Abstract
The present study aimed to investigate the regulatory mechanism of long non-coding RNA hypoxia-inducible factor 1α-anti-sense 1 (lncRNA HIF1α-AS1) in osteoblast differentiation as well as its targeting by sirtuin 1 (SIRT1), which may be inhibited by transforming growth factor (TGF)-β in bone marrow stromal cells (BMSCs). Real-time polymerase chain reaction (PCR), western blot analysis, lncRNA PCR arrays and chromatin immunoprecipitation were performed in order to examine the interference of SIRT1 expression by TGF-β, the effects of SIRT1 overexpression on lncRNA HIF1α-AS1 and the regulation of the expression of homeobox (HOX)D10, which promotes BMSC differentiation, by lncRNA HIF1α-AS1. The results showed that TGF-β interfered with SIRT1 expression. Furthermore, lncRNA HIF1α-AS1 was significantly downregulated following overexpression of SIRT1. In addition, low expression of HIF1α-AS1 was sufficient to block the expression of HOXD10. The present study further demonstrated that downregulation of HOXD10 by HIF1α-AS1 interfered with acetylation, and subsequently resulted in the inhibition of osteoblast differentiation. These results suggested that HIF1α-AS1 is an essential mediator of osteoblast differentiation, and may thus represent a gene-therapeutic agent for the treatment of human bone diseases.
Keywords: osteoblast differentiation, sirtuin 1, transforming growth factor-β, long non-coding RNA
Introduction
Osteoblasts are the major cellular components of bone and are cells with single nuclei that synthesize bone. They arise from bone marrow stromal cells (BMSCs) (1). BMSCs can differentiate into osteoblasts, adipocytes, chondrocytes and myoblasts. Sirtuin 1 (SIRT1) was shown to be able to de-acetylate β-catenin to promote its accumulation in the nucleus, which led to the transcription of genes associated with BMSC differentiation (2). Furthermore, SIRT1 has a key role in the differentiation of BMSCs into osteoblasts. SIRT1-activating and -inhibiting compounds increased and decreased, respectively, the expression of osteoblast markers and bone mineralization (3). In addition, SIRT1 has an important role in metabolism and in age-associated diseases. It has been used as a target for promoting bone formation in an anabolic approach for the treatment of osteoporosis (4).
Osteoblast differentiation is regulated by a number of growth factors, including transforming growth factor-β (TGF-β). TGF-β is produced by osteoblasts and osteocytes, and is abundantly deposited in bone matrices in a latent form (5). TGF-β is released from bone matrices through bone resorption and is activated by acids and matrix metalloproteinases secreted from osteoblast cells (6,7). TGF-β was shown to regulate the proliferation and differentiation of osteoblasts in vitro as well as in vivo (8). It stimulated proliferation and early osteoblast differentiation, while inhibiting terminal differentiation (9). TGF-β also suppressed osteoblastic differentiation of BMSCs (10). TGF-β was indicated to affect SIRT1 expression by mechanisms independent of methyl CpG binding protein 2 (11). TGF-β ligand activates TGF-β receptor-Smad3 signaling, which in turn collaboratively activates SIRT1 transcription (12). Homeobox (HOX)D10 has been reported to be a tumor suppressor and to regulate chondrocyte maturation as well as osteoblast differentiation (13,14).
Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts of >200 nucleotides (15). They are key regulators of diverse biological processes, including transcriptional regulation, cell growth and differentiation. Aberrant lncRNA expression and mutations have been linked to a diverse number of human diseases, including cancer, cardiovascular diseases and Alzheimer's disease (16). lncRNA-anti-differentiation ncRNA was shown to be an essential mediator of osteoblast differentiation (17). LncRNA hypoxia-inducible factor 1α-anti-sense 1 (HIF1α-AS1) has been shown to interact with BRG1, which is a key event in the proliferation and apoptosis of vascular smooth muscle cells in vitro (18). LncRNA HIF1α-AS1 is highly associated with cardiovascular diseases and is also over-expressed in advanced atherosclerosis tissues (19).
The aim of the present study was to explore the mechanisms of osteoblast differentiation with the goal of identifying novel regimens for treating osteoporosis, as well as providing protocols for optimal growth and differentiation of BMSCs into osteoblasts in vitro. First, real-time polymerase chain reaction (PCR) and western blot analyses were performed in order to explore the influence of TGF-β on the expression of SIRT1. Furthermore, an lncRNA PCR array was performed to assess the influence of SIRT1 overexpression on lncRNA HIF1α-AS1. Finally, the regulation of the expression of HOXD10, which is closely associated with cell differentiation, by HIF1α-AS1 was assessed.
Materials and methods
Cell culture
Human osteoblastic bone marrow stem cells (BMSCs) were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS; both from Hyclone Laboratories, Inc., Logan, UT, USA) at 37°C, in a humidified atmosphere of 5% CO2 in air. For osteoblastic differentiation, confluent cultures of BMSCs were maintained in low-serum medium, consisting of DMEM supplemented with 5% FBS and 200 ng/ml recombinant human bone morphogenic protein 2 (R&D Systems, Minneapolis, MN, USA). The medium was changed every 2–3 days. Human TGF-β was obtained from Sigma-Aldrich (St. Louis, MO, USA).
Treatment groups
To examine the influence of TGF-β on SIRT1 expression, four treatment groups of BMSCs were established: In the TGF-β group, BMSCs were incubated in DMEM containing 10% FBS and 200 µg/l TGF-β (Sigma-Aldrich) for 4 h; in the control group, BMSCs were cultured in DMEM supplemented with 10% FBS for 24 h; in the pirfenidone group, BMSCs were cultured in DMEM with 10% FBS and 200 µg/l pirfenidone (Marnac Inc., Dallas, TX, USA) for 4 h; in the dimethylsulfoxide (DMSO) group, BMSCs were cultured in DMEM containing 10% FBS and 200 µg/l DMSO (Sigma-Aldrich) for 4 h. Pirfenidone is an inhibitor of TGF-β at multiple steps: It reduces TGF-β promoter activity, TGF-β protein secretion, TGF-β-induced Smad2 phosphorylation and generation of reactive oxygen species (20). As pirfenidone was applied as a solution in DMEM, a positive control group treated with DMSO at a concentration of 200 µg/l, which is similar to the concentration of pirfenidone, was established in order to assess the effects of DMSO alone. Following incubation, reverse-transcription quantitative (RT-q)PCR was used to determine the expression of SIRT1 in BMSCs.
According to the SIRT1 gene sequence, the present study designed and synthesized small interfering (si)RNAs using the Silencer siRNA construction kit (Ambion; Invitrogen Life Technologies, Carlsbad, CA, USA) with the following sequences: siRNA-SIRT1, AAACCTGAGGCTGTTGTCTTCCTGTCTC and siRNA-negative control (NC), AAGGCTGTTTCACCTGAGTTCCTGTCTC. BMSCs were transfected using Lipofectamine 2000 (Invitrogen Life Technologies) and Opti-MEM (Invitrogen Life Technologies). To examine the effect of SIRT1 on lncRNA HIF1α-AS1 expression, the following experimental groups were established: The positive interference groups were transfected with the specific plasmid siRNA-SIRT1 (50 nM), SIRT1 inhibitor nicotinamide (Rugby Laboratories Inc., Norcross, GA, USA; 50 nM) or SIRT1 overexpression plasmid cDNA3.1(+)-SIRT1, which was constructed in our laboratory as previously described (21), while the negative interference group was transfected with non-specific plasmid siRNA-NC (50 nM) or non-specific vector pWPXL-SIRT1 (50 nM; Funeng Co., Ltd, Shanghai, China). RT-PCR was used to determine the relative mRNA levels of SIRT1 and HIF1α-AS1.
RT-qPCR
The total RNA was extracted and isolated from BMSCs using TRIzol reagent (Invitrogen Life Technologies, Inc.). RNA quality was assessed using a Nano Drop 1000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). RT-qPCR was performed using a QuantiTect Primer Assay (Qiagen GmbH, Hilden, Germany) and a QuantiTect SYBR Green RT-PCR kit (Qiagen GmbH) on a LightCycler 480 instrument (Roche Diagnostics, Mannheim, Germany). First, reverse-transcription was performed at 55°C for 30 min using random primers (cat no. 3801; Takara Bio, Inc, Otsu, Japan) and a Moloney Murine Leukemia Virus Reverse Transcriptase kit (Invitrogen Life Technologies, Inc.). The cDNA was added to a 2X Taq PCR Master mix (Tiangen Biotech Co., Ltd., Beijing, China) containing 10 pmol/l of each of the corresponding primer pairs. β-actin was used as an internal control. In the PCR amplification reaction, the thermocycling conditions were as follows: Initial activation for 15 min at 95°C, followed by 40 cycles of denaturation at 94°C for 15 sec, annealing for 30 sec at 55°C and extension for 30 sec at 72°C. The fluorescence was measured during the extension step. The relative expression of the target genes was determined using the 2−ΔΔCt method (22). Primers were obtained from Takara Bio Inc. with the following sequences: SIRT1 forward, 5′-GAGAATTCAGGACTTTCTTCGAGATC-3′ and reverse, 5′-CTGAATTCTCATTGTGGACTCGGC-3′; HIF1α-AS1 forward, 5′-ATGGTTCGCGTTGTATCTTTAA3′ and reverse, 5′-TCAGTTAATCCTGCCGCATTGA-3′; HoxD10 forward, 5′-TCTGTTATCTTTATTGGAGGAGTTTG-3′ and reverse, 5′-TTGAAACACCCATAGGACTGACT-3′; β-actin forward, 5′-CGAGCGCGGCTACAGCT-3′ and reverse, 5′-TCCTTAATGTCACGCACGATTT-3′.
Western blot analysis
Western blot analysis of SIRT1 and HOXD10 was performed as previously described (23). Cells were collected after transfection for 72 h, washed twice with cold phosphate-buffered saline and lysed in buffer 5A containing 10 mM HEPES (pH 7.0; Sigma-Aldrich), 10 mM NaCl, 1.5 mM MgCl2, 0.75 mM dithiothreitol (Sigma-Aldrich), 0.2 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich) and protease inhibitors (Roche, Basel, Switzerland) for 30 min. The mixture was centrifuged at 3,000 × g for 10 min after boiling for 10 min, and supernatants were collected. Protein concentrations were determined using a bicinchoninic acid assay (Pierce Biotechnology, Inc., Rockford, IL, USA). The cell lysate was separated by 12% SDS-PAGE and 15 ml protein was loaded onto nitrocellulose membranes. After blocking non-specific binding sites with 5% non-fat milk, the membrane was incubated with primary antibodies for 1.5 h at room temperature. The primary antibodies used were as follows: Monoclonal mouse anti-human SRT1 (cat. no. AP52838; Abgent, San Diego, CA, USA), monoclonal rabbit anti-human HoxD10 (cat. no. ab138508; Abcam, Cambridge, MA, USA) and monoclonal β-actin (cat. no. 12262; Cell Signaling Technology, Inc., Beverly, MA, USA). The membrane was then washed with Tris-buffered saline containing Tween 20 three times for 10 min each and incubated for 2 h with SIRT1 goat anti-mouse and HoxD10 goat anti-rabbit IgG secondary antibodies (cat. nos. ASS1021 and ASS1006, respectively; Abgent). Images of the blots were captured and densitometric analysis was performed using an LAS-3000 and Multi Gauge version 3.0 (Fujifilm, Tokyo, Japan).
lncRNA PCR array
The PCR-array was performed in a high-throughput manner in an RT-PCR system. The lncRNA PCR array kit (cat no. 20130123a) was purchased from Funeng Co. Ltd and all experiments were performed according to manufacturer's instructions. A 96-well PCR microarray plate (Funeng Co., Ltd) was prepared with a different primer-probe in each well. RNA was respectively converted into double-stranded cDNA by reverse transcription using a cDNA synthesis kit (Superscript Choice System; Invitrogen Life Technologies) with an oligo(dT)24 primer containing a T7 RNA polymerase promoter site at the 3′ end of the poly T (Genset, La Jolla, CA, USA). After mixing the ligated samples into a single tube, PCR amplification was performed using an adaptor primer and a gene-specific primer. The experimental procedure was performed according to a previously published procedure (24). The PCR cycling conditions were set as follows: Initial denaturation at 95°C for 10 min followed by 40 cycles of 94°C for 30 sec, 50°C for 30 sec and 72°C for 60 sec, followed by an incubation at 72°C for 20 min. BMSC cells without transfection were used as the source of the standard, relative to which the relative expression levels were determined. Each reaction mixture contained ten portions of standard cDNA with the shortest adaptor and two portions of standard cDNA with the second shortest adaptor. Amplified products were separated on an ABI 3100 DNA analyzer (Applied Biosystems, Thermo Fisher Scientific). Calibration curves were generated from data points of the two standards, which were used for determining the relative expression levels following conversion to a logarithmic scale. Finally, for comparison of gene expression in tumor (T) and surrounding non-tumor (NT) tissues, T versus NT (T/NT) ratios were estimated for each gene using the following formula: Log2(T/NT) = log2(T) − log2(NT). Hierarchical cluster analysis using the Ward method and principal component analysis were performed using GeneMaths 2.0 software (Applied Maths, Inc., Austin, TX, USA).
Chromatin immunoprecipitation
Cells were transfected with HIF1α-AS1 siRNA1 or HIF1α-AS1 siRNA2. Acetylhistone levels in cells were determined by RT-qPCR using the lncRNA PCR array kit (cat no. 20130123a; Funeng Co., Ltd) according to manufacturer's instructions. Chromatin immuno-precipitation was performed using the EZ ChIP™ Chromatin Immunoprecipitation kit (Millipore, Billerica, MA, USA) according to the manufacturer's instructions. Cross-linked chromatin was sonicated at 30 kHZ for 30 min for fragmentation. The chromatin was immunoprecipitated using the anti-acetylated histone 3 (cat no. sc-8654; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and rabbit polyclonal anti-histone H4 (acetyl K5) antibody (cat no. ab114146; Abcam). Normal human immunoglobulin (Ig)G was used as a negative control.
Statistical analysis
All values are expressed as the mean ± standard deviation from at least three separate experiments. Differences between groups were analyzed using Student's t-test in SPSS 10.0 software (SPSS, Inc., Chicago, IL, USA). Differences were considered statistically significant at P<0.05.
Results
TGF-β interferes with SIRT1 expression
The present study examined whether TGF-β was able to interfere with the expression of SIRT1. As expected (Fig. 1A), RT-qPCR analysis showed that the expression of SIRT1 in BMSCs was significantly decreased in the TGF-β group compared with that in the control group and the DMSO positive control. By contrast, the expression of SIRT1 was significantly increased in the group treated with pirfenidone, which added as an inhibitor of TGF-β, compared with that in the control group and the DMSO positive control.
Figure 1.
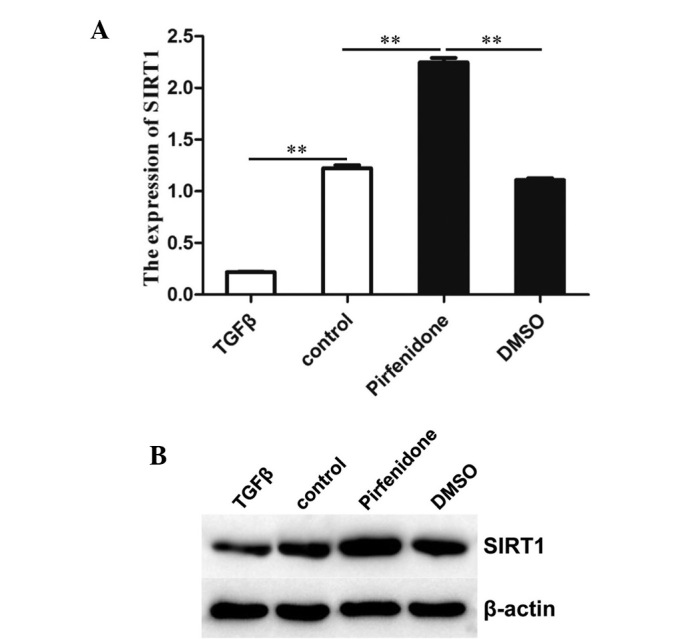
Interference of TGF-β with the expression of SIRT1. (A) SIRT1 mRNA levels in BMSCs. Total RNA was extracted and subjected to reverse transcription quantitative polymerase chain reaction to analyze the expression levels of SIRT1. β-actin was used as a reference. Values are expressed as the mean ± standard deviation (n=3). **P<0.01 compared with the control. (B) Western blot analysis of SIRT1 protein levels in BMSCs. β-actin was used as an internal control. TGF, transforming growth factor; SIRT1, sirtuin 1; DMSO, dimethyl sulfoxide; BMSCs, bone marrow stromal cells.
In analogy with the PCR results, western blot analysis showed that addition of TGF-β decreased the SIRT1 protein content in BMSCs (Fig. 1B). These results indicated that TGF-β inhibited SIRT1 expression in BMSCs.
Overexpression of SIRT1 results in downregulation of lncRNA HIF1α-AS1
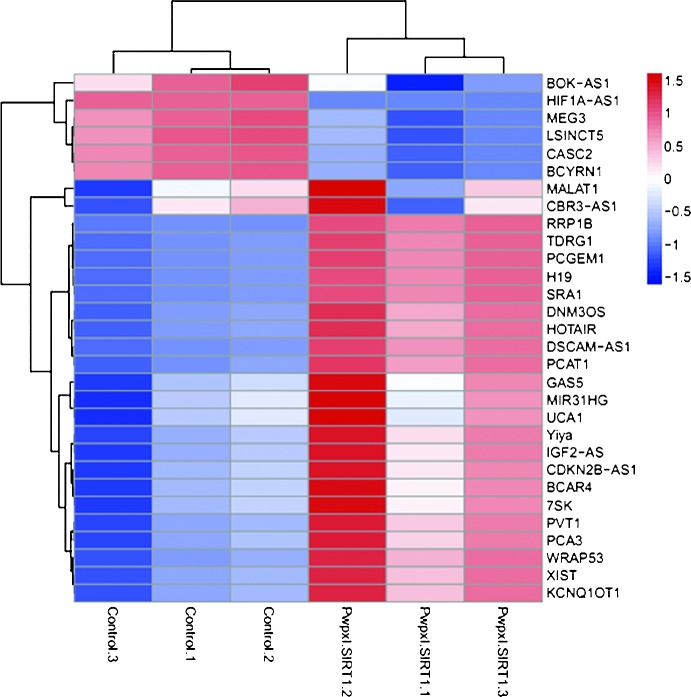
SIRT1 has a key role in the differentiation of BMSCs into osteoblasts, while lncRNA HIF1a-AS1 is important in the proliferation and apoptosis of tumor cells. Therefore, the present study explored the influence of SIRT1 on lncRNA HIF1a-AS1. To investigate the role of SIRT1 in the regulation of lncRNA HIF1α-AS1, the expression of a series of genes, including HIF1α-AS1, in cells treated with pWPXL-SIRT1 was assessed by lncRNA PCR array. The gene expression heatmap (Fig. 2) shows that pWPXL-SIRT1 suppressed lncRNA HIF1α-AS1 expression. The gene expression heatmap indicated that overexpression of SIRT1 downregulated the expression of lncRNA HIF1α-AS1.
Figure 2.
Heatmap of total RNA expression levels, including HIF1α-AS1 treated with pWPXL. SIRT1, assayed by polymerase chain reaction. pWPXL. SIRT1, SIRT1 overexpression vector.
Furthermore, RNA expression levels of SIRT1 and HIF1α-AS1 in cells respectively treated with small interfering (si) RNA-SIRT1, SIRT1 inhibitor nicotinamide or pWPXL-SIRT1 were assessed by PCR array. As shown in Fig. 3, siRNA-SIRT1 promoted HIF1α-AS1 expression and obviously resulted in increases in lncRNA HIF1α-AS1 expression. Conversely, pWPXL-SIRT1 markedly suppressed HIF1α-AS1 expression and significantly decreased HIF1α-AS1 expression. Inhibition of SIRT1 expression using nicotinamide resulted in the highest expression of HIF1α-AS1 among the three experimental groups and two control groups.
Figure 3.
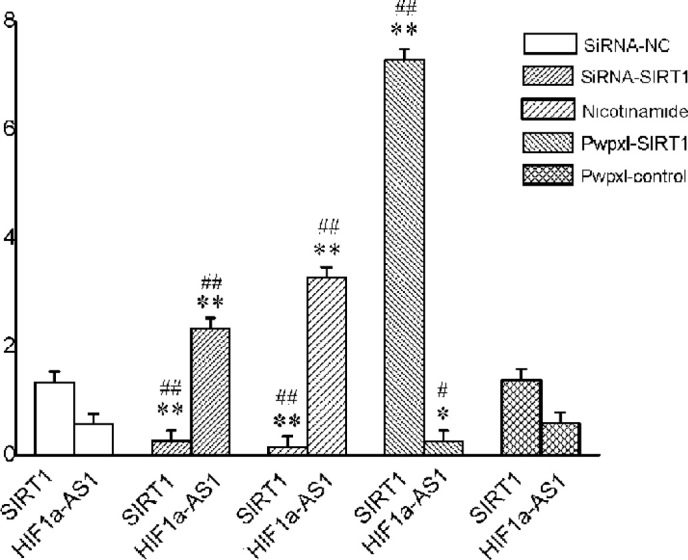
RNA expression levels of SIRT1 and HIF1α-AS1 in bone marrow stromal cells. Total RNA was extracted and subjected to polymerase chain reaction array to analyze the RNA expression levels. Nicotinamide was used as an SIRT1 inhibitor. Values are expressed as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 compared with the expression of SIRT1 in the siRNA-NC group; #P<0.05, ##P<0.01 compared with the expression of HIF1α-AS1 in the siRNA-NC group. SIRT1, SIRT1 overexpression vector; siRNA, small interfering RNA; NC, scrambled control.
The PCR array suggested that overexpression of SIRT1 downregulates the expression of lncRNA HIF1α-AS1, while downregulation or inhibition of SIRT1 upregulated the expression of lncRNA HIF1α-AS1.
HIF1α-AS1 decreases the expression of HOXD10
To investigate the biological role of lncRNA HIF1α-AS1 in the regulation of HOXD10, the HOXD10 expression levels in BMSCs treated with HIF1α-AS1 siRNA were assessed by RT-qPCR. Fig. 4A shows that HIF1α-AS1 siRNA1 as well as HIF1α-AS1 siRNA2 suppressed the mRNA expression of HOXD10. In analogy to the PCR results, western blot analysis demonstrated that HIF1α-AS1 siRNA blocked the expression of HOXD10 protein expression in BMSCs (Fig. 4B). These observations suggested that suppression of HIF1α-AS1 contributed to the downregulation of HOXD10.
Figure 4.
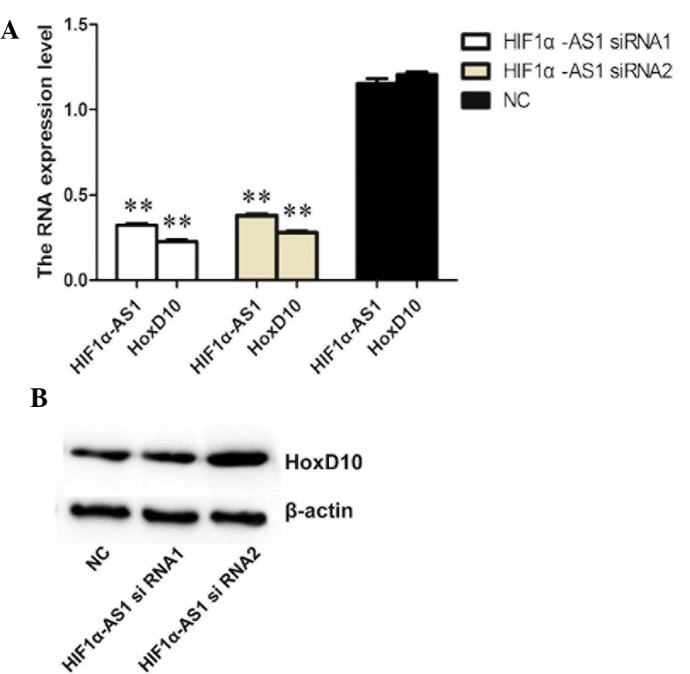
HOXD10 expression levels in cells treated with lncRNA HIF1α-AS1 siRNA. (A) mRNA expression levels in BMSCs. Total RNA was extracted and subjected to reverse transcription quantitative polymerase chain reaction to analyze the mRNA expression levels. β-actin was used as a reference. Values are expressed as the mean ± standard deviation (n=3). **P<0.01 compared with the siRNA-NC group (B) Western blot analysis of HOXD10 protein levels in BMSCs. β-actin was used as an internal controls. HOX, homeobox; NC, negative control; siRNA, small interfering RNA.
Acetylation-induced downregulation of HOXD10 by HIF1α-AS1 interference
The chromatin immunoprecipitation arrays showed that HIF1α-AS1 siRNA1 as well as HIF1α-AS1 siRNA2 interfered with acetylation and resulted in decreases in the enrichment of acetylhistone (Fig. 5). These results suggested that lncRNA HIF1α-AS1 enhances HOXD10 expression by promoting acetylation.
Figure 5.

Interference of HIF1α-AS1 with acetylhistone levels. Chromatin immunoprecipitation arrays were conducted using the indicated antibodies. Enrichment was determined relative to the input controls. Values are expressed as the mean ± standard deviation (n=3). **P<0.01 compared with NC group. NC, negative control; siRNA, small interfering RNA; IgG, immunoglobulin G.
Discussion
The results of the present study showed that the expression of SIRT1, which is known to have a key role in the differentiation of BMSCs into osteoblasts, was decreased by TGF-β. TGF-β is a secreted protein that regulates proliferation, differentiation and death of various cell types (25). It is also a potent inhibitor of skeletal muscle differentiation (26). TGF-β-induced repression of core-binding factor alpha 1 (CBFA1) by Smad3 decreases CBFA1 and osteocalcin expression and inhibits osteoblast differentiation (8). It is required for normal differentiation of cartilage in vivo. Loss of responsiveness to TGF-β has been demonstrated to promote terminal differentiation of chondrocytes and to result in development of degenerative joint disease resembling osteoarthritis in humans (27).
In the present study, the overexpression of SIRT1 down-regulated lncRNA HIF1α-AS1 expression. SIRT1 is a stress response factor that promotes longevity by regulating various cell survival pathways (28). It is increasingly recognized as a critical regulator of stress responses, replicative senescence, inflammation, metabolism and aging (29). SIRT1 regulates adipocyte formation during osteoblast differentiation; the efficiency of this process is further increased by the simultaneous action of SIRT1-mediated pathways (30). SIRT1 has also been reported to inhibit the proliferation and differentiation of pig pre-adipocytes through repression of adipocyte genes (31). These previous findings led to the conclusion that changes in SIRT1 expression are associated with osteoblast differentiation.
LncRNAs, the less intensively studied siblings of microRNA, have been suggested to have a robust role in developmental and adult tissue regulation similarly to that of microRNA (32). In recent years, the yeast germ-cell differentiation process was proven to be a powerful biological system for the identification and study of several lncRNAs that have a key role in regulating cellular differentiation by directly acting on chromatin (33). Bertani et al (34) uncovered that the lncRNA Mira had a role in epigenetic activation and cell differentiation by recruiting the epigenetic activator mixed-lineage leukemia 1 to chromatin. Transition from progenitor cells into highly differentiated cells involved tightly controlled gene-regulatory changes. A growing number of lncRNAs has been implicated in such processes (35). A fraction of them were shown to be functionally important in the differentiation of diverse cells and tissues. lncRNA has also been reported to be involved in gene expression (36). For instance, lncRNA has been shown to be implicated in the regulation of epidermal growth factor homology domain-1 (37), which may explain the result of the present study that gene HOXD10 was regulated by lncRNA HIF1α-AS1.
According to the results of the present study, HOXD10 expression was increased by lncRNA HIF1α-AS1. HOX genes are master regulators of organ morphogenesis and cell differentiation during embryonic development and continue to be expressed throughout post-natal life (38). HOXD10 is a member of the abdominal-B homeobox family and encodes a sequence-specific transcription factor with a homeobox DNA-binding domain (39). It was shown to have a key role in regulating cortical stromal-cell differentiation during kidney development (40). HOXD10 expression was found to be low in poorly-differentiated gastric cancer cell lines compared with that in well-differentiated gastric cancer cell lines (41). furthermore, HOXD10 has been demonstrated to have an important role in cell differentiation and morphogenesis during development (39).
The present study demonstrated that the expression of HOXD10 was increased by lncRNA HIF1α-AS1 via the promotion of acetylation; therefore, the present study hypothesized that HOXD10 is able to promote osteoblast differentiation.
In conclusion, the results of the present study suggested that TGF-β inhibits SIRT1 expression in BMSCs and the resulting low levels of SIRT1 lead to the upregulation of lncRNA HIF1α-AS1, which then enhances HOXD10 expression by promoting acetylation. As HOXD10 expression has a central role in promoting osteoblast differentiation, TGF causes the differentiation of BMSCs into osteoblasts via SIRT1 and lncRNA HIF1α-AS1. The present study provided a novel regulatory mechanism of osteoblast differentiation, which may be harnessed for the development of novel treatments of bone and joint diseases. In particular, lncRNA HIF1α-AS1 may represent a novel therapeutic agent against osteoarthritis.
Acknowledgments
This study was financially supported by Scientific Research Foundation of Shanghai Municipal Health Bureau (no. 20114266).
References
- 1.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 2.Simic P, Zainabadi K, Bell E, Sykes DB, Saez B, Lotinun S, Baron R, Scadden D, Schipani E, Guarente L, et al. SIRT1 regulates differentiation of mesenchymal stem cells by deacetylating β-catenin. EMBO mol Med. 2013;5:430–440. doi: 10.1002/emmm.201201606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäckesjö CM, Li Y, Lindgren U, Haldosén LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21:993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- 4.Cohen-Kfir E, Artsi H, Levin A, Abramowitz E, Bajayo A, Gurt I, Zhong L, D'Urso A, Toiber D, Mostoslavsky R, Dresner-Pollak R. Sirt1 is a regulator of bone mass and a repressor of Sost encoding for sclerostin, a bone formation inhibitor. Endocrinology. 2011;152:4514–4524. doi: 10.1210/en.2011-1128. [DOI] [PubMed] [Google Scholar]
- 5.Pfeilschifter J, Bonewald L, Mundy GR. Characterization of the latent transforming growth factor beta complex in bone. J Bone Miner Res. 1990;5:49–58. doi: 10.1002/jbmr.5650050109. [DOI] [PubMed] [Google Scholar]
- 6.Guise TA, Chirgwin JM. Transforming growth factor-beta in osteolytic breast cancer bone metastases. Clin Orthop Relat Res. 2003;(Suppl 415):S32–S38. doi: 10.1097/01.blo.0000093055.96273.69. [DOI] [PubMed] [Google Scholar]
- 7.Pfeilschifter J, Mundy GR. Modulation of type beta transforming growth factor activity in bone cultures by osteotropic hormones. Proc Natl Acad Sci USA. 1987;84:2024–2028. doi: 10.1073/pnas.84.7.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonewald LF, Dallas SL. Role of active and latent transforming growth factor beta in bone formation. J Cell Biochem. 1994;55:350–357. doi: 10.1002/jcb.240550312. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi K, Abe M, Hiasa M, Oda A, Amou H, Kido S, Harada T, Tanaka O, Miki H, Nakamura S, et al. Tgf-Beta inhibition restores terminal osteoblast differentiation to suppress myeloma growth. PLoS One. 2010;5:e9870. doi: 10.1371/journal.pone.0009870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volkmann I, Kumarswamy R, Pfaff N, Fiedler J, et al. MicroRNA-mediated epigenetic silencing of sirtuin1 contributes to impaired angiogenic responses. Circ Res. 2013;113:997–1003. doi: 10.1161/CIRCRESAHA.113.301702. [DOI] [PubMed] [Google Scholar]
- 12.Warburton D, Shi W, Xu B. TGF-β-Smad3 signaling in emphysema and pulmonary fibrosis: An epigenetic aberration of normal development? Amer J Physiol Lung Cell Mol Physiol. 2013;304:L83–L85. doi: 10.1152/ajplung.00258.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X, Lu Y, Nie Y, Fan D. MicroRNAs as critical regulators involved in regulating epithelial-mesenchymal transition. Curr Cancer Drug Targets. 2013;13:935–944. doi: 10.2174/15680096113136660099. [DOI] [PubMed] [Google Scholar]
- 14.Barham G, Clarke NM. Genetic regulation of embryological limb development with relation to congenital limb deformity in humans. J Child Orthop. 2008;2:1–9. doi: 10.1007/s11832-008-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo C, Wang Z, Lu H, Dai Z, Liu X, Cui L. Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Mol Med Rep. 2013;8:463–467. doi: 10.3892/mmr.2013.1540. [DOI] [PubMed] [Google Scholar]
- 17.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Zhang X, Yuan Y, Tan M, et al. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. Eur J Cardiothorac Surg. 2015;47:439–446. doi: 10.1093/ejcts/ezu215. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Feng G, Wang Y, Yue Y, Zhao W. Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: Implications for TAA pathogenesis. Int J Clin Exp Pathol. 2014;7:7643–7652. [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagita M. Inhibitors/antagonists of TGF-β system in kidney fibrosis. Nephrol Dialysis Transplant. 2012;27:3686–3691. doi: 10.1093/ndt/gfs381. [DOI] [PubMed] [Google Scholar]
- 21.Wu SQ, Wang M, Liu Q, Zhu YJ, Suo X, Jiang JS. Construction of DNA vaccines and their induced protective immunity against experimental Eimeria tenella infection. Parasitol Res. 2004;94:332–336. doi: 10.1007/s00436-004-1185-6. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Xu N, Shen C, Luo Y, Xia L, Xue F, Xia Q, Zhang J. Upregulated miR-130a increases drug resistance by regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell. Biochem Biophys Res Commun. 2012;425:468–472. doi: 10.1016/j.bbrc.2012.07.127. [DOI] [PubMed] [Google Scholar]
- 24.Kurokawa Y, Matoba R, Nakamori S, Takemasa I, Nagano H, Dono K, Umeshita K, Sakon M, Monden M, Kato K. PCR-array gene expression profiling of hepatocellular carcinoma. J Exp Clin Cancer Res. 2004;23:135–141. [PubMed] [Google Scholar]
- 25.Moustakas A, Pardali K, Gaal A, Heldin CH. Mechanisms of TGF-beta signaling in regulation of cell growth and differentiation. Immunol Lett. 2002;82:85–91. doi: 10.1016/S0165-2478(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 26.Liu D, Black BL, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001;15:2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serra R, Johnson M, Filvaroff EH, LaBorde J, Sheehan DM, Derynck R, Moses HL. Expression of a truncated, kinase-defective TGF-beta type II receptor in mouse skeletal tissue promotes terminal chondrocyte differentiation and osteoarthritis. J Cell Biol. 1997;139:541–552. doi: 10.1083/jcb.139.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qureshi IA, Mehler MF. Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol Med. 2011;17:337–346. doi: 10.1016/j.molmed.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H, Verdin E. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2010;2:415–431. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava S, Bedi U, Roy P. Synergistic actions of insulin-sensitive and Sirt1-mediated pathways in the differentiation of mouse embryonic stem cells to osteoblast. Mol Cell Endocrinol. 2012;361:153–164. doi: 10.1016/j.mce.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Bai L, Pang WJ, Yang YJ, Yang GS. Modulation of Sirt1 by resveratrol and nicotinamide alters proliferation and differentiation of pig preadipocytes. Mol Cell Biochem. 2008;307:129–140. doi: 10.1007/s11010-007-9592-5. [DOI] [PubMed] [Google Scholar]
- 32.Tye CE, Gordon JA, Martin-Buley LA, Stein JL, Lian JB, Stein GS. Could lncRNAs be the missing links in control of mesenchymal stem cell differentiation? J Cell Physiol. 2015;230:526–534. doi: 10.1002/jcp.24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiriart E, Verdel A. Long noncoding RNA-based chromatin control of germ cell differentiation: A yeast perspective. Chromosome Res. 2013;21:653–663. doi: 10.1007/s10577-013-9393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Kretz M. TINCR, staufen1 and cellular differentiation. RNA Biol. 2013;10:1597–1601. doi: 10.4161/rna.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tajiri N, Acosta SA, Shahaduzzaman M, Ishikawa H, Shinozuka K, Pabon M, Hernandez-Ontiveros D, Kim DW, Metcalf C, Staples M, et al. Intravenous transplants of human adipose-derived stem cell protect the brain from traumatic brain injury-induced neurodegeneration and motor and cognitive impairments: Cell graft biodistribution and soluble factors in young and aged rats. J Neurosci. 2014;34:313–326. doi: 10.1523/JNEUROSCI.2425-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, Chun CZ, Samant GV, Zhao B, Garnaas MK, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010;115:133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orr KS, Shi Z, Brown WM, O'Hagan KA, Lappin TR, Maxwell P, Percy MJ. Potential prognostic marker ubiquitin carboxyl-terminal hydrolase-L1 does not predict patient survival in non-small cell lung carcinoma. J Exp Clin Cancer Res. 2011;30:79. doi: 10.1186/1756-9966-30-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu X, Chen D, Cui Y, Li Z, Huang J. Targeting microRNA-23a to inhibit glioma cell invasion via HOXD10. Sci Rep. 2013;3:3423. doi: 10.1038/srep03423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yallowitz AR, Hrycaj SM, Short KM, Smyth IM, Wellik DM. Hox10 genes function in kidney development in the differentiation and integration of the cortical stroma. PLoS One. 2011;6:e23410. doi: 10.1371/journal.pone.0023410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Wang G, Scott SA, Capecchi MR. Hoxc10 and Hoxd10 regulate mouse columnar, divisional and motor pool identity of lumbar motoneurons. Development. 2008;135:171–182. doi: 10.1242/dev.009225. [DOI] [PubMed] [Google Scholar]