Abstract
In this issue of Blood, Zhu et al and Bougie et al provide the first evidence for a drug to alter the conformation of a drug-dependent antiplatelet antibody, thereby adding a new dimension to the mechanism of drug-induced immune thrombocytopenia (DITP).1,2
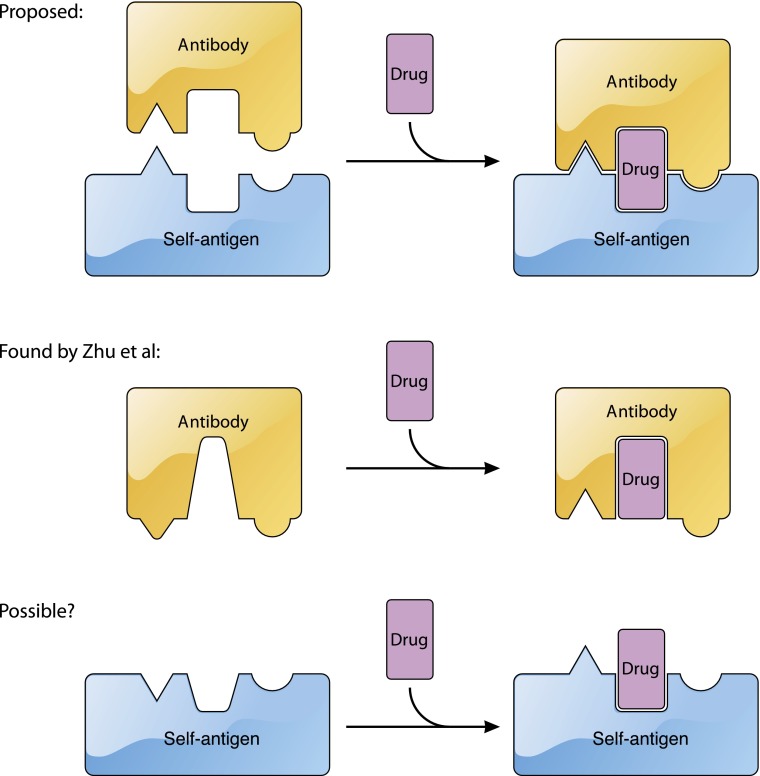
The current model for the recognition of platelet antigens by quinine-dependent antibodies. Current sandwich model (top). Summary of the findings of Zhu et al1 in which quinine is mostly embedded in the quinine-dependent antibody and quinine binding alters the conformation of the paratope (middle). Possibility that under certain circumstances quinine may also alter the conformation of the epitope in a platelet antigen (bottom). The figure has been adapted from Figure 5 in the article by Zhu et al1 that begins on page 2138. Professional illustration by Patrick Lane, ScEYEnce Studios.
In DITP, a drug somehow induces a tight interaction of antibody with one’s own platelet antigen, leading to drug-dependent depletion of platelets.3 In the absence of the drug, the antibody does not bind to the platelet with sufficient affinity to cause pathological damage. As the drug is often noncovalently associated with the antibody/antigen complex, to stop taking the drug is a simple but effective treatment. Thus, understanding of the molecular mechanism of DITP may not significantly impact the clinical outcome of individual patients, but if it can help to predict the likelihood of a drug to induce the depletion of platelets4 the benefit may be potentially enormous. DITP is characterized by its heterogeneity. Many drugs have been documented to cause DITP, although sharing little structural similarity.5 On the other hand, the same drug can facilitate binding of different antibodies to different epitopes on different platelet antigens.6-8 Therefore, the challenge is to glean common principles from studies of prototypical cases.
Quinine is such a prototypical drug. Studies of human sera containing quinine-dependent antibodies have produced most of the mechanistic insights on DITP, including the sandwich model,9 in which quinine becomes a part of the binding interface sandwiched between antibody and antigen. The detail of the quinine-dependent interface is lacking, although it has been implied, evident in all the cartoons depicting the sandwich model,3,5,9 that the conformation of antibody or antigen is not altered by quinine but only supplemented by it. It is also not clear why quinine, compared with most other drugs, is more likely to induce or facilitate the antibody/antigen interaction. To answer these questions, it will be important to elucidate the antibody/antigen-binding interface and to define the location of quinine relative to this interface.
Taking a critical step forward, Zhu et al has provided the first high-resolution glimpse of the interface by determining the crystal structures of quinine/antibody complexes.1 This impressive work follows the earlier, equally impressive, procurement of 2 monoclonal antibodies (mAbs) that bind tightly to the propeller domain of human integrin αIIb in the presence of quinine.10 The authors found that both mAbs, named 314.1 and 314.3, bound tightly to quinine. Crystal structures of the quinine/314-antibody complexes reveal that quinine is largely embedded in the antibody, surrounded by the complementarity-determining region (CDR) loops. It is likely that only a small portion of quinine may be in contact with the antigen. Importantly, comparison of the 314-antibody structure with the quinine/314-antibody complex structure reveals that upon binding, quinine alters the conformation of a CDR loop that is part of the paratope, thereby providing a new and critical element that is not covered by the sandwich model (see figure). The quinine-induced conformational change appears to be the molecular basis for the quinine-facilitated binding of 314 antibodies, as Bougie et al reported that the binding affinity of 314.1 antibody for purified integrin αIIbβ3 in the presence of quinine was 5 times tighter than that in the absence of quinine.2 The quinine/314-antibody structures also point to an interesting possibility regarding the origin of drug-dependent antibodies; quinine may fit, perhaps by chance, into a preexisting antibody to either greatly improve its affinity for a platelet antigen or enable its recognition of a different antigen.
Bougie et al2 reported that 314.1 antibody with quinine bound 2 copies of αIIbβ3 on the platelet, as its apparent affinity for the platelet was orders of magnitude higher than that for the purified monovalent αIIbβ3. This avidity effect, similar to the differential affinities of some anti-GPIbα antibodies for the platelet and for purified GPIb-IX complex,11 is enabled by the high expression level of αIIbβ3 in the platelet. Combining the aforementioned difference between monovalent affinities in the presence and absence of quinine (5 times) and the high antibody/quinine affinity for the platelet (0.15 nM) would produce an antibody affinity for the platelet in the absence of quinine at 3.75 nM. Bougie et al2 did not report the 314.1 affinity for the platelet without quinine, but did report binding of 314.1 to the platelet without quinine that could be inhibited by competing antibodies.
The work of Zhu et al1 stopped short of a structure of the quinine/antibody/antigen ternary complex, presumably due to some technical difficulties. Without a high-resolution structure showing the antibody/antigen interface in the presence of quinine, it would be difficult to discuss the interplay between quinine and antigen. Although a large body of evidence supports a direct interaction of quinine with antigen, a specific and stoichiometric interaction of quinine with its platelet antigen, primarily αIIbβ3 or GPIb-IX, has not been found. Paradoxically, quinine appears to prefer certain regions in the αIIbβ3 and GPIb-IX,6,8 suggesting that there may be an element other than chance recognition at play. Although providing no evidence on the quinine/antigen interaction, the quinine/antibody complex structures reported by Zhu et al1 raise a possibility that has not been explored vigorously. If quinine can alter the conformation of an antibody, is it also possible for quinine to alter the conformation of an antigen?12 Although the 314.1 antibody binds to the same region of αIIbβ3 in the absence and presence of quinine,2 it remains unclear whether the conformation of αIIbβ3 at the binding site is altered by the bound antibody. In the quinine/antibody structures, Zhu et al1 have showcased the usefulness of quinine-dependent mAbs. Perhaps an additional, ingenious characterization of these unique antibodies in the presence of αIIbβ3 and quinine would yield more mind-opening insights on the pathology of DITP.
Footnotes
Conflict-of-interest disclosure: The author declares no competing financial interests.
REFERENCES
- 1.Zhu J, Zhu J, Bougie DW, Aster RH, Springer TA. Structural basis for quinine-dependent antibody binding to platelet integrin αIIbβ3. Blood. 2015 doi: 10.1182/blood-2015-04-639351. 126(18):2138-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bougie DW, Peterson J, Rasmussen M, Aster RH. Mechanism of quinine-dependent monoclonal antibody binding to platelet glycoprotein IIb/IIIa. Blood. 2015 doi: 10.1182/blood-2015-04-643148. 126(18):2146-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aster RH, Bougie DW. Drug-induced immune thrombocytopenia. N Engl J Med. 2007;357(6):580–587. doi: 10.1056/NEJMra066469. [DOI] [PubMed] [Google Scholar]
- 4.Reese JA, Li X, Hauben M, et al. Identifying drugs that cause acute thrombocytopenia: an analysis using 3 distinct methods. Blood. 2010;116(12):2127–2133. doi: 10.1182/blood-2010-03-276691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aster RH, Curtis BR, McFarland JG, Bougie DW. Drug-induced immune thrombocytopenia: pathogenesis, diagnosis, and management. J Thromb Haemost. 2009;7(6):911–918. doi: 10.1111/j.1538-7836.2009.03360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess JK, Lopez JA, Berndt MC, Dawes I, Chesterman CN, Chong BH. Quinine-dependent antibodies bind a restricted set of epitopes on the glycoprotein Ib-IX complex: characterization of the epitopes. Blood. 1998;92(7):2366–2373. [PubMed] [Google Scholar]
- 7.Asvadi P, Ahmadi Z, Chong BH. Drug-induced thrombocytopenia: localization of the binding site of GPIX-specific quinine-dependent antibodies. Blood. 2003;102(5):1670–1677. doi: 10.1182/blood-2002-07-2175. [DOI] [PubMed] [Google Scholar]
- 8.Peterson JA, Nelson TN, Kanack AJ, Aster RH. Fine specificity of drug-dependent antibodies reactive with a restricted domain of platelet GPIIIA. Blood. 2008;111(3):1234–1239. doi: 10.1182/blood-2007-09-112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bougie DW, Wilker PR, Aster RH. Patients with quinine-induced immune thrombocytopenia have both “drug-dependent” and “drug-specific” antibodies. Blood. 2006;108(3):922–927. doi: 10.1182/blood-2006-01-009803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bougie DW, Birenbaum J, Rasmussen M, Poncz M, Aster RH. Quinine-dependent, platelet-reactive monoclonals mimic antibodies found in patients with quinine-induced immune thrombocytopenia. Blood. 2009;113(5):1105–1111. doi: 10.1182/blood-2008-09-177279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang X, Russell SR, Estelle S, et al. Specific inhibition of ectodomain shedding of glycoprotein Ibα by targeting its juxtamembrane shedding cleavage site. J Thromb Haemost. 2013;11(12):2155–2162. doi: 10.1111/jth.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R. A hypothesis that explains the heterogeneity of drug-induced immune thrombocytopenia. Blood. 2010;115(4):914. doi: 10.1182/blood-2009-09-242297. [DOI] [PMC free article] [PubMed] [Google Scholar]