Abstract
Quinolone antimicrobials are synthetic and widely used in clinical medicine. Resistance emerged with clinical use and became common in some bacterial pathogens. Mechanisms of resistance include two categories of mutation and acquisition of resistance-conferring genes. Resistance mutations in one or both of the two drug target enzymes, DNA gyrase and DNA topoisomerase IV, are commonly in a localized domain of the GyrA and ParE subunits of the respective enzymes and reduce drug binding to the enzyme-DNA complex. Other resistance mutations occur in regulatory genes that control the expression of native efflux pumps localized in the bacterial membrane(s). These pumps have broad substrate profiles that include quinolones as well as other antimicrobials, disinfectants, and dyes. Mutations of both types can accumulate with selection pressure and produce highly resistant strains. Resistance genes acquired on plasmids can confer low-level resistance that promotes the selection of mutational high-level resistance. Plasmid-encoded resistance is due to Qnr proteins that protect the target enzymes from quinolone action, one mutant aminoglycoside-modifying enzyme that also modifies certain quinolones, and mobile efflux pumps. Plasmids with these mechanisms often encode additional antimicrobial resistances and can transfer multidrug resistance that includes quinolones. Thus, the bacterial quinolone resistance armamentarium is large.
Keywords: topoisomerase, efflux pumps, plasmids, quinolone, DNA gyrase
Introduction
Quinolones have been a widely used class of synthetic antimicrobials.1, 2 The initial member of the class, nalidixic acid, was identified as a byproduct of chloroquine synthesis in 1962 and had limited clinical use because it was only sufficient for treatment of urinary tract infections and because of the early emergence of resistance.3 Chemical modifications of the core quinolone and related chemical scaffolds were, however, widely explored and generated compounds with greater potency, broader spectra of activity, improved pharmacokinetics, and lower frequency of development of resistance.4 A key modification of a fluorine substituent at position 8 led to the development of many members of what became known as the fluoroquinolone class with the introductions of norfloxacin in 1986 and ciprofloxacin in 1987 that exhibited substantially greater potency against gram-negative bacteria. Subsequently other fluoroquinolones, such as levofloxacin and moxifloxacin, were developed with enhanced activity against gram-positive bacteria. Because of their potency, spectrum of activity, oral bioavailability, and generally good safety profile, fluoroquinolones were used extensively for multiple clinical indications throughout the world. Although still clinically valuable, fluoroquinolone use has become limited in some clinical settings, as bacterial resistance has emerged over time. In the sections that follow we review the range of molecular mechanisms that underlie quinolone resistance.
Quinolone resistance due to mutation in chromosomal genes
Alterations in target enzymes
Quinolones target two essential bacterial type II topoisomerase enzymes, DNA gyrase and DNA topoisomerase IV.5 Each enzyme is a heterotetramer, with gyrase composed of 2 GyrA and 2 GyrB subunits and topoisomerase IV composed of 2 ParC and 2 ParE subunits. GyrA is homologous to ParC, and GyrB to ParE.6 Both enzymes act by catalyzing a DNA double-strand break, passing another DNA strand through the break, and resealing the break.7 The enzymes’ DNA strand-passing domains are localized in GyrA and ParC, and the enzymes’ ATPase activity, which drives the catalytic cycle, is localized in domains of GyrB and ParE. Quinolones block the resealing of the DNA double-strand break and in so doing inhibit enzyme activity as well as stabilize catalytic intermediate covalent complexes of enzyme and DNA that serve as a barrier to movement of the DNA replication fork and can be converted to double-strand DNA breaks, which correlate with quinolone bactericidal activity.8–10
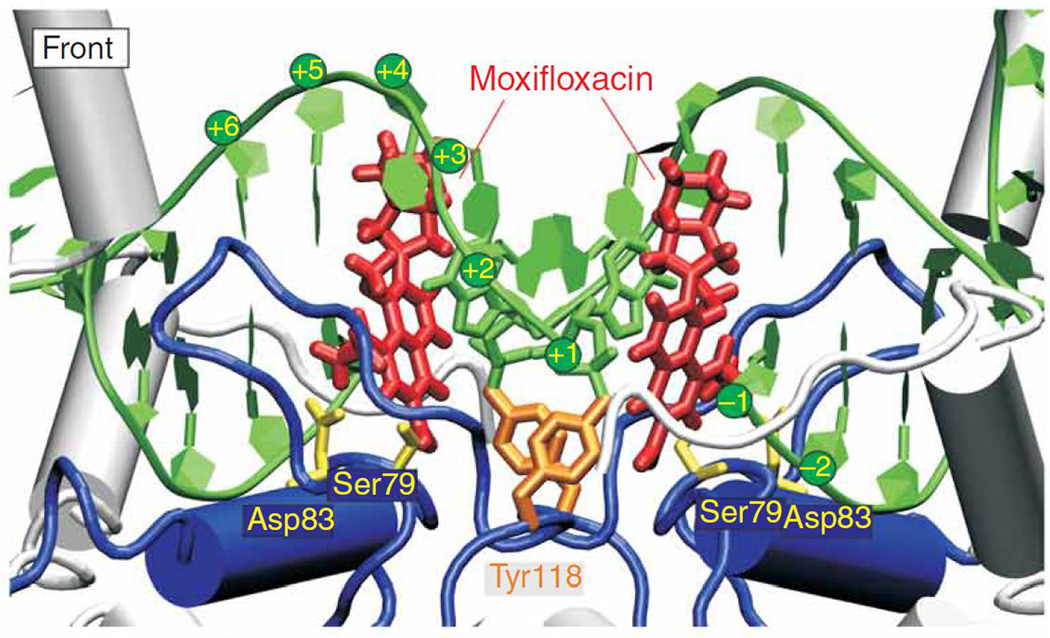
Single amino acid changes in either gyrase or topoisomerase IV can cause quinolone resistance. These resistance mutations have most commonly been localized to the amino terminal domains of GyrA (residues 67 to 106 for Escherichia coli numbering) or ParC (residues 63 to 102) and are in proximity to the active site tyrosines (Tyr122 for GyrA, Tyr120 for ParC), which are covalently linked to DNA in an enzyme intermediate, in both enzymes.11–14 This domain has been termed the quinolone resistance determining region (QRDR) of GyrA and ParC.15 The most common site of mutation in GyrA of E. coli is at Ser83 followed by Asp87, with similar predominance of mutations at equivalent positions in other species.7, 8, 16 There is conservation of an equivalent Ser and another acidic residue separated by four amino acids for GyrA in other species as well as for ParC, and likewise it is mutation in these residues that is most often present in resistant strains.7 Ser83Trp and Ser83Leu mutations of E. coli GyrA have been associated with reduced binding of the quinolone norfloxacin and enoxacin to gyrase-DNA complexes.17–19 Competition experiments with quinazolinediones and quinolones also suggest that the equivalent Ser81Phe resistance mutation in ParC of Bacillus anthracis causes selective decrease in quinolone affinity for the enzyme-DNA complex.20 Ser mutations in GyrA appear to have little effect on the E. coli gyrase catalytic efficiency, but mutations in the adjacent Asp87 (or other equivalently positioned acidic residues in other species) decrease overall catalytic efficiency five- to tenfold.7, 21 A crystal structure of moxifloxacin with topoisomerase IV of Streptococcus pneumoniae (Fig. 1) positioned the quinolone in proximity with the Ser and nearby acidic residues but not sufficiently close to determine binding directly.13 A subsequent structure of fused ParC-ParE fragments of topoisomerase IV of Acinetobacter baumannii with moxifloxacin, however, found positioning of the quinolone with a magnesium ion coordinating direct water interactions with Ser84 and Glu88, suggesting bridged contacts between drug and these conserved amino acids, contacts that are presumably disrupted when these amino acids are mutated.7, 12
Figure 1.
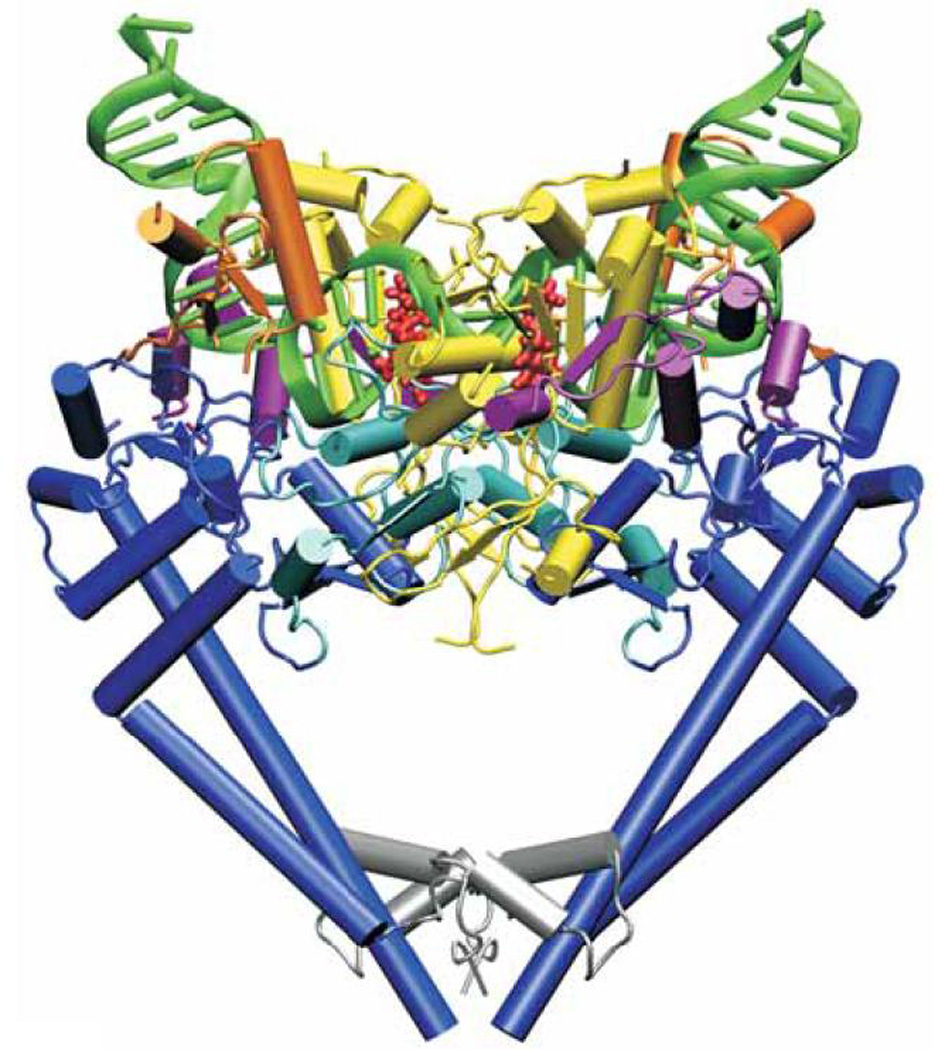
Structure of Streptococcus pneumoniae topoisomerase IV-DNA-moxifloxacin complex. From reference 13.
Mutations in specific domains of GyrB and ParE have also been shown to cause quinolone resistance,22, 23 although they are substantially less common in resistant clinical bacterial isolates than mutations in GyrA or ParC. GyrB resistance mutations have also been shown to have reduced binding of enoxacin to enzyme-DNA complexes.17 The QRDR of GyrB (or ParE) appears to be distant from the QRDR of GyrA (or ParC) based on the x-ray crystallographic structure of the homologous enzyme, topoisomerase II of yeast.24 Crystal structures of yeast topoisomerase II, however, identified other enzyme conformations in which the regions homologous to the QRDRs of GyrA and GyrB are in proximity,25 and the C7 basic substituents of ciprofloxacin and moxifloxacin were shown to be facing the GyrB subunit and could be cross-linked to GyrB Cys466.26 In addition, in the crystal structure of moxifloxacin and topoisomerase IV of A. baumannii, the quinolone C7 basic substituent is in proximity to Arg418, which is equivalent to Lys447 in E. coli.12 Notably mutations in acidic residues in this domain of GyrB in E. coli (Asp426Asn) and other species as well as in ParE have been shown to confer quinolone resistance, suggesting that drug-enzyme contacts in this region may be mediated by charge interactions.12 Thus, it appears that mutations in the QRDRs of both GyrA/ParC and GyrB/ParE act by reducing the affinity of quinolones for the enzyme-DNA complex. Although there are no direct quantitative data on quinolone binding to complexes of wild-type and mutant topoisomerase IV with DNA, the conservation of key resistance residues and the similarity of structures between gyrase and topoisomerase IV predict that resistance is also mediated by reduced drug affinity for the topoisomerase IV-DNA complex as it is for the gyrase-DNA complex.
The magnitude of resistance caused by single amino acid changes in the subunits of gyrase or topoisomerase IV varies by bacterial species and by quinolone.27, 28 The phenotype of a given resistance mutation is determined in part by the relative sensitivities of DNA gyrase and topoisomerase IV to a given quinolone. Because quinolone interaction with either target enzyme-DNA complex is sufficient to block cell growth and trigger cell death,9 the level of susceptibility of a wild-type bacterium is determined by the more sensitive of the two target enzymes. For many quinolones in clinical use, gyrase is the more sensitive enzyme in gram-negative bacteria, and topoisomerase IV is the more sensitive enzyme in gram-positive bacteria, but exceptions occur.28, 29 Target mutations occurring from first-step selection with quinolones are generally in the more sensitive target enzyme, constituting a genetic definition of the primary drug target enzyme.23, 30, 31 The magnitude of the increase in resistance from such a first-step mutation can be determined by either the magnitude of the effect of the mutation on enzyme sensitivity or the intrinsic level of sensitivity of the secondary target enzyme. Thus, the sensitivity of the secondary target can set a ceiling on the magnitude of resistance conferred by mutation in the primary target enzyme. This property implies that quinolones that have highly similar activities against both gyrase and topoisomerase IV of a given species may require mutations in both enzymes before the mutant bacterium exhibits a substantial resistance phenotype.32–34 For fluoroquinolones currently in clinical use, which generally have differences in potency between the two target enzymes, single target mutations typically result in an eight- to 16-fold increase in resistance.
Sequential mutations in both target enzymes have been shown to provide increasing levels of quinolone resistance. In many species high-level quinolone resistance is often associated with mutations in both gyrase and topoisomerase IV.35 There are also several species, Mycobacterium tuberculosis, Helicobacter pylori, and Treponema pallidum, for which genome sequencing has revealed the absence of genes for topoisomerase IV,16 indicating that for these organisms gyrase is the only quinolone target. Thus, selection of mutations with substantial resistance phenotypes is predicted to occur readily in these pathogens, an inference that is supported by clinical data indicating the frequent occurrence of resistance with clinical use of quinolones without use of other active agents to treat patients with infections with M. tuberculosis and H. pylori.36, 37
Altered drug permeation
Because gyrase and topoisomerase IV are cytoplasmic enzymes, quinolones must traverse the bacterial envelope to reach their targets, and mutations that result in reductions in cytoplasmic drug concentrations can confer resistance. This reduction is accomplished by active transport of quinolones out of the cell, reduced quinolone uptake, or a combination of the two. In Gram-positive bacteria active efflux transporters are the principal means of reducing cytoplasmic drug concentrations, and reduced diffusion across the cytoplasmic membrane has not been demonstrated as a mechanism of resistance. In contrast, in Gram-negative bacteria reduction in outer membrane porin diffusion channels, through which quinolones enter the periplasmic space, can contribute to resistance and act in concert with basal or increased expression of efflux transporters.38 Quinolones themselves in general do not induce expression of efflux pumps. Acquired quinolone resistance by altered drug permeation occurs largely by mutations in genes encoding regulatory proteins that control the transcription of efflux pump or porin genes.39 Less often mutations in efflux pump structural genes have been associated with changes in pump substrate profiles that include quinolones.40
Altered permeation in Gram-positive bacteria
In Gram-positive bacteria, quinolone resistance by increased efflux has been most extensively studied in Staphylococcus aureus.38, 41 Overexpression of each of three efflux pumps, NorA,42, 43 NorB,44 and NorC45 has been shown to cause four- to eightfold increases in resistance to quinolones, with some variations in substrate profiles among the three pumps. All three pumps are members of the major facilitator superfamily (MFS) of transporters that are secondary transporters powered by the proton gradient across the cytoplasmic membrane. NorA expression confers resistance to hydrophilic quinolones, such as norfloxacin and ciprofloxacin, whereas NorB and NorC expression each confers resistance to hydrophilic quinolones and hydrophobic quinolones, such as sparfloxacin and moxifloxacin; 43–45 these pumps also have substrate profiles extending beyond quinolones, in keeping with broad substrate profiles of many MFS transporters.
Regulation of expression of these transporters is complex and mediated by an interplay of several regulatory proteins. MgrA, has been most extensively studied, and it acts as a positive regulator of norA expression and a negative regulator of norB and norC expression.44, 46 Post-translational phosphorylation of MgrA by the PknB kinase results in the loss of the ability of MgrA dimers to bind the norA promoter and an increase in their binding to the norB promoter.47, 48 Acidic conditions alter the proportions of phosphorylated and unphosphorylated MgrA, and oxidative and aeration conditions also affect dimerization and promoter binding.49–51 Thus, relative levels of expression of NorA, NorB, and NorC are modified in response to a variety of environmental conditions. Particularly notable is the increased expression of norB in an abscess environment in response to low-free iron conditions relative to growth in laboratory media and the contribution of NorB to fitness and bacterial survival in abscesses,52 a common clinical manifestation of S. aureus infection. These findings imply that NorB, and likely NorA and NorC pumps, have natural substrates other than quinolones, which are synthetic agents. They also imply that susceptibility and response to quinolones may differ at sites of infection in vivo relative to standard clinical laboratory predictive susceptibility criteria, which are based on tests in vitro.
Other regulators such as NorG, a member of the GntR-like transcriptional regulators, can modulate pump expression and levels of quinolone resistance; it is a direct activator of norA and norB expression but a direct repressor of norC expression.53, 54 ArlRS, a two-component regulatory system, has been shown to affect expression of norA.55, 56 There are often hierarchies in regulatory networks, and other regulators can also affect expression of MgrA and NorG. Thus, there are additional complexities to the sum of various regulatory network contributors to what determines Nor pump expression under different conditions.
Other transporters in Gram-positive bacteria have also been shown to have effects on susceptibility to quinolones, but have been less extensively studied than the Nor pumps. In S. aureus overexpression of MFS transporters MdeA (norfloxacin, ciprofloxacin),57 SdrM (norfloxacin),58 QacB(III) (norfloxacin, ciprofloxacin),59 and LmrS (gatifloxacin)60 has also been shown to reduce susceptibility to quinolones. One member of the Multiple Antibiotic and Toxin Extrusion (MATE) family of secondary transporters, MepA, also confers resistance to norfloxacin, ciprofloxacin, moxifloxacin, and sparfloxacin in addition other antimicrobials and dyes.61 MepA is negatively regulated by MepR, and pentamidine, a MepA substrate, reduces MepR binding to the mepA promoter thereby increasing mepA expression. 62, 63 Thus, exposure to other agents may also affect quinolone susceptibility by upregulating broad-spectrum pumps. MFS transporters in other Gram-positive bacteria have also been shown to include quinolones in their substrate profiles. These transporters include those in the MFS group, Bmr, Bmr3, and Blt of Bacillus subtilis;64, 65 PmrA66 of Streptococcus pneumoniae; LmrP67 of Lactococcus lactis, and Lde68 of Listeria monocytogenes as well as those in the ABC transporter group, which are energized by ATP hydrolysis, PatAB69 of S. pneumoniae, SatAB70 of S. suis, and LmrA71 of L. lactis. In L. monocytogenes in addition, the FepA pump of the MATE family is overexpressed in quinolone-resistant strains and is regulated by the FepR transcriptional regulator of the TetR family, mutation in which accounted for pump overexpression and the resistance phenotype.72
Altered permeation in Gram-negative bacteria
In Gram-negative bacteria, the expression levels of a number of efflux pumps, most in the Resistance-Nodulation-Division (RND) superfamily, have been shown to confer increased quinolone resistance.73 The RND pumps have three structural components, a pump protein localized in the cytoplasmic membrane, an outer membrane channel protein, and a membrane fusion protein that links the pump and the outer membrane protein.74 Some outer membrane components may link to more than one pump-fusion protein pair.38 This structure allows for export of substrates across both inner and outer membranes that is coupled to movement of protons in the opposite direction, termed antiport exchange. Best studied has been the AcrAB-TolC pump complex of E. coli. Crystal structures of the complex have revealed a trimer of AcrB pump monomers that rotate around a central axis perpendicular to the membrane, with each monomer assuming a different conformation associated with substrate binding and extrusion through the channel as its rotation position changes.75 The drug access point is the periplasmic space between the inner and outer membranes or the outer leaflet of the inner membrane. Binding sites for ciprofloxacin and other substrates of diverse chemical types have been identified in the central cavity of the periplasmic domain of AcrB,76–78 accounting for the multidrug resistance properties of this pump. Fluoroquinolones as zwitterionic compounds are presumed to cross the outer membrane through porin diffusion channels OmpF and OmpC, and downregulation of these channels or mutation in their structural genes may also contribute as a resistance mechanism. Notably quinolone resistance mutations in the MarR regulator result in both an increase in acrB expression as well as a decrease in ompF expression.79 Thus, reduced quinolone influx through porin channels acts in concert with increased effux to generate a resistance phenotype. In addition to the Mar regulon, mutations in the E. coli SoxRS80, 81 and Rob82 regulons can also effect resistance to fluoroquinolones in part related to reductions in OmpF and in a manner that is dependent on AcrAB-TolC, similar to what occurs in mar mutants. Although initially quinolone and other antimicrobial resistances conferred by AcrAB-TolC were the phenotype most studied, this pump complex also confers resistance to bile salts and its expression is induced by bile salts,83 suggesting that one of its natural functions is to facilitate the ability of E. coli to thrive in its natural habitat, the lower gastrointestinal tract.
In Pseudomonas aeruginosa the OprF porin channel has permeability two orders of magnitude lower than that of OmpF in E. coli,84 accounting in part for its intrinsic relative resistance to quinolones and other antimicrobial agents. In addition, the MexAB-OprM efflux pump, an RND pump similar to AcrAB-TolC, is expressed in wildtype strains and acts in concert with the low permeability OprF to increase further the intrinsic level of resistance to fluoroquinolones, which is higher in P. aeruginosa than in E. coli.85 Both mexA and oprM structural gene mutants exhibit increased uptake of norfloxacin and increased susceptibility to fluoroquinolones.86 Overexpression of MexAB-OprM due to mutations in the MexR negative regulator causes increased resistance to ciprofloxacin and nalidixic acid, and mexR mutants can be selected with exposure to fluoroquinolones.87 P. aeruginosa also has three other efflux pump systems that include quinolones in their substrate profiles, MexCD-OprJ, MexEF-OprN, and MexXY-OprM.88 These pumps have limited or variable expression in wildtype strains expressing MexAB-OprM,89 but mutants overexpressing these pumps can be selected with fluoroquinolones and other antimicrobial substrates.90 Mutation in the NfxB repressor, which is encoded upstream of the mexCD-oprJ operon, results in increased expression of MexCD-OprJ and increased resistance to fluoroquinolones.91 MexEF-OprN expression varies inversely with the level of expression of MexAB-OprM, as does MexCD-OprJ expression. 89 Mutation in nfxC results in overexpression of MexEF-OprN, but details of the regulatory mechanism remain to be elucidated.92 Mutations in the global regulator MvaT, which affects quorum sensing and virulence, also causes increased expression of mexEF-oprM and resistance to norfloxacin. 93 Mutations in the MexZ repressor cause overexpression of MexXY-OprM and resistance to fluoroquinolones in addition to resistance to aminoglycosides and other pump substrates.94, 95 Notably, specific quinolones differ in which mutations they most commonly select. 90 Quinolones with a fluorine at position 6 and a positively charged substituent at position 7 (e.g., norfloxacin, ciprofloxacin, levofloxacin, moxifloxacin), which characterizes most quinolones currently in clinical use, tend to select nfxB-type mutants. In contrast, quinolones lacking a positive charge at position 7 (e.g., nalidixic acid) tend to select mexR and nfxC-type mutants, differences presumably reflecting differences in the relative efficiencies of efflux of different quinolones by the pumps overexpressed by a given mutation.
Other less extensively studied efflux pump systems that can confer quinolone resistance have been identified in many Gram-negative bacteria.38 In E. coli, EmrAB-TolC, a MFS pump that functions in tripartite structure like the RND pumps, is negatively regulated by EmrR and can confer resistance to nalidixic acid but not fluoroquinolones.96 MdfA, another MFS pump that was originally termed CmlA because of its ability to confer resistance to chloramphenicol, also confers resistance to fluoroquinolones.97 In Klebsiella pneumoniae, the OqxAB-TolC RND pump has been found on the chromosomes of most strains.98 Although originally identified on plasmids in E. coli isolated from pigs due to its ability to cause resistance to olaquindox, a growth promotant used in swine production, it also confers resistance to quinolones (see section on plasmid-mediated quinolone resistance below). Both Salmonella spp. 99 and Enterobacter aerogenes100 have AcrAB homologs the increased expression of which has been associated with quinolone resistance. The CmeABC RND pump of Campylobacter jejuni has been shown to contribute to the resistance of enrofloxacin-selected mutants.101, 102 The NorM MATE family pump can confer quinolone resistance in Vibrio parahaemolyticus.103 The NorA pump104 of Bacteroides fragilis and the BexA pump105 of B. thetaiotaomicron have also been shown to efflux fluoroquinolones.
Among non-enteric bacteria, in A. baumannii the AdeIJK RND pump106 is constitutively expressed and confers resistance to a large number of agents, including fluoroquinolones. In addition, overexpression of the AdeABC and AdeFGH RND pumps due to mutation in their respective regulators, AdeRS, a two-component sensor-regulator system, and AdeL, a LysR family regulator, can also confer a similarly broad resistance profile. Notably pump-overexpressing mutants exhibited decreased ability to form biofilms and accept plasmid DNA transfer.107, 108 In Stenotrophomonas maltophilia the SmeDEF RND pump109, 110 has been shown to contribute to resistance based on knock-out mutants with increased susceptibility and resistant strains with increased expression as well as its ability to confer quinolone resistance when overexpressed in E. coli.
In addition, there have been other examples in both Gram-positive and Gram-negative bacteria in which a relevant pump, its regulator, or a specific mutation have not been identified specifically but in which there is evidence of efflux in quinolone-resistant isolates determined by either reduction in resistance with addition of a broad efflux pump inhibitor or reduced quinolone accumulation in resistant cells.38, 75 Information on efflux mechanisms and resistance in over 50 bacterial species has recently been extensively reviewed and is beyond the scope of this review.38 Thus, efflux-mediated resistance to quinolones and many other antimicrobials is widespread, and since most efflux pumps effecting quinolone resistance have broad substrate profiles, efflux generally links quinolone resistance to multidrug resistance, as often also occurs with plasmid-mediated quinolone resistance discussed in the next section.
Plasmid-mediated quinolone resistance
Plasmid-mediated quinolone resistance was discovered inadvertently while studying β-lactam resistance produced by a multiresistance plasmid on transfer to a porin-deficient strain of K. pneumoniae. Ciprofloxacin resistance was evaluated as a control with the unexpected finding that it increased from 4 to 32 µg/ml on plasmid acquisition.111 The increase in resistance was much less marked in E. coli or K. pneumoniae with intact porins, but the plasmid was readily transferred and decreased quinolone susceptibility in strains of Citrobacter, Salmonella, and even P. aeruginosa. The responsible resistance gene was named qnr, later amended to qnrA, as additional alleles were discovered. Investigation of a qnrA plasmid from Shanghai that conferred more than the expected level of ciprofloxacin resistance resulted in the discovery of a second plasmid-mediated mechanism: modification of certain quinolones by a particular aminoglycoside acetyltransferase, AAC(6 ′)-Ib-cr.112 A third mechanism of plasmid-mediated quinolone resistance (PMQR) was added with the discovery of plasmid-mediated quinolone efflux pumps QepA 113, 114 and OqxAB.115 In the last decade PMQR genes have been found in bacterial isolates worldwide. They reduce bacterial susceptibility to quinolones, usually not to the level of clinical nonsusceptibility, but facilitate the selection of mutants with higher level quinolone resistance and promote treatment failure.
Qnr structure and function
Cloning and sequencing qnrA disclosed that it coded for a protein of 218 amino acids with a tandem repeat unit of five amino acids indicating membership in the large (more than 1000 member) pentapeptide repeat family of proteins.116 Knowledge of the sequence allowed search for qnrA by PCR, and it was soon discovered in E. coli, K. pneumoniae, and S. enterica strains from around the world.117–121 qnrA was followed by the discovery of plasmid-mediated qnrS,122 qnrB,123 qnrC,124 qnrD,125 and most recently qnrVC.126, 127 These qnr genes generally differ in sequence by 35% or more from qnrA and from each other. Allelic variants differing by 10% or less have also been described in almost every family: currently a single allele for qnrC, 2 for qnrD, 7 each for qnrA and qnrVC, 9 for qnrS, and 78 for qnrB.128
The first pentapeptide repeat protein to have its structure determined by X-ray crystallography was MfpA, encoded by the chromosome of mycobacterial species, where its deletion increases and its overexpression decreases fluoroquinolone susceptibility.129 MfpA is a dimer linked C-terminus to C-terminus and folded into a right-handed quadrilateral β helix with size, shape, and charge mimicking the B-form of DNA.130 The 5 unit repeat occupies one face of the quadrilateral with each of the 8 helical coils of the MfpA monomer thus consisting of 20 residues. The central, usually hydrophobic, amino acid (i) of the pentapeptide repeat and the first polar or hydrophobic residue (i-2) generally point inward forming the core of the molecule, while the remaining amino acids (i-1, i+1, i+2) are oriented outward, presenting an anionic surface. Hydrogen bonding between backbone atoms of neighboring coils stabilizes the helix, which is just the size to fit into the cationic G segment DNA binding saddle of DNA gyrase and topoisomerase IV.130
The three-dimensional structure of three Qnr proteins has been determined by x-ray crystallography: chromosomally-encoded EfsQnr from Enterococcus faecalis, 131 chromosomally encoded AhQnr from Aeromonas hydrophila,132 and plasmid-mediated QnrB1.133 All are rod-like dimers (Fig. 1). The monomers of QnrB1 and AhQnr have projecting loops of 8 and 12 amino acids that are important for their activity. Deletion of the smaller A loop reduces quinolone protection, while deletion of the larger B loop or both loops destroys protective activity. Deletion of even a single amino acid in the larger loop compromises protection.134 Other essential residues in QnrB are found in pentapeptide repeat positions i and i-2 where alanine substitution for the native amino acid eliminates protection as does deletion of more than 10 amino acids at the N-terminus or as few as 3 amino acids from the dimerization module at the C-terminus.134 MfpA and EfsQnr lack loops, but EfsQnr differs from MfpA in having an additional β-helical rung, a capping peptide, and a 25-amino acid flexible extension that interacts with a lengthwise grove along the β-helix and is required for full protective activity.131
Although quinolones can bind gyrase alone in some species,135 DNA enhances and increases the binding specificity to the enzyme-DNA complex.136, 137 Thus, a molecule like MfpA that mimics and competes with DNA can decrease quinolone susceptibility by reducing the number of lethal double stranded breaks that result from quinolone stabilization of the cleavage complex. It lacks a protective effect against ciprofloxacin and only inhibits DNA gyrase in vitro.130, 138 In contrast QnrA,116, 139 QnrB,123, 134, 138 QnrS,140 AhQnr,132 and EfsQnr131 have been shown to protect purified DNA gyrase from quinolone inhibition . Protection occurs at low concentrations of Qnr relative to quinolone. For DNA gyrase inhibited by 6 µM (2 µg/ml) ciprofloxacin, half protection required only 0.5 nM QnrB1, and some protective effect was seen with as little at 5 pM.123 At high Qnr concentrations (25–30 µM) gyrase inhibition is observed. 123, 138 EfsQnr is intermediate in effect. It partially protects E. coli gyrase against ciprofloxacin inhibition but also inhibits ATP-dependent supercoiling activity of gyrase with an IC50 of 1.2 µM.131 Evidently added structural features (loops, N-terminal extension) of Qnr proteins allow interactions with regions of gyrase besides the DNA binding groove132 and could allow more specific binding to and destabilization of the topoisomerase-DNA-quinolone cleavage complex.133
Qnr Origin
Qnr homologs can be found on the chromosome of many γ-Proteobacteria, Firmicutes, and Actinomycetales, including species of Bacillus, Enterococcus, Listeria, and Mycobacterium, as well as anaerobes such as Clostridium difficile and Clostridium perfringens.141–144 Nearly 50 allelic variants have been found on the chromosome of S. maltophilia.141, 145–148 Aquatic bacteria are especially well represented, including species of Aeromonas, Photobacterium, Shewanella, and Vibrio.149–151 QnrA1 is 98% identical to the chromosomally determined Qnr of Shewanella algae.151 QnrS1 is 83% identical to Qnr from Vibrio splendidus,152 and QnrC is 72% identical to chromosomal Qnr in V. orientalis or V. cholerae.124 QnrB homologs, on the other hand, are encoded by the chromosome of members of the Citrobacter freundii complex of both clinical153 and environmental154 origin. The small, nonconjugative plasmids that carry qnrD can be found in other Enterobacteriaceae but are especially likely to be found in Proteeae, such as Proteus mirabilis, Proteus vulgaris, and Providencia rettgeri 155 and may have originated there.156, 157
The worldwide distribution of qnr suggests an origin well before quinolones were discovered Indeed, qnrB genes and pseudogenes have been discovered on the chromosome of C. freundii strains collected in the 1930s.158
Qnr Plasmids
PMQR genes have been found on plasmids varying in size and incompatibility specificity (Table 1), indicating that the spread of multiple plasmid types has been responsible for the dissemination of this resistance around the world. Such plasmid heterogeneity also indicates that plasmid acquisition of qnr and other quinolone resistance determinants occurred independently multiple times. qnr genes are almost invariably associated with a mobile or transposable element, especially ISCR1 and IS26 (Table 1). qnrD and qnrS2 are located within mobile insertion cassettes, elements with bracketing inverted repeats but lacking a transposase,157, 159 while qnrVC is so far the only qnr gene located in a cassette with a linked attC site.159a
Table 1.
Plasmids and mobilizing elements associated with PMQR genes
| PMQR gene | Plasmid Inc groups | Mobilizing element |
References |
|---|---|---|---|
| qnrA1 | A/C2, FII, HI2, I1, L/M, N | ISCR1 | 111, 116, 215–217 |
| qnrA3 | N | ISCR1, IS26 | 216, 218 |
| qnrA6 | A/C | ISCR1 | 219 |
| qnrB1 | FIIK, H family, L/M | Orf1005, IS26 | 123, 199, 216, 220, 221 |
| qnrB2 | FIA, FII, L/M, N | ISCR1 | 199, 216, 222–224 |
| qnrB4 | FIA, FIIAs, L/M, R | ISCR1 | 216, 225, 226 |
| qnrB6 | FIIAs | ISCR1 | 216, 227 |
| qnrB10 | UTa | ISCR1 | 199, 228 |
| qnrB19 | ColE, L/M, N | ISEcp1, IS26 | 199, 216, 223, 228, 229 |
| qnrB20 | Orf1005, IS26 | 230 | |
| qnrS1 | ColE, FI, HI1, HI2, I1, L/M, N, NT, R, UT, X1, X2 | IS2, IS26, ISEcl2 | 122, 199, 215, 223, 224, 228, 231–234 |
| qnrS2 | Q, U | micb | 216, 235 |
| qnrC | ISPmi1 | 124 | |
| qnrD1 | UT | mic | 125, 155, 157, 236 |
| qnrVC1 | attC | 126,159a | |
| qnrVC4 | ISCR1 | 237 | |
| aac(6’)-Ib-cr | ColE, FII, L/M, N, R | IS26, attC | 175, 216, 226, 238–240 |
| oqxAB | F, FII, HI2, N, X1 | IS26 | 190, 195, 240–242 |
| qepA1 | FII, HI2 | IS26, ISCR3C | 113, 228, 243, 244 |
| qepA2 | FI | ISCR3C | 216, 243 |
UT= untypable
mic = mobile insertion cassette
qnr genes are usually found in multiresistance plasmids linked to other resistance determinants. β-lactamase genes, including genes for extended spectrum β-lactamases (ESBLs), AmpC enzymes, and carbapenemases, have been conspicuously common, (reviewed in Ref. 160). qnrB alleles are also frequently found in plasmids linked to variable portions of the operons for psp (phage shock protein) and sap (peptide ABC transporter, ATP-binding protein) genes. These genes flank qnrB on the chromosome of several Citrobacter spp., and their co-acquisition with qnrB is one of the arguments for Citrobacter as the source of qnrB alleles.153
Spread of qnr plasmids
PMQR genes have been found in a variety of Enterobacteriaceae, especially E. coli and species of Enterobacter, Klebsiella, and Salmonella (reviewed in Ref. 160). They have been conspicuously rare in non-fermenters but have occasionally been reported in P. aeruginosa, other Pseudomonas spp., A. baumannii, and S. maltophilia. qnr genes are found in a variety of Gram-positive organisms but are chromosomal and not plasmid-mediated. Of the various qnr varieties, qnrB seems somewhat more common than qnrA or qnrS, which are more common than qnrD.161–163 Only a single isolate of qnrC is known.124 The earliest known qnr outside of Citrobacter spp., dates from 1988.164 Studies in the last decade suggest that qnr detection is increasing but is still usually less than 10% in unselected clinical isolates with the exception of a qnr prevalence of 39%, which was reached in an unselected sample of E. cloacae isolates at one hospital in China.165 Higher frequencies result if samples are preselected for ESBL or other resistance phenotypes.163, 165, 166
Although most prevalence studies have surveyed hospital isolates, animals have not been neglected. PMQR genes have been found in a great variety of wild and domestic animals, including samples from birds, cats, cattle, chickens, dogs, ducks, fish, geese, horses, pigs, reptiles, sheep, turkeys, and zoo animals (reviewed in Ref.160).
Regulation of qnr
Environmental conditions affect expression of qnr genes and may offer clues concerning the native function of these genes. Expression of the qnrA gene of S. algae, an organism adapted to growth at low temperature, is stimulated up to 8-fold by cold shock but not by other conditions such as DNA damage, oxidative or osmotic stress, starvation, or heat shock. 167 Expression of qnrB alleles, on the other hand, is augmented up to 9-fold by exposure to DNA damaging agents such as ciprofloxacin or mitomycin C via an upstream LexA binding site and the classical SOS system.168, 169 qnrD and the chromosomal qnr of S. marcescens are similarly regulated.170 Expression of plasmid-mediated qnrS1 or the related chromosomal qnrVS1 of V. splendidus is also stimulated by ciprofloxacin up to 30-fold, but by a mechanism independent of the SOS system. No LexA binding site is found upstream from these qnr genes, but upstream sequence is required for quinolone induction to occur.171 Some naturally occurring quinolone-like compounds such as quinine, 2-hydroxyquinoline, 4-hydroxyquinoline, or the Pseudomonas quinolone signal for quorum sensing also induce qnrS1, but not qnrVS1.172
AAC(6′)-Ib-cr
AAC(6′)-Ib-cr is a bifunctional variant of a common acetyltransferase active on such aminoglycosides as amikacin, kanamycin, and tobramycin but also able to acetylate those fluoroquinolones with an amino nitrogen on the piperazinyl ring, such as ciprofloxacin and norfloxacin.112 Compared to other AAC(6′)-Ib enzymes, the –cr variant has two unique amino acid substitutions: Trp102Arg and Asp179Tyr, both of which are required for quinolone acetylating activity. Models of enzyme action suggest that the Asp179Tyr replacement is particularly important in permitting π-stacking interactions with the quinolone ring to facilitate quinolone binding. The role of Trp102Arg is to position the Tyr face for optimal interaction173 or to hydrogen bond to keto or carboxyl groups of the quinolone to anchor it in place.174
The aac(6′)-Ib-cr gene is usually found in a cassette as part of an integron in a multiresistance plasmid, which may contain other PMQR genes. Association with ESBL CTX-M-15 is particularly common. A mobile genetic element, especially IS26, is often associated.175 aac(6′)-Ib-cr may also be chromosomal.176, 177 The gene has been found world-wide in a variety of Enterobacteriaceae and even in P. aeruginosa.178 It is more prevalent in E. coli than other Enterobacteriaceae,179–182 and is more common than qnr alleles in some samples.183 184
QepA and OqxAB
QepA is a plasmid-mediated efflux pump in the major facilitator (MFS) family that decreases susceptibility to hydrophilic fluoroquinolones, especially ciprofloxacin and norfloxacin.113, 114qepA has often been found on plasmids also encoding aminoglycoside ribosomal methylase rmtB.114, 185–187 Substantial differences in quinolone resistance produced by different qepA transconjugants suggest variability in the level of qepA expression, by mechanisms as yet to be defined. 186
OqxAB is an efflux pump in the RND family that was initially recognized on transmissible plasmids responsible for resistance to olaquindox used for growth enhancement in pigs.188, 189 It has a wide substrate specificity, including chloramphenicol, trimethoprim, and quinolones such as ciprofloxacin, norfloxacin, and nalidixic acid.115 oqxAB has been found on plasmids in clinical isolates of E. coli and K. pneumoniae and in the chromosome and on plasmids of S. enteritis flanked in both locations by IS26-like elements.190–195 In E. coli isolates from farms in China where olaquindox was in use, oqxAB was found on transmissible plasmids in 39% of isolates from animals and 30% of isolates from farm workers.192 Linkage of oqxAB with genes for CTX-M-14 and other plasmid-mediated CTX-M alleles has been noted.196 It is common (usually 75% or more) on the chromosome of K. pneumoniae isolates, where up to 20-fold variation in expression implies the presence of regulatory control.191, 194, 197–199 In K. pneumoniae overexpression of the nearby rarA gene is associated with increased oqxAB expression, while increased expression of adjacent oqxR gene down regulates OqxAB production.200, 201
Resistance produced by PMQR determinants
Table 2 shows the minimum inhibitory concentration (MIC) produced in an E. coli strain by PMQR genes. qnr genes produce about the same resistance to ciprofloxacin and levofloxacin as single mutations in gyrA, but have less effect on susceptibility to nalidixic acid. Thus, reduced susceptibility to fluoroquinolones combined with susceptibility to nalidixic acid is a clue to the presence of PMQR and potentially resistance to other agents because of their linkage to qnr.202, 203 aac(6′)-Ib-cr and qepA give lower levels of resistance, which is confined to ciprofloxacin and norfloxacin in the case of aac(6′)-Ib-cr because of its substrate specificity. All provide a decrease in susceptibility that does not reach the clinical breakpoint for even intermediate resistance, but PMQR genes are important because they facilitate the selection of higher levels of quinolone resistance.
Table 2.
Effect of different quinolone resistance mechanisms on quinolone susceptibility ofE. coli
| E. coli Strain | MIC (µg/ml) | ||
|---|---|---|---|
| Ciprofloxacin | Levofloxacin | Nalidixic acid | |
| J53 | 0.008 | 0.015 | 4 |
| J53 gyrA (S83L) | 0.25 | 0.5 | ≥256 |
| J53 pMG252 (qnrA1) | 0.25 | 0.5 | 16 |
| J53 pMG298 (qnrB1) | 0.25 | 0.5 | 16 |
| J53 pMG306 (qnrS1) | 0.25 | 0.38 | 16 |
| J53 pMG320 (aac(6’)-Ib-cr) | 0.06 | 0.015 | 4 |
| J53 pAT851 (qepA) | 0.064 | 0.032 | 4 |
| CLSI susceptibility breakpoint | ≤1.0 | ≤2.0 | ≤16 |
If E. coli J53 pMG252 is exposed to increasing concentrations of ciprofloxacin, a diminishing number survives until a concentration of more than 1 µg/ml ciprofloxacin is reached. This limiting concentration has been termed the mutant prevention concentration (MPC), and the concentration between the MIC and MPC at which mutants are selected is the mutant selection window.204 PMQR genes exert their influence by widening the mutant selection window and elevating the MPC, as shown for qnr,205, 206 aac(6′)-Ib-cr,112, 207 and oqxAB.207 Surprisingly, in qnr-harboring E. coli gyrA resistance mutants are rarely selected,208 although resistance produced by qnr and gyrA is additive.209–211 Rather higher level ciprofloxacin resistant derivatives of E. coli J53 pMG252 (qnrA1) have mutations in regulatory genes marR or soxR leading to increased expression of the AcrAB pump or mutations in rfaD or rfaE associated with defects in lipopolysaccharide biosynthesis.212
It should be noted that higher levels of quinolone resistance are seen if a plasmid or strain carries two or more genes for quinolone resistance, such as both qnr and aac(6′)-Ib-cr , and that ciprofloxacin MICs of 2 µg/ml can be reached with qnrA in E. coli overexpressing the AcrAB multi-drug efflux pump.213 A fully resistant E. coli with a ciprofloxacin MIC of 4 µg/ml has been reported with plasmid-mediated qnrS1 and oqxAB as well as overexpression of AcrAB and other efflux pumps.214
Areas for future study
Much has been learned about the mechanisms of quinolone resistance over many years, but a number of areas await further studies. Because quinolones are synthetic compounds, those efflux pumps and plasmid-encoded proteins that confer resistance, although advantageous to the bacterium in the presence of quinolone use in humans and animals, likely have functions in addition to resistance mediation in Nature. Further understanding of their natural functions, the determinants of their mobilization, and the regulation of their expression should better inform the links between bacterial physiology, adaptation to environmental conditions, and virulence with antimicrobial resistance, an understanding that will be important for future strategies for optimizing antimicrobial use.
Figure 2.
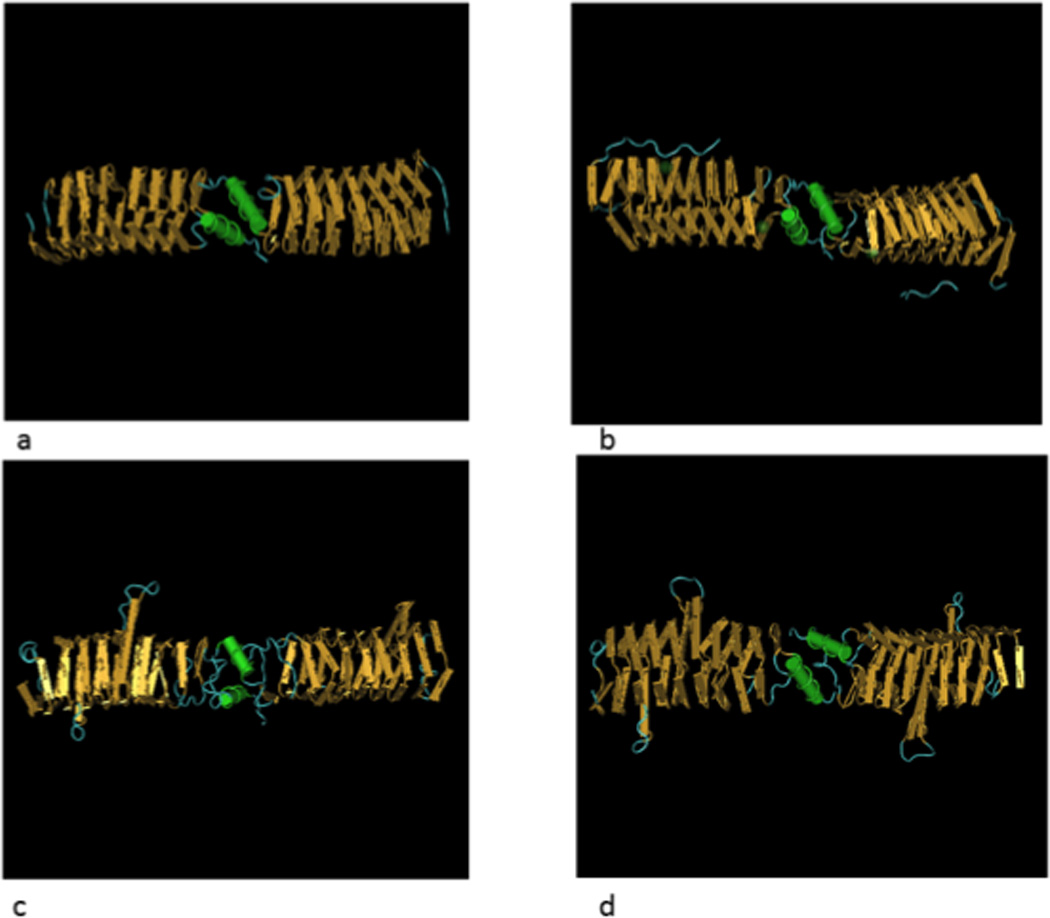
3-D representation of pentapeptide repeat proteins. (A) MfpA from M. tuberculosis (PDB ID: 2BM6), (B) EfsQnr from E. faecalis (PDB ID: 2W7Z), (C) AhQnr from A. hydrophila (PDB ID: 3PSS) and (D) plasmid-mediated QnrB1 (PDB ID: 2XTW)
ACKNOWLEDGMENTS
This work was supported by grants R01 AI057576 (to D.C.H and G.A.J.), R37 AI023988 (D.C.H.), and P01 AI083214 (D.C.H.) from the US Public Health Service, National Institutes of Health.
Reference List
- 1.Owens RC, Jr, Ambrose PG. Clinical use of the fluoroquinolones. Med. Clin. N. Amer. 2000;84:1447–1469. doi: 10.1016/s0025-7125(05)70297-2. [DOI] [PubMed] [Google Scholar]
- 2.Kim ES, Hooper DC. Clinical importance and epidemiology of quinolone resistance. Infect Chemother. 2014;46:226–238. doi: 10.3947/ic.2014.46.4.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lesher GY, Forelich ED, Gruet MD, et al. 1,8-Naphthyridine derivatives. A new class of chemotherapeutic agents. J Medicinal Pharm Chem. 1962;5:1063–1068. doi: 10.1021/jm01240a021. [DOI] [PubMed] [Google Scholar]
- 4.Domagala JM, Hagen SE. Structure-activity relationships of the quinolone antibacterials in the new millennium: some things change and some do not. In: Hooper DC, Rubinstein E, editors. Quinolone antimicrobial agents. 3rd. Washington, D.C: ASM Press; 2003. pp. 3–18. [Google Scholar]
- 5.Hooper DC. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin. Infect. Dis. 1997;27:S54–S63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 6.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 7.Aldred KJ, Kerns RJ, Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drlica K, Hiasa H, Kerns R, et al. Quinolones: action and resistance updated. Curr. Top. Med. Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drlica K, Zhao XL. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiasa H, Yousef DO, Marians KJ. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J. Biol. Chem. 1996;271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- 11.Morais Cabral JH, Jackson AP, Smith CV, et al. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 12.Wohlkonig A, Chan PF, Fosberry AP, et al. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 2010;17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 13.Laponogov I, Sohi MK, Veselkov DA, et al. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat Struct Mol Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 14.Laponogov I, Veselkov DA, Crevel IM, et al. Structure of an ‘open’ clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport. Nucleic Acids Res. 2013;41:9911–9923. doi: 10.1093/nar/gkt749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida H, Bogaki M, Nakamura M, et al. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper DC. Mechanisms of quinolone resistance. In: Hooper DC, Rubinstein E, editors. Quinolone antimicrobial agents. 3rd. Washington, D.C: ASM Press; 2003. pp. 41–67. [Google Scholar]
- 17.Yoshida H, Nakamura M, Bogaki M, et al. Mechanism of action of quinolones against Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 1993;37:839–845. doi: 10.1128/aac.37.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willmott CJ, Maxwell A. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 1993;37:126–127. doi: 10.1128/aac.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willmott CJ, Critchlow SE, Eperon IC, et al. The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J. Mol. Biol. 1994;242:351–363. doi: 10.1006/jmbi.1994.1586. [DOI] [PubMed] [Google Scholar]
- 20.Aldred KJ, McPherson SA, Wang P, et al. Drug interactions with Bacillus anthracis topoisomerase IV: biochemical basis for quinolone action and resistance. Biochemistry. 2012;51:370–381. doi: 10.1021/bi2013905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiasa H. The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry. 2002;41:11779–11785. doi: 10.1021/bi026352v. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H, Bogaki M, Nakamura M, et al. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breines DM, Ouabdesselam S, Ng EY, et al. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of parE gene encoding a subunit of topoisomerase IV. Antimicrob. Agents Chemother. 1997;41:175–179. doi: 10.1128/aac.41.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger JM, Gamblin SJ, Harrison SC, et al. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 25.Fass D, Bogden CE, Berger JM. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nature Struct. Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 26.Mustaev A, Malik M, Zhao X, et al. Fluoroquinolone-gyrase-DNA complexes: two modes of drug binding. J. Biol. Chem. 2014;289:12300–12312. doi: 10.1074/jbc.M113.529164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fournier B, Zhao X, Lu T, et al. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 2000;44:2160–2165. doi: 10.1128/aac.44.8.2160-2165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan XS, Fisher LM. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanche F, Cameron B, Bernard FX, et al. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob. Agents Chemother. 1996;40:2714–2720. doi: 10.1128/aac.40.12.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trucksis M, Wolfson JS, Hooper DC. A novel locus conferring fluoroquinolone resistance in Staphylococcus aureus. J. Bacteriol. 1991;173:5854–5860. doi: 10.1128/jb.173.18.5854-5860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng EY, Trucksis M, Hooper DC. Quinolone resistance mutations in topoisomerase IV: relationship of the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan XS, Fisher LM. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: overexpression, purification, and differential inhibition by fluoroquinolones. Antimicrob. Agents Chemother. 1999;43:1129–1136. doi: 10.1128/aac.43.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan XS, Fisher LM. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strahilevitz J, Hooper DC. Dual targeting of topoisomerase IV and gyrase to reduce mutant selection: direct testing of the paradigm by using WCK-1734, a new fluoroquinolone, and ciprofloxacin. Antimicrob. Agents Chemother. 2005;49:1949–1956. doi: 10.1128/AAC.49.5.1949-1956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz FJ, Jones ME, Hofmann B, et al. Characterization of grlA grlB gyrA and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MICAntimicrob. Agents Chemother. 1998;42:1249–1252. doi: 10.1128/aac.42.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukamura M, Nakamura E, Yoshii S, et al. Therapeutic effect of a new antibacterial substance ofloxacin (DL8280) on pulmonary tuberculosis. Am. Rev. Resp. Dis. 1985;131:352–356. doi: 10.1164/arrd.1985.131.3.352. [DOI] [PubMed] [Google Scholar]
- 37.Mégraud F. Epidemiology and mechanism of antibiotic resistance in Helicobacter pylori. Gastroenterology. 1998;115:1278–1282. doi: 10.1016/s0016-5085(98)70101-5. [DOI] [PubMed] [Google Scholar]
- 38.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blair JM, Bavro VN, Ricci V, et al. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3511–3516. doi: 10.1073/pnas.1419939112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindler BD, Frempong-Manso E, DeMarco CE, et al. Analyses of Multidrug Efflux Pump-Like Proteins Encoded on the Staphylococcus aureus Chromosome. Antimicrob Agents Chemother. 2015;59:747–748. doi: 10.1128/AAC.04678-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ubukata K, Itoh-Yamashita N, Konno M. Cloning and expression of the norA gene for fluoroquinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1989;33:1535–1539. doi: 10.1128/aac.33.9.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu JL, Grinius L, Hooper DC. NorA functions as a multidrug efflux protein in both cytoplasmic membrane vesicles and reconstituted proteoliposomes. J. Bacteriol. 2002;184:1370–1377. doi: 10.1128/JB.184.5.1370-1377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truong-Bolduc QC, Dunman PM, Strahilevitz J, et al. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 2005;187:2395–2405. doi: 10.1128/JB.187.7.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truong-Bolduc QC, Strahilevitz J, Hooper DC. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50:1104–1107. doi: 10.1128/AAC.50.3.1104-1107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingavale S, Van Wamel W, Luong TT, et al. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 2005;73:1423–1431. doi: 10.1128/IAI.73.3.1423-1431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truong-Bolduc QC, Ding Y, Hooper DC. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 2008;190:7375–7381. doi: 10.1128/JB.01068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truong-Bolduc QC, Hooper DC. Phosphorylation of MgrA and Its Effect on Expression of the NorA and NorB Efflux Pumps of Staphylococcus aureus. J. Bacteriol. 2010;192:2525–2534. doi: 10.1128/JB.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Truong-Bolduc QC, Bolduc GR, Okumura R, et al. Implication of the NorB efflux pump in the adaptation of Staphylococcus aureus to growth at acid pH and in resistance to moxifloxacin. Antimicrob. Agents Chemother. 2011;55:3214–3219. doi: 10.1128/AAC.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Truong-Bolduc QC, Liao C-H, Villet R, et al. Reduced aeration affects the expression of the NorB efflux pump of Staphylococcus aureus by posttranslational modification of MgrA. J. Bacteriol. 2012;194:1823–1834. doi: 10.1128/JB.06503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen PR, Bae T, Williams WA, et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus Nature Chemical Biology. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 52.Ding Y, Onodera Y, Lee JC, et al. NorB, an efflux pump in Staphylococcus aureus MW2, contributes to bacterial fitness in abscesses. J. Bacteriol. 2008;190:7123–7129. doi: 10.1128/JB.00655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Truong-Bolduc QC, Dunman PM, Eidem T, et al. Transcriptional profiling analysis of the global regulator NorG, a GntR-like protein of Staphylococcus aureus. J. Bacteriol. 2011;193:6207–6214. doi: 10.1128/JB.05847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Truong-Bolduc QC, Hooper DC. Transcriptional regulators NorG and MgrA modulate resistance to both quinolones and β-lactams in Staphylococcus aureus. J. Bacteriol. 2007;189:2996–3005. doi: 10.1128/JB.01819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fournier B, Klier A, Rapoport G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 2001;41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 56.Fournier B, Hooper DC. A new two-component regulatory system involved in adhesion autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 2000;182:3955–3964. doi: 10.1128/jb.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J, O’Toole PW, Shen W, et al. Novel chromosomally encoded multidrug efflux transporter MdeA in Staphylococcus aureus. Antimicrob. Agents Chemother. 2004;48:909–917. doi: 10.1128/AAC.48.3.909-917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada Y, Hideka K, Shiota S, et al. Gene cloning and characterization of SdrM, a chromosomally-encoded multidrug efflux pump, from Staphylococcus aureus. Biol Pharm. Bull. 2006;29:554–556. doi: 10.1248/bpb.29.554. [DOI] [PubMed] [Google Scholar]
- 59.Nakaminami H, Noguchi N, Sasatsu M. Fluoroquinolone efflux by the plasmid-mediated multidrug efflux pump QacB variant QacBIII in Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54:4107–4111. doi: 10.1128/AAC.01065-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Floyd JL, Smith KP, Kumar SH, et al. LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54:5406–5412. doi: 10.1128/AAC.00580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaatz GW, DeMarco CE, Seo SM. MepR, a repressor of the Staphylococcus aureus MATE family multidrug efflux pump MepA, is a substrate-responsive regulatory protein. Antimicrob. Agents Chemother. 2006;50:1276–1281. doi: 10.1128/AAC.50.4.1276-1281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindler BD, Patel D, Seo SM, et al. Mutagenesis and modeling to predict structural and functional characteristics of the Staphylococcus aureus MepA multidrug efflux pump. J. Bacteriol. 2013;195:523–533. doi: 10.1128/JB.01679-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumaraswami M, Schuman JT, Seo SM, et al. Structural and biochemical characterization of MepR, a multidrug binding transcription regulator of the Staphylococcus aureus multidrug efflux pump MepA. Nucleic Acids Res. 2009;37:1211–1224. doi: 10.1093/nar/gkn1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohki R, Murata M. bmr3 a third multidrug transporter gene of Bacillus subtilis. J. Bacteriol. 1997;179:1423–1427. doi: 10.1128/jb.179.4.1423-1427.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klyachko KA, Schuldiner S, Neyfakh AA. Mutations affecting substrate specificity of the Bacillus subtilis multidrug transporter Bmr. J. Bacteriol. 1997;179:2189–2193. doi: 10.1128/jb.179.7.2189-2193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gill MJ, Brenwald NP, Wise R. Identification of an efflux pump gene pmrA associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1999;43:187–189. doi: 10.1128/aac.43.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolhuis H, Poelarends G, Van Veen HW, et al. The lactococcal lmrP gene encodes a proton motive force-dependent drug transporter. J. Biol. Chem. 1995;270:26092–26098. doi: 10.1074/jbc.270.44.26092. [DOI] [PubMed] [Google Scholar]
- 68.Godreuil S, Galimand M, Gerbaud G, et al. Efflux pump Lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob. Agents Chemother. 2003;47:704–708. doi: 10.1128/AAC.47.2.704-708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boncoeur E, Durmort C, Bernay B, et al. PatA and PatB form a functional heterodimeric ABC multidrug efflux transporter responsible for the resistance of Streptococcus pneumoniae to fluoroquinolones. Biochemistry. 2012;51:7755–7765. doi: 10.1021/bi300762p. [DOI] [PubMed] [Google Scholar]
- 70.Escudero JA, San MA, Gutierrez B, et al. Fluoroquinolone efflux in Streptococcus suis is mediated by SatAB and not by SmrA. Antimicrob. Agents Chemother. 2011;55:5850–5860. doi: 10.1128/AAC.00498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Veen HW, Margolles A, Müller M, et al. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J. 2000;19:2503–2514. doi: 10.1093/emboj/19.11.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guerin F, Galimand M, Tuambilangana F, et al. Overexpression of the novel MATE fluoroquinolone efflux pump FepA in Listeria monocytogenes is driven by inactivation of its local repressor FepR. PLoS. ONE. 2014;9:e106340. doi: 10.1371/journal.pone.0106340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015;28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Du D, Wang Z, James NR, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu EW, Aires JR, Nikaido H. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 2003;185:5657–5664. doi: 10.1128/JB.185.19.5657-5664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu EW, Aires JR, McDermott G, et al. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 79.Alekshun MN, Levy SB. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 1999;7:410–413. doi: 10.1016/s0966-842x(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 80.Chou JH, Greenberg JT, Demple B. Postranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J. Bacteriol. 1998;175:1026–1031. doi: 10.1128/jb.175.4.1026-1031.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller PF, Gambino L, Sulavik MC, et al. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 1994;38:1773–1779. doi: 10.1128/aac.38.8.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jair KW, Yu X, Skarstad K, et al. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rosenberg EY, Bertenthal D, Nilles ML, et al. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 2003;48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 84.Nikaido H, Nikaido K, Harayama S. Identification and characterization of porins in Pseudomonas aeruginosa. J. Biol. Chem. 1991;266:770–779. [PubMed] [Google Scholar]
- 85.Li XZ, Zhang L, Poole K. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000;45:433–436. doi: 10.1093/jac/45.4.433. [DOI] [PubMed] [Google Scholar]
- 86.Poole K, Tetro K, Zhao QX, et al. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Poole K, Krebes K, McNally C, et al. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Masuda N, Sakagawa E, Ohya S, et al. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000;44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li XZ, Barré N, Poole K. Influence of the MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and MexE-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000;46:885–893. doi: 10.1093/jac/46.6.885. [DOI] [PubMed] [Google Scholar]
- 90.Köhler T, Michea-Hamzehpour M, Plesiat P, et al. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Poole K, Gotoh N, Tsujimoto H, et al. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB- type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 92.Köhler T, Michea-Hamzehpour M, Henze U, et al. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 93.Westfall LW, Carty NL, Layland N, et al. mvaT mutation modifies the expression of the Pseudomonas aeruginosa multidrug efflux operon mexEF-oprN. FEMS Microbiol. Lett. 2006;255:247–254. doi: 10.1111/j.1574-6968.2005.00075.x. [DOI] [PubMed] [Google Scholar]
- 94.Hay T, Fraud S, Lau CH, et al. Antibiotic inducibility of the mexXY multidrug efflux operon of Pseudomonas aeruginosa: involvement of the MexZ anti-repressor ArmZ. PLoS. ONE. 2013;8:e56858. doi: 10.1371/journal.pone.0056858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matsuo Y, Eda S, Gotoh N, et al. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol. Lett. 2004;238:23–28. doi: 10.1016/j.femsle.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 96.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang S, Clayton SR, Zechiedrich EL. Relative contributions of the AcrAB, MdfA and NorE efflux pumps to quinolone resistance in Escherichia coli. J. Antimicrob. Chemother. 2003;51:545–556. doi: 10.1093/jac/dkg126. [DOI] [PubMed] [Google Scholar]
- 98.Kim HB, Wang M, Park CH, et al. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baucheron S, Imberechts H, Chaslus-Dancla E, et al. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar typhimurium phage type DT204. Microb. Drug Resist. 2002;8:281–289. doi: 10.1089/10766290260469543. [DOI] [PubMed] [Google Scholar]
- 100.Pradel E, Pagès JM. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob. Agents Chemother. 2002;46:2640–2643. doi: 10.1128/AAC.46.8.2640-2643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luo N, Sahin O, Lin J, et al. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 2003;47:390–394. doi: 10.1128/AAC.47.1.390-394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin J, Michel LO, Zhang QJ. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 2002;46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morita Y, Kataoka A, Shiota S, et al. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J. Bacteriol. 2000;182:6694–6697. doi: 10.1128/jb.182.23.6694-6697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miyamae S, Nikaido H, Tanaka Y, et al. Active efflux of norfloxacin by Bacteroides fragilis. Antimicrob. Agents Chemother. 1998;42:2119–2121. doi: 10.1128/aac.42.8.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miyamae S, Ueda O, Yoshimura F, et al. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 2001;45:3341–3346. doi: 10.1128/AAC.45.12.3341-3346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fernando DM, Xu W, Loewen PC, et al. Triclosan can select for an AdeIJK-overexpressing mutant of Acinetobacter baumannii ATCC 17978 that displays reduced susceptibility to multiple antibiotics. Antimicrob. Agents Chemother. 2014;58:6424–6431. doi: 10.1128/AAC.03074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yoon EJ, Courvalin P, Grillot-Courvalin C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 2013;57:2989–2995. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yoon EJ, Chabane YN, Goussard S, et al. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio. 2015;6:e00309–e00315. doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang L, Li XZ, Poole K. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2001;45:3497–3503. doi: 10.1128/AAC.45.12.3497-3503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alonso A, Martinez JL. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2000;44:3079–3086. doi: 10.1128/aac.44.11.3079-3086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 112.Robicsek A, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006;12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 113.Yamane K, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 2007;51:3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Périchon B, Courvalin P, Galimand M. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 2007;51:2464–2469. doi: 10.1128/AAC.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hansen LH, et al. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother. 2007;60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- 116.Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. U S A. 2002;99:5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang M, et al. Plasmid-mediated quinolone resistance in clinical isolates of. Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 2003;47:2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cheung TK, et al. Plasmid-mediated resistance to ciprofloxacin and cefotaxime in clinical isolates of Salmonella enterica serotype Enteritidis in Hong Kong. J. Antimicrob. Chemother. 2005;56:586–589. doi: 10.1093/jac/dki250. [DOI] [PubMed] [Google Scholar]
- 119.Jeong JY, et al. Detection of qnr in clinical isolates of Escherichia coli from Korea. Antimicrob. Agents Chemother. 2005;49:2522–2524. doi: 10.1128/AAC.49.6.2522-2524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jonas D, et al. Plasmid-mediated quinolone resistance in isolates obtained in German intensive care units. Antimicrob. Agents Chemother. 2005;49:773–775. doi: 10.1128/AAC.49.2.773-775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mammeri H, et al. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 2005;49:71–76. doi: 10.1128/AAC.49.1.71-76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hata M, et al. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 2005;49:801–803. doi: 10.1128/AAC.49.2.801-803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jacoby GA, et al. qnrB another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 2006;50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang M, et al. New plasmid-mediated quinolone resistance gene qnrC found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 2009;53:1892–1897. doi: 10.1128/AAC.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cavaco LM, et al. qnrD a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 2009;53:603–608. doi: 10.1128/AAC.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fonseca EL, Vicente AC. Epidemiology of qnrVC alleles and emergence out of the Vibrionaceae family. J. Med. Microbiol. 2013;62:1628–1630. doi: 10.1099/jmm.0.062661-0. [DOI] [PubMed] [Google Scholar]
- 127.Pons MJ, Gomes C, Ruiz J. QnrVC, a new transferable Qnr-like family. Enfermedades infecciosas y microbiologia clinica. 2013;31:191–192. doi: 10.1016/j.eimc.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 128.Jacoby G, et al. qnr gene nomenclature. Antimicrob. Agents Chemother. 2008;52:2297–2299. doi: 10.1128/AAC.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Montero C, et al. Intrinsic resistance of Mycobacterium smegmatis to fluoroquinolones may be influenced by new pentapeptide protein MfpA. Antimicrob. Agents Chemother. 2001;45:3387–3392. doi: 10.1128/AAC.45.12.3387-3392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hegde SS, et al. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 131.Hegde SS, et al. Structural and biochemical analysis of the pentapeptide repeat protein EfsQnr, a potent DNA gyrase inhibitor. Antimicrob. Agents Chemother. 2011;55:110–117. doi: 10.1128/AAC.01158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xiong X, et al. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a gram-negative bacterium. Nucleic Acids Res. 2011;39:3917–3927. doi: 10.1093/nar/gkq1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vetting MW, et al. Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J. Biol. Chem. 2011;286:25265–25273. doi: 10.1074/jbc.M111.226936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jacoby GA, et al. Mutational analysis of quinolone resistance protein QnrB1. Antimicrob. Agents Chemother. 2013;57:5733–5736. doi: 10.1128/AAC.01533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kumar R, Madhumathi BS, Nagaraja V. Molecular basis for the differential quinolone susceptibility of mycobacterial DNA gyrase. Antimicrob. Agents Chemother. 2014;58:2013–2020. doi: 10.1128/AAC.01958-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Willmott CJ, Maxwell A. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 1993;37:126–127. doi: 10.1128/aac.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shen LL, et al. Mechanism of quinolone inhibition of DNA gyrase. Appearance of unique norfloxacin binding sites in enzyme-DNA complexes. J. Biol. Chem. 1989;264:2973–2978. [PubMed] [Google Scholar]
- 138.Mérens A, et al. The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex. J. Bacteriol. 2009;191:1587–1594. doi: 10.1128/JB.01205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 2005;49:118–125. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tavio MM, Jacoby GA, Hooper DC. QnrS1 structure-activity relationships. J. Antimicrob. Chemother. 2014;69:2102–2109. doi: 10.1093/jac/dku102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sánchez MB, et al. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol. 2008;8:148–161. doi: 10.1186/1471-2180-8-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jacoby GA, Hooper DC. Phylogenetic analysis of chromosomally determined Qnr and related proteins. Antimicrob. Agents Chemother. 2013;57:1930–1934. doi: 10.1128/AAC.02080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arsène S, Leclercq R. Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob. Agents Chemother. 2007;51:3254–3258. doi: 10.1128/AAC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rodríguez-Martínez JM, et al. Qnr-like pentapeptide repeat proteins in gram-positive bacteria. J. Antimicrob. Chemother. 2008;61:1240–1243. doi: 10.1093/jac/dkn115. [DOI] [PubMed] [Google Scholar]
- 145.Shimizu K, et al. Smqnr, a new chromosome-carried quinolone resistance gene in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2008;52:3823–3825. doi: 10.1128/AAC.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sánchez MB, Martínez JL. SmQnr contributes to intrinsic resistance to quinolones in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2010;54:580–581. doi: 10.1128/AAC.00496-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gordon NC, Wareham DW. Novel variants of the Smqnr family of quinolone resistance genes in clinical isolates of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2010;65:483–489. doi: 10.1093/jac/dkp476. [DOI] [PubMed] [Google Scholar]
- 148.Zhang R, et al. Detection of the Smqnr quinolone protection gene and its prevalence in clinical isolates of Stenotrophomonas maltophilia in China. J. Med. Microbiol. 2012;61:535–539. doi: 10.1099/jmm.0.037309-0. [DOI] [PubMed] [Google Scholar]
- 149.Poirel L, Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front. Microbiol. 2012;3:24. doi: 10.3389/fmicb.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Poirel L, et al. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 2005;56:1118–1121. doi: 10.1093/jac/dki371. [DOI] [PubMed] [Google Scholar]
- 151.Poirel L, et al. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 2005;49:3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cattoir V, et al. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 2007;51:2650–2651. doi: 10.1128/AAC.00070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jacoby GA, Griffin CM, Hooper DC. Citrobacter spp. as a source of qnrB alleles. Antimicrob Agents Chemother. 2011;55:4979–4984. doi: 10.1128/AAC.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang R, et al. High Prevalence of qnr and aac(6’)-Ib-cr genes in both water-borne environmental bacteria and clinical isolates of Citrobacter freundii in China. Microbes Environ. 2011 doi: 10.1264/jsme2.ME11308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang S, et al. Prevalence and plasmid characterization of the qnrD determinant in Enterobacteriaceae isolated from animals, retail meat products, and humans. Microb. Drug. Resist. 2013;19:331–335. doi: 10.1089/mdr.2012.0146. [DOI] [PubMed] [Google Scholar]
- 156.Guillard T, et al. Description of a 2,683-base-pair plasmid containing qnrD in two Providencia rettgeri isolates. Antimicrob. Agents Chemother. 2012;56:565–568. doi: 10.1128/AAC.00081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guillard T, et al. Mobile insertion cassette elements found in small non-transmissible plasmids in Proteeae may explain qnrD mobilization. PLoS One. 2014;9:e87801. doi: 10.1371/journal.pone.0087801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Saga T, et al. Characterization of qnrB-like genes in Citrobacter species of the American Type Culture Collection. Antimicrob. Agents Chemothe.r. 2013;57:2863–2866. doi: 10.1128/AAC.02396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Picão RC, et al. Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J. Antimicrob. Chemother. 2008;62:948–950. doi: 10.1093/jac/dkn341. [DOI] [PubMed] [Google Scholar]
- 159a.Fonseca EL, et al. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg. Infect. Dis. 2008;14:1129–1131. doi: 10.3201/eid1407.080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol. Spectrum. 2014;2 doi: 10.1128/microbiolspec.PLAS-0006-2013. pPLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jeong HS, et al. Prevalence of plasmid-mediated quinolone resistance and its association with extended-spectrum beta-lactamase and AmpC beta-lactamase in Enterobacteriaceae. Korean J. Lab. Med. 2011;31:257–264. doi: 10.3343/kjlm.2011.31.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Silva-Sanchez J, et al. Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-lactamase-producing Enterobacteriaceae isolates in Mexico. Microb. Drug Resist. 2011;17:497–505. doi: 10.1089/mdr.2011.0086. [DOI] [PubMed] [Google Scholar]
- 163.Jlili Nel H, et al. Trend of plasmid-mediated quinolone resistance genes at the Children’s Hospital in Tunisia. J. Med. Microbiol. 2014;63:195–202. doi: 10.1099/jmm.0.062216-0. [DOI] [PubMed] [Google Scholar]
- 164.Jacoby GA, et al. Temporal appearance of plasmid-mediated quinolone resistance genes. Antimicrob. Agents Chemother. 2009;53:1665–1666. doi: 10.1128/AAC.01447-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao X, et al. Decreased quinolone susceptibility in high percentage of Enterobacter cloacae clinical isolates caused only by Qnr determinants. Diagn. Microbiol. Infect. Dis. 2010;67:110–113. doi: 10.1016/j.diagmicrobio.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 166.Robicsek A, et al. Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob. Agents Chemother. 2005;49:3001–3003. doi: 10.1128/AAC.49.7.3001-3003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim HB, et al. Cold shock induces qnrA expression in Shewanella algae. Antimicrob. Agents Chemother. 2011;55:414–416. doi: 10.1128/AAC.00991-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang M, et al. SOS regulation of qnrB expression. Antimicrob. Agents Chemother. 2009;53:821–823. doi: 10.1128/AAC.00132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Da Re S, et al. The SOS response promotes qnrB quinolone-resistance determinant expression. EMBO Rep. 2009;10:929–933. doi: 10.1038/embor.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Briales A, et al. Exposure to diverse antimicrobials induces the expression of qnrB1, qnrD and smaqnr genes by SOS-dependent regulation. J. Antimicrob Chemother. 2012;67:2854–2859. doi: 10.1093/jac/dks326. [DOI] [PubMed] [Google Scholar]
- 171.Okumura R, et al. Quinolone Induction of qnrVS1 in Vibrio splendidus and plasmid-carried qnrS1 in Escherichia coli, a mechanism independent of the SOS system. Antimicrob. Agents Chemother. 2011;55:5942–5945. doi: 10.1128/AAC.05142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kwak YG, Jacoby GA, Hooper DC. Induction of plasmid-carried qnrS1 in Escherichia coli by naturally occurring quinolones and quorum-sensing signal molecules. Antimicrob. Agents Chemother. 2013;57:4031–4034. doi: 10.1128/AAC.00337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vetting MW, et al. Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6’)-Ib and its bifunctional fluoroquinolone-active AAC(6’)-Ib-cr variant. Biochemistry. 2008;47:9825–9835. doi: 10.1021/bi800664x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Maurice F, et al. Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance. EMBO Rep. 2008;9:344–349. doi: 10.1038/embor.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ruiz E, et al. New genetic environments of aac(6’)-Ib-cr gene in a multiresistant Klebsiella oxytoca strain causing an outbreak in a pediatric intensive care unit. Diagn. Microbiol. Infect. Dis. 2011;69:236–238. doi: 10.1016/j.diagmicrobio.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 176.Ruiz E, et al. qnr, aac(6’)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp: genetic environments and plasmid and chromosomal location. J. Antimicrob. Chemother. 2012;67:886–897. doi: 10.1093/jac/dkr548. [DOI] [PubMed] [Google Scholar]
- 177.Musumeci R, et al. Prevalence of plasmid-mediated quinolone resistance genes in uropathogenic Escherichia coli isolated in a teaching hospital of northern Italy. Microb. Drug Resist. 2012;18:33–41. doi: 10.1089/mdr.2010.0146. [DOI] [PubMed] [Google Scholar]
- 178.Ogbolu DO, et al. High levels of multidrug resistance in clinical isolates of Gram-negative pathogens from Nigeria. Int. J. Antimicrob. Agents. 2011;37:62–66. doi: 10.1016/j.ijantimicag.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 179.Park CH, et al. Prevalence in the United States of aac(6’)Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 2006;50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pitout JD, et al. Surveillance for plasmid-mediated quinolone resistance determinants in Enterobacteriaceae within the Calgary Health Region. Canada: the emergence of aac(6’)-Ib-cr. J. Antimicrob. Chemother. 2008;61:999–1002. doi: 10.1093/jac/dkn068. [DOI] [PubMed] [Google Scholar]
- 181.Kim ES, et al. Prevalence of aac(6’)-Ib-cr encoding a ciprofloxacin-modifying enzyme among Enterobacteriaceae blood isolates in Korea. Antimicrob. Agents Chemother. 2009;53:2643–2645. doi: 10.1128/AAC.01534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kim HB, et al. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob. Agents Chemother. 2009;53:639–645. doi: 10.1128/AAC.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Guillard T, et al. aac(6’)-IB-cr is the major plasmid-mediated quinolone resistance determinant in extended-spectrum β-lactamase-producing. Escherichia coli in eastern France. J. Glob. Antimicrob. Resist. 2014;2:111–113. doi: 10.1016/j.jgar.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 184.Piekarska K, et al. Co-existence of plasmid-mediated quinolone resistance determinants and mutations in gyrA and parC among fluoroquinolone-resistant clinical Enterobacteriaceae isolated in a tertiary hospital in Warsaw, Poland. Int. J. Antimicrob. Agents. 2015;45:238–243. doi: 10.1016/j.ijantimicag.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 185.Yamane K, et al. Plasmid-mediated qepA gene among. Escherichia coli clinical isolates from Japan. Antimicrob. Agents Chemother. 2008;52:1564–1566. doi: 10.1128/AAC.01137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu JH, et al. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6’)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob. Agents Chemother. 2008;52:2992–2993. doi: 10.1128/AAC.01686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Périchon B, et al. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob. Agents Chemother. 2008;52:2581–2592. doi: 10.1128/AAC.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sorensen AH, et al. Conjugative plasmid conferring resistance to olaquindox. Antimicrob. Agents Chemother. 2003;47:798–799. doi: 10.1128/AAC.47.2.798-799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hansen LH, et al. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob. Agents Chemother. 2004;48:3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Norman A, et al. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid. 2008;60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 191.Kim HB, et al. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhao J, et al. Prevalence and dissemination of oqxAB in Escherichia coli Isolates from animals, farmworkers, and the environment. Antimicrob. Agents Chemother. 2010;54:4219–4224. doi: 10.1128/AAC.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wong MH, Chen S. First detection of oqxAB in Salmonella spp. isolated from food. Antimicrob. Agents Chemother. 2013;57:658–660. doi: 10.1128/AAC.01144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rodríguez-Martínez JM, et al. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2013;68:68–73. doi: 10.1093/jac/dks377. [DOI] [PubMed] [Google Scholar]
- 195.Li L, et al. Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J. Antimicrob. Chemother. 2013;68:2263–2268. doi: 10.1093/jac/dkt209. [DOI] [PubMed] [Google Scholar]
- 196.Liu BT, et al. Dissemination and characterization of plasmids carrying oqxAB-bla CTX-M genes in Escherichia coli isolates from food-producing animals. PLoS One. 2013;8:e73947. doi: 10.1371/journal.pone.0073947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yuan J, et al. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and. Escherichia coli clinical isolates. J. Antimicrob. Chemother. 2012;67:1655–1659. doi: 10.1093/jac/dks086. [DOI] [PubMed] [Google Scholar]
- 198.Perez F, et al. OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob. Agents Chemother. 2013;57:4602–4603. doi: 10.1128/AAC.00725-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Andres P, et al. Differential distribution of plasmid-mediated quinolone resistance genes in clinical enterobacteria with unusual phenotypes of quinolone susceptibility from Argentina. Antimicrob. Agents Chemother. 2013;57:2467–2675. doi: 10.1128/AAC.01615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Veleba M, et al. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob. Agents Chemothe.r. 2012;56:4450–4458. doi: 10.1128/AAC.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.De Majumdar S, et al. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2013;57:1603–1609. doi: 10.1128/AAC.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hopkins KL, Day M, Threlfall EJ. Plasmid-mediated quinolone resistance in Salmonella enterica United Kingdom. Emerg. Infect. Dis. 2008;14:340–342. doi: 10.3201/eid1402.070573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Campos MJ, et al. Prevalence of quinolone resistance determinants in non-typhoida. Salmonella isolates from human origin in Extremadura, Spain. Diagn. Microbiol. Infect. Dis. 2014;79:64–69. doi: 10.1016/j.diagmicrobio.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 204.Drlica K. The mutant selection window and antimicrobial resistance. J. Antimicrob Chemother. 2003;52:11–17. doi: 10.1093/jac/dkg269. [DOI] [PubMed] [Google Scholar]
- 205.Jacoby GA. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005;2(41 Suppl):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 206.Rodríguez-Martínez JM, et al. Mutant prevention concentrations of fluoroquinolones for. Enterobacteriaceae expressing the plasmid-carried quinolone resistance determinant qnrA1. Antimicrob. Agents Chemother. 2007;51:2236–2239. doi: 10.1128/AAC.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wong MH, et al. PMQR genes oqxAB and aac(6’)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front. Microbiol. 2014;5:521. doi: 10.3389/fmicb.2014.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cesaro A, et al. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J. Antimicrob. Chemother. 2008;61:1007–1015. doi: 10.1093/jac/dkn077. [DOI] [PubMed] [Google Scholar]
- 209.Martínez-Martínez L, et al. Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 2003;51:1037–1039. doi: 10.1093/jac/dkg157. [DOI] [PubMed] [Google Scholar]
- 210.Briales A, et al. In vitro effect of qnrA1, qnrB1, and qnrS1 genes on fluoroquinolone activity against isogenic Escherichia coli isolates with mutations in gyrA and parC. Antimicrob. Agents Chemother. 2011;55:1266–1269. doi: 10.1128/AAC.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Luo Y, et al. Joint effects of topoisomerase alterations and plasmid-mediated quinolone-resistant determinants in. Salmonella enterica. Typhimurium.. Microb. Drug Resist. 2011;17:1–5. doi: 10.1089/mdr.2010.0074. [DOI] [PubMed] [Google Scholar]
- 212.Vinué L, et al. Genetic analysis of enhanced quinolone resistance without gyrase mutations in Escherichia coli J53 pMG252 (qnrA1) In Abst. 54th Intersci. Conf. Antimicrob. Agents Chemother. 2014 abstr C-1429. [Google Scholar]
- 213.Jeong JY, et al. Effects of a plasmid-encoded qnrA1 determinant in Escherichia coli strains carrying chromosomal mutations in the acrAB efflux pump genes. Diagn. Microbiol. Infect. Dis. 2008;60:105–107. doi: 10.1016/j.diagmicrobio.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 214.Sato T, et al. Fluoroquinolone resistance mechanisms in an Escherichia coli isolate, HUE1, without quinolone resistance-determining region mutations. Front. Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00125. Doi: 10.3389/fmicb.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Poirel L, et al. Expanded-spectrum β-lactamase and plasmid-mediated quinolone resistance. Emerg. Infect. Dis. 2007;13:803–805. doi: 10.3201/eid1305.061293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Carattoli A. Resistance plasmid families in Enterobacteriaceae Antimicrob. Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Garza-Ramos U, et al. Transfer of quinolone resistance gene qnrA1 to Escherichia coli through a 50 kb conjugative plasmid resulting from the splitting of a 300 kb plasmid. J. Antimicrob. Chemother. 2012;67:1627–1634. doi: 10.1093/jac/dks123. [DOI] [PubMed] [Google Scholar]
- 218.Lascols C, et al. A plasmid-borne. Shewanella algae gene, qnrA3, and its possible transfer in vivo between Kluyvera ascorbata and Klebsiella pneumoniae. J. Bacteriol. 2008;190:5217–5223. doi: 10.1128/JB.00243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Arpin C, et al. Evolution of an incompatibility group IncA/C plasmid harboring bla CMY-16 and qnrA6 genes and its transfer through three clones of Providencia stuartii during a two-year outbreak in a Tunisian burn unit. Antimicrob. Agents Chemother. 2012;56:1342–1349. doi: 10.1128/AAC.05267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Villa L, et al. Complete sequencing of an IncH plasmid carrying the bla NDM-1 bla CTX-M-15 and qnrB1 genes. J. Antimicrob. Chemothe.r. 2012;67:1645–1650. doi: 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 221.Dolejska M, et al. Plasmid content of a clinically relevant Klebsiella pneumoniae clone from the Czech Republic producing CTX-M-15 and QnrB1. Antimicrob. Agents Chemothe.r. 2013;57:1073–1076. doi: 10.1128/AAC.01886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Fortini D, et al. Novel genetic environment of plasmid-mediated quinolone resistance gene qnrB2 in Salmonella Bredeney from poultry. J. Antimicrob. Chemother. 2009;64:1332–1334. doi: 10.1093/jac/dkp356. [DOI] [PubMed] [Google Scholar]
- 223.García-Fernández A, et al. Characterization of plasmids harbouring. qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 2009;63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 224.Jones-Dias D, et al. Assessing the molecular basis of transferable quinolone resistance in Escherichia coli and Salmonella spp. from food-producing animals and food products. Veterinary microbiology. 2013;167:523–531. doi: 10.1016/j.vetmic.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 225.Lee CH, et al. Spread of ISCR1 elements containing bla DHA-1 and multiple antimicrobial resistance genes leading to increase of flomoxef resistance in extended-spectrum-β-lactamase-producing. Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2011;55:4058–4063. doi: 10.1128/AAC.00259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Compain F, et al. Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2014;58:4207–4210. doi: 10.1128/AAC.02773-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ma J, et al. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6’)-Ib-cr and. qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob. Agents Chemother. 2009;53:519–524. doi: 10.1128/AAC.00886-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Fortini D, et al. Plasmid-mediated quinolone resistance and β-lactamase-producing. Klebsiella pneumoniae Escherichia coli from healthy animals from Nigeria. J. Antimicrob. Chemother. 2011;66:1269–1272. doi: 10.1093/jac/dkr085. [DOI] [PubMed] [Google Scholar]
- 229.Cattoir V, et al. ISEcp1-mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob. Agents Chemother. 2008;52:2929–2932. doi: 10.1128/AAC.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Teo JW, Ng KY, Lin RT. Detection and genetic characterisation of qnrB in hospital isolates of Klebsiella pneumoniae in Singapore. Int. J. Antimicrob. Agents. 2009;33:177–180. doi: 10.1016/j.ijantimicag.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 231.Chen YT, et al. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 2006;50:3861–3866. doi: 10.1128/AAC.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hu FP, et al. Coexistence of qnrB4 and qnrS1 in a clinical strain of Klebsiella pneumoniae. Acta pharmacologica Sinica. 2008;29:320–324. doi: 10.1111/j.1745-7254.2008.00757.x. [DOI] [PubMed] [Google Scholar]
- 233.Carattoli A, et al. Complete nucleotide sequence of the IncN plasmid pKOX105 encoding VIM-1, QnrS1 and SHV-12 proteins in Enterobacteriaceae from Bolzano, Italy compared with IncN plasmids encoding KPC enzymes in the USA. J. Antimicrob. Chemother. 2010;65:2070–2075. doi: 10.1093/jac/dkq269. [DOI] [PubMed] [Google Scholar]
- 234.Dolejska M, et al. Complete sequences of IncHI1 plasmids carrying bla CTX-M-1 and qnrS1 in equine Escherichia coli provide new insights into plasmid evolution. J. Antimicrob. Chemother. 2014;69:2388–2393. doi: 10.1093/jac/dku172. [DOI] [PubMed] [Google Scholar]
- 235.Cattoir V, et al. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental. Aeromonas spp. Emerg. Infect. Dis. 2008;14:231–237. doi: 10.3201/eid1402.070677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mazzariol A, et al. Description and plasmid characterization of qnrD determinants in Proteus mirabilis and Morganella morganii. Clin. Microbiol. Infect. 2012;18:E46–E48. doi: 10.1111/j.1469-0691.2011.03728.x. [DOI] [PubMed] [Google Scholar]
- 237.Xia R, et al. qnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong Province, China. Antimicrob. Agents Chemother. 2010;54:3471–3474. doi: 10.1128/AAC.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Woodford N, et al. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 2009;53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.de Toro M, et al. pMdT1, a small ColE1-like plasmid mobilizing a new variant of the aac(6’)-Ib-cr gene in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 2013;68:1277–1280. doi: 10.1093/jac/dkt001. [DOI] [PubMed] [Google Scholar]
- 240.Jiang HX, et al. Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int. J. Antimicrob. Agents. 2014;43:242–247. doi: 10.1016/j.ijantimicag.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 241.Dotto G, et al. High prevalence of oqxAB in Escherichia coli isolates from domestic and wild lagomorphs in Italy. Microb. Drug Resist. 2014;20:118–123. doi: 10.1089/mdr.2013.0141. [DOI] [PubMed] [Google Scholar]
- 242.Liu BT, et al. Characterization of plasmids carrying oqxAB in blaCTX-M-negative Escherichia coli isolates from food-producing animals. Microb. Drug Resist. 2014;20:641–650. doi: 10.1089/mdr.2014.0022. [DOI] [PubMed] [Google Scholar]
- 243.Cattoir V, Poirel L, Nordmann P. Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob Agents Chemother. 2008;52:3801–3804. doi: 10.1128/AAC.00638-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Périchon B, et al. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob. Agents Chemother. 2008;52:2581–2592. doi: 10.1128/AAC.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]