Abstract
Active DNA demethylation in mammals involves TET-mediated oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxycytosine (5caC). However, genome-wide detection of 5fC at single-base resolution remains challenging. Here we present a bisulfite-free method for whole-genome analysis of 5fC, based on selective chemical labeling of 5fC and subsequent C-to-T transition during PCR. Base-resolution 5fC maps reveal limited overlap with 5hmC, with 5fC-marked regions more active than 5hmC-marked ones.
The ten-eleven translocation (TET)-dependent generation and removal of oxidized derivatives of 5-methylcytosine (5mC), namely 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), uncovered a new paradigm of active DNA demethylation in mammalian genomes1–3. Besides acting as demethylation intermediates, these oxidized variants of 5mC may also play functional roles4. Emerging evidence has suggested 5hmC is a stable epigenetic modification implicated in many biological processes and various diseases4,5. 5fC and 5caC, further oxidation products of 5hmC, accumulate at distal regulatory elements as active DNA demethylation intermediates6–8 and can be removed through base excision repair by mammalian thymine DNA glycosylase (TDG)3,9.
5fC is found in many cell types and all major organs2,10, yet it is present at a level of 0.02 to 0.002% of cytosines, approximately 10–100 fold lower than that of 5hmC2,10. Therefore, highly sensitive and selective methods are required to allow genome-wide detection of 5fC. We and others have developed chemical-, enzyme- or antibody-based methods for enrichment of 5fC-containing genomic DNA6–8; yet such affinity-based approaches fall short with regard to resolution. More recent base-resolution methods all rely on bisulfite treatment8,11–13, which causes substantial DNA degradation. Harsh bisulfite treatment is required for effective deamination of 5fC14, which can result in further DNA degradation. Furthermore, due to its limited abundance, only partial genome has been investigated for 5fC in wild-type mouse embryonic stem cells (mESCs)11,13. Whole-genome mapping of 5fC using bisulfite-based methods requires unusually high sequencing depth and hence is cost-prohibitive15,16.
Here we present a bisulfite-free method that detects whole-genome 5fC signals in mESCs at single-base resolution. Friedländer synthesis utilizes 2-aminobenzaldehyde and ketones to form quinoline derivatives (Fig. 1a); such intramolecular cyclization inspired us to screen for chemicals that could react in similar ways with 5fC in DNA (Supplementary Fig. 1). We successfully identified several chemicals that react readily with 5fC (Supplementary Figs. 2, 3 and Supplementary Note 1). These chemicals formed the intended cyclization products involving the exocyclic amino group of 5fC; such products are read as “C” during PCR amplification (Supplementary Fig. 4a). Interestingly, the adduct between 5fC and 1,3-indandione (5fC-I) is read as a “T” instead of a “C” during PCR (Supplementary Fig. 4b–e). One notable difference between 5fC-I and other 5fC adducts is that the original 4-amino group of 5fC is no longer a competent proton donor in 5fC-I; thus, 5fC-I may fail to form a canonical base pair with dG (Supplementary Fig. 5 and Supplementary Note 2). Although the mechanism of the C-to-T transition awaits future investigations, we envisioned that such transition could be utilized as a direct readout of 5fC and hence would provide a simple solution for bisulfite-free and base-resolution sequencing of 5fC.
Figure 1. Cyclization labeling of 5fC and fC-CET.
(a) Scheme for Friedländer Reaction. (b) AI-mediated cyclization labeling of 5fC and subsequent conjugation of biotin via click chemistry. (c) Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) Mass Spectrometry characterization of the AI-mediated labeling of 5fC in a 9-mer DNA oligo. Calculated and observed MS are shown. (d) Sanger sequencing results of the 5fC-containing 76-mer DNA. AI labeling caused C-to-T transition during PCR (indicated with the red inverted triangle). (e) Schematic diagram of fC-CET. Genomic DNA was sequentially labeled with AI, conjugated to biotin for pull-down enrichment, washed with NaOH to remove 5fC-null strands, cleaved from beads with DTT and ligated to adaptors. Such DNA was then subjected to next generation sequencing, and C-to-T transitions were specifically searched for to define 5fC sites in the whole-genome. (f) Fold enrichment of spike-in controls using qPCR. Values represent fold enrichment over the input (n = 3), normalized to the C-Ref sequence (reference with only regular Cs). 5xC mix: PCR-amplified DNA with 70% dCTP, 15% dmCTP, 10% dhmCTP and 5% dcaCTP; Single 5fC: synthetic DNA with single 5fC site; 10% 5fC: PCR-amplified DNA with 10% dfCTP (within dCTP).
To enrich 5fC-containing genomic DNA, we synthesized an azido derivative of 1,3-indandione (AI) (Supplementary Note 3). AI completely converted 5fC to the 5fC-AI adduct under very mild conditions, without causing detectable DNA degradation and hence allowing high recovery of DNA (Fig. 1b, c and Supplementary Fig. 6). The reaction was also highly selective for 5fC among all modified cytosines (Supplementary Fig. 7). We then coupled a cleavable biotin to the AI-labeled 5fC via click chemistry (Fig. 1b–d and Supplementary Fig. 8). We also screened different polymerases to minimize PCR bias and washed away DNA strands that did not contain 5fC, aiming to maximize the C-to-T signals in the sequencing reads (Supplementary Fig. 9). Such cyclization-enabled C-to-T transition of 5fC (fC-CET) was utilized from the sequencing reads to obtain genome-wide maps of 5fC at single-base resolution (Fig. 1e).
We used several spike-in DNA sequences (Supplementary Table 1) to confirm the specificity and sensitivity of fC-CET by quantitative PCR. The results proved that AI showed no cross-reactivity to C, 5mC, 5hmC or 5caC (Fig. 1f). Moreover, our chemical-assisted pull-down demonstrated efficient enrichment for 5fC-containing DNAs (Fig. 1f); even for sequences with one single 5fC, fC-CET enriched the sequence by ~100 fold, showing little density bias commonly associated with antibody-based strategies.
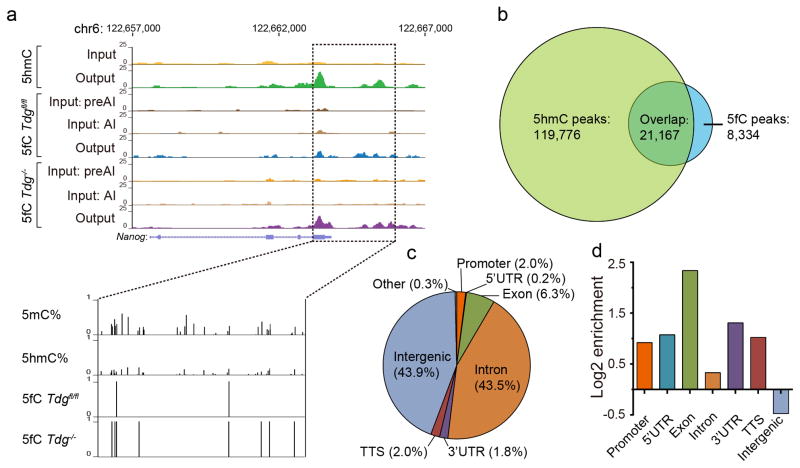
We then applied fC-CET to wild-type (Tdgfl/fl) and Tdg-depleted (Tdg−/−) mESCs, and readily identified 29,501 and 77,750 5fC-enriched regions in Tdgfl/fl and Tdg−/− mESCs, respectively (Fig. 2a, Supplementary Figs. 10 and 11a, b). The majority of the 5fC-enriched regions in Tdgfl/fl mESCs fall within those in Tdg−/− mESCs (Supplementary Fig. 11c), confirming extensive TDG-dependent active DNA demethylation. fC-CET detected 5fC-enriched regions are also in good agreement with those by fC-Seal8 (Supplementary Fig. 11d, e). Additionally, we compared 5fC-enriched regions with 5hmC-enriched regions17 and found that ~71.8% of 5fC-enriched regions overlap with those of 5hmC (Fig. 2b), consistent with our previous observations8. Taken together, we conclude that fC-CET is highly efficient in genome-wide 5fC profiling and can reliably recapitulate sequencing results from previous methods.
Figure 2. fC-CET reveals base-resolution 5fC maps in the whole-genome.
(a) Genome browser view of representative 5fC-enriched regions in the Nanog gene. The data shown here represents results from two biological replicates. Results from hmC-Seal in the same region were also plotted for comparison. (b) Venn diagram showing that fC-CET detected 5fC-marked regions largely overlap with hmC-Seal detected 5hmC regions. (c,d) Overall distribution of 5fC sites in genomic elements of wild-type mESCs (c) and their relative enrichment (d).
We next sought to detect the base-resolution map of 5fC in the whole genome of mESCs. Requiring positive hits in both replicates, we identified 32,685 and 139,027 high-confidence 5fC sites in Tdgfl/fl and Tdg−/− mESCs, respectively (Supplementary Fig. 12a). Previous base-resolution maps of 5fC were either obtained by reduced representation bisulfite sequencing from wild-type mESCs or by whole-genome bisulfite sequencing from Tdg−/− mESCs11,12. In comparison, fC-CET readily identified a comprehensive view of 5fC in the whole genome of mESCs. Using the Nanog gene as an example (Fig. 2a), fC-CET detected 5fC sites in Tdgfl/fl mESCs and additional 5fC sites in Tdg−/− mESCs, all of which were also previously identified as 5mC sites18. Moreover, a 5fC-enriched region can contain one or multiple 5fC sites (Fig. 2a and Supplementary Fig. 10b), demonstrating the sensitivity of fC-CET in detecting 5fC in both loosely- and densely-modified regions. In Tdgfl/fl mESCs, a large fraction of 5fC sites are located in intragenic regions, with particular enrichment in exons (Fig. 2c, d). A similar pattern of 5fC distribution was observed in the Tdg−/− mESCs (Supplementary Fig. 12b, c). Furthermore, we selected nine 5fC sites for loci-specific validation; five and nine 5fC sites were validated by fCAB-seq and fC-CET, respectively (Supplementary Fig. 13 and Supplementary Table 2).
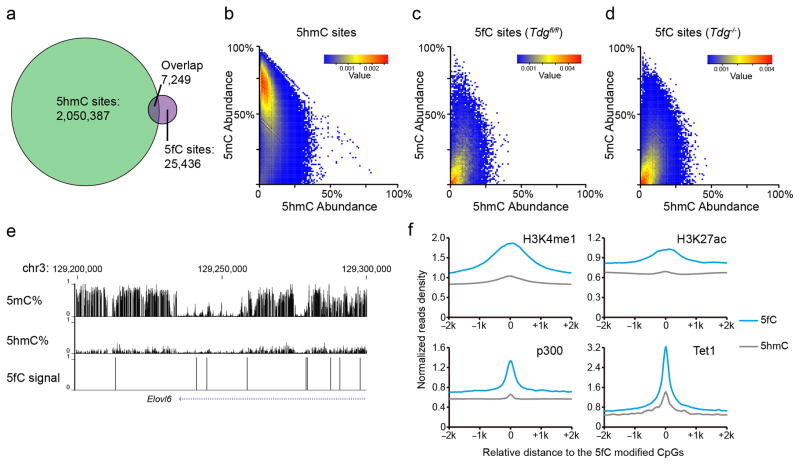
With base-resolution maps of 5fC and TAB-seq-detected 5hmC18, we next sought to investigate their spatial relationship. Although 5fC- and 5hmC-enriched regions largely overlap (Fig. 2b), 5fC and 5hmC sites share limited overlap on the single-base level: only ~22.2% 5fC sites were previously identified as 5hmC (Fig. 3a). These observations suggest that 5fC and 5hmC sites may have different steady-state features and that the degree of TET-mediated oxidation reactions may be subjected to further regulation. Given that 5fC and 5hmC may be recognized by different reader proteins19,20, the limited overlap between 5fC and 5hmC sites on the single-base level further hints their different biological roles.
Figure 3. 5fC represents a more active marker than 5hmC.
(a) Venn diagram of 5fC sites detected by fC-CET and 5hmC sites detected by TAB-seq in wild-type mESCs, showing limited overlap of 5fC and 5hmC on the single-base level. (b–d) Heatmaps of the abundance of 5hmC (horizontal) and 5mC (longitudinal) for the TAB-seq-detected 5hmC sites (b) and fC-CET-detected 5fC sites in wild-type (c) or TDG null (d) mESCs. (e) A representative view showing that 5fC-marked sites exhibit lower 5mC abundance compared to 5hmC. 5mC and 5hmC data were shown as mean value of two biological replicates; 5fC data represents results from two biological replicates. (f) Normalized read densities of 5fC (blue, fC-CET) and 5hmC (grey, hmC-Seal) at H3K4me1-, H3K27ac-, p300- and Tet1-enriched regions in wild-type mESCs.
To characterize the relationship of 5fC with 5mC and 5hmC, we calculated the abundance of 5hmC and 5mC at the TAB-seq-detected 5hmC sites (Fig. 3c) and fC-CET-detected 5fC sites (Fig. 3d), respectively. On the single-base level, 5hmC sites show high levels of 5mC (Fig. 3c). Previous profiling results have noticed a decrease of 5mC abundance in 5fC-marked regions7,8. We show herein that on the single-base level, 5fC sites are indeed very low in 5mC abundance (Fig. 3b,d) (mean value = 18.67% for Tdgfl/fl mESCs, compared to the mean value of 54.56% for 5hmC sites). Similar observations were also found in the Tdg−/− mESC (mean value = 18.78%) (Fig. 3e). The markedly lower abundance of 5mC in the 5fC-occupied sites suggests that 5fC-marked genomic regions may be more active than 5hmC-marked regions.
We calculated the ChIP-seq signals of active histone modification markers H3K4me1 and H3K27ac at the corresponding genomic regions. Enhancer marker H3K4me1 exhibits enrichment for both 5hmC and 5fC, while active enhancer marker H3K27ac is enriched for 5fC but exhibits only weak signals for 5hmC8. In fact, both H3K4me1 and H3K27ac signals are much higher for 5fC than 5hmC (Fig. 3f and Supplementary Fig. 14). Moreover, compared to 5hmC, 5fC-marked regions are more enriched for the transcriptional coactivator p300, Tet1, and DNase I hypersensitive regions, although CTCF-bound regions are similarly enriched (Fig. 3f and Supplementary Fig. 15). Taken together, our results reveal that 5fC marks distinct regulatory elements and represents a more active marker than 5hmC; the iterative oxidation of 5mC to 5hmC and 5fC also displays a gradient of reduced DNA methylation and increased enhancer activity.
In summary, we report a bisulfite-free and base-resolution sequencing method for 5fC in the whole genome of mESCs. fC-CET is achieved through selective chemical labeling of 5fC and subsequent C-to-T transition during PCR. This cyclization reaction demonstrated efficient enrichment, enabled robust C-to-T transition and exhibited minimal density bias. fC-CET has allowed detection of base-resolution 5fC maps in the whole genome of both Tdg−/− mESCs and Tdgfl/fl mESCs, the latter of which is here obtained for the first time. Such comprehensive views reveal on the single-base level that 5fC exhibits limited overlap with 5hmC, occupies genomic sites with low 5mC abundance and represents a more active marker than 5hmC. In addition, fC-CET exhibits no noticeable DNA degradation, indicating its potential in analyzing precious DNA including clinical-related samples. Furthermore, by combining selective conversion of 5mC/5hmC to 5fC with fC-CET, this bisulfite-free method could find wider applications in epigenome sequencing.
Online methods
Oligonucleotide synthesis and model DNA preparation
Oligonucleotides containing 5fC, 5mC, 5hmC or 5caC were synthesized using the ABI Expedite 8909 Nucleic Acid Synthesizer. The modified nucleotides were site-specifically incorporated at desired positions (Supplementary Table 1) with commercially available phosphoramidites (Glen Research). Subsequent deprotection and purification were carried out with Glen-Pak Cartridges (Glen Research) following the manufacturer’s instructions. Purified oligonucleotides were characterized by MALDI-TOF (< 40-mer). Regular oligonucleotides (and PCR primers) were purchased from Sangon Biotech.
Long duplex DNAs (Supplementary Table 1 and 2) were prepared through ligation of short duplexes (20–40 bp) with sticky overhangs21. In brief, the ligation-site oligonucleotides were phosphorylated with T4 polynucleotide kinase (NEB) and then annealed with the corresponding complementary strands. Annealed duplexes with sticky overhangs were mixed and ligated with T7 DNA ligase (NEB) at 16 °C for 4 h, followed by purification with native PAGE (10%).
“10% 5fC” dsDNA and “5xC mix” dsDNA used in the qPCR assay were prepared through PCR amplification as previously described18. All modified dCTPs were purchased from Trilink.
Cell lines and genomic DNA
To generate the TDG null (Tdg−/−) mESCs, the nucleus of a Tdg−/− iPS cell was transferred into an enucleated oocyte to produce a Tdg−/− embryo; the Tdg−/− mESC was then derived from inner cell mass of the Tdg−/− embryo. Wild-type (Tdgfl/fl) mESC was prepared in parallel. The genomic DNA was prepared by SDS/proteinase K digestion, followed by phenol/chloroform extraction and ethanol precipitation.
5fC cyclization labelling and click chemistry
Typically, the 5fC cyclization-labelling chemicals can be divided into two groups (Supplementary Figure 1). For 1,3-indandione (J&K) and AI (self-synthesized), the reaction was performed in a suspension of 1,3-indandione or AI in 100 mM MES buffer (pH 6.0). For diethyl malonate (J&K), methyl/ethyl acetoacetate (J&K) or ethyl 6-azido-3-oxohexanoate (self-synthesized), the reaction was performed in 100 mM NaOH methanol solution with 100 mM of the corresponding chemical. 4 μg oligonucleotide or model DNA per 100 μL reaction was used, and the reaction mixture was incubated in the thermomixer (Eppendorf, 850 rpm) in an Eppendorf tube at 37°C for 24 h. After the reaction, ethanol precipitation was used to purify the short DNAs with the help of glycogen (Invitrogen), while genomic DNA samples were purified with QIAquick PCR Purification Kit (QIAGEN). Click chemistry was performed by adding the DBCO-S-S-PEG3-Biotin (Click Chemistry Tools, Cat. No. A112-10) to a final concentration of 400 mM and incubating at 37°C for 2 h. Purification steps were performed with QIAquick PCR Purification Kit.
The precipitated DNA was again applied to Micro Bio-Spin P-6 Gel Columns (Bio-Rad) to remove any additional chemicals. Products were characterized with MALDI-TOF, and the 1,3-indandione and AI reaction products were enzymatically digested to nucleosides and further analyzed with HPLC17.
Sanger sequencing and TOPO cloning tests on model DNA
Chemically labelled model DNA were prepared as described above. Bisulfite treatment was performed with EpiTect Fast Bisulfite Conversion Kit (QIAGEN) according to the manufacturer’s instructions. PCR amplification was performed under common reaction conditions (Model-F and Model-Seq-R)18, except for the bisulfite-treated products, which were amplified (Model-BS-F and Model-Seq-R) with Hotstar Taq polymerase (QIAGEN). PCR products were purified with QIAquick PCR Purification Kit (QIAGEN) and Sanger-sequenced with unified Sequencing Primer, or used directly in TOPO cloning tests using the pEASY-T5 Zero Cloning Kit (TransGen) according to the manufacturer’s instructions. For oligonucleotides and primers, see Supplementary Table 1.
Single nucleotide primer extension assay
Templates and primer (Supplementary Table 1) were adapted from Obeid, S. et al22. Primer was labeled with [γ32P]-ATP according to the standard protocol. For each reaction, 100 nM of 32P-primer, 130 nM of template, 800 μM of one of the dNTPs and 0.5 U of Bsu DNA polymerase large fragment (NEB) were used in 10 μL 1 × NEB buffer 2 (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.9). Reaction mixtures were incubated at 37 °C for different amount of time and quenched by adding 22.5 μL stop solution (20 mM EDTA, 80% [v/v] formamide, 0.25% [w/v] xylene cyanol and 0.25% [w/v] bromophenol blue). The quenched reaction mixtures were analyzed by 12% denaturing PAGE containing 8M urea. Visualization of the gel was performed by phosphoimaging on a Typhoon FLA 7000 biomolecular imager (GE Healthcare).
AI-mediated enrichment of 5fC-containing DNA
AI-mediated labelling and purification were described above. Typically, 2 μg of model DNA or fragmented genomic DNA (~100–400 bp, with NEB dsDNA Fragmentase) was used per reaction. The Dynabeads MyOne Streptavidin C1 (Invitrogen) was used to pull-down the biotin-labelled DNA with minor modifications to the suggested immobilizing procedure for nucleic acids. Specifically, the 1× binding and washing buffer (B&W buffer, pH 7.5) was added with 0.1% Tween-20, and the canonical washing step was repeated for 5 times followed by 50 μL 1× SSC buffer (pH 7.0) washing. Then the beads were resuspended and incubated in freshly prepared 0.15 M NaOH at room temperature for 10 min. Beads with biotinylated DNA strand were then sequentially washed once with 50 μL 0.1 M NaOH, once with 50 μL of 1× B&W buffer and once with 50 μL 1× TE buffer (pH 7.5). Beads were then resuspended and incubated in freshly prepared 50 mM DTT at 37 °C for 2 h to release the 5fC-containing strand. Then the supernatant containing the desired DNA was purified with Micro Bio-Spin P-6 Gel Columns to remove DTT.
Dot blot assay
Chemical labeling of 76mer model DNA containing single 5fC or Tdg−/− mESC gDNA was performed as described above. For the dot blot assay, different amounts of model DNA or gDNA were denatured in advance with NaOH solution (final concentration of 0.15M), spotted on the Amersham Hybond-N+ membrane (GE Healthcare) and air-dried for 5 min. The membrane was UV crosslinked and then blocked with 5% nonfat milk in 1x TBST at room temperature for 2 h. The membrane was then incubated with anti-5fC antiserum (Active Motif, 61223, 1:2,500 dilution) overnight at 4 °C followed by 1× TBST washing three times. After incubation with HRP-conjugated anti-rabbit IgG secondary antibody (CW Biotech, CW0103, 1:10,00 dilution) at room temperature for 1 h and washing with 1× TBST for three times, the membrane was supplied with 1 mL SuperSignal West Chemiluminescent Substrate (Thermo Scientific) and then visualized by chemiluminescence exposure.
FspI restriction enzyme digestion
The FspI restriction site (TG5fC/GCA)-containing 70-mer oligonucleotide was prepared and chemically labelled as described above. Labelling product was PCR amplified (Model-F and Model-R, Supplementary Table 1) to introduce the C-to-T transition within the cutting site. PCR products were purified with QIAquick PCR Purification Kit and subjected to Fsp I restriction enzyme (NEB) digestion according to the manufacturer’s instructions. The digested products were analyzed with 4% agarose gel.
Pull-down specificity test with quantitative PCR
The model DNAs and primers for qPCR (Supplementary Table 2) were prepared as described above. 2 pg of each spike-in DNA was added per 1 μg of fragmented genomic DNA background. The qPCR test was run in triplicate with SYBR Premix Ex TaqTM II (TAKARA) according to the manufacturer’s instructions. Reactions were run on the ABI viiA7 Instrument. Three biological replicates were repeated to validate the results.
Library preparation and next generation sequencing of fC-CET-enriched DNA sample
The fC-CET-enriched genomic DNAs were used directly for library preparation using the TELP protocol23. A minor modification was the use of MightyAmp DNA Polymerase (TAKARA) for one round of on-bead primer extension before PCR amplification. The adaptor-ligated samples were then PCR amplified using NEBNext 2× PCR Master Mix (NEB) and indexed primers (NEB). Libraries were checked using the Agilent 2100 Bioanalyzer before loading onto the Illumina Hiseq 2500 platform. A single-end (100 bp or longer) sequencing mode was suggested for maximal data collection.
Two biological replicates of each mESCs were prepared and sequenced, which means that in parallel, two non-enriched input DNAs (Input: preAI), two AI labeled samples (Input: AI) and two pull-down output samples were sequenced simultaneously following the same procedure.
Validation of loci-specific 5fC sites
Nine 5fC sites (Supplementary Table 3) from Tdg−/− mESC genome, containing both previously identified sites (by fCAB-seq or MAB-seq)8,13 and novel sites identified by fC-CET, were chosen for validation. For fC-CET, different amounts of starting DNA material (1 μg or 100 ng) were tested. For fCAB-Seq, 1 μg input DNA was protected with 10 mM O-ethylhydroxylamine (Aldrich, 274992) in 100 μL of 100 mM MES buffer (pH 5.0) at 37 °C for 4 h, and then purified with ethanol precipitation. For MAB-Seq, 1 μg input DNA was treated by M.SssI (NEB) in 50 μL for four rounds (each round consists of a 2 h initial treatment [1.5 μL M.SssI and 1 μL SAM] and another 4 h treatment after adding 0.5 μL M.SssI and 1 μL SAM). Subsequent DNA purification was performed with phenol/chloroform/isoamyl alcohol (25:24:1) extraction followed by ethanol precipitation. Bisulfite treatment of normal, O-ethylhydroxylamine protected or M.SssI treated DNAs was performed using EpiTect Fast DNA Bisulfite Kit (QIAGEN) according to the manufacturer’s instructions, except that two thermo cycles were run. Bisulfite-treated or fC-CET-treated DNAs (genomic loci or model DNA) were PCR amplified with bisulfite primers or normal primers, respectively. The PCR-amplified samples were purified and then applied to Illumina library preparation using NEBNext Ultra DNA Library Prep Kit (NEB). The libraries were then pooled together and sequenced on Illumina MiSeq platform with singled-end reads of 150bp.
Data processing and analysis
Raw reads were first trimmed for Poly C sequence at the 3′ end. Reads shorter than 60bp were discarded. Processed reads were then mapped to the mouse genome (mm9) by bismark v0.8.324, using options -n 1 -l 40 -chunkmbs 512. The 5fC-enriched regions in each output sample were detected by using the model-based analysis of ChIP-seq (MACS) peak-calling algorithm25, with the corresponding input AI sample serving as the input control. The numbers of converted (NT) and unconverted cytosines (NC) were further extracted from each output dataset. CpGs with fewer than 10 reads or 2 converted reads were discarded. We used the binomial distribution with parameter N as (NT + NC) and r as normal cytosine conversion rate (evaluated by spike-in sequence, r = 1.87% & 1.41% for WT replicates, r = 2.01% & 1.42% for KO replicates) to calculate the probability of observing NT or greater C-to-T conversion by chance. To identify modified CpGs that were significantly enriched for 5fC, we considered CpGs with Holm-Bonferroni method-adjusted P < 0.05 in both replicate samples and located within 5fC-enriched regions to be genuine 5fC sites. Genome annotation analysis and reads visualization were performed by Homer software26. To plot the distribution of ChIP-Seq signals around 5fC sites, the sites located at repeat regions were further discarded.
External data
H3K4me1 (GSM881352) and H3K27ac (GSM881349) ChIP-Seq datasets were obtained from Xiao, S. et al27. Tet1 (GSM611192) dataset was obtained from Williams, K. et al28. P300 (GSM1019072) dataset was obtained from Song, C. X. et al8. MethylC-Seq and TAB-Seq were obtained from Hon, G. C. et al29.
Supplementary Material
Acknowledgments
The authors would like to thank R. Meng, S.T. Huang, J.Y. Liu, J.Y. Li, X.T. Shu, X.Y. Li and C.X. Zhu for technical assistance; X.X. Zhang and H.S. Guo (Peking University) for genomic DNA at the beginning of the project; C.F. Xia for synthetic suggestions; and O. Stovicek for editing the manuscript. This work was supported by the National Basic Research Foundation of China (No. 2014CB964900 to C.Y.), the National Natural Science Foundation of China (No. 31270838 and No. 21472009 to C.Y.), and US National Institutes of Health (R01 HG006827 to C.H.). C.H. is supported by the Howard Hughes Medical Institute.
Footnotes
Note: Any Supplementary Information is available in the online version of the paper.
Accession code
Sequencing data have been deposited into the Gene Expression Omnibus (GEO) under the accession number: GSE66144.
Author contributions
B.X. and C.Y. conceived the original idea, and designed the experiments with the help of X.L. and C.H.; B.X. performed the experiments with the help of X.L., H.Z., M.L., and X.J.; D.H. performed bioinformatics analysis; Z.S. and A.Z. synthesized the chemicals; Q.Y. and W.X. helped with the library preparation; C.H. and C.Y. supervised the project. B.X. and C.Y. wrote the manuscript with contributions from D.H., X.L. and C.H.
Competing financial interests
B.X., A.Z. and C.Y. are co-inventors on a filed patent (WO2015043493) for the labeling strategies and sequencing methods reported herein.
References
- 1.Tahiliani M, et al. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S, et al. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He YF, et al. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song CX, He C. Trends Biochem Sci. 2013;38:480–484. doi: 10.1016/j.tibs.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachman M, et al. Nat Chem. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raiber EA, et al. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen L, et al. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song CX, et al. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiti A, Drohat AC. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaffeneder T, et al. Angew Chem Int Ed. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 11.Booth MJ, Marsico G, Bachman M, Beraldi D, Balasubramanian S. Nat Chem. 2014;6:435–440. doi: 10.1038/nchem.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu X, et al. Cell Res. 2015 [Google Scholar]
- 13.Wu H, Wu X, Shen L, Zhang Y. Nat Biotechnol. 2014;32:1231–1240. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booth MJ, et al. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 15.Rivera CM, Ren B. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neri F, et al. Cell Rep. 2015 [Google Scholar]
- 17.Song CX, et al. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu M, et al. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iurlaro M, et al. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spruijt CG, et al. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, et al. Biochemistry. 2003;42:6747–6753. doi: 10.1021/bi034264k. [DOI] [PubMed] [Google Scholar]
- 22.Obeid S, et al. EMBO J. 2010;29:1738–1747. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng X, et al. Nucleic Acids Res. 2015;43:e35. doi: 10.1093/nar/gku818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krueger F, Andrews SR. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinz S, et al. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao S, et al. Cell. 2012;149:1381–1392. doi: 10.1016/j.cell.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams K, et al. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hon GC, et al. Mol Cell. 2014;56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.