Abstract
Dysregulation of miRNAs is a common feature in human cancers, but this phenomenon has not been studied extensively in hepatocellular carcinoma (HCC). miR-128 has been found to be downregulated in cancer. However its role in HCC remains unclear. miR-128 was underexpressed in HCC tissues and cell lines compared with their normal controls. Additionally, ITGA2 and ITGA5 were predicted as the target genes of miR-128. ITGA2 and ITGA5 were inversely correlated with the expression of miR-128 in HCC cells. Importantly, we demonstrate that the overexpression of miR-128 significantly inhibits HCC cell metastasis and stem-cell like properties via ITGA2 and ITGA5. Our results suggest the existence of a novel miR-128-ITGA pathway and indicate that miR-128 acts as a tumor suppressor during hepatocellular carcinogenesis. These results may provide a promising alternative strategy for the therapeutic treatment of HCC.
Keywords: miR-128, ITGA2, ITGA5, hepatocellular carcinoma
Introduction
HCC is one of the most common and deadly malignant primary tumors. Although new therapeutic approaches have been developed, there are still many common problems that need to be resolved, including therapy resistance to improve long-term survival for this cancer [1,2]. Recent advances in our understanding of the biological features of HCC offer opportunities for the design of a new therapeutic strategy by targeting essential signaling pathways.
MicroRNAs (miRNAs) are noncoding mRNA sequences containing around 22-nucleotides that act as important regulators of gene expression [3]. miRNAs can silence their cognate target genes by specifically binding and cleaving mRNAs or inhibiting their translation. Approximately half of all human miRNAs are located in cancer-associated genomic regions; thus, they can function as tumor-suppressor or oncogenic miRNAs, depending on their targets [4,5]. miRNAs have also been associated with HCC formation and growth. Recent advances demonstrated that a number of miRNAs have been reported to be aberrantly over-expressed or down-regulated during HCC progression [6]. miR-128 plays important roles in cancer progression and its function includes cell proliferation, apotosis, cell cycle, metastasis, DNA damage, drug resistance and so on [7-18]. However, the role of miR-128 in HCC is still unclear.
In this study, we investigated the potential roles of miR-128 in HCC. We examined the expression of miR-128 in human HCC cells and tissues. Cell growth, cell-cycle distribution, colony formation, migration, and invasion were also examined. We also investigated a potential role of miR-128 on HCC tumorigenesis in a murine model. We predicted ITGA2 and ITGA5 as potential target genes of miR-128 and this was verified in HCC cells. Our results suggest that miR-128 is a tumor suppressor which negatively regulates integrin signal pathway in progression of HCC.
Material and methods
Clinical specimens
Primary HCC biopsy specimens and normal biopsies were obtained from the patients of Southern Medical University (Guangzhou, China), who did not receive either radiotherapy or chemotherapy. Both tumor and normal tissues were histologically confirmed by H&E (hematoxylin and eosin) staining. Informed consent was obtained from each patient, and the research protocols were approved by the Ethics Committee of the hospital.
Cell culture
Human HCC cells HEL7702, HEL7404, 7721, HepG2 and PG5 were obtained primarily from the American Type Culture Collection (ATCC) were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/mL of penicillin and 100 µg/mL of streptomycin. Cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
miRNA and siRNA transfection
Lentiviral vectors mediated miR-128, ITGA2 and ITGA5 were constructed according to the protocol (Invitrogen). Lentivirus construction was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Lentivirus-mediated silencing of miR-128 was examined by qRT-PCR. ITGA2 and ITGA5 siRNAs were ordered from Sigma (need catalog number).
RNA isolation and real-time RT-PCR
Total RNA, following the manufacturer’s instructions, was isolated from the cells using Trizol reagent (Invitrogen). To measure mRNA expression, real-time RT-PCR was performed using a sequence detector (ABI-Prism, Applied Biosystems). Primers were purchased from Invitrogen (need the primer seuence). The relative expression levels were calculated by comparing Ct values of the samples with those of the reference, all data normalized to the internal control GAPDH.
MTT assay
MTT assay was employed to detect the growth of liver cancer cells and the growth curve was delineated. The cells were incubated at 37°C, 5% CO2 until cells cover the bottom of the well (a flat-bottom 96-well plate), and then the cells were cultured. 20 ul of the MTT solution was added to each well (5 mg/ml, 0.5% MTT) and the cells were continued to culture for 4 h. After the incubation, the supernatant was discarded and 150 ul dimethyl sulfoxide was added to each well, and the culture plate was shaked at low speed for 10 min until crystal dissolved completely. The ELISA reader was used to measure the absorbance at 570 nm.
Colony formation assay
Cells in logarithmic growth phase were digested in 0.5% trypsin/0.04% EDTA and single cell suspension was prepared. Then, these cells were added to 6-well plates (200 cells/well) followed by incubation at 37°C in an environment with saturated humidity and 5% CO2 for 24 h. Non-adherent cells were removed. After culture for 10-14 days, lcolonies were present. These cells were seeded into 96-well plates followed by incubation at 37°C in an environment with saturated humidity and 5% CO2. The colony formation efficiency and the morphology of colonies were observed.
Cells were harvested by trypsinization, washed in ice-cold PBS, fixed in ice-cold 80% ethanol in PBS, centrifuged at 4°C and resuspended in chilled PBS. Bovine pancreatic RNAase (Sigma-Aldrich) was added at a final concentration of 2 μg/ml, incubated at 37°C for 30 min, then 20 μg/ml propidium iodide (Sigma-Aldrich) was added and incubated for 20 min at room temperature. In each group, 50,000 cells were analyzed by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA, USA).
Western blot analysis
Cells were lysed in RIPA buffer containing 1 X protease inhibitor cocktail, and protein concentrations were determined using the Bradford assay (Bio-Rad, Philadelphia, PA). Proteins were separated by 12.5% SDS/PAGE and transferred to membranes (Millipore, Bedford, MA) at 55 V for 4 h at 4°C. After blocking in 5% nonfat dry milk in TBS, the membranes were incubated with primary antibodies at 1:1,000 dilution in TBS overnight at 4°C, washed three times with TBS-Tween 20, and then incubated with secondary antibodies conjugated with horseradish peroxidase at 1:5,000 dilution in TBS for 1 hour at room temperature. Membranes were washed again in TBS-Tween 20 for three times at room temperature. Protein bands were visualized on X-ray film using an enhanced chemiluminescence detection system.
Migration assay
For transwell migration assays, 1×105 pancreatic cancer cells were plated in the top chamber onto the noncoated membrane (24-well insert; pore size, 8 μm; Corning Costar) and allowed to migrate toward serum-containing medium in the lower chamber. Cells were fixed after 24 hours of incubation with methanol and stained with 0.1% crystal violet (2 mg/ml, Sigma-Aldrich). The number of cells invading through the membrane was counted under a light microscope (three random fields per well). Invasion assay is the same to migration assay except for the coated Matrigel on membrane.
Statistical analysis
Each experiment was performed independently at least three with similar results; one representative experiment was presented. All statistical analyses were performed using the SPSS 13.0 statistical software package (SPSS, Chicago, IL, USA). The significance of the data was determined using Student’s t test. One-way ANOVA was used to compare gene expression in different groups. All the statistical tests were two-sided, and a P value < 0.05 was considered significant.
Results
Decreased expression of miR-128 in HCC
In order to investigate the possible roles of miR-128 in HCC, we first examined the expression of miR-128 in HCC specimens by real time RT-PCR. As shown in Figure 1A, the expression levels of miR-128 in HCC samples were lower than those in normal samples. The clinical relevance of the downregulated miR-128 was associated HCC metastasis (Table 1) and miR-128 was related to HCC stage (Figure 1B). Similarly, miR-128 was lower in human HCC cells compared with normal cells (Figure 1C). These results indicated that miR-128 may play a suppressing miRNA in the development of HCC.
Figure 1.
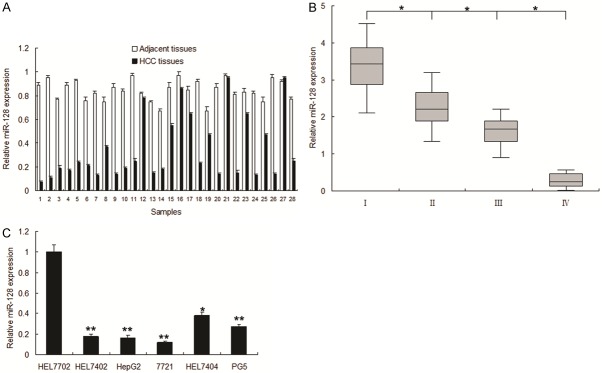
miR-128 expression decreased in HCC. A. The relative expression level of miR-128 was examined in HCC tissues (HCC) and their adjacent tissues (Adjacent tissues). miR-128 expression was examined by real time RT-PCR. B. miR-128 expression in different stages of HCC. C. The expression level of miR-128 was compared in HCC cell lines: HepG2, BEL-7402, BEL-7404, SMMC7721 and normal liver cells 7702. All experiments were repeated at least three times. Each bar represents the mean of three independent experiments. *p < 0.05. **p < 0.01.
Table 1.
Correlations between miR-128 expression and clinicopathologic characteristics of HCC patients
| Characteristics | Cases | miR-128 expression | Chi-square p-value | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| Gender | 0.091 | |||
| Male | 82 | 43 | 45 | |
| Female | 10 | 3 | 7 | |
| Age (years) | 0.308 | |||
| ≤ 60 | 51 | 25 | 26 | |
| > 60 | 41 | 19 | 22 | |
| Hepatitis history | 0.895 | |||
| Yes | 74 | 38 | 36 | |
| No | 18 | 10 | 8 | |
| Liver cirrhosis | 0.196 | |||
| Yes | 69 | 34 | 35 | |
| No | 22 | 10 | 12 | |
| Tumor size | 0.019 | |||
| ≤ 5 | 43 | 17 | 26 | |
| > 5 | 55 | 21 | 34 | |
| Clinical stage | 0.002 | |||
| I | 9 | 3 | 6 | |
| II | 33 | 15 | 18 | |
| III | 46 | 14 | 32 | |
| IV | 4 | 1 | 3 | |
| Vascular invasion | < 0.01 | |||
| Yes | 39 | 11 | 28 | |
| No | 53 | 26 | 27 | |
| AFP | 0.032 | |||
| ≤ 400 ug/L | 67 | 32 | 35 | |
| > 400 ug/L | 25 | 12 | 13 | |
miR-128 inhibits proliferation of HCC
To explore the effect of miR-128 on cell proliferation, BEL7404 (7404) and 7721 cells were infected with lentivirus vector with miR-128 (miR-128) or the miRNA control (miRNA-control) and miR-128 increased in the group of infection (Figure 2A). The results of MTT assay displayed that miR-128 inhibited cell proliferation in 7404 cells (Figure 2B) and 7721 cells (Figure 2C). The colony formation rate in the two cells with miR-128 overexpression increased compared with the controls (Figure 2D and 2E). Cell cycle is related to cell growth, to further observe miR-128 mediating growth suppression, cells were infected with miR-128 and cell-cycle distribution was examined in 7404 and 7721 cells. It was shown that G1 phase percentage in the cells with miR-128 expression got more than the control (Figure 2F and 2G). Cell proliferation and cell cycle associated regulatory proteins were detected by western blot. The data showed that PCNA, cyclinD1 decreased and p27 increased in the cells with miR-128 overexpression (Figure 2H). These results suggested that the proliferation inhibiting effect of miR-128 was partly due to a G1-phase arrest.
Figure 2.
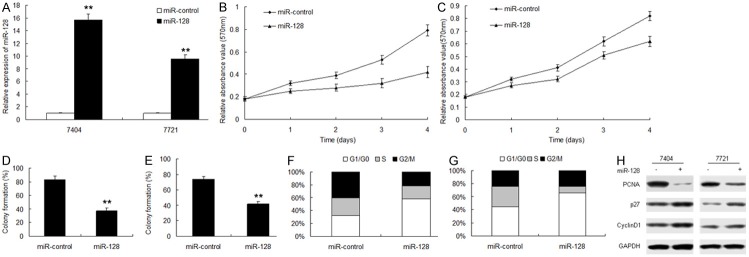
miR-128 inhibits HCC cell proliferation. A. Real-time PCR analysis of expression of miR-128 in 7404 and 7721 cells. U6 was used as a loading control. B and C. miR-128 caused cell proliferatiod inhibition in 7404 and 7721 cells by MTT assay. D and E. miR-128 caused cell proliferation inhibition in BEL7404 and 7721 cells by colony formation assay. F and G. Quantitative analysis of G0/G1 proportion in 7404 and 7721 cells. Cell cycle was examined by flow cytometry analysis. H. G0/G1 phase cell cycle related protein expression including p21, p27 and CyclinD1 were determined by western blot in 7404 and 7721 cells. The experiments were repeated three times. Data are mean ± SD. **p < 0.01.
miR-128 inhibits HCC metastasis
Above data from the clinic showed that lack of miR-128 was related to HCC metastasis. To investigate the role of miR-128 in HCC metastasis and stem cell-like properties, 7404 and 7721 cells were infected with miR-128, and the results showed that up-regulation of miR-128 could significantly suppressed cell migration (Figure 3A) and invasion (Figure 3B). Metastasis associated markers such as Vimentin and Twist1 decreased in the 7721 cells treated with miR-128 and were detected by Western blot analysis (Figure 3C).
Figure 3.
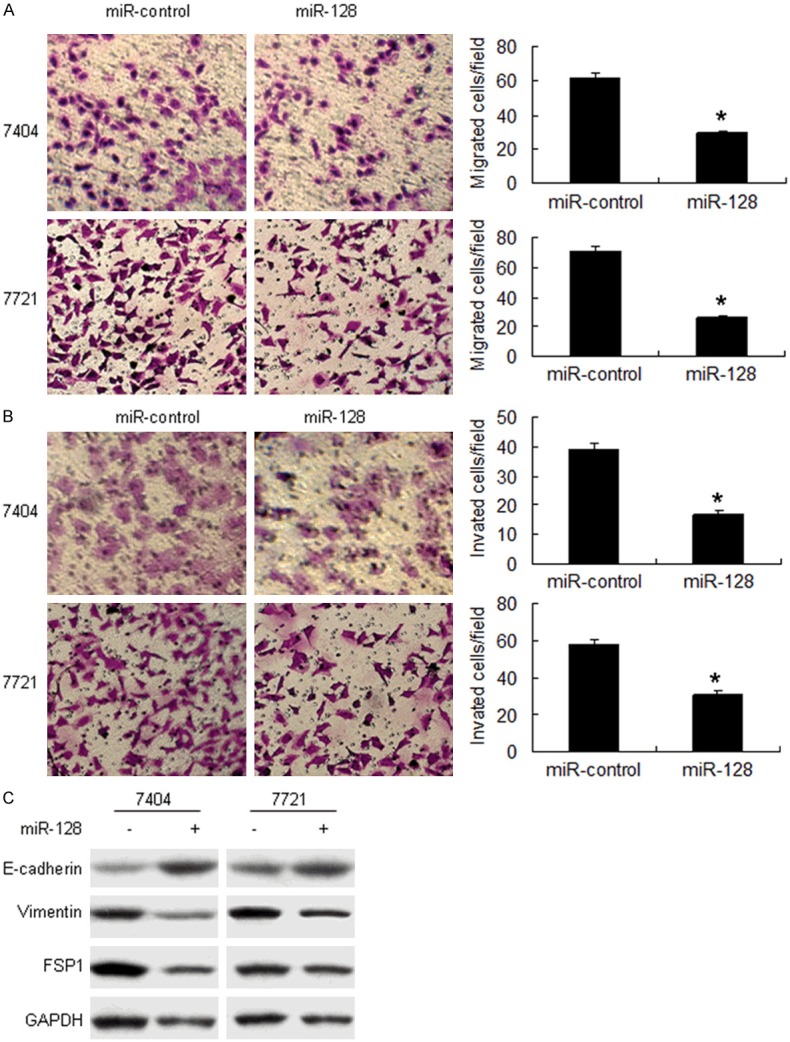
miR-128 inhibits HCC metastasis. A. miR-128 inhibits HCC cell migration. 7404 and 7721 cells were infected with miR-128 and assayed by transwell system. B. miR-128 inhibited cell invasion assayed by transwell system. C. miR-128 inhibited metastasis associated protein expression. The chambers have been coated with matrigel, which functions as the extracellular cell matrix. **p < 0.01.
ITGA2 and ITGA5 are target genes of miR-128 in HCC cells
We then investigated the mechanisms by which miR-128 inhibit HCC progression. Bioinformatic analysis showed that ITGA2 and ITGA5 were directly suppressed by miR-128 (Figure 4A). As shown in Figure 4B and 4C, the luciferase activity of wide typed ITGA2 and ITGA5 in 7721 cells was much lower than in control cells. The luciferase activity of mutated ITGA2 and ITGA5 was rescued in 7721 cells. We next examined whether miR-128 could regulate endogenous ITGA2 and ITGA5 expression in 7721 cells. Compared with control, endogenous ITGA2 and ITGA5 mRNA levels (Figure 4D and 4E) were down-regulated when 7404 and 7721 cells were transfected with miR-128. ITGA2 and ITGA5 protein increased in the cells with anti-miR-128 (Figure 4F).
Figure 4.

miR-128 downregulates ITGA2 and ITGA5 expression via directly targeting their 3’UTR. A. Sequence alignment of human ITGA2 and ITGA5 3’UTR with miR-128. B and C. miR-128 targeted the wild-type but not the mutant 3’UTR of ITGA2 and ITGA5. Luciferase activity assay of indicated 7404 and 7721 cells transfected with the pGL3-ITGA2-3’UTR or pGL3-ITGA5-3’UTR reporter with miR-128. D and E. Ectopic expression of miR-128 downregulated ITGA2 and ITGA5 mRNA expression in 7721 and 7404 cells as determined by real time RT-PCR. F. miR-128 decreased ITGA2 and ITGA5 protein level in 7721 and 7404 cells by western blot. Error bars represent from three independent experiments. **p < 0.01. *p < 0.05.
ITGA2 and ITGA5 promote HCC metastasis
To verify whether ITGA2 and ITGA5 play as oncogenes in HCC, ITGA2 and ITGA5 protein was examined in HCC tissues and cell lines. It was found that ITGA2 and ITGA5 protein was expressed higher in HCC than in their adjacent ones (Figure 5A). The result was verifed in HCC cell lines (Figure 5B). When ITGA2 and ITGA5 were knocked down, cell proliferation and invasion were suppressed in HCC cells (Figure 5C and 5D).
Figure 5.
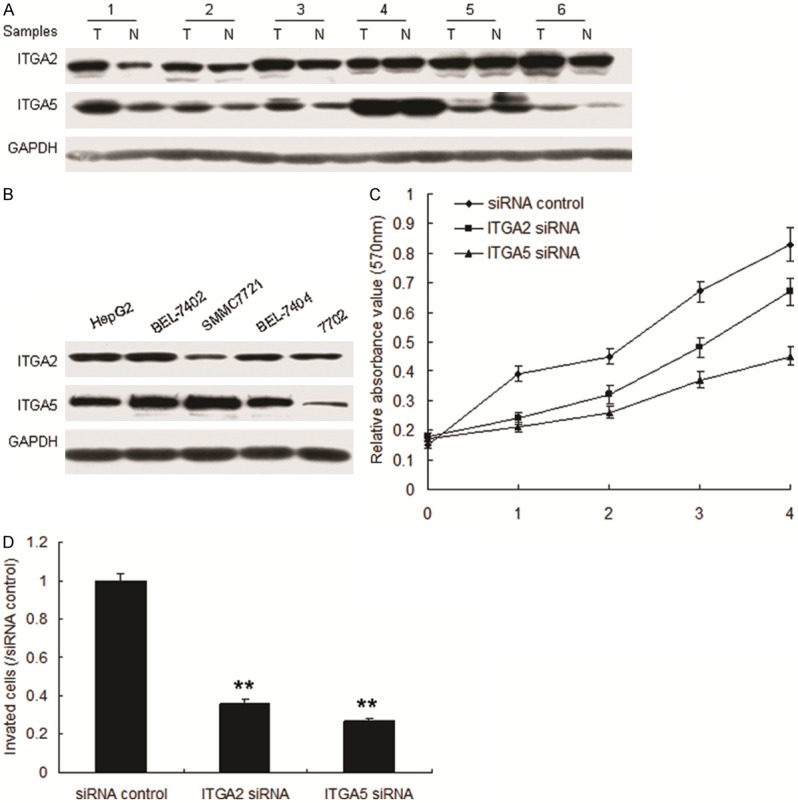
ITGA2 and ITGA5 promotes HCC metastasis. A. ITGA2 and ITGA5 was examined in HCC tissues by western blot. B. ITGA2 and ITGA5 was examined in HCC cell lines by western blot. C. Inhibition of ITGA2 or ITGA5 suppressed cell proliferation in 7404 cells. D. miR-128 inhibited cell invasion assayed by transwell system. The chambers have been coated with matrigel, which functions as the extracellular cell matrix. **p < 0.01.
miR-128 inhibits HCC by targeting ITGA2 and ITGA5
The above data suggested that miR-128 inhibits HCC by targeting integrin signal pathway in vitro. In order to investigate miR-128 mediating growth inhibition in vivo, HCC nude mice were set up using 7404-miR-128 and 7404-miR-control or ITGA2 and ITGA5, the results showed that miR-128 suppressed HCC growth via ITGA2 and ITGA5 (Figure 6A). The clinic data showed that miR-128 was negatively correlated with ITGA2 and ITGA5 protein in HCC (Figure 6B and 6C).
Figure 6.
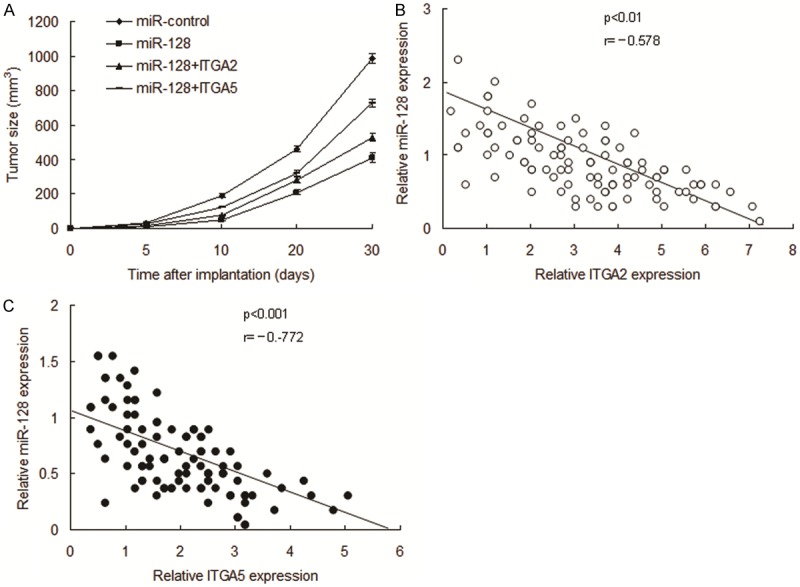
miR-128 inhibits HCC by targeting ITGA2 and ITGA5. A and B. HCC nude mice models were set up to analyze role of miR-128. B and C. miR-128 was negatively associated with ITGA2 and ITGA5 protein in HCC. The data presented are shown as means ± s.d. collected from three independent experiments.
Discussion
In the present study, we have identified miR-128 as a negative regulator of HCC cell migration, invasion and metastasis by targeting ITGA2 and ITGA5 expression. Our study is the first to explore the role of miR-128 in HCC metastasis. We have demonstrated the relevance of miR-128 in clinical malignancies in that low expression of miR-128 and high expression of ITGA2 and ITGA5 is significantly associated with advanced stages in patients with HCC. We demonstrated that miR-128 mediates inhibition of metastasis via modulating pathways directly affecting cell migration and invasion. miR-128 could be a metastasis suppressor in HCC depending on its target genes.
Recent studies showed that the differential expression of miR-128 in many type of cancers, such as lung cancer targeting VEGFC [7], glioma targeting SNAI1, SP1, EphB2 [8,13], prostate cancer targeting RPS6KB1/HIF-1α/PKM2 [11], ovarian cancer targeting CSF-1 [15], breast cancer targeting Bmi-1, ABCC5 [17]. However, a study showed that miR-128 is up-regulated in osteosarcoma and plays a role of oncogene [11]. Our data show that miR-128 is downregulated in advanced and metastatic HCC by targeting ITGA2 and ITGA5. Taken together with previous studies, it suggests that miR-128 can be multitasking by regulating different downstream effectors in different cancer contexts and it functions mainly as a metastasis suppressor in HCC.
Integrins are members of a family of cell-surface heterodimeric proteins that mediate cell-matrix and cell-cell interactions [19-22]. The 18 α-subunits and 8 β-subunits form together at least 25 different integrins, each pair being specific for a unique set of ligands. It has been demonstrated that integrins may play a crucial role in carcinogenesis, tumor behavior and metastasis [20-22]. The integrin, α2 gene (ITGA2) encodes the alpha subunit of a transmembrane receptor for collagens and related proteins. ITGA5 (integrin alpha 5) is anthor member of integrin family. We found that ITGA2 and ITGA5 were downstream targets of miR-128. Knockdown of ITGA2 or ITGA5 expression reduced cell proliferation and invasion of 7404 and 7721 cells. However, in mouse model studies, re-expression of ITGA2 or ITGA5 could significantly but not completely reverse the miR-128-imposed inhibition on tumor growth, which suggests that other potential targets of miR-128 may exist.
Our present study has identified a novel metastasis suppressor miRNA, miR-128, in HCC that can negatively regulate ITGA2 and ITGA5 in vitro and in vivo. Taken together with clinical observations, our finding suggests that miR-128 and ITGA2 and ITGA5 are significant biomarkers for metastasis and could be targets for the development of antimetastasis strategy in the treatment of HCC.
Disclosure of conflict of interest
None.
References
- 1.Yang JD, Roberts LR. Epidemiology and management of hepatocellular carcinoma. Infect Dis Clin North Am. 2010;24:899–919. doi: 10.1016/j.idc.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morise Z, Kawabe N, Tomishige H, Nagata H, Kawase J, Arakawa S, Yoshida R, Isetani M. Recent advances in the surgical treatment of hepatocellular carcinoma. World J Gastroenterol. 2014;20:14381–14392. doi: 10.3748/wjg.v20.i39.14381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Z, Zhang X, Wang G, Zheng H. Role of MicroRNAs in Hepatocellular Carcinoma. Hepat Mon. 2014;14:e18672. doi: 10.5812/hepatmon.18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callegari E, Gramantieri L, Domenicali M, D’Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22:46–57. doi: 10.1038/cdd.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scaggiante B, Kazemi M, Pozzato G, Dapas B, Farra R, Grassi M, Zanconati F, Grassi G. Novel hepatocellular carcinoma molecules with prognostic and therapeutic potentials. World J Gastroenterol. 2014;20:1268–88. doi: 10.3748/wjg.v20.i5.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma L, Chua MS, Andrisani O, So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World J Gastroenterol. 2014;20:333–45. doi: 10.3748/wjg.v20.i2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J, Cheng Y, Li Y, Jin Z, Pan Y, Liu G, Fu S, Zhang Y, Feng K, Feng Y. microRNA-128 plays a critical role in human non-small cell lung cancer tumourigenesis, angiogenesis and lymphangiogenesis by directly targeting vascular endothelial growth factor-C. Eur J Cancer. 2014;50:2336–50. doi: 10.1016/j.ejca.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Dong Q, Cai N, Tao T, Zhang R, Yan W, Li R, Zhang J, Luo H, Shi Y, Luan W, Zhang Y, You Y, Wang Y, Liu N. An axis involving SNAI1, microRNA-128 and SP1 modulates glioma progression. PLoS One. 2014;9:e98651. doi: 10.1371/journal.pone.0098651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao T, Li G, Dog Q, Liu D, Liu C, Han D, Huang Y, Chen S, Xu B, Chen M. Loss of SNAIL inhibits cellular growth and metabolism through the miR-128-mediated RPS6KB1/HIF-1α/PKM2 signaling pathway in prostate cancer cells. Tumour Biol. 2014;35:8543–50. doi: 10.1007/s13277-014-2057-z. [DOI] [PubMed] [Google Scholar]
- 10.Seca H, Lima RT, Almeida GM, Sobrinho-Simoes M, Bergantim R, Guimaraes JE, Vasconcelos MH. Effect of miR-128 in DNA damage of HL-60 acute myeloid leukemia cells. Curr Pharm Biotechnol. 2014;15:492–502. doi: 10.2174/1389201015666140519122524. [DOI] [PubMed] [Google Scholar]
- 11.Shen L, Chen XD, Zhang YH. MicroRNA-128 promotes proliferation in osteosarcoma cells by downregulating PTEN. Tumour Biol. 2014;35:2069–74. doi: 10.1007/s13277-013-1274-1. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Fu W, Wo L, Shu X, Liu F, Li C. miR-128 and its target genes in tumorigenesis and metastasis. Exp Cell Res. 2013;319:3059–64. doi: 10.1016/j.yexcr.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Lin L, Chen X, Peng X, Zhou J, Kung HF, Lin MC, Jiang S. MicroRNA-128 promotes cell-cell adhesion in U87 glioma cells via regulation of EphB2. Oncol Rep. 2013;30:1239–48. doi: 10.3892/or.2013.2596. [DOI] [PubMed] [Google Scholar]
- 14.Peruzzi P, Bronisz A, Nowicki MO, Wang Y, Ogawa D, Price R, Nakano I, Kwon CH, Hayes J, Lawler SE, Ostrowski MC, Chiocca EA, Godlewski J. MicroRNA-128 coordinately targets Polycomb Repressor Complexes in glioma stem cells. Neuro Oncol. 2013;15:1212–24. doi: 10.1093/neuonc/not055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo HH, László CF, Greco S, Chambers SK. Regulation of colony stimulating factor-1 expression and ovarian cancer cell behavior in vitro by miR-128 and miR-152. Mol Cancer. 2012;11:58. doi: 10.1186/1476-4598-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi ZM, Wang J, Yan Z, You YP, Li CY, Qian X, Yin Y, Zhao P, Wang YY, Wang XF, Li MN, Liu LZ, Liu N, Jiang BH. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One. 2012;7:e32709. doi: 10.1371/journal.pone.0032709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, Gong C, Chen J, Su F, Zhang Y, Song E. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17:7105–15. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
- 18.Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JC, Gill RM, Huillard E, Liu C, Zong H, Rowitch DH, Barres BA, Verma IM, Kosik KS. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene. 2012;31:1884–95. doi: 10.1038/onc.2011.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu BH, Wu ZZ, Qin J. Effects of integrins on laminin chemotaxis by hepatocellular carcinoma cells. Mol Biol Rep. 2010;37:1665–70. doi: 10.1007/s11033-009-9790-1. [DOI] [PubMed] [Google Scholar]
- 20.Fu BH, Wu ZZ, Qin J. Effects of integrins on laminin chemotaxis by hepatocellular carcinoma cells. Mol Biol Rep. 2010;37:1665–70. doi: 10.1007/s11033-009-9790-1. [DOI] [PubMed] [Google Scholar]
- 21.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 22.Parise LV, Lee J, Juliano RL. New aspects of integrin signaling in cancer. Semin Cancer Biol. 2000;10:407–414. doi: 10.1006/scbi.2000.0337. [DOI] [PubMed] [Google Scholar]


