Abstract
Melanoma is the leading cause of skin cancer death owing to its highly metastatic nature and resistance to chemotherapy. It may account for 80% of the deaths relating to skin cancers. Once it progressed to metastatic stage, no current effective treatment is available for melanoma. Therefore, in-depth understanding of the mechanism underlying the metastatic process is imperative and would be of great help for improving the treatment of melanoma. Here, wedemonstrate that RING finger protein 11 (RNF11) disruption by insertional mutagenesis impairs the metastatic potential of murine melanoma B16F10 cells. The requirement of RNF11 in the migration of melanoma cells is further confirmed by gene knockdown and overexpression experiments in vitro. Together, our findings suggest a novel role of RNF11 in promoting the metastasis of melanoma cells which may potentially be useful for the treatment of melanoma by developing a new intervention target.
Keywords: RNF11, melanoma, metastasis, gene knockdown, overexpression
Introduction
Melanoma is originated from malignant transformation of melanocytes which are pigment-producing cells derived from skin neural crest [1]. Though melanoma is only the third most common skin cancer, it accounts for 80% of the mortality relating to skin cancers with a 5-year survival rate less than 5% [2]. Early stage thin lesions of melanoma can be cured by surgical resection. However, it is always fatal once it developed to metastatic stage [3,4]. Even though many metastasis promotive and suppressive factors have been identified to be involved in the progression of melanoma, the molecular mechanisms underlying this process are still obscure [5-7]. Hence, further study to reveal the molecular mechanism underlying the complex metastatic process is urgently required to find new potential targets for the treatment of melanoma. In the present study, insertional mutagenesis was employed by using a specifically designed retroviral vector pDisrup 8 that can randomly disrupt genes in genome to isolate genes involved in the metastasis of melanoma cells [8,9]. As a result, RING finger protein 11 (RNF11) was found to be involved in melanoma metastasis.
RNF11 is an evolutionally conserved 154-aa-containing RING-H2 E3 ligase that was initially isolated from breast tumor cells [10,11]. It harbors two functional modules: an N-terminal PPPY motif that interacts with WW-domain containing proteins, such as AIP4/Itch, Nedd4, Smurf1/2, and Cullin, and a C-terminal RING-H2 domain that functions as a scaffold for the coordinated transfer of ubiquitin to substrate proteins [12]. RNF11 is highly expressed in several human tumor tissues, including breast, pancreatic and colon [10,11]. However, the function of RNF11 in cancer progression remains largely undetermined.
In the present study, a novel role for RNF11 was identified by insertional mutagenesis. Disruption of RNF11 expression drastically reduced the metastatic potential of murine melanoma B16F 10 cells. The role of RNF11 in the migration of melanoma cells in vitro was further conformed by gene knockdown and overexpression. More convincingly, knockdown of RNF11 expression in melanoma B16F10 cells significantly impaired their metastasis in vivo.
Materials and methods
Cell culture and reagents
Murine melanoma B16F10, B16F0 cells, and NIH 3T3 cells were purchased from American Type Culture Collection (ATCC). Cells were routinely cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal calf serum (FCS) (Invitrogen, USA), and 1% penicillin/streptomycin mix (Invitrogen, USA) and maintained at 37°C in a humidified atmosphere containing 5% CO2.
Plasmids and DNA constructs
The pDisrup 8 vector was kindly provided by Dr. Han Jiahuai (Xiamen University, China). A sequence specifically targeted to mouse RNF11 gene was selected to produce small interfering RNA (siRNA) (5’-AGG AGT TTA TGA CCC TGGA-3’). Target and scrambled control oligonucleotides duplexes were cloned into pSilencer 3.1-U6 vector (Ambion, USA) according to the manufacturer’s instructions. The cDNA of RNF11 were cloned into pXJ-40-Myc vector containing a Myc-tag with gene specific primers (forward: 5’-CG CTCGAG ATG GGG AAC TGC CTC AAA TC-3’; reverse: 5’-GA CCCGGG TCA ATT AGT CTC ATA GGA TG-3’). Transfection was carried out with Lipofectamine 2000 (Invitrogen, USA) according to the manufacture’s instruction. For pDisrup 8 clone selections, cells were selected with Blasticidin S. HCl at 25 μg/ml (Invitrogen, USA).
Western blot
Cells were washed twice with PBS (3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl and 140 mM NaCl, pH 7.4) and extracted with cold lysis buffer (20 mM Tris, 100 mM NaCl, 5 mM EDTA, 1 mM EGTA, 5 mM MgCl2, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM b-glycerolphosphate, 1 mM Na3VO4, 1 mM PMSF, and Roche complete protease inhibitors) following centrifugation at 15,000 g for 15 min at 4°C. The supernatant was collected and protein concentration was determined with Bradford assay (Biorad, USA). For western blot, samples were separated by electrophoresis on 8-16% SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Millipore, USA). After blocking with 5% skimmed milk for 1 h, membranes were incubated with different specific primary antibodies in 5% skimmed milk (anti-Myc-tag, anti-RNF11, anti-actin from Sigma). The membranes were developed with Pierce’s West Pico chemiluminescence substrate (Millipore, USA) after incubation with corresponding horseradish peroxidase-conjugated secondary antibodies.
In vitro invasion and migration assay
Invasion of melanoma cells was evaluated by BD BioCoatTM MatrigelTM Invasion Chamber (BD Biosciences, USA) assay in vitro following the manufacturer’s instructions with some modifications. In brief, a suspension of 1×105 cells in 500 μl serum-free medium was plated into the top chamber and 750 μl of NIH-3T3 fibroblast conditioned medium was added into the bottom chamber as chemo-attractant. After 24 h incubation at 37°C, the non-invasive cells in the top surface were carefully removed with a cotton swab and the invasive cells that had traversed to the bottom surface were fixed and stained with 0.5% crystal violet for 30 min. To quantify the traversed cells, cell counting was carried out by photographing 20 random fields under microscope at 20× magnification. The migration assay was performed in a similar strategy with chamber membrane without coating with matrigel.
Animals and experimental metastasis assay
Female C57BL/6J mice at 6-8 weeks old (15-20 g) were purchased from the Vital River Laboratories (China). Procedures for handling mice were approved by Ethic Committee of Xinxiang Medical University. For experimental metastasis analysis, 12 animals were randomly divided into two experimental groups. Single cell suspensions (5×105) in 100 μl PBS were injected into lateral tail vein of mice. 2 weeks after inoculation, mice were euthanized and all organs were examined for the presence of macroscopic metastases. Lung metastatic nodules were determined under a dissecting microscope.
Statistical analysis
Data were expressed in means ± SD. Statistical analysis was performed using student’s t-test or one way ANOVA for groupwise comparisons. Differences were considered statistically significant at p<0.05.
Results
RNF11 disruption impairs the metastatic potential of murine melanoma cells
To isolate genes involved in the metastasis of melanoma cells, murine melanoma B16F10 cells were transfected with a specifically designed retroviral vector pDisrup 8 that can randomly generate disruption in genome, followed by selection with Blasticidin. Then wound healing assay and 3’-RACE were performed to identify genes disrupted by pDisrup 8 vector (data not shown). As a result, RNF11 was found to be disrupted and this cell clone was designated as RNF11mut which demonstrated reduced migratory potential. To verify whether RNF11 is indeed disrupted in melanoma B16F10 cells, western blot was carried out to determine the expression of RNF11. As shown in Figure 1A, the expression of RNF11 was drastically reduced in RNF11mut cells compared to the control cells. Then transwell migration and invasion assays were performed to assess the cell migratory potential. As expected, the number of RNF11mut cells migrated across chamber membrane was much less than the wild type cells (Figure 1B and 1C), indicating that RNF11 disruption significantly impaired the metastatic potential of B16F10 cells.
Figure 1.
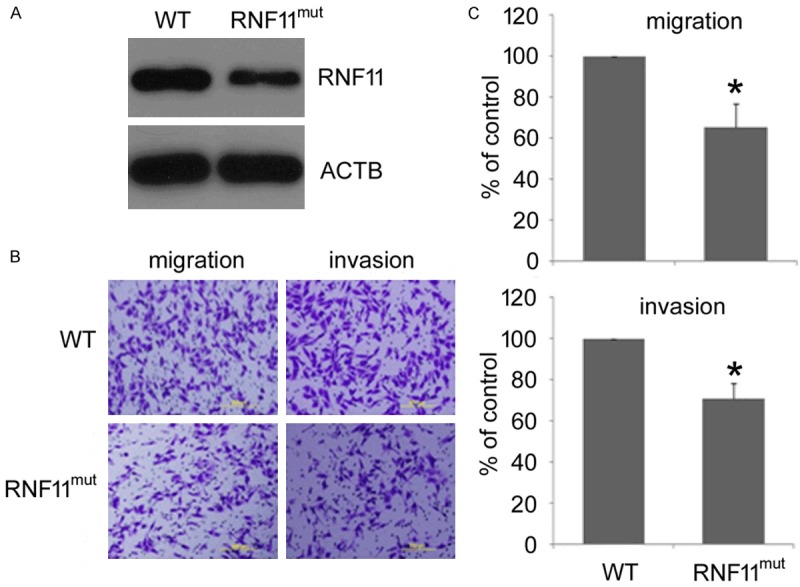
RNF11 disruption reduces the metastatic potential of murine melanoma B16F10 cells. A. RNF11 expression in RNF11mut cells was analyzed by western blot. Cell migration and invasion were evaluated by transwell migration and matrigel invasion assays respectively. B. Representative pictures were taken after staining with crystal violet. Scale bars: 200 mm. C. Cells migrated across traswell membrane were quantified (upper panel, migration; lower panel, invasion). Data are collected from three independent experiments and are average ± SD. values. *p<0.05, compared to wild type (WT) cells.
Confirmation of the role of RNF11 in melanoma migration by gene knockdown and overexpression
To confirm the gene isolated is indeed involved in decreased migration of B16F10 cells, whether the effect of RNF11 disruption on migratory potential of melanoma cells could be repeated by gene knockdown with siRNA was assessed. RNF11 expression in melanoma B16F10 and B16F0 cells were first knocked down by using a siRNA-incorporated plasmid targeting a specific site of mouse RNF11 mRNA. As shown in Figure 2A, RNF11 expression in cells transfected with siRNA plasmid (siRNF11) was greatly reduced compared with the cells transfected with scrambled siRNA (siCTL). Then transwell migration and invasion assay were performed to evaluate cell migration and invasion. Consistently, results from transwell assay also showed that the cell number of siRNF11 cells moved across the membrane was much fewer than the siCTL cells (Figure 2B). Taken together, these results indicated that RNF11 knock down could reproduce the effect of RNF11 disruption and significantly decreased the metastatic potential of melanoma cells.
Figure 2.
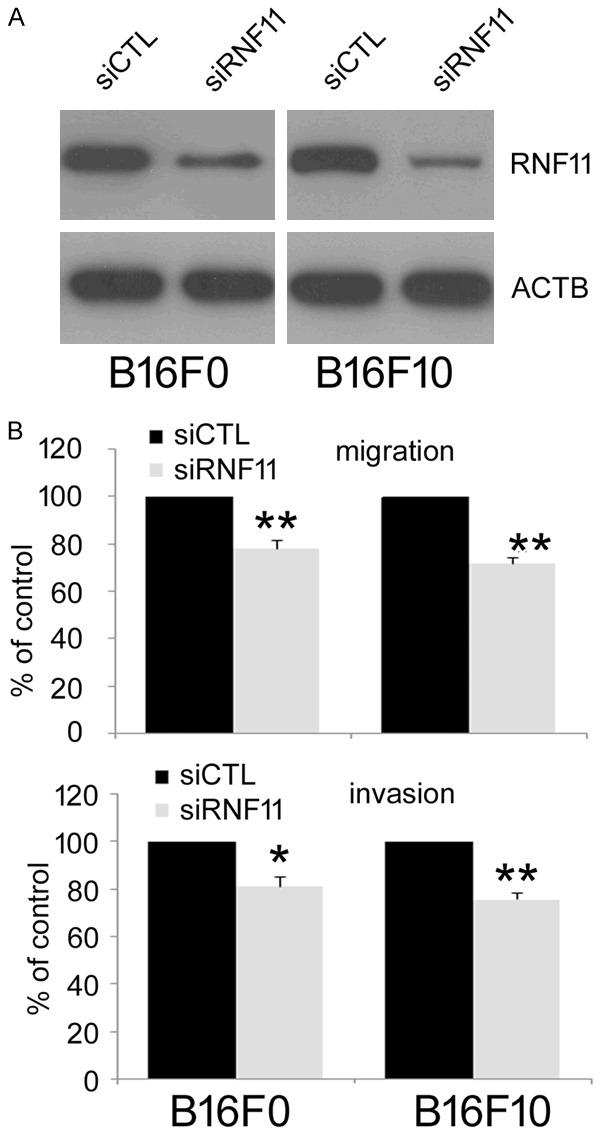
RNF11 gene knockdown decreases the migration and invasion of melanoma B16F10 and B16F0 cells. A. pSilence3.1-U6 vectors carrying siRNF11 or siCTL were transfected into melanoma B16F10 and B16F0 cells separately. The expression of RNF11 was assessed by western blot. B. Cell migration and invasion were evaluated by transwell migration (upper panel) and matrigel invasion (lower panel) assays. Data are from three repeated experiments and are average ± SD. values. *p<0.05, **p<0.01, compared to control cells.
To further assess the role of RNF11 in melanoma metastasis, RNF11 was cloned into pXJ-40/myc vector and transfected it into melanoma B16F10 and B16F0 cells. The overexpression of RNF11 was conformed by western blot with myc-tag and RNF11 antibodies (Figure 3A). The migration and invasion of RNF11-overexpressing cells was then examined by transwell migration and invasion assay. As shown in Figure 3B, there were more RNF11-overexpressing cells migrated across the membrane. Collectively, these results suggest that RNF11 overexpression increases the migratory potential of melanoma cells.
Figure 3.
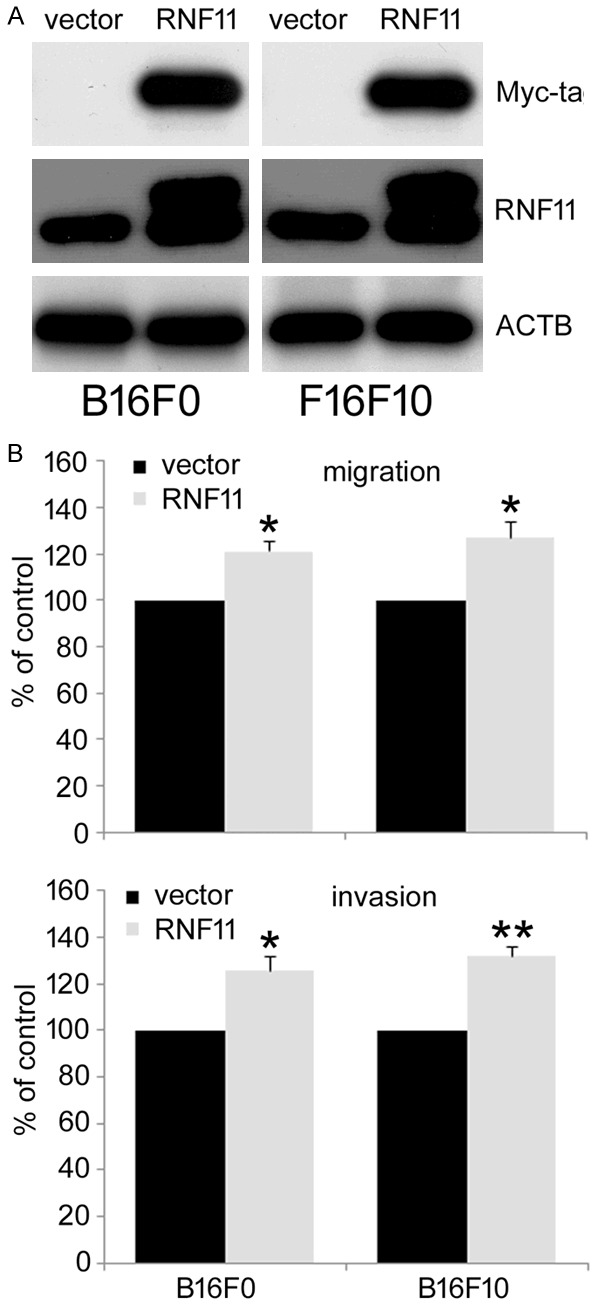
RNF11 overexpression promotes the migration and invasion of murine melanoma B16F10 and B16F0 cells. A. RNF11 was cloned into pXJ-40 vector and transfected into murine melanoma B16F10 and B16F0 cells. The cells transfected with an empty vector were used as control. The expression of RNF11 was determined by western blot with antibodies against Myc-tag and RNF11. B. Cell migration and invasion was evaluated by transwell assays. Data are from three independent experiments and are average ± SD. values. *p<0.05, **p<0.01, compared to control cells.
RNF11 knockdown reduces the metastasis of murine melanoma cells in vivo
To further validate the involvement of RNF11 in the metastasis of melanoma cells in vivo, an experimental metastasis assay was performed. Tumor metastasis in mice was mainly observed in the lungs as previously reported [13]. As shown in Figure 4B, injection of B16F10 control cells formed much more lung colonies (median, 345; range, 287-489) than mice injected with RNF11 knockdown cells (median, 123; range, 88-175, p<0.01). Moreover, control cells generated nodules that occupied a higher percentage of the total lung area compared with RNF11 knockdown cells (Figure 4A). These results further prove that RNF11 knockdown impairs the metastasis of melanoma cells in vivo.
Figure 4.

RNF11 gene knockdown reduces metastasis of murine melanoma B16F10 cells in vivo. A. Representative pictures of lungs from mice injected with siCTL and siRNF11 cells were taken after 2 weeks of injection. B. Numbers of lung metastasis were quantified and showed by each data point, **p<0.01, compared to control cells.
Discussion
Among the three major types of skin cancers, malignant melanoma has the highest risk of mortality for its highly metastatic potential [14]. In recent years, both incidence and mortality rate of melanoma continue to increase [15]. However, there is currently no effective treatment once melanoma progressed to metastatic stage partly because of the complex mechanism underlying its metastasis. In this study, a novel role of RNF11 promoting the metastasis of murine melanoma cells was identified by insertional mutagenesis. It was found that RNF11 potentiates the metastasis of murine melanoma cells in vitro and in vivo. Our results may give rise to a new target for intervention in the melanoma treatment and may improve the future treatment of melanoma.
RNF11 was initially isolated from breast tumor cells and its expression was enhanced in several human tumor tissues, such as breast, pancreatic and colon [10,11]. However, the role of RNF11 in cancer is still obscure. In this work, a novel function for RNF11 in the metastasis of melanoma cells firstly reported. It was found that the metastatic potential of murine melanoma B16F10 cells was significantly reduced when the RNF11 gene was disrupted by insertional mutagenesis. Further investigation with gene knockdown of RNF11 demonstrated that the migratory and invasive ability of melanoma cells was significantly decreased as revealed by the transwell migration and invasion assay. In contrast, overexpression of RNF11 in either murine melanoma B16F10 or B16F0 cells greatly enhanced the migration and invasion of melanoma cells. More convincingly, RNF11 gene knockdown markedly impaired the lung metastasis of murine melanoma B16F10 cells in vivo. All these data presented that RNF11 is involved in the metastasis of melanoma cells, but it is unclear how RNF11 regulates the metastasis of melanoma cells.
It was speculated that the mechanism underlying RNF11-promoted melanoma metastasis involves its ability to enhance TGF-β signaling. RNF11 has been demonstrated to directly enhance Smad4-dependant TGF-β signaling and R-Smad-Smad4 signals through association and stabilization of Smad4, the common Smad for TGF-β, activin and BMP signaling, in the presence of TGF-β [16]. RNF11 has also been reported to indirectly enhance TGF-β signaling by interacting with Smurf2 to abrogate Smurf2-mediated ubiquitination of TGF-β receptor and degradation of AMSH (associated molecule with the SH3 domain of STAM), a de-ubiquitination enzyme that increases TGF-β signaling [12]. TGF-β signaling enhances the migratory and invasive properties of cancer by inducing epithelial-mesenchymal transition (EMT) through a combination of Smad-dependent transcriptional events and inhibition of TGF-β signaling with SD-208, a small molecule inhibitor of TGF-β receptor I kinase, prevent the development of melanoma metastases [17,18]. It is possible that RNF11 promotes the metastasis of melanoma through upregulating TGF-β signaling.
Considering the critical function of RNF11 in the metastasis of melanoma cells, it is worthwhile to investigate the precise molecular mechanisms underlying RNF11-promoted the metastasis of melanoma cells in the future work.
Acknowledgements
This work was financially supported by Scientific Research Fund of Xinxiang Medical University (2013QZ107) and the Joint Fund for Fostering Talents of National Natural Science Foundation of China (U1404816). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Miller AJ, Mihm MC Jr. Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 3.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 4.Vidwans SJ, Flaherty KT, Fisher DE, Tenenbaum JM, Travers MD, Shrager J. A melanoma molecular disease model. PLoS One. 2011;6:e18257. doi: 10.1371/journal.pone.0018257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rofstad EK, Mathiesen B, Henriksen K, Kindem K, Galappathi K. The tumor bed effect: increased metastatic dissemination from hypoxia-induced up-regulation of metastasis-promoting gene products. Cancer Res. 2005;65:2387–2396. doi: 10.1158/0008-5472.CAN-04-3039. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24:448–456. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Stafford LJ, Vaidya KS, Welch DR. Metastasis suppressors genes in cancer. Int J Biochem Cell Biol. 2008;40:874–891. doi: 10.1016/j.biocel.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Zarubin T, Jing Q, New L, Han J. Identification of eight genes that are potentially involved in tamoxifen sensitivity in breast cancer cells. Cell Res. 2005;15:439–446. doi: 10.1038/sj.cr.7290312. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Ono K, Kim SO, Kravchenko V, Lin SC, Han J. Metaxin is required for tumor necrosis factor-induced cell death. EMBO Rep. 2001;2:628–633. doi: 10.1093/embo-reports/kve135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seki N, Hattori A, Hayashi A, Kozuma S, Sasaki M, Suzuki Y, Sugano S, Muramatsu MA, Saito T. Cloning and expression profile of mouse and human genes, Rnf11/RNF11, encoding a novel RING-H2 finger protein. Biochim Biophys Acta. 1999;1489:421–427. doi: 10.1016/s0167-4781(99)00190-6. [DOI] [PubMed] [Google Scholar]
- 11.Subramaniam V, Li H, Wong M, Kitching R, Attisano L, Wrana J, Zubovits J, Burger AM, Seth A. The RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligase. Br J Cancer. 2003;89:1538–1544. doi: 10.1038/sj.bjc.6601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azmi P, Seth A. RNF11 is a multifunctional modulator of growth factor receptor signalling and transcriptional regulation. Eur J Cancer. 2005;41:2549–2560. doi: 10.1016/j.ejca.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Liu R, Zhang S, Xia YY, Yang HJ, Guo K, Zeng Q, Feng ZW. Neural cell adhesion molecule potentiates invasion and metastasis of melanoma cells through CAMP-dependent protein kinase and phosphatidylinositol 3-kinase pathways. Int J Biochem Cell Biol. 2011;43:682–690. doi: 10.1016/j.biocel.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Jemal A, Saraiya M, Patel P, Cherala SS, Barnholtz-Sloan J, Kim J, Wiggins CL, Wingo PA. Recent trends in cutaneous melanoma incidence and death rates in the United States, 1992-2006. J Am Acad Dermatol. 2011;65:S17–25. e11–13. doi: 10.1016/j.jaad.2011.04.032. [DOI] [PubMed] [Google Scholar]
- 15.Giblin AV, Thomas JM. Incidence, mortality and survival in cutaneous melanoma. J Plast Reconstr Aesthet Surg. 2007;60:32–40. doi: 10.1016/j.bjps.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Azmi PB, Seth AK. The RING finger protein11 binds to Smad4 and enhances Smad4-dependant TGF-beta signalling. Anticancer Res. 2009;29:2253–2263. [PubMed] [Google Scholar]
- 17.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 18.Mohammad KS, Javelaud D, Fournier PG, Niewolna M, McKenna CR, Peng XH, Duong V, Dunn LK, Mauviel A, Guise TA. TGF-beta-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 2011;71:175–184. doi: 10.1158/0008-5472.CAN-10-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]


