Abstract
The actin cytoskeleton is a key target of numerous microbial pathogens, including protozoa, fungi, bacteria and viruses. In particular, bacterial pathogens produce and deliver virulence effector proteins that hijack actin dynamics to enable bacterial invasion of host cells, allow movement within the host cytosol, facilitate intercellular spread or block phagocytosis. Many of these effector proteins directly or indirectly target the major eukaryotic actin nucleator, the Arp2/3 complex, by either mimicking nucleation promoting factors or activating upstream small GTPases. In contrast, this review is focused on a recently identified class of effector proteins from Gram-negative bacteria that function as direct actin nucleators. These effector proteins mimic functional activities of formins, WH2-nucleators and Ena/VASP assembly promoting factors demonstrating that bacteria have coopted the complete set of eukaryotic actin assembly pathways. Structural and functional analyses of these nucleators have revealed several motifs and/or mechanistic activities that are shared with eukaryotic actin nucleators. However, functional effects of these proteins during infection extend beyond plain actin polymerization leading to interference with other host cell functions such as vesicle trafficking, cell cycle progression and cell death. Therefore, their use as model systems could not only help in the understanding of the mechanistic details of actin polymerization but also provide novel insights into the connection between actin dynamics and other cellular pathways.
Keywords: actin, nucleation, bacterial virulence, effector protein, secretion system
In this review, the authors discuss the mechanisms and function of the emerging group of bacterial virulence factors that act as actin nucleators.
Graphical Abstract Figure.
In this review, the authors discuss the mechanisms and function of the emerging group of bacterial virulence factors that act as actin nucleators.
INTRODUCTION
The actin cytoskeleton is found essentially in all eukaryotic cells. It is fundamental to mediate or control cell migration, cell shape, cell division, intracellular membrane trafficking or muscle contraction. Actin monomers (G-actin) polymerize to yield polarized microfilaments (F-actin), in which G-actin addition is faster at the plus end (barbed end) and slower at the minus end (pointed end). The process is very dynamic, such that controlled actin polymerization and depolymerization are constantly occurring within living cells, and is regulated by different types of actin-binding proteins. Because assembly of two actin monomers leads to unstable dimers, the rate-limiting step of actin filament polymerization is nucleation, consisting in the formation of a stable nucleus of three actin subunits (Sept and McCammon 2001). In addition to the formation of this actin nucleus being kinetically unfavourable, the majority of the G-actin pool in the cell is bound to profilin, which sequesters actin and further inhibits spontaneous nucleation. To initiate the formation of actin filaments in a temporally and spatially controlled manner, eukaryotic cells use proteins or protein complexes known as actin nucleators. Since the identification of the major eukaryotic actin nucleator, the Arp2/3 complex (Welch, Iwamatsu and Mitchison 1997), two other families of nucleators have arisen: formins and tandem-monomer-binding proteins (Firat-Karalar and Welch 2011).
The actin cytoskeleton and its regulators are a major target of bacterial pathogens, which modulate actin dynamics for invasion or exit of host cells, for inhibition of phagocytosis or for intracellular motility and cell-to-cell spread. In particular, through different protein secretion systems, bacterial effector proteins gain access to the host cell cytosol where they have been shown to usurp the function of the majority of the eukaryotic actin nucleators, by modulating directly or indirectly their activity (Welch and Way 2013; Truong, Copeland and Brumell 2014). Furthermore, bacterial toxins can also directly interfere with actin polymerization. For example, some toxins ADP-ribosylate actin monomers and in this way inhibit the addition of the latter to the fast growing ends of actin filaments (e.g. Clostridium botulinum exotoxin C2), whereas others modify actin by enzymatic crosslinking (e.g. Vibrio cholerae MARTX; reviewed in Aktories et al. 2011). A remarkable example of bacterial manipulation of the actin cytoskeleton is the polymerization of actin tails at one of the poles of bacterial cells mediated, e.g. by ActA of Listeria monocytogenes and IcsA of Shigella flexneri, that propels the bacteria in the host cell cytosol and promotes bacterial cell-to-cell spread. ActA and IcsA do not mediate polymerization actin directly, but instead act as activators of the Arp2/3 complex, i.e. they are nucleation-promoting factors (NPFs) (see below), and there are many excellent reviews discussing their mode of action and function (Cossart 2000; Travier and Lecuit 2014). Here, we will focus on the role of the emerging group of bacterial effectors that function as direct actin nucleators.
Eukaryotic actin nucleators
The Arp2/3 complex is formed by seven subunits. Two of them, Arp2 and Arp3, form a structural mimic of an actin dimer, serving as template for the formation of a new filament. This complex is a weak nucleator by itself and needs to be activated by NPFs, such as WASP (Wiskott–Aldrich syndrome protein; Fig. 1A) and related proteins (N-WASP, WAVE, WHAMM and WASH). These NPFs, upon activation, usually by small GTPases of the Rho/Rac/Cdc42 family, interact with Arp2/3 via a central and an acidic domain and recruit G-actin to the barbed end of the filament using a ∼18 amino acid WH2 (WASP-Homology 2) motif (Figs 1 and 2), and profilin-actin via proline-rich regions. The NPF-activated Arp2/3 complex usually binds to the sides of existing filaments in a way that new filaments grow at an angle of ∼70° relatively to the mother filament, giving rise to a branched actin network (Fig. 2A). Noteworthy, exceptions exist, as Arp2/3 activated by the Schizosaccharomyces pombe Dip1 NPF forms linear, unbranched actin filaments (Wagner et al. 2013).
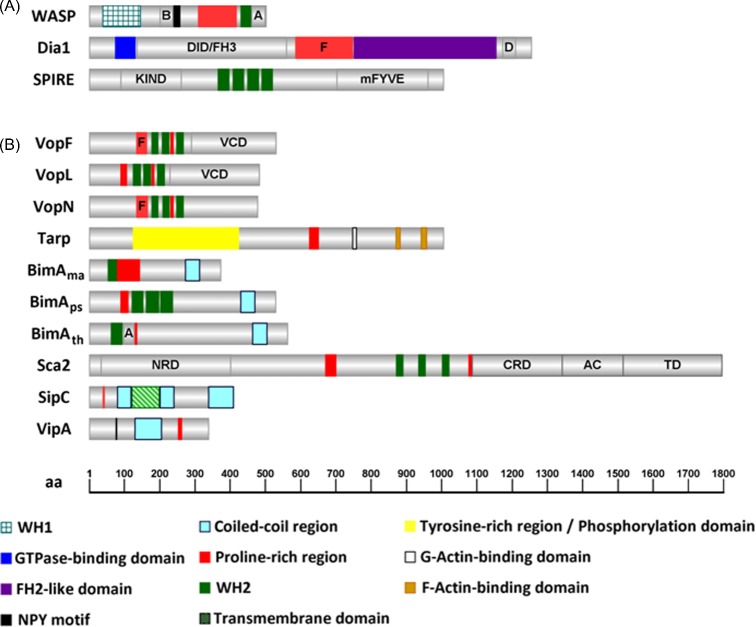
Figure 1.
Graphical representation of the domain organization of proteins involved in actin nucleation. (A) Eukaryotic actin nucleators: WASP, Dia1 and SPIRE (Homo sapiens; Sarkar et al. 2014; Goode and Eck 2007; Qualmann and Kessels 2009). (B) Bacterial actin nucleators: VopN and VopF (V. cholerae; Tam et al. 2010); VopL (V. parahaemolyticus, Yu et al. 2011); Tarp (C. trachomatis; Jiwani et al. 2013); BimAma (B. mallei; Benanti et al. 2015), BimAps (B. pseudomallei; Benanti et al. 2015), BimAth (B. thailandensis, Benanti et al. 2015), Sca2 (R. conorii; Madasu et al. 2013); SipC (S. Typhimurium; Hayward and Koronakis 1999); and VipA (L. pneumophila; Franco, Shohdy and Shuman 2012). The number of amino acids of the proteins and the approximate position of regions of interest are indicated (bottom). B, basic region; A, Arp2/3-binding central and acidic motifs; DID/FH3, diaphanous inhibitory domain/Formin homology 3; F, FH1-like domain; D, diaphanous auto-regulatory domain; KIND, kinase noncatalytic C-lobe domain; mFYVE, modified zinc finger motif; NPY, asparagine, proline, tyrosine; VCD, Vop C-terminal domain; NRD, N-terminal repeat domain; CRD, C-terminal repeat domain; AC, auto-chaperone domain; TD, translocator domain. The figure was generated using IBS (Illustrator of Biological Sequences) v1.0 software.
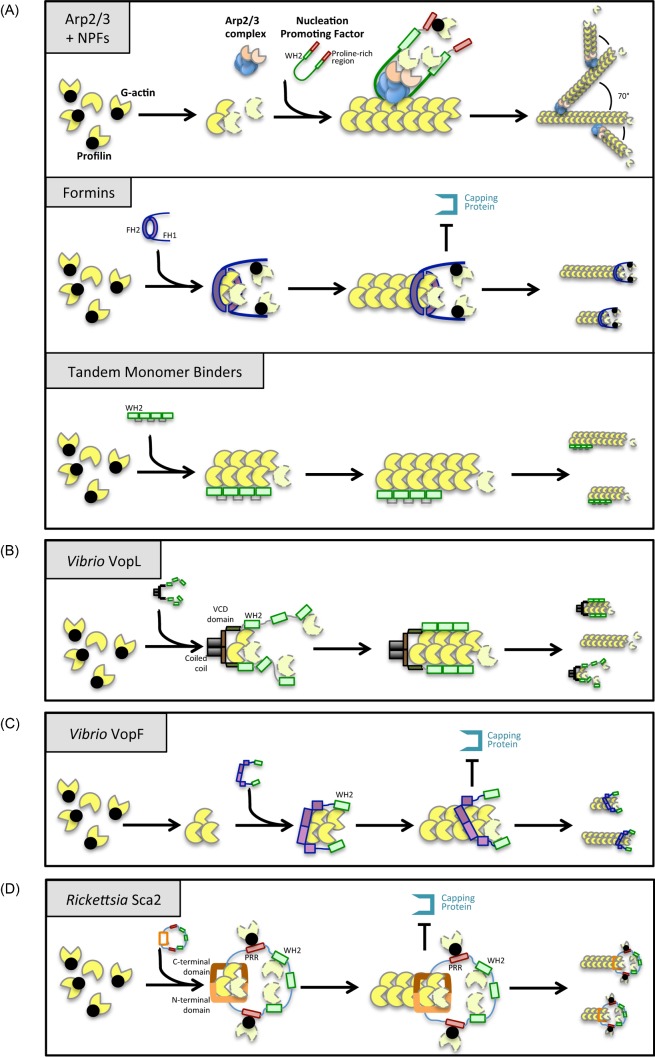
Figure 2.
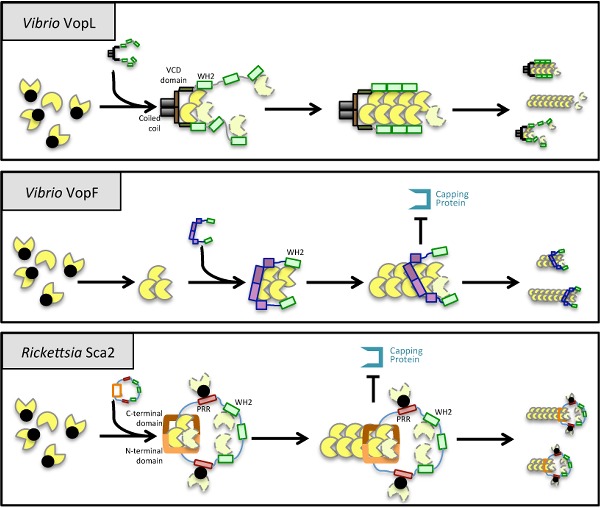
Models for the mechanism of actin nucleation and elongation by eukaryotic and bacterial nucleators. (A) After activation by NPFs, the Arp2/3 complex binds to the side of pre-existing actin filaments. WH2 domains and proline-rich regions (PRRs) within NPFs recruit actin monomers and profilin-actin, respectively. New filaments elongate at a 70° angle, originating a branched actin network with Arp2/3 bound at the pointed end. Formins act as homodimers that induce nucleation by stabilizing two actin subunits through their FH2 domains. As the filament elongates, formins remain bound to the barbed end to which the FH1 domains recruit profilin-actin. Nucleators belonging to the tandem monomer binding family contain tandem motifs that bind G-actin (generally WH2 motifs) that act as a scaffold that brings together actin subunits to form a nucleus (Firat-Karalar and Welch 2011). (B) In V. parahaemolyticus VopL, the VCD contributes to nucleation by stabilizing the pointed end of an actin nucleus and by allowing the dimerization of the WH2 domains that recruit G-actin. After nucleation, VopL dissociates from the growing filament but might remain bound to the pointed-end subunits, initiating a new cycle of polymerization (Namgoong et al. 2011). (C) Each V. cholerae VopF monomer contains three WH2 domains, but only the first and the third WH2 (the only WH2 represented in the figure for simplification) bind actin monomers. Nucleation involves dimerization of the WH2 domains and formation of a tetrameric actin nucleus where each terminal WH2 binds one barbed-end actin subunit. Processive elongation of the filament is accomplished by continuous displacement and re-association of the WH2 to newly added barbed-end subunits (Pernier et al. 2013). (D) Rickettsia conorii Sca2 monomers recruit profilin-actin via PRRs and G-actin via its WH2 domains. Interaction between its N- and C-terminal regions is thought to allow Sca2 to move processively during elongation, remaining at the barbed end and preventing filament capping (Madasu et al. 2013).
In contrast to the Arp2/3 complex, formins nucleate the formation of long unbranched F-actin, for which the presence of characteristic Formin-homology 1 and Formin-homology 2 (FH1 and FH2) domains is required (Paul and Pollard 2009; Chesarone, DuPage and Goode 2010) (Fig. 1A). While FH1 domains are proline-rich regions that recruit profilin-actin and thereby aid in elongation of actin filaments, FH2 domains are involved in the initial nucleation phase. As the filament grows, FH2 domains remain bound to the barbed end, acting as a ‘leaky capper’, which allows the addition of actin monomers and simultaneously prevents the binding of a capping protein that would lead to termination of polymerization (Fig. 2A).
A third family of eukaryotic actin nucleators comprises tandem-monomer-binding proteins, such as Spire, Cordon-bleu and leiomodin (Qualmann and Kessels 2009). These proteins typically contain tandem repeats of WH2 domains, which act as a scaffold to bring together actin monomers (Fig. 1). Unlike formins, but similarly to the Arp2/3 complex, these nucleators remain bound to the pointed end of the filament and create actin clusters (Fig. 2A). Importantly, there is also cross-talk between proteins belonging to the three different families of nucleators, revealing important cooperation that fine-tune actin dynamics in the cell (Firat-Karalar and Welch 2011).
Actin nucleation mediated by bacterial virulence proteins
Vibrio VopF, VopL and VopN: disruption of the intestinal epithelial barrier
Pathogenic Vibrio spp. are associated with gastroenteritis and, less frequently, septicemia by the infection of open wounds. These bacteria are typically found in saline water habitats and are transmitted by the ingestion of contaminated water (V. cholerae) or undercooked seafood (V. parahaemolyticus). Three Vibrio outer membrane proteins (Vops), secreted by a type III secretion system (T3SS), are involved in the characteristic disruption of the intestinal barrier and function as actin nucleators: V. cholerae VopF and VopN, and V. parahaemolyticus VopL (Table 1). In addition to sharing 57% amino acid identity, VopF and VopL contain a C-terminal dimerizing region, three WH2-like domains required for actin nucleation, as well as profilin-binding FH1-like regions containing poly-proline repeats (Dziejman et al. 2005) (Fig. 1B). In spite of these resemblances, the mechanistic activities of VopL and VopF on actin filaments diverge, as shown by functional assays and determination of the three-dimensional structure of their domains in complex with actin (Rebowski et al. 2010; Namgoong et al. 2011; Yu et al. 2011; Pernier et al. 2013) (Fig. 2).
Table 1.
Bacterial actin nucleators.
| Protein | Organism | Secretion System | Functions in infection | Activities on actin | Additional eukaryotic binding partners | References |
|---|---|---|---|---|---|---|
| Tarp | Chlamydia spp. | T3SS | Invasion, host cell survival | Nucleation, bundling, Arp2/3-mediated polymerization (via recruitment of FAK/Cdc42 and activation of Rac) | Rac GEFs (Vav2, Sos1); PI3K; tyrosine Kinases (Src family, Abl, Yes, Fyn); human adaptor protein SHC-1; focal adhesion kinase | Lane et al. (2008); Jewett et al. (2008); Mehlitz et al. (2008, 2010); Jiwani et al. (2013); Thwaites et al. (2014) |
| VopL | Vibrio parahaemolyticus | T3SS (T3SS2) | Formation of stress fibres, disruption of actin homeostasis | Nucleation, pointed-end binding | Dziejman et al. (2005); Liverman et al. (2007); Rebowski et al. (2010); Namgoong et al. (2011); Yu et al. (2011); Zahm et al. (2013) | |
| VopF | Vibrio cholerae | T3SS | Formation of filopodia, alteration in tight junctions, inhibition of apoptosis, alteration of cell cycle, cytotoxicity | Nucleation, severing, barbed-end binding, CapZ dissociation | Cyclophilin G (PPIG); breast cancer associated gene 3 (BCA3/AKIP); copper metabolism domain containing proteins | Tam et al. (2007, 2010); Pernier et al. (2013); Tripathi et al. (2013); Avvaru, Pernier and Carlier (2015) |
| VopN | Vibrio cholerae | T3SS | Disruption of cell polarity and tight junctions, inhibition of apoptosis and alteration of cell cycle | Nucleation | Filamin A and B | Tam et al. (2010) |
| Sca2 | Rickettsia conorii Rickettsia rickettsi Rickettsia parkeri | T5SS | Intra- and intercellular motility, adherence and invasion of host cell | Nucleation, elongation, barbed-end binding, CapZ dissociation | Host cell receptor? | Cardwell and Martinez (2009); Haglund et al. (2010); Kleba et al. (2010); Madasu et al. (2013); Reed et al. (2014) |
| Burkholderia pseudomallei | T5SS | Motility, host cell fusion | Nucleation, elongation, bundling, barbed-end binding, CapZ dissociation | Stevens, Ulrich and Taylor (2005); Stevens et al. (2005); Sitthidet et al. (2010); Benanti et al. (2015) | ||
| BimA | Burkholderia mallei | T5SS | Motility, host cell fusion | Nucleation, elongation, bundling, barbed-end binding, CapZ dissociation | ||
| Burkholderia thailandensis | T5SS | Motility, host cell fusion | Nucleation, Arp2/3 activation | Arp2/3 | ||
| SipC | Salmonella enterica serovarTyphimurium | T3SS (SPI-1) | Invasion; formation of the SPI-1 T3SS translocon; recruitment of LAMP-1 and exocytic vesicles | Nucleation, bundling | Syntaxin6, Exo70 | Hayward and Koronakis (1999); Scherer et al. (2000); Nichols and Casanova (2010); Cain, Hayward and Koronakis (2008) |
| VipA | Legionella pneumophila | T4BSS (Icm/Dot) | Vesicle trafficking | Nucleation | Disruption of vacuolar protein trafficking in yeast | Shohdy et al. (2005); Franco, Shohdy and Shuman (2012) |
VopL WH2 domains capture actin monomers due to their high affinity for G-actin, but as a consequence of their inherent flexibility, they are unable to organize these molecules into microfilaments. This action is accomplished by the dimerization region, which has low affinity for actin, but in turn is able to enforce a filament-like organization for these monomers originating a productive nucleus (Fig. 2B). As a result, VopL relies on the synergistic organizer/recruiter activities of the dimerization and WH2 domains to drive filament nucleation and promote fast cycles of nucleation and detachment from the pointed end (Namgoong et al. 2011).
The mechanism of microfilament nucleation and elongation by VopF also involves a fundamental role of the dimerization and WH2 domains (Pernier et al. 2013) (Fig. 2C). Taken together, structural data suggest that VopF binds actin monomers in a non-nucleating, sequestered conformation, and actin nucleation occurs only upon dimerization of two opposite WH2 domains from two VopF protomers (Fig. 2C). However, in contrast with VopL, VopF-mediated elongation is presumed to involve processive filament growth, with sequential cycles of partial dissociation of one WH2 from the recently added actin monomer allowing insertion of a new actin subunit, followed by new association of the same WH2 domain with this newly added actin (Pernier et al. 2013) (Fig. 2C). This process involves also exclusion of capping protein from the growing barbed ends of a microfilament, similarly to many formins. In fact, the fundamental role of VopF in filopodia formation and its localization at filopodia tips strongly suggests that its main activities might be the uncapping of actin filaments and regulation of barbed-end growth. These activities are thought to impair the WASP and Arp2/3-mediated formation of branched actin meshworks (which require barbed-end cappers) and in contrast favour the effect of VopF on the host cell cycle and intestinal colonization by V. cholerae (Pernier et al. 2013).
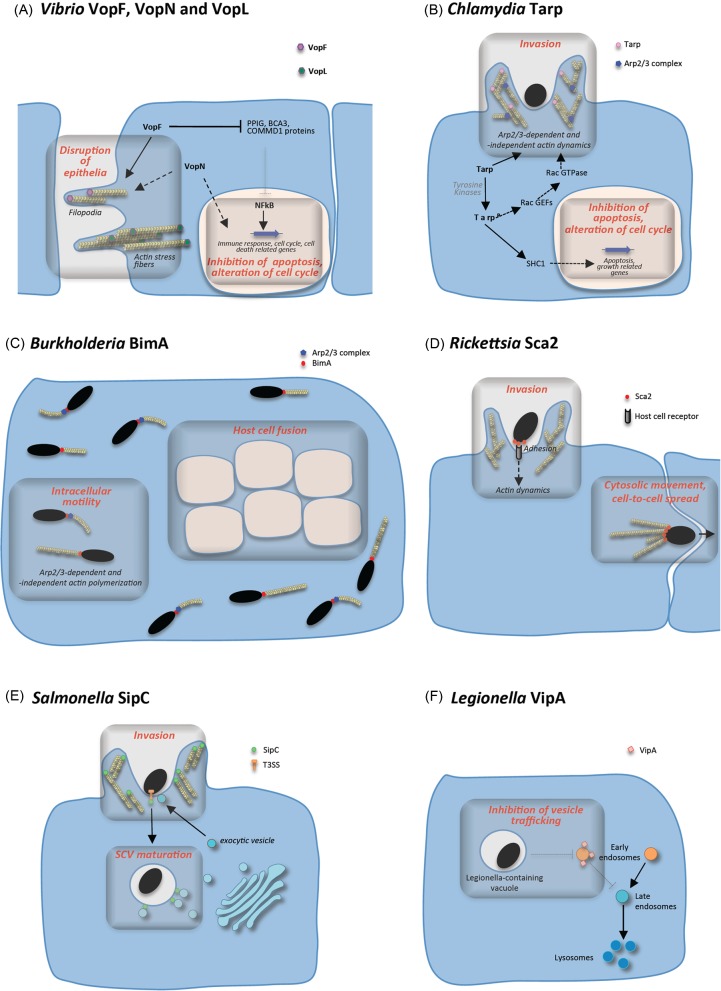
The activity of the VopL and VopF actin nucleators has been linked to enterotoxic effects during infection. Vibrio parahaemolyticus VopL disrupts actin homeostasis by inducing the formation of large stress fibres in infected cells (Liverman et al. 2007), while VopF mediates the formation of aberrant protrusions on the surface and periphery of the cells and localizes at the tip of these protrusions, bound to the barbed end of the growing microfilaments (Tam et al. 2007) (Fig. 3). These cytoskeletal rearrangements have implications in the alteration of actin homeostasis during infection, leading to a decrease in transepithelial resistance and to the increase in diarrhoea. In addition to its modulation of actin dynamics, a multifunctional role for VopF in the gut epithelia is reinforced by its effect on the host cell cycle and cell death (Tam et al. 2010) likely by targeting of several binding partners: cyclophilin G (PPIG), a nuclear protein involved in regulation of the cell cycle; two repressors of NF-kB-dependent transcription, breast cancer associated gene 3 (BCA3/AKIP) and copper metabolism domain containing proteins (COMMD1) (Tam et al. 2010). In fact, VopF inhibits TNF-α-dependent NF-kB activity, preventing the activation of genes driving cell cycle and cell death (Fig. 3A). During infection, these effects of VopF are thought to stimulate bacterial colonization by inhibiting cell renewal and extending the lifespan of the intestinal epithelia, and as a result affecting epithelial integrity.
Figure 3.
Function of bacterial actin nucleators during infection. (A) Vibrio VopF, VopN and VopL contribute to the disruption of intestinal epithelial integrity by inducing the formation of aberrant protrusions, filopodia and altering tight junctions. VopF further contribute to cell alterations by inhibiting apoptosis and arresting the cell cycle, by relieving the inhibition of NF-kB regulated genes—a function also exerted by VopN by an unknown mechanism. (B) Chlamydia Tarp contributes to host cell invasion by rapidly nucleating actin filaments, and indirectly by triggering the Rac signalling cascade that leads to activation of the Arp2/3 complex. Phosphorylated Tarp also has a role in mediating host cell survival by binding the eukaryotic SHC-1 adaptor protein. (C) BimA nucleates actin filaments from the Burkholderia cell pole allowing bacterial motility in the cytosol and host cell fusion. Burkholderia thailandensis BimA also recruits and activates the Arp2/3 complex, leading to the formation of curved actin tails. (D) Rickettsia Sca2 promotes invasion of the host cell by mediating adhesion to an unknown receptor and inducing actin remodelling. After uptake and escape into the cytosol, Sca2 induces the formation of actin comet tails from the bacterial cell pole, thus allowing intracellular movement and spread to neighbouring cells. (E) Salmonella SipC is part of the T3SS translocon complex and is thought to nucleate actin filaments to facilitate invasion. Additionally, SipC is involved in the recruitment of exocytic vesicles to the site of entry, presumably to provide membrane to the invasion process. SipC might also contribute to the maturation of the SCVs by recruiting Golgi-derived vesicles enriched in LAMP1. (F) Legionella VipA interferes with eukaryotic vesicle trafficking in the MVB pathway and associates with early endosomes during macrophage infection, possibly contributing to the avoidance by the LCV of the endocytic machinery.
Similarly to VopF, VopN also interferes with host cell cycle and cell death (Tam et al. 2010). Nevertheless, in contrast with VopF, VopN localizes to actin stress fibres. The divergent localization of these two homologous proteins is determined by their distinct N-termini, which in the case of VopN interacts with host cell Filamin A and Filamin B, F-actin crosslinkers that regulate intracellular communication and signalling (Tam et al. 2010). A vopN null-mutant strain, however, shows no phenotypic defect in colonization in a mouse model of infection and so far the function of this nucleator during infection has mostly remained elusive (Tam et al. 2010). Thus, in spite of their divergent mechanisms of action on actin dynamics and other cell processes, VopL, VopF and VopN converge in their role in altering the intestinal epithelia homeostasis.
Chlamydia Tarp: a multifunctional signalling hub that triggers invasion
Chlamydiae are a large group of obligate intracellular bacteria that includes human (e.g. Chlamydia trachomatis or C. pneumoniae) and animal pathogens. Chlamydia trachomatis is a particular clinical and public health concern, causing human preventable blindness (serovars A-C) and sexually transmitted diseases (serovars D-L) (Wright, Turner and Taylor 2008; Bebear and de Barbeyrac 2009). Among sexually transmitted C. trachomatis, serovars A-G cause infections restricted to the genital epithelium while serovars L1-L3 are more invasive and cause lymphogranuloma venereum (LGV). The Chlamydia developmental cycle involves the interconversion between an infectious and non-replicative form, the elementary body (EB), and a replicative but non-infectious form, the reticulate body (RB) (Abdelrahman and Belland 2005). During this cycle, Chlamydia resides and multiplies intracellularly within a membrane-bound compartment termed inclusion, and translocates effector proteins into host cells using a T3SS. One of these effectors, the Translocated actin-recruiting phosphoprotein (Tarp; Table 1) is delivered into host cells upon attachment of EBs to host cells and triggers immediate actin recruitment at the site of entry and thereby contributes to invasion (Clifton et al. 2004; Lane et al. 2008; Jewett et al. 2010).
Chlamydia trachomatis Tarp can be divided into two functional regions of ∼500 amino acids in length: the N-terminal region contains several repeats of ∼50 residues each highly enriched in tyrosines, whose phosphorylation by host kinases has been associated both with activation of the Arp2/3 complex and interference with intracellular signalling to promote host cell survival; the C-terminal region encompasses determinants for interaction and polymerization of actin, and of Tarp oligomerization (Fig. 1B). Tarp orthologues are present in all chlamydial pathogenic species identified wherein recruitment of actin to the entry site occurs. Nonetheless, they display relatively low levels of amino acid sequence identity and the tyrosine-rich repeats are only present in Tarp of C. trachomatis (Jewett et al. 2010). Even within C. trachomatis, different serovars show significant differences in the amino acid sequence of Tarp that might imply distinct mechanisms of actin polymerization (Jewett et al. 2010). In fact, differences between Tarp of C. trachomatis LGV strains (serovars L1-L3) and ocular strains (serovars A-C) correlate with the number of tyrosine-rich domains and actin-binding domains, putting forward Tarp as possibly involved in adaptation to specific niches in the host and contributing to C. trachomatis tissue-tropism (Lutter et al. 2010).
Binding of C. trachomatis serovar L2 Tarp to actin monomers is mediated by a single C-terminal WH2-like domain, which alone is insufficient to induce polymerization of filaments. To achieve actin nucleation, a proline-rich region is required to prompt oligomerization of Tarp into large multimeric complexes (>800 kDa), which presumably nucleate microfilaments by bringing multiple actin molecules into apposition (Jewett et al. 2006). In addition to its actin nucleating activity, Tarp is also able to bundle microfilaments via two regions within the C-terminal half of the protein, the F-actin binding domains 1 and 2 (FAB1 and FAB2), both conserved among C. trachomatis serovars (Jiwani et al. 2013) (Fig. 1B). However, the involvement of C. trachomatis Tarp in microfilament formation at the entry site extends beyond the nucleating and bundling activities mediated by its C-terminal region. In fact, the N-terminal half of C. trachomatis Tarp is implicated in an elaborate, and still not completely defined, signalling process that involves multiple tyrosine phosphorylations and binding partners. Lane et al. (2008) found that Tarp activates the Rac GTPase-dependent signalling cascade by regulating two Rac guanine nucleotide exchange factors (GEFs): the complex Sos1/Eps8/Abi1 and Vav2. Interaction with these GEFs requires Tarp phosphorylation and additionally the presence of the phophatidylinositol 3,4,5-triphosphate (PIP3) in the case of Vav2. Abolishment of binding of Tarp to these GEFs decreases the recruitment of Rac and, consequently, negatively affects invasion of non-phagocytic cells by Chlamydia. More recently, a different pathway of Tarp-mediated Arp2/3 activation was revealed, via a signalling cascade involving focal adhesion kinase and Cdc42 (Thwaites et al. 2014). Therefore, actin remodelling during Chlamydia invasion seems to engage a combined action of Arp2/3-dependent and -independent actin nucleation mechanisms, both elicited by Tarp (Fig. 3B).
Another level of signalling involving Tarp has been revealed by the identification of Tarp as a substrate of multiple host tyrosine kinases (Src family members, Syk and Abl) (Elwell et al. 2008; Jewett et al. 2008; Mehlitz et al. 2008). However, in C. trachomatis, entry of EBs into host cells is not dependent on tyrosine phosphorylation of Tarp (Jewett et al. 2008), and the tyrosine-rich repeats at the N-terminal region are absent in Tarp from other chlamydial species, which are nevertheless proficient in invasion of host cells. Therefore, the relevance of the activity of these host tyrosine kinases on Tarp is still unclear. Nevertheless, the determination of the Tarp phospho-interactome revealed a significant increase in high-affinity interactions triggered by phosphorylation, including the human adaptor protein SHC1. This protein is involved in the regulation of apoptosis and growth-related genes, and binding to Tarp promotes host cell survival during early stages of infection (Mehlitz et al. 2010) (Fig. 3B).
In summary, the direct and indirect action of Tarp on actin led to its establishment as a key regulator of actin dynamics required for invasion. Still, recent identification of multiple eukaryotic interactants is contributing to recognizing Tarp also as a signalling hub (Mehlitz et al. 2010), anticipating the discovery of further pathways targeted by this effector during infection.
Burkholderia BimA: bacterial motility, host cell fusion
The Burkholderia genus comprises species pathogenic for humans or other animals (e.g. Burkholderia pseudomallei, B. mallei or B. cepacia) but also closely related non-human pathogens with unknown hosts (B. thailandensis). After entry into host cells, Burkholderia escapes the newly formed vacuolar compartment and induces the formation of actin tails to propel itself in the cytosol. The force generated by this movement is thought to bring membranes of infected cells into close contact with adjacent cells, facilitating cell fusion and leading to the formation of giant multinucleated cells, a process essential for Burkholderia virulence (Kespichayawattana et al. 2000; Galyov, Brett and DeShazer 2010; Toesca, French and Miller 2014). Actin-based motility requires the Burkholderia intracellular motility A protein (BimA; Table 1). BimA is an autotransporter (i.e. a type V secretion system; T5SS) that after secretion is localized at bacterial cell poles, from where it nucleates actin (Fig. 3C). BimA orthologues are encoded in the genomes of B. pseudomallei (BimAps), B. mallei (BimAma) and B. thailandensis (BimAth). They are all potent actin nucleators but differ on their mechanisms of action as a consequence of significant differences in the composition and organization of their functional domains (Fig. 1B), which also impacts the efficiency of cell fusion and the outcome of infection (Benanti, Nguyen and Welch 2015). In addition to being an actin nucleator, BimAth is an NPF that binds to the Arp2/3 complex via its acidic domain (Sitthidet et al. 2010). BimAth-mediated activation of Arp2/3 promotes branched filament assembly, a feature absent in BimAma and BimAps (Sitthidet et al. 2010). Conversely, BimAma and BimAps are both able to bind filament barbed ends and exclude capping protein, as well as to processively elongate and simultaneously bundle filaments. These activities are typical features of actin elongators belonging to the ENA/Vasp family (Sitthidet et al. 2011; Benanti, Nguyen and Welch 2015).
There is a correlation between the differences in the functional domains and mechanisms of action of BimAth, BimAma and BimAps and features of filament organization, actin-based motility and efficiency of host cell fusion (Benanti, Nguyen and Welch 2015). While BimAth induces the formation of curved actin tails consisting of a dense network of filaments, consistent with Arp2/3 activation, BimAma and BimAps promote long and straight tails composed of bundled F-actin (Benanti, Nguyen and Welch 2015). This results in two distinct types of bacterial movement in the host cell cytosol (Benanti, Nguyen and Welch 2015) (Fig. 3C). Notwithstanding, the most predominant effect in influencing cell fusion is the frequency of actin tail formation at the bacterial pole. In fact, in BimAps and BimAth the higher number of intracellular Burkholderia polymerizing actin tails leads to larger multinucleated cells (Benanti, Nguyen and Welch 2015). Hence, the evolution of different mechanisms of actin polymerization in closely related species seems to reflect different host cell features. Burkholderia pseudomallei is an environmental saprophyte that infects a wide range of mammals, and consequently BimAps might have developed the ability to function in different mammalian hosts, whereas B. mallei is a clonal descendent that evolved to be an obligate intracellular pathogen of horses, and so BimAma may have been optimized for specific infection of equine cells requiring additional host actin regulators that are present in equine cells, but absent from human cells (Benanti, Nguyen and Welch 2015).
Rickettsia Sca2: adhesion to host cells and bacterial movement
The Rickettsia genus comprises several pathogenic species classified into different groups according to their biology, genetic and antigenic features. The spotted fever group includes Rickettsia rickettsii, R. conorii and R. parkeri, the etiological agents of Rocky Mountain, Mediterranean and mild-to-moderate spotted fever, respectively (Walker and Ismail 2008). These obligate intracellular bacterial pathogens are transmitted to the human host by arthropod vectors and their life cycle involves a stage within the host cell cytosol. Similarly to other cytosolic pathogens, Rickettsia induces its own intra- and intercellular movement by the polymerization of actin comet tails, thereby spreading from cell to cell and escaping lysosomal degradation and immune detection (Heinzen et al. 1999; Stevens, Galyov and Stevens 2006; Ray et al. 2009). Essential for these processes is the Surface cell antigen 2 (Sca2), a strong formin-like nucleator (Table 1). Similarly to Burkholderia BimA, Sca2 is a member of the autotransporter family that remains anchored to the outer membrane at the bacterial cell pole after secretion, from where it mediates the formation of microfilaments (Fig. 3D).
Although Sca2 homologues are present in many different Rickettsia, only members belonging to the spotted fever group possess WH2, FH1 and proline-rich regions in the bacterial cell surface exposed passenger domain (Fig. 1B) (Kleba et al. 2010). Functional and structural studies performed in Sca2 from R. rickettsii (Haglund et al. 2010) and R. parkeri (Madasu et al. 2013) have revealed Sca2 as a unique example of a bacterial formin-like nucleator. Similarities with formins include its role in enhancing nucleation of microfilaments, a high affinity for barbed ends, where it competes with capping protein binding and induction of profilin-dependent barbed-end elongation (Haglund et al. 2010; Madasu et al. 2013) (Fig. 2D). However, these similarities with formins remain mostly at the functional level. First, Sca2 sequence identity is restricted to other Sca2 homologues. Second, full-length Sca2 protein is active and all protein regions are involved in nucleation and elongation, in contrast to formins that are activated only when repression imposed by the autoinhibitory domain is relieved. Moreover, where formins are active as dimers, Sca2 is a monomer that uses the interaction between its N- and the C-terminal domains to bring proline-rich regions together, thus allowing incorporation of G-actin during processive elongation (Fig. 2D) (Madasu et al. 2013). In summary, Sca2 functionally mimics formins activities on actin but this is accomplished by different molecular and structural mechanisms.
Sca2 has different functionalities at different times of infection (Fig. 3D). It is required and sufficient for the critical stage of adherence to and invasion of host epithelial and endothelial cells by R. conorii, presumably by binding to a still unknown mammalian membrane receptor through a region also involved in actin nucleation (Cardwell and Martinez 2012). Inside the eukaryotic cell, it induces the formation of actin comet tails that drive the propulsion of bacteria intracellularly. Interestingly, at earlier stages of infection, bacterial movement is triggered by another effector, RickA, an NPF that recruits Arp2/3 to the bacterial pole. The global result is an initial RickA and Arp2/3-mediated slow and meandering movement resulting from short, curved actin tails, which is subsequently replaced by a more directionally steady and faster propulsion driven by nucleation of longer filaments by Sca2 (Reed et al. 2014).
Salmonella SipC: invasion and vacuole maturation
Salmonella enterica serovar Typhimurium (S. Typhimurium) causes mild gastroenteritis and severe infections in humans and domestic animals, normally by ingestion of contaminated food or water. This facultative intracellular pathogen is able to invade non-phagocytic epithelial cells and to replicate within a membrane-bound compartment, the Salmonella-containing vacuole (SCV) that interacts selectively with the endocytic pathway (Haraga, Ohlson and Miller 2008). The virulence of S. Typhimurium is dependent on two distinct T3SSs at least partially functioning during different stages of infection. The Salmonella pathogenicity island 1 (SPI-1) T3SS translocates effector proteins across the plasma membrane and is essential for bacterial invasion, while SPI-2 T3SS effectors traverse the SCV membrane and allow intracellular bacterial replication (Haraga, Ohlson and Miller 2008; Agbor and McCormick 2011). Salmonella invasion protein C (SipC; Table 1) was the first described bacterial actin nucleator (Hayward and Koronakis 1999). SipC is a SPI-1 T3SS substrate forming a translocon complex in the host cell plasma membrane through which SPI-1 effectors reach the host cell cytosol (Scherer, Cooper and Miller 2000). In addition, SipC has also an effector role and is possibly also directly involved in Salmonella invasion of non-phagocytic cells. Nucleation of microfilaments and their bundling by SipC leads to cytoskeletal rearrangements in tissue cultured cells that result in membrane ruffles underneath the site of bacterial attachment (Chang, Chen and Zhou 2005; Myeni and Zhou 2010) (Fig. 3E). These activities involve also the cooperation of the effector SPI-1 T3SS effector SipA, which stimulates the action of SipC (McGhie, Hayward and Koronakis 2001).
SipC can be divided into three functional domains (Fig. 1B). The C-terminus has a fundamental role in effector translocation (Chang, Chen and Zhou 2005) and, together with the central dimerization region, mediates nucleation of microfilaments (Hayward and Koronakis 1999). The role of the N-terminal domain is still a matter of debate. This region was suggested as the one mediating F-actin bundling, while the C-terminal was responsible for actin nucleation (Hayward and Koronakis 1999; Chang, Chen and Zhou 2005; Chang et al. 2007). In spite of this, later work performed by Myeni and Zhou (2010) demonstrated that the SipC C-terminus contains the F-actin binding site and is sufficient for its F-actin bundling activity. More recently, novel interactions of SipC with host cell proteins have suggested roles in interference with vesicle trafficking. In particular, SipC can bind Exo70, a component of the exocyst complex involved in the tethering of exocytic vesicles to the plasma membrane (Nichols and Casanova 2010). During infection, exocyst components are indeed enriched at the plasma membrane around attached Salmonella and this localized targeting of vesicles is presumed to provide additional membrane in bacterial uptake (Nichols and Casanova 2010). Additionally, SipC binds SNARE Syntaxin 6 (Madan et al. 2012). It has been suggested that through its binding to Syntaxin 6, SipC promotes the recruitment of vesicles that stabilize the SCV and help avoiding lysosome fusion (Madan et al. 2012).
Legionella pneumophila VipA: vesicular trafficking
Legionnaires’ disease is a frequently fatal type of pneumonia caused by the facultative intracellular human pathogen Legionella pneumophila. In nature, this Gram-negative bacterium is found in soil and freshwater, where it infects unicellular protozoan hosts. In man-made habitats such as cooling towers, spas and air-conditioning systems, the balance between bacteria and protozoa can be shifted towards a rapid proliferation of the pathogen, which can lead to disease upon inhalation of contaminated aerosols (McDade et al. 1977; Muder, Yu and Woo 1986). After phagocytic uptake by human alveolar macrophages, the bacterium prevents phagosomal maturation and triggers the biogenesis of the Legionella-containing vacuole (LCV), a niche suitable for bacterial survival and replication (Franco, Shuman and Charpentier 2009; Isberg, O'Connor and Heidtman 2009). This escape from host bactericidal mechanisms is greatly due to the translocation of more than 300 bacterial effectors via a T4SS. One of the substrates of this T4SS of Legionella is VipA (Vacuolar protein sorting inhibitor protein A), discovered using a pathogen effector protein screening in yeast (Shohdy et al. 2005). VipA was found to impair vacuolar protein trafficking in Saccharomyces cerevisiae, specifically interfering with the multivesicular body (MVB) pathway, a function that seems to be linked to its interaction with actin (Shohdy et al. 2005; Franco, Shohdy and Shuman 2012) (Fig. 3F). During infection of THP-1 macrophages, translocated VipA associates with F-actin patches and early endosomes, a distribution similar to the one observed when ectopically expressed in mammalian cells. Functional studies of truncated versions of the protein implicate its C-terminal region in mediating actin binding and polymerization, as well as interference with organelle trafficking in yeast (Bugalhão et al., submitted for publication). Analysis of the VipA amino acid sequence revealed the presence of a proline-rich region within its C-terminus, but similarly to Tarp and SipC no WH2 motifs seem to be present in the protein (Fig. 1B). Although the assessment of the role of VipA during infection has been hindered by the usual absence of phenotype in L. pneumophila single knockout mutants, its function as a link between the host actin cytoskeleton and organelle trafficking allows to anticipate a new activity amongst bacterial actin nucleators.
CONCLUDING REMARKS
Bacterial virulence proteins able to directly nucleate actin are encoded in the genomes of many Gram-negative bacteria. They access the cytosol of host cells via different protein secretion systems (e.g. T3SS, T4SS or T5SS). These bacterial actin nucleators possess specific structural domains involved in actin dynamics often found in eukaryotic proteins: for example, G-actin binding WH2 domains and proline-rich regions involved in the recruitment of profilin-actin (Fig. 1). The presence of these two regions enables bacterial actin nucleators to functionally mimic eukaryotic nucleators. In particular, the mechanism of action of some bacterial nucleators is similar to the one of formins (e.g. Rickettsia Sca2) or of tandem monomer binders (e.g. Vibrio VopL, Chlamydia Tarp) (Fig. 2). While the mechanism of action of Vibrio VopF is also similar to formins (and to Sca2), as VopF remains bound to and promotes filament polymerization at the barbed end, it is also different from formins (and from Sca2) because VopF does not require binding to profilin for its activity of enhancement of actin elongation at the barbed end. Some of these bacterial nucleators also display functional analogies to eukaryotic proteins of the ENA/VASP family that promote filament elongation and bundling (e.g. B. mallei and B. pseudomallei BimA), or that have an NPF-like function by direct activation of the Arp2/3 complex (e.g. B. thailandensis BimA) or by indirect activation of the Arp2/3 complex through the modulation of the activity of Rac GEFs (e.g. Tarp). Therefore, bacterial nucleators display a plethora of mechanisms enabling them to hijack the host cell actin cytoskeleton, ranging from direct and indirect filament nucleation to mediation of elongation and F-actin crosslinking.
In spite of their common function as actin nucleators, these bacterial virulence proteins show a variety of roles during infection. While Tarp, Sca2 and SipC are used by intracellular pathogens to trigger actin polymerization and promote bacterial invasion of non-phagocytic cells, extracellular Vibrio use Vop nucleators to disturb the integrity of intestinal epithelia. BimA is involved in the formation of actin tails that mediate intra- and intercellular movement, a function also present in Sca2, and in the classical examples of Listeria ActA and Shigella IcsA. Finally, VipA and SipC interfere with vesicle trafficking, and effects of Tarp, VopF and VopN in apoptosis and cell cycle have been described, although the mechanisms used to achieve these activities are still unclear.
The fact that even organisms belonging to closely related phylogenetically groups, such as different C. trachomatis serovars or Burkholderia spp., express slightly distinct nucleators indicates that these proteins have been fine-tuned during evolution to fit their specific host cell. Therefore, further mechanistic and functional studies on these (and new) bacterial actin nucleators might not only provide a deeper insight on the crosstalk between pathogen and host, but also reveal new eukaryotic pathways that are regulated by actin dynamics.
FUNDING
Work in the laboratory of IF has been funded by grants PTDC/BIA-MIC/2821/2012 from Fundação para a Ciência e Tecnologia (FCT) and PCOFUND-GA-2009-246542 from the European Union Seventh Framework Programme FP7/2007-2013 (to IF) and UID/Multi/04378/2013 from FCT (to UCIBIO).
Conflict of interest. None declared.
REFERENCES
- Abdelrahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29:949–59. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Agbor TA, McCormick BA. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol. 2011;13:1858–69. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktories K, Lang AE, Schwan C, et al. Actin as target for modification by bacterial protein toxins. FEBS J. 2011;278:4526–43. doi: 10.1111/j.1742-4658.2011.08113.x. [DOI] [PubMed] [Google Scholar]
- Avvaru BS, Pernier J, Carlier MF. Dimeric WH2 repeats of VopF sequester actin monomers into non-nucleating linear string conformations: An X-ray scattering study. J Struct Biol. 2015;190:192–9. doi: 10.1016/j.jsb.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Bebear C, de Barbeyrac B. Genital Chlamydia trachomatis infections. Clin Microbiol Infec. 2009;15:4–10. doi: 10.1111/j.1469-0691.2008.02647.x. [DOI] [PubMed] [Google Scholar]
- Benanti EL, Nguyen CM, Welch MD. Virulent burkholderia species mimic host actin polymerases to drive actin-based motility. Cell. 2015;161:348–60. doi: 10.1016/j.cell.2015.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain RJ, Hayward RD, Koronakis V. Deciphering Interplay between Salmonella Invasion Effectors. PLoS Pathog. 2008;4:e1000037. doi: 10.1371/journal.ppat.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell MM, Martinez JJ. Identification and characterization of the mammalian association and actin-nucleating domains in the Rickettsia conorii autotransporter protein, Sca2. Cell Microbiol. 2012;14:1485–95. doi: 10.1111/j.1462-5822.2012.01815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell MM, Martinez JJ. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect Immun. 2009;77:5272–80. doi: 10.1128/IAI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Chen J, Zhou D. Delineation and characterization of the actin nucleation and effector translocation activities of Salmonella SipC. Mol Microbiol. 2005;55:1379–89. doi: 10.1111/j.1365-2958.2004.04480.x. [DOI] [PubMed] [Google Scholar]
- Chang J, Myeni SK, Lin TL, et al. SipC multimerization promotes actin nucleation and contributes to Salmonella-induced inflammation. Mol Microbiol. 2007;66:1548–56. doi: 10.1111/j.1365-2958.2007.06024.x. [DOI] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Bio. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Clifton DR, Fields KA, Grieshaber SS, et al. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. P Natl Acad Sci USA. 2004;101:10166–71. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell Microbiol. 2000;2:195–205. doi: 10.1046/j.1462-5822.2000.00053.x. [DOI] [PubMed] [Google Scholar]
- Dziejman M, Serruto D, Tam VC, et al. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. P Natl Acad Sci USA. 2005;102:3465–70. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Ceesay A, Kim JH, et al. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 2008;4:e1000021. doi: 10.1371/journal.ppat.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco IS, Shohdy N, Shuman HA. The Legionella pneumophila effector VipA is an actin nucleator that alters host cell organelle trafficking. PLoS Pathog. 2012;8:e1002546. doi: 10.1371/journal.ppat.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco IS, Shuman HA, Charpentier X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 2009;11:1435–43. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- Galyov EE, Brett PJ, DeShazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol. 2010;64:495–17. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CT, et al. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12:1057–63. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999;18:4926–34. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen RA, Grieshaber SS, Van Kirk LS, et al. Dynamics of actin-based movement by Rickettsia rickettsii in vero cells. Infect Immun. 1999;67:4201–7. doi: 10.1128/iai.67.8.4201-4207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Dooley CA, Mead DJ, et al. Chlamydia trachomatis tarp is phosphorylated by src family tyrosine kinases. Biochem Bioph Res Co. 2008;371:339–44. doi: 10.1016/j.bbrc.2008.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Fischer ER, Mead DJ, et al. Chlamydial TARP is a bacterial nucleator of actin. P Natl Acad Sci USA. 2006;103:15599–604. doi: 10.1073/pnas.0603044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Miller NJ, Dooley CA, et al. The conserved Tarp actin binding domain is important for chlamydial invasion. PLoS Pathog. 2010;6:e1000997. doi: 10.1371/journal.ppat.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiwani S, Alvarado S, Ohr RJ, et al. Chlamydia trachomatis Tarp harbors distinct G and F actin binding domains that bundle actin filaments. J Bacteriol. 2013;195:708–16. doi: 10.1128/JB.01768-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kespichayawattana W, Rattanachetkul S, Wanun T, et al. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect Immun. 2000;68:5377–84. doi: 10.1128/iai.68.9.5377-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleba B, Clark TR, Lutter EI, et al. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun. 2010;78:2240–7. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane BJ, Mutchler C, Al Khodor S, et al. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 2008;4:e1000014. doi: 10.1371/journal.ppat.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liverman AD, Cheng HC, Trosky JE, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. P Natl Acad Sci USA. 2007;104:17117–22. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter EI, Bonner C, Holland MJ, et al. Phylogenetic analysis of Chlamydia trachomatis Tarp and correlation with clinical phenotype. Infect Immun. 2010;78:3678–88. doi: 10.1128/IAI.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade JE, Shepard CC, Fraser DW, et al. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Eng J Med. 1977;297:1197–203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- McGhie EJ, Hayward RD, Koronakis V. Cooperation between actin-binding proteins of invasive Salmonella: SipA potentiates SipC nucleation and bundling of actin. EMBO J. 2001;20:2131–9. doi: 10.1093/emboj/20.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan R, Rastogi R, Parashuraman S, et al. Salmonella acquires lysosome-associated membrane protein 1 (LAMP1) on phagosomes from Golgi via SipC protein-mediated recruitment of host Syntaxin6. J Biol Chem. 2012;287:5574–87. doi: 10.1074/jbc.M111.286120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madasu Y, Suarez C, Kast DJ, et al. Rickettsia Sca2 has evolved formin-like activity through a different molecular mechanism. P Natl Acad Sci USA. 2013;110:E2677–86. doi: 10.1073/pnas.1307235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlitz A, Banhart S, Hess S, et al. Complex kinase requirements for Chlamydia trachomatis Tarp phosphorylation. FEMS Microbiol Lett. 2008;289:233–40. doi: 10.1111/j.1574-6968.2008.01390.x. [DOI] [PubMed] [Google Scholar]
- Mehlitz A, Banhart S, Maurer AP, et al. Tarp regulates early Chlamydia-induced host cell survival through interactions with the human adaptor protein SHC1. J Cell Biol. 2010;190:143–57. doi: 10.1083/jcb.200909095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muder RR, Yu VL, Woo AH. Mode of transmission of Legionella pneumophila. A critical review. Arch Intern Med. 1986;146:1607–12. [PubMed] [Google Scholar]
- Myeni SK, Zhou D. The C terminus of SipC binds and bundles F-actin to promote Salmonella invasion. J Biol Chem. 2010;285:13357–63. doi: 10.1074/jbc.M109.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S, Boczkowska M, Glista MJ, et al. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat Struct Mol Biol. 2011;18:1060–7. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CD, Casanova JE. Salmonella-directed recruitment of new membrane to invasion foci via the host exocyst complex. Curr Biol. 2010;20:1316–20. doi: 10.1016/j.cub.2010.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskel. 2009;66:606–17. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernier J, Orban J, Avvaru BS, et al. Dimeric WH2 domains in Vibrio VopF promote actin filament barbed-end uncapping and assisted elongation. Nat Struct Mol Biol. 2013;20:1069–76. doi: 10.1038/nsmb.2639. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM. New players in actin polymerization–WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19:276–85. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Ray K, Marteyn B, Sansonetti PJ, et al. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–40. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- Rebowski G, Namgoong S, Boczkowska M, et al. Structure of a longitudinal actin dimer assembled by tandem w domains: implications for actin filament nucleation. J Mole Biol. 2010;403:11–23. doi: 10.1016/j.jmb.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SC, Lamason RL, Risca VI, et al. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol. 2014;24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar K, Sadhukhan S, Han SS, et al. Disruption of hSWI/SNF complexes in T cells by WAS mutations distinguishes X-linked thrombocytopenia from Wiskott-Aldrich syndrome. Blood. 2014;124:3409–19. doi: 10.1182/blood-2014-07-587642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer CA, Cooper E, Miller SI. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol Microbiol. 2000;37:1133–45. doi: 10.1046/j.1365-2958.2000.02066.x. [DOI] [PubMed] [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–74. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohdy N, Efe JA, Emr SD, et al. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. P Natl Acad Sci USA. 2005;102:4866–71. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitthidet C, Korbsrisate S, Layton AN, et al. Identification of motifs of Burkholderia pseudomallei BimA required for intracellular motility, actin binding, and actin polymerization. J Bacteriol. 2011;193:1901–10. doi: 10.1128/JB.01455-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitthidet C, Stevens JM, Field TR, et al. Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. J Bacteriol. 2010;192:5249–52. doi: 10.1128/JB.00608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- Stevens JM, Ulrich RL, Taylor LA. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J Bacteriol. 2005;187:7857–62. doi: 10.1128/JB.187.22.7857-7862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MP, Stevens JM, Jeng RL, et al. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol Microbiol. 2005;56:40–53. doi: 10.1111/j.1365-2958.2004.04528.x. [DOI] [PubMed] [Google Scholar]
- Tam VC, Serruto D, Dziejman M, et al. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe. 2007;1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Tam VC, Suzuki M, Coughlin M, et al. Functional analysis of VopF activity required for colonization in Vibrio cholerae. mBio. 2010;1:e00289-10. doi: 10.1128/mBio.00289-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thwaites T, Nogueira AT, Campeotto I, et al. The Chlamydia effector TarP mimics the mammalian leucine-aspartic acid motif of paxillin to subvert the focal adhesion kinase during invasion. J Biol Chem. 2014;289:30426–42. doi: 10.1074/jbc.M114.604876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toesca IJ, French CT, Miller JF. The Type VI secretion system spike protein VgrG5 mediates membrane fusion during intercellular spread by pseudomallei group Burkholderia species. Infect Immun. 2014;82:1436–44. doi: 10.1128/IAI.01367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travier L, Lecuit M. Listeria monocytogenes ActA: a new function for a ‘classic’ virulence factor. Curr Opin Microbiol. 2014;17:53–60. doi: 10.1016/j.mib.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Tripathi R, Kaithwas V, Dureja C, et al. Alanine-scanning mutagenesis of WH2 domains of VopF reveals residues important for conferring lethality in a Saccharomyces cerevisiae model. Gene. 2013;525:116–23. doi: 10.1016/j.gene.2013.04.071. [DOI] [PubMed] [Google Scholar]
- Truong D, Copeland JW, Brumell JH. Bacterial subversion of host cytoskeletal machinery: hijacking formins and the Arp2/3 complex. BioEssays. 2014;36:687–96. doi: 10.1002/bies.201400038. [DOI] [PubMed] [Google Scholar]
- Wagner AR, Luan Q, Liu SL, et al. Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr Biol. 2013;23:1990–8. doi: 10.1016/j.cub.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008;6:375–86. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–9. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Welch MD, Way M. Arp2/3-mediated actin-based motility: a tail of pathogen abuse. Cell Host Microbe. 2013;14:242–55. doi: 10.1016/j.chom.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright HR, Turner A, Taylor HR. Trachoma. Lancet. 2008;371:1945–54. doi: 10.1016/S0140-6736(08)60836-3. [DOI] [PubMed] [Google Scholar]
- Yu B, Cheng HC, Brautigam CA, et al. Mechanism of actin filament nucleation by the bacterial effector VopL. Nat Struct Mol Biol. 2011;18:1068–74. doi: 10.1038/nsmb.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahm JA, Padrick SB, Chen Z, et al. The bacterial effector VopL organizes actin into filament-like structures. Cell. 2013;155:423–34. doi: 10.1016/j.cell.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]