Abstract
Escherichia coli is one of the first causes of Gram-negative orthopedic implant infections (OII), but little is known about the pathogenicity of this species in such infections that are increasing due to the ageing of the population. We report how this pathogen interacts with human osteoblastic MG-63 cells in vitro, by comparing 20 OII E. coli strains to two Staphylococcus aureus and two Pseudomonas aeruginosa strains. LDH release assay revealed that 6/20 (30%) OII E. coli induced MG-63 cell lysis whereas none of the four control strains was cytotoxic after 4 h of coculture. This high cytotoxicity was associated with hemolytic properties and linked to hlyA gene expression. We further showed by gentamicin protection assay and confocal microscopy that the non-cytotoxic E. coli were not able to invade MG-63 cells unlike S. aureus strains (internalization rate <0.01% for the non-cytotoxic E. coli versus 8.88 ± 2.31% and 4.60 ± 0.42% for both S. aureus). The non-cytotoxic E. coli also demonstrated low adherence rates (<7%), the most adherent E. coli eliciting higher IL-6 and TNF-α mRNA expression in the osteoblastic cells. Either highly cytotoxic or slightly invasive OII E. coli do not show the same infection strategies as S. aureus towards osteoblasts.
Keywords: E. coli, human osteoblasts, cytotoxicity, internalization, alpha-hemolysin, TNF-α
This study describes the interaction of Escherichia coli clinical strains with an osteoblastic cell line, showing no internalization unlike Staphylococcus aureus, but a strong cytotoxicity of the Hly-A producing strains.
Graphical Abstract Figure.
This study describes the interaction of Escherichia coli clinical strains with an osteoblastic cell line, showing no internalization unlike Staphylococcus aureus, but a strong cytotoxicity of the Hly-A producing strains.
INTRODUCTION
Despite serious technological advances in orthopedic surgery and improvement in clinical preventive practice, orthopedic implant infections (OII) continue to be a problem, with an incidence increasing with the boom of prosthetic joint implantations (Kurtz et al. 2012; Lima et al. 2013). Approximately 6–23% of all OII are caused by Gram-negative bacilli, and Escherichia coli is the most frequently isolated microorganism in these cases (Zmistowski et al. 2011; Peel et al. 2012; Rodriguez-Pardo et al. 2014).
Progress has been made over the past decade in the understanding of the physiopathogenesis of staphylococcal OII. Based on research in this field, bacterial adherence and biofilm formation on the implant surface represent major factors involved in the pathogenesis process, providing protection against antimicrobial therapy and host immune responses (Montanaro et al. 2011; Ribeiro, Monteiro and Ferraz 2012; Molina-Manso et al. 2013). Regarding bone infection, Staphylococcus aureus has been shown to invade osteoblasts, persist intracellularly and induce the secretion of key proinflammatory mediators and potent stimulators of osteoclastogenesis and bone resorption (Alexander et al. 2003; Marriott 2004; Marriot et al. 2005; Somayaji et al. 2008; Wright and Nair 2010; Ning et al. 2011; Hamza et al. 2013). Several virulence determinants, such as specific microbial surface components recognizing adhesive matrix molecules with in particular the fibronectin-binding proteins (FnBPs), the surface-bound protein A (SpA) or a class of pore-forming toxins named the phenol-soluble modulins (PSMs), have been demonstrated to play a role in S. aureus invasiveness and bone destruction (Wright and Nair 2010; Cassat et al. 2013; Claro et al. 2013; Rasigade et al. 2013).
Escherichia coli is known as a harmless commensal of the intestinal flora, but through virulence and fitness genes gain, evolves as a highly diverse and adapted pathogen (Köhler and Dobrindt 2011; Croxen et al. 2013). Very few data currently exist on the pathophysiological mechanisms involved in E. coli OII. As previously demonstrated by our group (Crémet et al. 2012), most of E. coli strains recovered from peri-implant tissues show a high virulence potential, but no molecular pathogenic signature distinguishes these strains from other extraintestinal pathogenic E. coli (ExPEC) like uropathogenic strains (UPEC). Furthermore, we showed that only a small number of OII E. coli forms strong biofilms on inert surfaces in experimental conditions (Crémet et al. 2012).
Little is known about the interaction of E. coli with bone. Major insights into osteoblasts and osteoclasts responses to E. coli infection come from studies of bone cells stimulated with the bacterial cell-wall component LPS (lipopolysaccharide) (Suda et al. 2004; Inada et al. 2006; Ochi et al. 2010; Yan et al. 2010; Gao et al. 2013; Guo et al. 2014; Nakao et al. 2014). Thus, in the present study, we used a well-characterized panel of 20 clinical OII E. coli strains to investigate whether this species can infect and survive within a human osteoblastic cell line and whether this infection elicits the secretion of proinflammatory mediators and promotes bone destruction (Crémet et al. 2012).
MATERIALS AND METHODS
Bacterial strains and growth conditions
A total of 20 clinical strains of E. coli (Ec1 to Ec20) involved in hip (14 strains) or knee (6 strains) OII were selected for this study. All OII E. coli were obtained from cultures of intraoperative tissue specimens of 20 patients, who displayed typical clinical signs of OII with acute presentation, and fulfilled diagnostic criteria for OII (Crémet et al. 2012). In five cases (Ec1, Ec3, Ec5, Ec14, Ec17), blood cultures were positive with the same E. coli, and in four patients, E. coli isolates were recovered from polymicrobial infections (Ec4, Ec8, Ec10, Ec12). Only one isolate per patient was included. The genetic relatedness of the 20 OII E. coli was studied by MLST analysis according to the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli). The strains were also investigated for the most common O-serotypes of UPEC and 20 established or putative virulence factors, by PCR (Table 1) (Li et al. 2010; Crémet et al. 2012). For comparison, we used the well-characterized osteomyelitis strain of S. aureus ATCC 49230 (Sa49230), the Pseudomonas aeruginosa laboratory reference strain PaO1 and two clinical strains from our collection: one S. aureus (Saclin) involved in infection of a total hip prosthesis and one P. aeruginosa (Paclin) recovered from an infected locking compression plate used to treat a tibia–fibula fracture. Two isogenic variants of the E. coli strain A0 34/86 [ZKLR+ (HlyA+) and Zhly– (ΔhlyA mutant)] were also introduced as controls in some experiments (Table 1) (Sheshko et al. 2006).
Table 1.
Virulence profiles of the 20 OII E. coli and 2 E. coli control strains studied.
| Site of | Phylogenetic | Virulence factorsb: | ||||
|---|---|---|---|---|---|---|
| Isolatea | infection | group | MLST | Serotype | fimH, papC, papGII, papGIII, sfa/foc, hra, tsh, iucC, hlyA, traT, sat, cnf1, usp, cdt, vat, ibeA, ompT, neuB, csgA and pgaA | Reference |
| Ec1 | Knee | B2 | ST95 | O2 | fimH, papC, papGII, iucC, traT, usp, vat, ompT, neuB, csgA and pgaA | This study |
| Ec2 | Knee | B2 | ST80 | O75 | fimH, sfa/foc, hra, hlyA, cnf1, usp, vat, ibeA, ompT, neuB, csgA and pgaA | This study |
| Ec3 | Hip | B2 | ST537 | O75 | fimH, papC, papGIII, sfa/foc, hra, hlyA, cnf1, usp, cdt, vat, ibeA, ompT, csgA and pgaA | This study |
| Ec4 | Knee | B2 | ST126 | - | fimH, papC, papGIII, sfa/foc, hra, hlyA, cnf1, usp, cdt, vat, ibeA, ompT, csgA and pgaA | This study |
| Ec5 | Hip | B2 | ST141 | O2 | fimH, papC, papGIII, sfa/foc, hra, hlyA, cnf1, usp, vat, ompT, neuB, csgA and pgaA | This study |
| Ec6 | Hip | B2 | ST404 | O75 | fimH, papGII, iucC, sat, usp, vat, ompT, csgA and pgaA | This study |
| Ec7 | Hip | B2 | ST73 | - | fimH, sfa/foc, hra, hlyA, cnf1, usp, vat, ompT, csgA and pgaA | This study |
| Ec8 | Hip | B2 | ST73 | O6 | fimH, papC, papGII, sfa/foc, iucC, hlyA, sat, usp, vat, ompT, csgA and pgaA | This study |
| Ec9 | Hip | B2 | ST550 | O75 | fimH, sfa/foc, iucC, sat, usp, vat, ompT, csgA and pgaA | This study |
| Ec10 | Hip | B2 | ST95 | O1 | fimH, papC, papGII, iucC, traT, usp, vat, ompT, neuB, csgA and pgaA | This study |
| Ec11 | Knee | B2 | ST141 | O2 | fimH, papC, papGIII, sfa/foc, hra, hlyA, cnf1, usp, vat, ompT, neuB, csgA and pgaA | This study |
| Ec12 | Hip | B2 | ST1618 | O4 | fimH, sfa/foc, usp, cdt, vat, ompT, csgA and pgaA | This study |
| Ec13 | Hip | B2 | ST95 | O1 | fimH, papC, papGII, iucC, traT, usp, vat, ompT, neuB, csgA and pgaA | This study |
| Ec14 | Hip | D | ST354 | O1 | fimH, iucC, traT, usp, ibeA and csgA | This study |
| Ec15 | Knee | D | ST405 | O2 | fimH, papC, hra, iucC, hlyA, traT, sat, ompT, csgA and pgaA | This study |
| Ec16 | Hip | D | ST420 | – | fimH, usp, vat, ibeA, ompT, neuB, csgA and pgaA | This study |
| Ec17 | Hip | A | ST361 | – | fimH, traT, csgA and pgaA | This study |
| Ec18 | Knee | A | ST88 | O8 | fimH, papC, hra, iucC, csgA and pgaA | This study |
| Ec19 | Hip | A | ST10 | – | fimH, papC, hra, iucC, traT, ompT, csgA and pgaA | This study |
| Ec20 | Hip | B1 | ST1167 | O21 | fimH, iucC, ompT, csgA and pgaA | This study |
| ZKLR± | – | B2 | – | O83 | fimH, papC, papGII, papGIII, sfa/foc, hra, tsh, hlyA, cnf1, usp, vat, ibeA, ompT, csgA and pgaA | (Sheshko et al. 2006) |
| Zhly_ | – | B2 | – | O83 | fimH, papC, papGII, papGIII, sfa/foc, hra, tsh, cnf1, usp, vat, ibeA, ompT, csgA and pgaA | (Sheshko et al. 2006) |
aE. coli strains that induced osteoblasts lysis in vitro are underlined.
bVirulence factors significantly associated with the osteoblasts lysis potential are shown in bold type.
Before infection of the osteoblast cultures, the strains were grown overnight at 37°C in 10 mL of Luria-Bertani (LB) broth, harvested by centrifugation for 10 min at 800 g and washed once in 5 mL of phosphate-buffered saline (PBS). The pellets were resuspended in 5 mL of Dulbecco's Modified Eagle's Medium (DMEM; Lonza, Belgium). Bacterial suspensions were adjusted with a nephelometer to obtain 1 × 108 CFU mL−1 and then diluted in DMEM.
Human osteoblast cultures
The human osteoblast-like osteosarcoma cell line MG-63 purchased from ATCC was cultured in a 5% CO2 atmosphere at 37°C in DMEM supplemented with 5% of fetal bovine serum (Hyclone Perbio, France). One day before infection, the cells were seeded at 105 cells/wells in 24-well culture plates to obtain confluent monolayers.
Lactate dehydrogenase (LDH) release assays
Cytotoxicity was determined by quantifying LDH release into MG-63 cell culture supernatants, after 2 or 4 h of infection with the different strains at a multiplicity of infection (MOI) of 10:1. The enzymatic activity of LDH was measured with the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega), as described by the manufacturer. Cells mortality was calculated relative to that of uninfected cells (set at 0%), and cells lysed with 1% Triton X-100 (positive control, 100%).
Hemolysis assays
The hemolytic activity of the strains was quantified in defibrinated horse blood (bioMérieux, Marcy l'Etoile, France) diluted to 10% (v/v) in PBS with Ca2+ and Mg2+. For these experiments, 200 μL of each bacterial suspension at a concentration of 108 CFU mL–1 in PBS with Ca2+ and Mg2+ was added to 200 μL of the erythrocytes suspension. Following 2, 6, or 24 h of incubation at 37°C, the unlysed erythrocytes were pelleted by centrifugation at 1500 × g for 2 min, and the optical density (OD) of 150 μL of the cell-free supernatants was measured at 450 nm. In each assay, a negative control (0% lysis) and a positive control (100% lysis) were obtained by addition of 200 μL of PBS with Ca2+ and Mg2+ or 1% Triton X-100 in place of the bacterial suspension, respectively. The percentage of hemolysis was calculated as follows: (OD450nm supernatants – OD450nm negative control)/(OD450nm positive control – OD450nm negative control) × 100.
Measurement of hlyA and cnf1 mRNA expression
The expression of the hlyA and cnf1 genes was analyzed in a relative quantification assay by real-time PCR, using the E. coli gapA housekeeping gene as a reference. The strains were grown overnight in LB broth, then adjusted to a density of 0.5 MacFarland units and diluted one-tenth in 10 mL of LB broth. The diluted suspensions were cultured for an additional 3 h in a 37°C water bath, and the total RNA from 1 mL of each suspension was then extracted using the High Pure RNA Isolation Kit (Roche) following the manufacturer's instructions. The RNA yield was assessed by UV absorbance, and 100 ng of total RNA was reverse transcribed and amplified using a One Step SYBR® PrimeScript RT-PCR Kit II (TaKaRa) with primers listed in Table 2.
Table 2.
Primer sequences used in RT-PCR analysis.
| Primer | Nucleotide sequence (5′-3′) |
|---|---|
| gapA-F | ACGAAGTTGGTGTTGACGTTG |
| gapA-R | ATAACCACTTTCTTCGCACCA |
| hlyA-F | AGCAGGACAAAGCACGAAA |
| hlyA-R | GTAATCGCCGTGCCATTCTT |
| cnf1-F | ATGAAGCCCCGGTTGAAGTA |
| cnf1-R | TCCCCGTCCTTTAAGCACAT |
| β-actin-F | CCCAGCCATGTACGTTGCTA |
| β-actin-R | AGGGCATACCCCTCGTAGATG |
| h-IL-6-F | TCCACAAGCGCCTTCGGTCCAG |
| h-IL-6-R | CTCAGGGCTGAGATGCCGTCG |
| h-TNF-α-F | CAGCCTCTTCTCCTTCCTGAT |
| h-TNF-α-R | GCCAGAGGGCTGATTAGAGA |
Analysis of the chromosomal region upstream of the hlyC gene
Primer pairs previously developed from sequences of PAI I (81f/r) and PAI II (72f/r) from the UPEC strain 536 were used for amplification and sequencing of the region upstream of the hlyC gene (Burgos and Beutin 2010). Primers ops-hly-f 5′ ACATGAGCAAAACGGATGGG 3′and ops-hly-r 5′ ATCCCAGCACGCAAAAGAAG 3′ were used for amplification and sequencing of the ops element (operon polarity suppressor) upstream of the hlyCABD genes.
Internalization and adherence assays
For bacterial internalization experiments, the MG-63 monolayers were infected during 2 h (MOI of 10:1). The number of bacteria present in the inocula was verified by plating dilutions on Tryptic Soy Agar (TSA) plates. After 2 h of infection, the MG-63 cells were washed twice with PBS and incubated for further 4 h in 1 mL of DMEM containing either 100 μg mL−1 gentamicin for cells infected with the S. aureus or E. coli strains or 1 mg mL−1 gentamicin for cells infected with the P. aeruginosa strains, to kill the remaining extracellular bacteria (all strains were susceptible to gentamicin with the VITEK2® system). The MG-63 monolayers were then washed three times and lysed with 0.1% Triton X-100. The lysates were serially diluted, and plated on TSA plates to determine the number of intracellular bacteria.
To measure the number of adherent bacteria, the MG-63 monolayers were infected as for internalization experiments, without the use of gentamicin to kill extracellular bacteria. After washing three times with PBS, MG-63 cells were lysed as described above, and lysates were serially diluted and plated on TSA plates.
Adherence and invasion efficiencies were calculated as the ratio of the number of cell-associated bacteria or cell-internalized bacteria, respectively, and the number of bacteria used to infect MG-63 monolayers.
Bacterial internalization assessment by confocal microscopy
MG-63 cells were seeded onto glass cover slips and infected with three clinical strains from OII (Saclin, Paclin and Ec2), as described above for the internalization assays. Following incubation with gentamicin, the MG-63 cells were washed three times, fixed with 4% paraformaldehyde and then permeabilized and blocked with 0.05% Triton X-100 + 1% bovine serum albumin. FITC-labeled rabbit anti-E. coli, S. aureus or P. aeruginosa antibodies (Thermo Fisher Scientific) were used to visualize the bacteria, along with DRAQ5 far-red fluorescent DNA dye (Eurobio) and Alexa Fluor 546® phalloidin (Life Technologies) for labeling F-actin. Slides were acquired on a Nikon A1R-Si confocal microscope, and pictures were analyzed with the Fiji software.
Measurement of IL-6 and TNF-α mRNA expression by relative quantitative RT-PCR
Following 4 or 6 h of infection (MOI of 100:1), the MG-63 monolayers were washed twice with PBS and RNA was isolated using the High Pure RNA Isolation Kit (Roche) following the manufacturer's instructions. The RNA yield was assessed by UV absorbance, and 2 μg of total RNA was reverse transcribed using the SuperScript III Reverse Transcriptase (Life technologies) with random primers (Promega), as recommended by the manufacturers. Real-time amplification of the cDNA was performed using the SYBR® Premix Ex Taq (TaKaRa) and primers were listed in Table 2. The expression of the lL-6 and TNF-α mRNAs was normalized against β-actin mRNA and quantified using the comparative 2−ΔCT method.
IL-6 and TNF-α secretion assays
The release of IL-6 and TNF-α in culture supernatants of uninfected and infected MG63 cells (5 × 105 cells/wells in 6-well culture plates, MOI of 1:100) was quantified by ELISA, according to the manufacturer's instructions (eBioscience, Paris, France), after 24 h of incubation or 72 h, including a treatment with 100 μg mL−1 gentamicin at 24 h post-infection.
Statistical analysis
All assays were performed on three separate occasions. Results were expressed as means ± SD. Fisher's exact test and Kruskal–Wallis test with Dunn's multiple comparison post test were used to calculate P values. P values less than 0.05 were considered statistically significant.
RESULTS
Some OII E. coli strains induce osteoblasts lysis in vitro
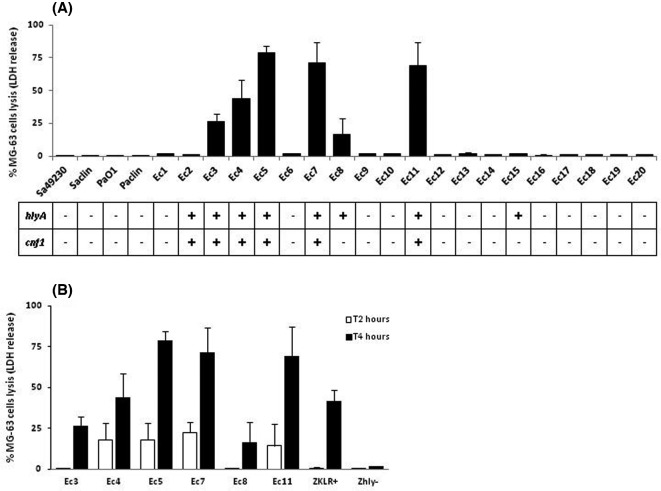
Viability of the MG-63 cells was assessed following infection with the 20 E. coli and 6 control strains. Osteoblasts were not lysed following 2 or 4 h of infection with the S. aureus or P. aeruginosa control strains. By contrast, 6/20 OII E. coli induced cytotoxicity following 4 h (Fig. 1A), and cell lysis could already be observed at 2 h for four strains (Fig. 1B). No cytotoxic effect was detected when the strains were heat inactivated at 100°C for 10 min before infection.
Figure 1.
LDH release assays. In vitro lytic effects on MG-63 cells of the 20 OII E. coli and six control strains. (A) Percentage of MG-63 cells lysis after 4 h of infection (MOI of 10:1), and correlation with the detection of two virulence genes (hlyA and cnf1), by PCR. (B) Levels of mortality obtained after 2 and 4 h of infection (MOI of 10:1) for the six cytotoxic E. coli strains of our panel, and both E. coli control strains: ZKLR+, positive for the hlyA gene, and Zhly–, the ΔhlyA mutant of ZKLR+.
Cytotoxicity, hemolytic activity and alpha-hemolysin (HlyA) expression are linked
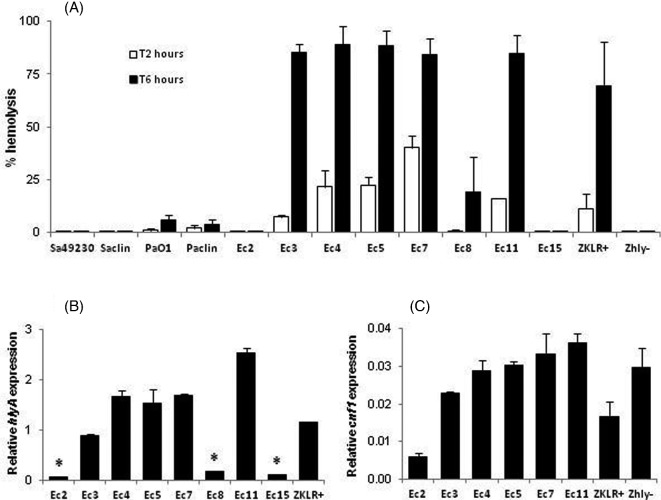
Investigation of the virulence profiles of the 20 E. coli strains showed that the cytolytic potential was significantly associated with two virulence genes (hlyA and cnf1, P = 0.0007 and 0.0022, respectively; Fisher's exact test) (Table 1, Fig. 1A). The six strains that induced cytotoxicity after 4 h of infection were positive for hlyA, and experiments with both E. coli ZKLR+ (hlyA+) and Zhly– (ZKLR+ ΔhlyA mutant) confirmed the link between the hlyA gene and MG-63 cells lysis (Fig. 1B). Since two hlyA-positive strains (Ec2 and Ec15) did not induce lysis (Fig. 1A), we analyzed the hemolytic activity of the eight hlyA-positive strains of our panel and studied the hlyA gene expression in these strains (Fig. 2), to characterize the influence of alpha-hemolysin production on MG-63 cells lysis. In accordance with the results shown above, two hlyA-positive strains (Ec2 and Ec15) were non-hemolytic. The six cytotoxic E. coli strains and the ZKLR+ control, but not the Zhly– control, caused hemolysis with some differences, the maximum effect being obtained after 6 h of incubation with erythrocytes (Fig. 2A). The observed differences were correlated with the levels of hlyA gene expression (Fig. 2B), and with the lytic effects on MG-63 cells (some variations observed for Ec3 being probably due to differences in response between the two cell types). Genetic analysis of the regions located upstream of the hlyCABD genes revealed that the eight hly operons were encoded on pathogenicity islands (PAI) II, and that the sequences of the respective ops elements, involved in transcriptional activation of the hlyCABD genes, were identical. The cnf1 gene was less strongly expressed than the hlyA gene, and while both genes were weakly expressed in the non-cytotoxic Ec2 strain, results obtained with the Zhly– E. coli confirmed that this virulence factor was not responsible for the MG-63 cells lysis (Figs 1B and 2C).
Figure 2.
Analysis of the hemolytic potential of hlyA-positive strains of E. coli. (A) Hemolytic activity of the eight hlyA-positive E. coli of our panel, and the six control strains of our study, after 2 and 6 h of incubation with horse erythrocytes. The 12 hlyA-negative E. coli of our panel did not induce hemolysis and are not shown. (B) Relative expression of the hlyA gene in the eight hlyA-positive strains, and the ZKLR+ E. coli control strain. * P < 0.01 versus Ec3, Ec4, Ec5, Ec7, Ec11 or ZKLR+ (Kruskal–Wallis test followed by a Dunn's post test). (C) Relative expression of the cnf1 gene in the six cnf1-positive strains, and both ZKLR+ and Zhly– E. coli control strain.
OII E. coli strains show fairly inefficient internalization in osteoblasts
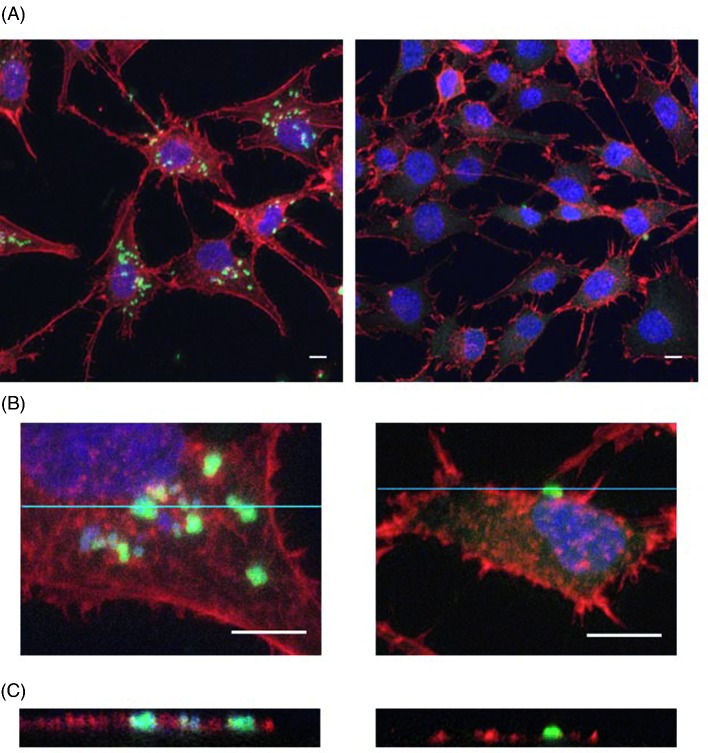
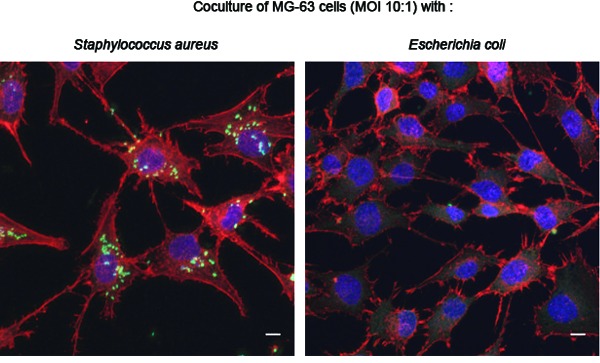
Evaluation of bacterial internalization was performed with the S. aureus and P. aeruginosa control strains and the 16 E. coli strains that did not induce cytotoxicity following 2 h of infection. All E. coli strains were less invasive than the control strains towards osteoblasts (mean percentage of internalized bacteria <0.01% for the 16 E. coli strains versus 8.88 ± 2.31% and 4.60 ± 0.42% for Saclin and Sa49230, respectively, or 0.57 ± 0.17% and 0.04 ± 0.03% for Paclin and PaO1, respectively). Determination of bacterial subcellular localization by confocal microscopy confirmed the intracellular presence of S. aureus or P. aeruginosa and nearly no internalization for E. coli (Fig. 3).
Figure 3.
Bacterial internalization assessment by confocal microscopy. Immunofluorescence stainings of F-actin (red, phalloidin), nuclei (blue, DRAQ5) and bacteria (green, polyclonal anti S. aureus or E. coli), performed after 2 h of coculture of MG-63 cells with the Saclin strain or with the OII E. coli strain Ec2 (MOI of 10:1) and 4 h of incubation with 100 μg mL−1 gentamicin. (A) The image on the left shows several S. aureus cells associated with the MG-63 cells, whereas very few E. coli cells were detected (image on the right). (B) Cropped images of the stacks analyzed in A confirm the presence of aggregates of S. aureus cells (image on the left) and a single E. coli cell (image on the right) in close contact with the MG-63 cells. (C) Orthogonal views derived from the blue lines of the crops analyzed in B show the intracellular location of the S. aureus cells (image on the left, green signals framed with red), whereas the E. coli cell was extracellular (image on the right, no red frame around the green signal). Scale bars: 10 μm.
OII E. coli strains show differences in adherence to osteoblasts
The adherence assays revealed differences among the 16 non-cytotoxic strains of E. coli (Fig. 4). Only two strains (Ec6 and Ec12) showed an adherence rate higher than 4%, and eight showed an adherence rate lower than 1%. No significant association was found between specific virulence genes and the ability of the strains to adhere to MG-63 cells in vitro.
Figure 4.
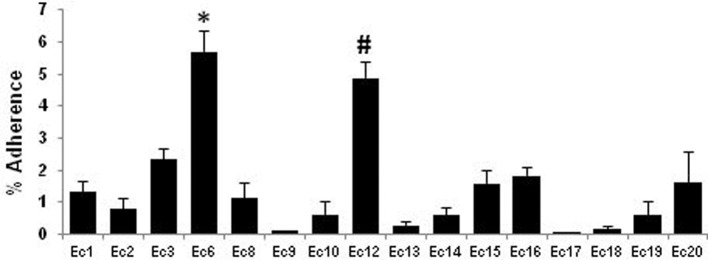
Adherence efficiencies of the OII E. coli strains. Percentage of adherent bacteria after 2 h of coculture of MG-63 cells with the 16 non-cytotoxic E. coli strains (MOI of 10:1). The four E. coli strains (Ec4, Ec5, Ec7 and Ec11) that induced osteoblasts lysis after 2 h of infection were not tested. The Ec6 and Ec12 strains were tested statistically with the Kruskal–Wallis test followed by a Dunn's post test. *, P < 0.05 versus Ec2, Ec9, Ec10, Ec13, Ec14, Ec17, Ec18 or Ec19; #, P < 0.05 versus Ec9, Ec10, Ec13, Ec14, Ec17, Ec18 or Ec19.
Adherence of OII E. coli strains to osteoblasts elicits weak induction of IL-6 and TNF-α responses
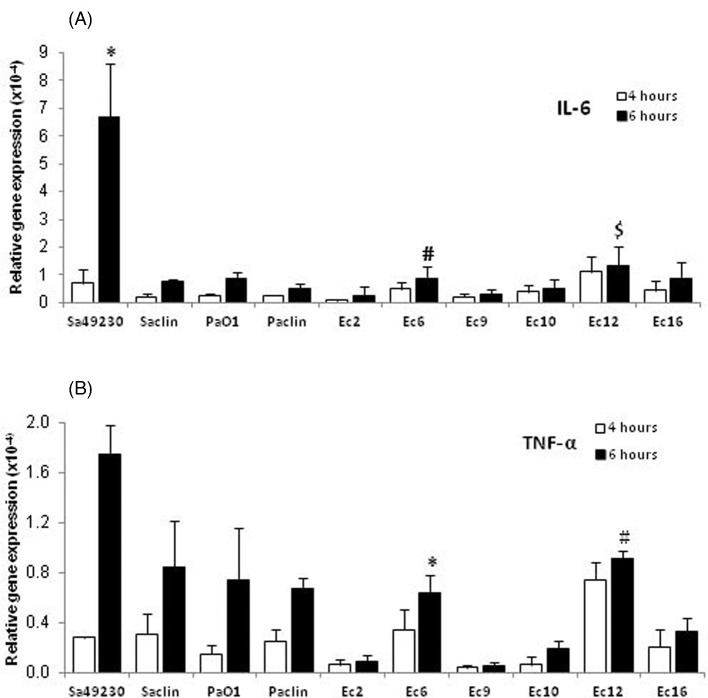
We examined the effects of the S. aureus and P. aeruginosa control strains and six E. coli strains on IL-6 and TNF-α mRNAs expression in MG-63 cells. Both most adherent E. coli (Ec6 and Ec12), and four other E. coli strains [including three (Ec2, Ec9 and Ec10) that showed adherence rates < 1%] were studied. As shown in Fig. 5, the strain Sa49230 induced much higher level of IL-6 mRNA than any other strain, 6 h post-infection, and was the only strain to induce IL-6 secretion 24 and 72 h post-infection (54.08 ± 26.73 and 131.71 ± 43.25 pg mL−1, respectively). Both most adherent Ec6 and Ec12 strains induced significantly higher levels of IL-6 mRNA than the less adherent Ec2 and Ec9 strains. Similarly, induction of TNF-α mRNA expression was correlated with the adherence rates of the E. coli strains. Thus, the levels of TNF-α mRNA expressed after 6 h by the Ec6 or Ec12-infected MG-63 cells were comparable to those of the Saclin, PaO1, or Paclin-infected cells, but were significantly higher than those of the Ec2, Ec9 or Ec10-infected cells. TNF-α was not detected in supernatants of the infected cells 24 or 72 h post-infection, even for the strain Sa49230, which was the most efficient to induce TNF-α mRNA expression.
Figure 5.
Relative expression of IL-6 and TNF-α mRNAs in MG-63 cells. The cells were infected with the four control strains or six selected OII E. coli strains, for 4 or 6 h (MOI of 100:1), before total RNA extraction. Results are expressed as relative gene expression after normalization using the β-actin housekeeping gene. (A) IL-6 mRNA expression. The Sa49230, Ec6 and Ec12 strains were tested statistically with the Kruskal–Wallis test followed by a Dunn's post test. *, P < 0.05 versus Saclin, PaO1, Paclin, Ec2, Ec6, Ec9, Ec10 or Ec16 (6 h of infection); #, P < 0.05 versus Sa49230, Ec2 or Ec9 (6 h of infection); $, P < 0.05 versus Paclin, Ec2, Ec9 or Ec10 (6 h of infection). (B) TNF-α mRNA expression. The Ec6 and Ec12 strains were tested statistically with the Kruskal–Wallis test followed by a Dunn's post test. *, P < 0.05 versus Sa49230, Ec2, Ec9 or Ec10 (6 h of infection); #, P < 0.05 versus Ec2, Ec9, Ec10 or Ec16 (6 h of infection).
DISCUSSION
Several subgroups of ExPEC through production of large arrays of virulence factors have been shown to adhere to and cross different epithelial barriers (e.g. intestine, bladder), triggering severe tissue damages and proinflammatory responses (Croxen et al. 2013; Ulett et al. 2013). In this investigation, we wanted to determine if such virulence mechanisms are developed by OII E. coli towards human osteoblasts. For this purpose, we used a panel of 20 clinical strains of E. coli from OII. These strains belonged to different sequence types and serotypes, most of them presenting the characteristics of UPEC (serotypes and PAI-encoded virulence genes of UPEC).
The osteoblasts, derived from mesenchymal bone marrow precursors, control the bone remodeling process by synthesizing the components of bone matrix, and by modulating the activity of the bone-resorbing osteoclasts. These cells have also been shown to produce different soluble inflammatory mediators [IL-6, IL-1, TNF-α, receptor activator of NF-κB ligand (RANKL)], following S. aureus infection (Marriott 2004; Somayaji et al. 2008; Wright and Nair 2010; Ning et al. 2011; Claro et al. 2013). Thus, the bone destruction induced by S. aureus exposure results mainly from the inflammatory reaction elicited by the infection, even if some S. aureus lineages that overexpress the newly studied family of PSM peptides share cytolytic properties following invasion of osteoblasts (Rasigade et al. 2013). The present study provides pieces of evidence that 30% of the E. coli strains of our panel are also strongly cytotoxic towards osteoblasts. Interestingly, we observed that most of these cytotoxic strains at an MOI as low as 10:1 reduced osteoblasts cells viability by approximately 20% after 2 h of infection, whereas both S. aureus strains of our study were non-cytotoxic. Other authors reported cytolytic effects for S. aureus infection of the same MG-63 cell line, at higher MOI (100:1) and after 24 h of infection (Rasigade et al. 2013).
In our study, two virulence factors, namely an alpha-hemolysin (HlyA) and a toxin named cytotoxic necrotizing factor (CNF1), were significantly associated with the cytolytic activity on MG63 cells. This cytolytic activity was also confirmed on human mesenchymal stem cells (derived from bone marrow) that are precursors of osteoblasts (data not shown). HlyA is an extracellular pore-forming cytolysin from the RTX (repeats-in-toxin) family, while the protein CNF1 belongs to a family of factors activating intracellular Rho GTPases (Landraud et al. 2003; Burgos and Beutin 2010; Garcia et al. 2013; Ulett et al. 2013). Both virulence factors are known to induce apoptosis or necrosis/lysis, depending on concentration, in several cell types, including macrophages, epithelial cells and neutrophils (Burgos and Beutin 2010; Garcia et al. 2013; Ulett et al. 2013). In UPEC -producing CNF1, the cnf1 gene has been shown to be coupled to the hemolysin operon (hlyCABD) in the PAI II (Landraud et al. 2003). Transcription of both hlyCABD operon and cnf1 gene is enhanced by the regulator RfaH which binds on the ops DNA site located downstream from the hlyCABD promoter, and suppresses transcription polarity (Leeds and Welch 1997; Landraud et al. 2003). In the present study, all cytotoxic E. coli strains exhibited a hemolytic activity linked to the expression of the hlyA gene. We observed that non-hemolytic strains were not cytotoxic, regardless of whether or not they carried a cnf1 gene. We found, in fact, that the cnf1 gene was less expressed than the hlyA gene, and that its expression was well correlated with that of hlyA gene. Accordingly, all hlyCABD operons were found encoded on PAIs II, with an ops element upstream of the hlyC gene. Several polymorphisms were found in the upstream sequences of the hlyC genes, but no nucleotide deletions and/or substitutions was detected in the ops elements of the strains that weakly expressed the hlyA gene. Thus, while no explanation was found for the low hlyA expression in some E. coli, our results highlight the cytolytic effect of a functioning hemolysin on osteoblasts, and are in accordance with those of Island et al. (1998), who found that CNF1 does not affect the cytotoxicity of hemolytic isolates towards human bladder cells in vitro. In fact, the Zhly– control (ZKLR+ ΔhlyA mutant) that expressed cnf1 did not induce a cytolytic effect in our experimental conditions.
Detailed analyses of the interaction between S. aureus and bone revealed that the S. aureus surface-associated proteins FnBPs and SpA mediate attachment of the bacterium to fibronectin and tumor necrosis factor receptor-1 (TNFR-1) on osteoblasts, and leads to bacterial uptake via an integrin α5β1-mediated mechanism (Claro et al. 2013). Consistent with this model, both S. aureus strains of our study showed the highest internalization capacity (approximately 5 and 9% of internalized bacteria). The percentage of internalized P. aeruginosa was comparatively 10-fold to 100-fold lower, and all strains of E. coli presented an extremely low internalization rate (<0.01%). The poor uptake of E. coli could be reasonably attributed to the low ability of the strains to adhere to osteoblasts, since the adherence rates of our OII E. coli were not higher than 7%, half of the strains showing an adherence rate lower than 1%. Interestingly, these results are very similar to those from Fiedler et al. (2013) that recently investigated the adherence and internalization in adipose-derived mesenchymal stem cells of one E. coli obtained from wound infection (i.e. 1.4% of adherent bacteria and 0.008% of internalized bacteria). As far as we know, there is not any animal model showing histopathological evidence of E. coli adherence to osteoblasts during an experimental infection. Nonetheless, E. coli is well known to express various adhesins, including several FnBPs. The amyloid fibers curli, involved in biofilm formation, are the most important and were previously identified in all OII E. coli studied (Barnhart and Chapman 2006; Henderson et al. 2011; Crémet et al. 2012). Approximately 60% (12/20) of our E. coli strains demonstrated a strong affinity for the curli-binding dye, congo red (data not shown), including both most adherent strains, Ec6 and Ec12.
Finally, a major component of the outer membrane of E. coli, namely the LPS, has been proposed to be a potent stimulator of bone destruction in several bone inflammatory diseases (Ochi et al. 2010; Yan et al. 2010). Thus, following binding to Toll-like receptor 4 (TLR4) on osteoblasts, LPS promotes synthesis of proinflammatory mediators, among which IL-6, TNF-α, RANKL and PGE2 inhibit osteoblast differentiation, promote osteoblast apoptosis and stimulate osteoclastogenesis directly or indirectly (Suda et al. 2004; Inada et al. 2006; Ochi et al. 2010; Yan et al. 2010; Gao et al. 2013; Guo et al. 2014). However, LPS is a surrogate marker of Gram-negative bacteria, which do not sufficiently reflect whole bacteria and only partly mimics osteoblasts infection, with the risk of non-physiological stimuli. There are very few experimental models of osteoblasts infection by Gram-negative organisms, the existing studies being essentially focused on the intracellular pathogens Salmonella and Brucella (Alexander et al. 2001; Marriott 2004; Delpino, Fossati and Baldi 2009; Scian et al. 2011). Furthermore, these studies are often based on small numbers of clinical strains, even in the case of S. aureus infections, where the osteomyelitis clinical strain ATCC 49230 has been the most studied (Alexander et al. 2001, 2003; Marriott et al. 2005; Somayaji et al. 2008; Ning et al. 2011).
In the present study, in accordance with the current knowledge on osteoblasts responses to S. aureus, we found that infection with the strain Sa49230 elicited the secretion of IL-6 from MG-63 cells (Ning et al. 2011). This secretion seemed to be strain dependent, since stimulation of osteoblasts with the other S. aureus elicited a lower IL-6 response and did not result in IL-6 secretion. Moreover, we show that infection with both S. aureus induced TNF-α mRNA expression, but did not induce TNF-α release from MG-63 cells. These results are consistent with previous data that either showed an endogenous cell-surface TNF-α expression of MG-63 cells or established that S. aureus- or Brucella-infected osteoblasts do not secrete detectable levels of TNF-α in vitro (Bu et al. 2003; Delpino, Fossati and Baldi 2009; Scian et al. 2011).
Differences in IL-6 or TNF-α mRNA expression were observed between the E. coli strains, and adherence to osteoblasts seemed to contribute to the induction of IL-6 or TNF-α mRNA expression. Our findings indicate that the most adherent OII E. coli strains elicit IL-6 or TNF-α responses, which are equivalent to those elicited by S. aureus or P. aeruginosa strains recovered in the same clinical context. It would be now interesting to extend those findings to other proinflammatory mediators and to study the molecular mechanisms mediating osteoblasts immune response to infection by these different bacterial species. Furthermore, even though the low adherence seemed correlated to a poor inflammatory response, further experiments could be helpful to rule out a subversion of immune response by the bacteria, as previously described in ExPEC isolates (Cirl et al. 2008).
In conclusion, our study is the first to investigate the effects of 20 clinical strains of E. coli from OII on human osteoblastic cells. Even if we did not find that OII E. coli shared common virulence properties towards osteoblasts, our data provide evidence that the HlyA-producing strains, known to be highly virulent, are associated with cytolytic phenotypes. The others are poorly internalized into osteoblasts, the most adherent of them eliciting IL-6 or TNF-α responses in the same range as those induced by some strains of S. aureus or P. aeruginosa from OII.
Acknowledgments
We thank Dr J. Holubova and Pr P. Sebo (Laboratory of Molecular Biology of Bacterial Pathogens, Institute of Microbiology of the Academy of Sciences of the Czech Republic, Prague, Czech Republic) for kindly providing the ZKLR+ and Zhly– E. coli control strains used in this study.
FUNDING
This work was supported by the French ‘Ministère de l'Enseignement Supérieur et de la Recherche’.
Conflict of interest. None declared.
REFERENCES
- Alexander EH, Bento JL, Hughes FM, Jr, et al. Staphylococcus aureus and Salmonella enterica serovar Dublin induce tumor necrosis factor-related apoptosis-inducing ligand expression by normal mouse and human osteoblasts. Infect Immun. 2001;69:1581–6. doi: 10.1128/IAI.69.3.1581-1586.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander EH, Rivera FA, Marriott I, et al. Staphylococcus aureus-induced tumor necrosis factor-related apoptosis-inducing ligand expression mediates apoptosis and caspase-8 activation in infected osteoblasts. BMC Microbiol. 2003;3:5. doi: 10.1186/1471-2180-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–47. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu R, Borysenko CW, Li Y, et al. Expression and function of TNF-family proteins and receptors in human osteoblasts. Bone. 2003;33:760–70. doi: 10.1016/j.bone.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Burgos Y, Beutin L. Common origin of plasmid encoded alpha-hemolysin genes in Escherichia coli. BMC Microbiol. 2010;10:193. doi: 10.1186/1471-2180-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Hammer ND, Campbell JP, et al. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe. 2013;13:759–72. doi: 10.1016/j.chom.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirl C, Wieser A, Yadav M, et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- Claro T, Widaa A, McDonnell C, et al. Staphylococcus aureus protein A binding to osteoblast tumour necrosis factor receptor 1 results in activation of nuclear factor kappa B and release of interleukin-6 in bone infection. Microbiology. 2013;159:147–54. doi: 10.1099/mic.0.063016-0. [DOI] [PubMed] [Google Scholar]
- Crémet L, Corvec S, Bémer P, et al. Orthopædic-implant infections by Escherichia coli: molecular and phenotypic analysis of the causative strains. J Infect. 2012;64:169–75. doi: 10.1016/j.jinf.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Croxen MA, Law RJ, Scholz R, et al. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26:822–80. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpino MV, Fossati CA, Baldi PC. Proinflammatory response of human osteoblastic cell lines and osteoblast-monocyte interaction upon infection with Brucella spp. Infect Immun. 2009;77:984–95. doi: 10.1128/IAI.01259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler T, Salamon A, Adam S, et al. Impact of bacteria and bacterial components on osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Exp Cell Res. 2013;18:2883–92. doi: 10.1016/j.yexcr.2013.08.020. [DOI] [PubMed] [Google Scholar]
- Gao A, Kantarci A, Herrera BS, et al. A critical role for suppressors of cytokine signaling 3 in regulating LPS-induced transcriptional activation of matrix metalloproteinase-13 in osteoblasts. Peer J. 2013;1:e51. doi: 10.7717/peerj.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia TA, Ventura CL, Smith MA, et al. Cytotoxic necrotizing factor 1 and hemolysin from uropathogenic Escherichia coli elicit different host responses in the murine bladder. Infect Immun. 2013;81:99–109. doi: 10.1128/IAI.00605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yuan L, Wang JG, et al. Lipopolysaccharide (LPS) induces the apoptosis and inhibits osteoblast differentiation through JNK pathway in MC3T3-E1 cells. Inflammation. 2014;37:621–31. doi: 10.1007/s10753-013-9778-9. [DOI] [PubMed] [Google Scholar]
- Hamza T, Dietz M, Pham D, et al. Intra-cellular Staphylococcus aureus alone causes infection in vivo. Eur Cell Mater. 2013;25:341–50. doi: 10.22203/ecm.v025a24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B, Nair S, Pallas J, et al. Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol Rev. 2011;35:147–200. doi: 10.1111/j.1574-6976.2010.00243.x. [DOI] [PubMed] [Google Scholar]
- Inada M, Matsumoto C, Uematsu S, et al. Membrane-bound prostaglandin E synthase-1-mediated prostaglandin E2 production by osteoblast plays a critical role in lipopolysaccharide-induced bone loss associated with inflammation. J Immunol. 2006;177:1879–85. doi: 10.4049/jimmunol.177.3.1879. [DOI] [PubMed] [Google Scholar]
- Island MD, Cui X, Foxman B, et al. Cytotoxicity of hemolytic, cytotoxic necrotizing factor 1-positive and -negative Escherichia coli to human T24 bladder cells. Infect Immun. 1998;66:3384–9. doi: 10.1128/iai.66.7.3384-3389.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler CD, Dobrindt U. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol. 2011;301:642–7. doi: 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Kurtz SM, Lau E, Watson H, et al. Economic burden of periprosthetic joint infection in the United States. J Arthroplasty. 2012;27:61–5e1. doi: 10.1016/j.arth.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Landraud L, Gibert M, Popoff MR, et al. Expression of cnf1 by Escherichia coli J96 involves a large upstream DNA region including the hlyCABD operon, and is regulated by the RfaH protein. Mol Microbiol. 2003;47:1653–67. doi: 10.1046/j.1365-2958.2003.03391.x. [DOI] [PubMed] [Google Scholar]
- Leeds JA, Welch RA. Enhancing transcription through the Escherichia coli hemolysin operon, hlyCABD: RfaH and upstream JUMPStart DNA sequences function together via a postinitiation mechanism. J Bacteriol. 1997;179:3519–27. doi: 10.1128/jb.179.11.3519-3527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu B, Chen M, et al. A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J Microbiol Meth. 2010;82:71–7. doi: 10.1016/j.mimet.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Lima AL, Oliveira PR, Carvalho VC, et al. Periprosthetic joint infections. Interdiscip Perspect Infect Dis. 2013;2013:542796–802. doi: 10.1155/2013/542796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott I. Osteoblast responses to bacterial pathogens. Immunol Res. 2004;30:291–308. doi: 10.1385/IR:30:3:291. [DOI] [PubMed] [Google Scholar]
- Marriott I, Gray DL, Rati DM, et al. Osteoblasts produce monocyte chemoattractant protein-1 in a murine model of Staphylococcus aureus osteomyelitis and infected human bone tissue. Bone. 2005;37:504–12. doi: 10.1016/j.bone.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Molina-Manso D, del Prado G, Ortiz-Pérez A, et al. In vitro susceptibility to antibiotics of staphylococci in biofilms isolated from orthopædic infections. Int J Antimicrob Ag. 2013;41:521–3. doi: 10.1016/j.ijantimicag.2013.02.018. [DOI] [PubMed] [Google Scholar]
- Montanaro L, Speziale P, Campoccia D, et al. Scenery of Staphylococcus implant infections in orthopedics. Future Microbiol. 2011;6:1329–49. doi: 10.2217/fmb.11.117. [DOI] [PubMed] [Google Scholar]
- Nakao J, Fujii Y, Kusuyama J, et al. Low-intensity pulsed ultrasound (LIPUS) inhibits LPS-induced inflammatory responses of osteoblasts through TLR4-MyD88 dissociation. Bone. 2014;58:17–25. doi: 10.1016/j.bone.2013.09.018. [DOI] [PubMed] [Google Scholar]
- Ning R, Zhang X, Guo X, et al. Staphylococcus aureus regulates secretion of interleukin-6 and monocyte chemoattractant protein-1 through activation of nuclear factor kappaB signaling pathway in human osteoblasts. Braz J Infect Dis. 2011;15:189–94. doi: 10.1016/s1413-8670(11)70173-8. [DOI] [PubMed] [Google Scholar]
- Ochi H, Hara Y, Tagawa M, et al. The roles of TNFR1 in lipopolysaccharide-induced bone loss: dual effects of TNFR1 on bone metabolism via osteoclastogenesis and osteoblast survival. J Orthop Res. 2010;28:657–63. doi: 10.1002/jor.21028. [DOI] [PubMed] [Google Scholar]
- Peel TN, Cheng AC, Buising KL, et al. Microbiological ætiology, epidemiology, and clinical profile of prosthetic joint infections: are current antibiotic prophylaxis guidelines effective? Antimicrob Agents Ch. 2012;56:2386–91. doi: 10.1128/AAC.06246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasigade JP, Trouillet-Assant S, Ferry T, et al. PSMs of hypervirulent Staphylococcus aureus act as intracellular toxins that kill infected osteoblasts. PLoS One. 2013;8:e63176. doi: 10.1371/journal.pone.0063176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M, Monteiro FJ, Ferraz MP. Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions. Biomatter. 2012;2:176–94. doi: 10.4161/biom.22905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pardo D, Pigrau C, Lora-Tamayo J, et al. Gram-negative prosthetic joint infection: outcome of a debridement, antibiotics, and implant retention approach. A large multicenter study. Clin Microbiol Infect. 2014;20:O911–9. doi: 10.1111/1469-0691.12649. [DOI] [PubMed] [Google Scholar]
- Scian R, Barrionuevo P, Giambartolomei GH, et al. Granulocyte-macrophage colony-stimulating factor- and tumor necrosis factor alpha-mediated matrix metalloproteinase production by human osteoblasts and monocytes after infection with Brucella abortus. Infect Immun. 2011;79:192–202. doi: 10.1128/IAI.00934-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheshko V, Hejnova J, Rehakova Z, et al. HlyA knock out yields a safer Escherichia coli A0 34/86 variant with unaffected colonization capacity in piglets. FEMS Immunol Med Micr. 2006;48:257–66. doi: 10.1111/j.1574-695X.2006.00140.x. [DOI] [PubMed] [Google Scholar]
- Somayaji SN, Ritchie S, Sahraei M, et al. Staphylococcus aureus induces expression of receptor activator of NF-kappaB ligand and prostaglandin E2 in infected murine osteoblasts. Infect Immun. 2008;76:5120–6. doi: 10.1128/IAI.00228-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda K, Udagawa N, Sato N, et al. Suppression of osteoprotegerin expression by prostaglandin E2 is crucially involved in lipopolysaccharide-induced osteoclast formation. J Immunol. 2004;172:2504–10. doi: 10.4049/jimmunol.172.4.2504. [DOI] [PubMed] [Google Scholar]
- Ulett GC, Totsika M, Schaale K, et al. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr Opin Microbiol. 2013;16:100–7. doi: 10.1016/j.mib.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Wright JA, Nair SP. Interaction of staphylococci with bone. Int J Med Microbiol. 2010;300:193–204. doi: 10.1016/j.ijmm.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Cao J, Wu M, et al. Suppressor of cytokine signaling 3 inhibits LPS-induced IL-6 expression in osteoblasts by suppressing CCAAT/enhancer-binding protein β activity. J Biol Chem. 2010;285:37227–39. doi: 10.1074/jbc.M110.132084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmistowski B, Fedorka CJ, Sheehan E, et al. Prosthetic joint infection caused by Gram-negative organisms. J Arthroplasty. 2011;26(6 Suppl):104–8. doi: 10.1016/j.arth.2011.03.044. [DOI] [PubMed] [Google Scholar]