Abstract
Francisella tularensis subspecies tularensis is a highly virulent intracellular bacterial pathogen, causing the disease tularemia. However, a safe and effective vaccine for routine application against F. tularensis has not yet been developed. We have recently constructed the deletion mutants for the DsbA homolog protein (ΔdsbA/FSC200) and a hypothetical protein IglH (ΔiglH/FSC200) in the type B F. tularensis subsp. holarctica FSC200 strain, which exerted different protection capacity against parental virulent strain. In this study, we further investigated the immunological correlates for these different levels of protection provided by ΔdsbA/FSC200 and ΔiglH/FSC200 mutants. Our results show that ΔdsbA/FSC200 mutant, but not ΔiglH/FSC200 mutant, induces an early innate inflammatory response leading to strong Th1-like antibody response. Furthermore, vaccination with ΔdsbA/FSC200 mutant, but not with ΔiglH/FSC200, elicited protection against the subsequent challenge with type A SCHU S4 strain in mice. An immunoproteomic approach was used to map a spectrum of antigens targeted by Th1-like specific antibodies, and more than 80 bacterial antigens, including novel ones, were identified. Comparison of tularemic antigens recognized by the ΔdsbA/FSC200 post-vaccination and the SCHU S4 post-challenge sera then revealed the existence of 22 novel SCHU S4 specific antibody clones.
Keywords: tularemia, cytokines, antibody response, protection, immunoproteomics
This work is focused on characterization of immune response during in vivo infection of two attenuated Francisella tularensis mutant (ΔdsbA and ΔiglH) strains; importantly, the ΔdsbA mutant, but not the ΔiglH mutant, induced an early innate inflammatory response leading to strong Th1-like antibody response.
Graphical Abstract Figure.
This work is focused on characterization of immune response during in vivo infection of two attenuated Francisella tularensis mutant (ΔdsbA and ΔiglH) strains; importantly, the ΔdsbA mutant, but not the ΔiglH mutant, induced an early innate inflammatory response leading to strong Th1-like antibody response.
INTRODUCTION
Tularemia is a severe disease caused by the intracellular pathogenic bacterium Francisella tularensis (F. tularensis). Human infections are most commonly acquired through direct contact with infected material (usually animals) or through vector-borne transmission, such as bites by infected insects. By infection through skin, the ulceroglandular tularemia form develops, which represents approximately 90% of all tularemia cases (Tarnvik and Berglund 2003). A more severe form of tularemia may be caused by respiratory infections after inhalation of aerosols containing as little as 10 bacteria of subsp. tularensis, which may result in 30–60% mortality if untreated (Evans et al. 1985). The potential risk of F. tularensis to be misused as a biological weapon led to this bacterium being classified as a category A agent by Centers for Disease Control and Prevention, USA (Oyston, Sjostedt and Titball 2004).
In general, tularemia is treated with antibiotics where streptomycin is recommended as the drug of first choice with tetracyclines serving as potential alternatives (Russell et al. 1998; Dennis et al. 2001; Johansson et al. 2001). However, the successful antibiotic therapy requires prompt diagnosis which is still a serious problem in some countries, and therefore, the development of a safe vaccine is urgently needed. Currently, tularemia vaccine development focuses on improvement of existing attenuated Francisella live vaccine strain (LVS) or on construction of new attenuated mutant strains for genes that are involved in pathogenic mechanisms of tularemic microbe (Marohn and Barry 2013). Compared to these two approaches, designing a subunit vaccine represents much more difficult task because of the current lack of knowledge of suitable immunodominant antigens. Up to now, immunoproteomics exploiting immune sera for identification of new immunoreactive antigens has been the easiest way to acquire information about candidates for protective antigens (Kilmury and Twine 2010).
Previously, we constructed two attenuated type B F. tularensis strains, one with deletion in gene encoding a homolog to the protein family of disulfide oxidoreductases DsbA (FTS_1067) and the second one with deletion in gene encoding the FPI protein IglH (FTS_0106/FTS_1134) (Straskova et al. 2009, 2012). Both mutants showed attenuated phenotype and protective potential against subsequent subcutaneous challenge with parental European clinical isolate of subsp. holarctica strain, denoted as FSC200 strain. While immunization with ΔdsbA/FSC200 led to complete protection of BALB/c mice against the FSC200 strain challenge, administration of the ΔiglH/FSC200 mutant provided only partial dose-dependent protection with maximal protective effect when a dose of more than 3 × 107 CFUs was applied (Straskova et al. 2009, 2012).
In this study, we investigated the immunological parameters which might be responsible for differential protection capacity of the ΔdsbA/FSC200 and the ΔiglH/FSC200 mutant strains. We found that the ability of in vivo induction of early innate inflammatory response and the Th1-like antibody response clearly differ between both mutants. Furthermore, we demonstrated that immune response induced by the ΔdsbA/FSC200 mutant is also sufficient for protection against challenge with Francisella type A strain SCHU S4. Finally, using an immunoproteomic approach, we defined the profile of Francisella membrane proteins recognized by post-vaccination and post-challenge sera and their comparison enabled the determination of novel immunoreactive SCHU S4 antigens.
MATERIALS AND METHODS
Animals
Female BALB/c mice were purchased from Velaz, s.r.o. (Unetice, Czech Republic) and entered experiments at 6–8 weeks of age. All procedures using mice were performed in accordance with guidelines of Animal Care and Use Ethical Committee of the Faculty of Military Health Sciences, University of Defence, Czech Republic. At USAMRIID, research was conducted under an IACUC approved protocol in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
Bacteria and culture conditions
Wild-type F. tularensis subsp. tularensis SCHU S4 strain (Collection of Animal Pathogenic Microorganisms, No. 5600, Veterinary Research Institute, Brno, Czech Republic or USAMRIID strain collection) and F. tularensis subsp. holarctica FSC200 strain were used. Generation of mutant strains with the in frame deletion of the iglH gene in the FSC200 strain (ΔiglH/FSC200) and with the deletion of the dsbA gene in FSC200 (ΔdsbA/FSC200) strain has been described previously (Straskova et al. 2009, 2012). Bacterial stocks of each strain were grown on McLeod agar supplemented with bovine hemoglobin (Becton Dickinson San Jose, CA) and IsoVitalex (Becton Dickinson) for 24 h at 37°C and 5% CO2. Before each experiment, bacteria were grown for 24 h at 37°C and 5% CO2 on McLeod agar plates and thereafter suspended in PBS (phosphate-buffered saline, pH 7.4) to an OD600 = 1, which is approximately 3 × 109 bacteria mL−1. Studies involving F. tularensis SCHU S4 strain were conducted at the BSL-3 facility at the Faculty of Military Health Sciences following appropriate biosafety requirements.
Animal infection, cytokine and antibody assays
For immunological assays, groups of BALB/c mice (n = 3) were subcutaneously (s.c.) infected with 102 CFU/mouse of F. tularensis strain FSC200 and with 107 CFU/mouse of the ΔiglH/FSC200 or the ΔdsbA/FSC200 mutant strain. After 1, 3, 5, 7, 14, 21 and 28 days post-infection, mice were killed and sera together with livers and spleens were collected. Blood was obtained from vena cava and pooled for each strain from three mice per treatment. Sera were then separated from blood, filtered through a 0.22-μm filter and stored at −80°C until needed. Individual livers and spleens were aseptically removed from each mouse, homogenized in PBS and stored frozen at −20°C until needed. Organ homogenates and sera samples were used undiluted and analyzed for levels of cytokines and antibodies using Custom Quantibody Array technology (RayBiotech, Inc., Norcross GA, USA) following the manufacturer´s protocol. The cytokine/antibody concentrations were calculated against the standards using software H20 OV Q-Analyzer v8.10.4 (Raybiotech, Inc., Norcross, GA).
To determine bacterial burden in targeted organs, BALB/c mice (n = 3 for each treatment) were infected with 102 CFU/mouse of the F. tularensis FSC200 parental strain or with 107 CFU/mouse of the ΔdsbA/FSC200 mutant. Control group of mice was inoculated with sterile saline solution only. After 1, 3, 5, 7, 14, 21 and 28 days of infection, livers, spleens and lungs were aseptically removed, homogenized in 2 mL of PBS, serially diluted and plated on McLeod agar plates enriched with 100 U mL−1 of penicillin to minimize unwanted contamination. After 3 days of incubation at 37°C in 5% CO2, the bacterial colonies were enumerated and CFUs per organ were calculated.
For in vivo subcutaneous protection studies, groups of BALB/c mice (n = 5) were s.c. inoculated with 10, 102, 103, 104, 105 and 107 CFU/mouse of the ΔdsbA/FSC200 mutant or with 107 CFU/mouse of the ΔiglH/FSC200 mutant strain. After 3 weeks of immunization, mice were challenged s.c. with 102 CFU/mouse of virulent SCHU S4 strain. For intranasal protection studies, groups of BALB/c mice (n = 10) were vaccinated intranasally (i.n.) with 10, 102, 103, 104, 105 or 106 CFU/mouse of the ΔdsbA/FSC200 mutant. After 4 weeks, mice were challenged i.n. using 102 CFU/mouse of the F. tularensis SCHU S4 strain. In both studies, mice were monitored daily for morbidity and mortality. The study endpoint was euthanasia when moribund or survival to 21 days following exposure.
For immunoproteomic studies, BALB/c mice (n = 10) were s.c. vaccinated with 107 CFU/mouse of the ΔdsbA/FSC200 mutant strain. After 21 days, five ΔdsbA/FSC200-vaccinated mice were killed to obtain sera. The remaining ΔdsbA/FSC200-immunized mice were further s.c. challenged with 102 CFU/mouse of highly virulent SCHU S4 strain. After 21 days of infection, sera were collected and stored at −80°C until needed.
Detergent-enriched fraction preparation
The detergent-enriched fraction was prepared using the Triton X-114 phase separation similar to those described by Shimizu, Kida and Kuwano (2005). Briefly, F. tularensis SCHU S4 was grown in chemically defined Chamberlain medium until up to an OD600nm of 0.8. Culture was then pelleted by centrifugation and washed twice with cold PBS. The cell pellet was resuspended in ice-cold PBS supplemented with proteases inhibitors cocktail Complete EDTA-free (Roche, A.G., Switzerland) and disintegrated by French Pressure Cell Press. Then, the whole cell lysate was ultracentrifuged at 100 000 × g for 1 h at 4°C to pellet membrane-associated proteins. Pellets were resuspended in ice-cold PTX buffer (PBS supplemented with 350 mm NaCl, 2% Triton X-114, protease inhibitor mixture) and incubated at 4°C for 1 h under end-over-end rotation. Samples were centrifuged at 12 000 rpm 4°C for 30 min, and the supernatants were kept at 37°C for 10 min to induce detergent phase separation. Following centrifugation at 14 000 rpm for 10 min at room temperature, the upper aqueous phase was discarded and replaced with the same volume of PBS supplemented with 350 mm NaCl. This phase separation was repeated three times, and the final detergent phase was resuspended in PBS to the original volume. Protein concentration in the suspension was measured with a BCA protein assay kit (Sigma-Aldrich, St. Louis, MO, USA).
Pilin protein-enriched fraction preparation
Francisella tularensis SCHU S4 was grown on McLeod plates for 48 h and then the bacteria were harvested from the plates and suspended in PBS. The supernatant enriched for the pili was acquired by vortexing the suspension at the maximum speed for 2 min. The bacteria were pelleted by centrifugation at 13 000 rpm for 10 min and the supernatant was collected, which was then heated at 65°C for 2 h to eliminate any remaining bacteria. The pili were left to aggregate on an orbital shaker at 4°C for 18 h. The suspension was then ultracentrifuged at 150 000 × g 4°C for 1 h, the pellets were resuspended in PBS and the protein concentration was quantified using a BCA protein assay kit (Sigma-Aldrich).
Two-dimensional polyacrylamide gel electrophoresis (2D PAGE), Western Blotting and MALDI TOF/TOF protein identification
Protein samples were precipitated with cold acetone, solubilized in ASB-D buffer (7 M urea, 2 M thiourea, 40 mM Tris, 1% Triton X-100, 1% ASB-14, 0.5% bromphenol blue and 1.2% DeStreak) and separated using immobilized pH gradient strips (IPG), non-linear pH 3–10, 18 cm or linear pH 6–11, 18 cm (GE Healthcare, Uppsala, Sweden) using 150 or 200 μg of protein/gel for western blots and 1 mg of protein/gel for Coomassie blue staining (Colloidal Blue Stain Kit, Invitrogen). Following IEF, the IPG strips were equilibrated for 15 min in equilibration buffer (2% SDS, 50 mM Tris-HCl (pH 8.8), 6 M urea, 30% glycerol and 1% DTT) followed by a second 15 min equilibration step (2% SDS, 50 mM Tris-HCl (pH 8.8), 6 M urea, 30% glycerol and 14% iodoacetamide). Approximately 9–16% gels were used for second dimension separations. Proteins from the gels were transferred onto BioTrace PVDF membrane (Pall Corporation, Pensacola, FL) and subjected to immunoblotting using the pooled sera either from the ΔdsbA/FSC200 vaccinated or SCHU S4-challenged immunized BALB/c mice. As a secondary antibody, the polyclonal goat anti-mouse immunoglobulins/HRP (Dako, Denmark), which recognizes IgG, IgA and IgM isotypes, was used. Chemiluminiscence detection was performed using a BM chemiluminiscence blotting substrate POD according to the manufacturer's instructions (Roche Applied Science). For these experiments, three biological replicates of detergent and pilin protein-enriched fractions were prepared.
Alignments of immunoreactive spots on 2D blots with Coomassie blue-stained gels were done manually.
Protein spots corresponding to immunoreactive spots on western blots were excised from Coomassie blue-stained 2D-gels and in-gel tryptically digested as described elsewhere (Balonova et al. 2010). The mass spectra were recorded in positive MS and MS/MS modes on a 4800 MALDI-TOF/TOF mass analyzer (AB Sciex, Forster City, CA). Internal calibration of mass spectra was performed using tryptic autolytic peptides. Acquired data were processed using GPS Explorer software version 3.6 (AB Sciex) cooperating with the Mascot search algorithm version 2.2 and the search was done against a Francisella tularensis SCHU S4 database (NC_006570.2). Trypsin was selected as the proteolytic enzyme and one missed cleavage was allowed. Carbamidomethylation of cysteine residues and methionine oxidation was set as a variable and fixed modification, respectively. Proteins were considered identified with the confidence when GPS protein score confidence interval was 100% and at least two peptides per protein were identified.
STATISTICS
Differences in cytokine levels were compared by two-way ANOVA followed by Tukey´s multiple-comparison post-test as appropriate, using GraphPad Prizm 5 software. In all cases, differences were considered significant at P < 0.05, where the group of wt FSC200 infected mice and the ΔdsbA/FSC200 vaccinated mice were compared and the group of wt FSC200 strain and the ΔiglH/FSC200 mutant were compared. Each experiment was independently repeated two times.
RESULTS
In vivo cytokine immune responses elicited in BALB/c mice after ΔdsbA/FSC200 and ΔiglH/FSC200 s.c. vaccination
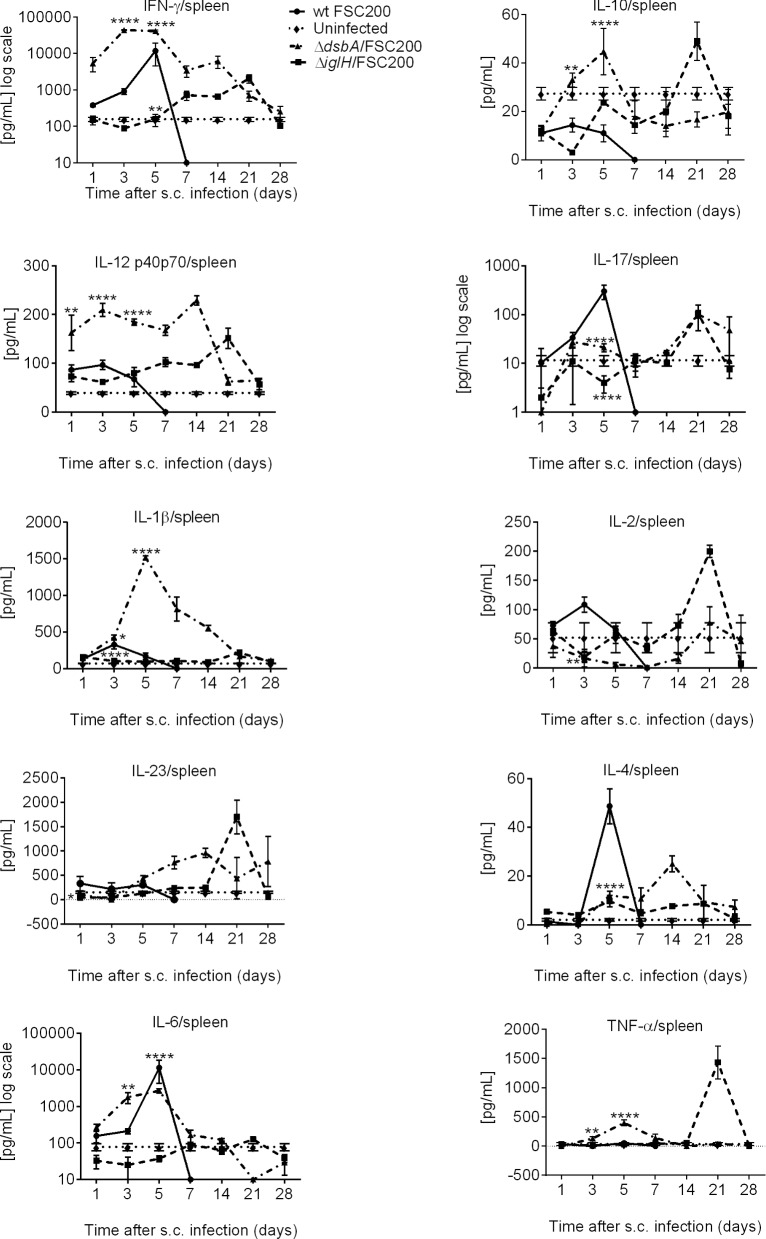
To determine whether the ΔdsbA/FSC200 mutant or the ΔiglH/FSC200 mutant elicited different immune responses in BALB/c mice, the levels of IFN-γ, IL-10, IL-12, IL-17, IL-1β, IL-2, IL-23, IL-4, IL-6 and TNF-α were measured in spleens, livers and sera on days 1, 3, 5, 7, 14, 21 and 28 post-vaccination.
Immunization of BALB/c mice with the ΔdsbA/FSC200 mutant led to significantly upregulated levels of IFN-γ and IL-12 in spleen compared to the group infected with wt strain (Fig. 1; Fig. S1, Supporting Information). The increased levels of these cytokines started very early on day 3 after vaccination and persisted till day 14 (Fig. 1). Similarly, IL-6 production increased significantly on day 3, which was maintained until day 5 but then declined by day 7. Day 5 after ΔdsbA/FSC200 immunization was also characterized by steep production of IL-1β and TNF-α. In contrast to TNF-α, the increased level of IL-1β in spleen declined more slowly and persisted till day 21 (Fig. 1; Fig. S1, Supporting Information). Cytokine profile after 2 weeks of ΔdsbA/FSC200 infection is associated with the peaks of IL-4 and IL-23 production (Fig. 1). The immunization with the ΔiglH/FSC200 mutant influenced expression of IFN-γ, IL-2, IL-17, IL-10, IL-23 and IL-12 cytokines in spleen; nevertheless, their levels appeared with a delay of more than 3 weeks in comparison to the ΔdsbA/FSC200 mutant (Fig. 1; Fig. S1, Supporting Information).
Figure 1.
In vivo cytokine immune responses elicited in BALB/c mice spleens after ΔdsbA/FSC200 and ΔiglH/FSC200 vaccination. Groups of BALB/c mice (n = 3) were s.c. inoculated either with 102 CFU/mouse of wt FSC200 strain (circles) or with 107 CFU/mouse of ΔdsbA/FSC200 (triangles) or 107 CFU/mouse of ΔiglH/FSC200 (squares) mutant strains. Individual spleen was removed at given time interval and analyzed for cytokine levels using cytokine arrays. Statistical comparison was done between groups vaccinated with ΔdsbA/FSC200 mutant and wt FSC200 strain and between groups vaccinated with ΔiglH/FSC200 mutant and wt FSC200 strain. Results represent means ± standard errors, where P < 0.05 was considered to be significant. The results shown are representatives of two separate experiments.
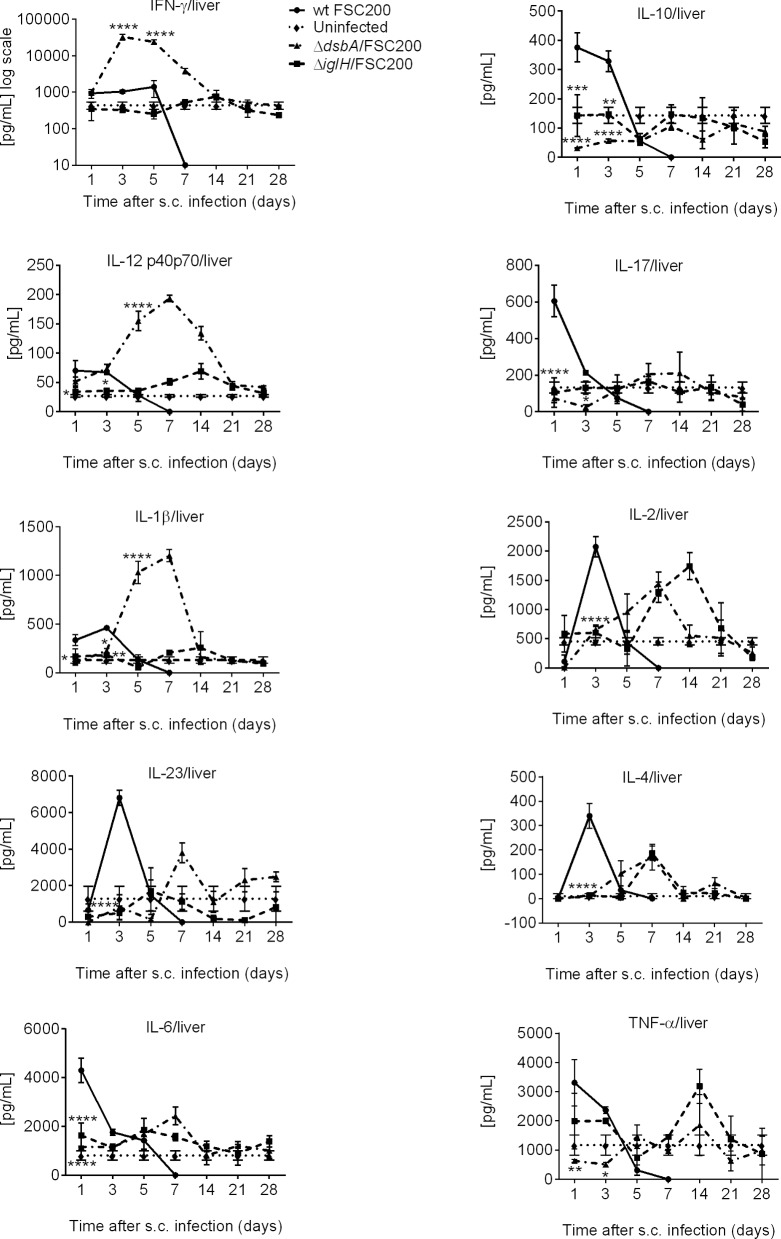
Likewise in spleen, the ΔdsbA/FSC200 mutant was also able to induce strong upregulation of IFN-γ, IL-1β and IL-12 levels in liver (Fig. 2; Fig. S2, Supporting Information). The increased production of IL-6 was shifted to day 7 after infection in liver, and on day 7 also the upregulation of IL-17 production occurred. Both ΔdsbA/FSC200 and ΔiglH/FSC200 mutants then stimulated secretion of IL-2 and IL-4 on day 7 in liver and TNF-α on day 14 after immunization (Fig. 2; Fig. S2, Supporting Information).
Figure 2.
In vivo cytokine immune responses elicited in BALB/c mice livers after ΔdsbA/FSC200 and ΔiglH/FSC200 vaccination. Groups of BALB/c mice (n = 3) were s.c. inoculated either with 102 CFU/mouse of FSC200 strain (circles) or with 107 CFU/mouse of ΔdsbA/FSC200 (triangles) or 107 CFU/mouse of ΔiglH/FSC200 (squares) mutant strains. At selected time intervals after infection, individual livers were removed and further analyzed for cytokine levels using cytokine arrays. Statistical comparison was done between groups vaccinated with ΔdsbA/FSC200 mutant and wt FSC200 strain and between groups vaccinated with ΔiglH/FSC200 mutant and wt FSC200 strain. Results represent means ± standard errors, where P < 0.05 was considered to be significant. The results shown are representatives of two independent experiments.
Investigation of cytokine patterns in sera of vaccinated BALB/c mice confirmed that the ΔdsbA/FSC200 mutant can upregulate the early IFN-γ, IL-12 and IL-6 responses (Fig. 3; Fig. S3, Supporting Information). Furthermore, there was strong increase of IL-1β, IL-4, TNF-α and IL-10, but in these cases the response was divided in two phases, one on day 5 and the second 3 weeks after infection (Fig. 3; Fig. S3, Supporting Information). The complicated kinetics exhibited production of IL-2 with three maxima on days 1, 5 and 14. Late time responses are associated with increased levels of IL-17 and IL-23. Like in spleen and liver, the ΔiglH/FSC200 mutant induced only a weak inflammatory cytokine response in serum (Fig. 3; Fig. S3, Supporting Information). The only exception was TNF-α production, but even in this case the ΔdsbA/FSC200 mutant was more efficient than the ΔiglH/FSC200 mutant (Fig. 3; Fig. S3, Supporting Information).
Figure 3.
In vivo cytokine immune responses elicited in BALB/c mice sera after ΔdsbA/FSC200 and ΔiglH/FSC200 vaccination. Groups of BALB/c mice (n = 3) were s.c. inoculated either with 102 CFU/mouse of FSC200 strain (circles) or with 107 CFU/mouse of ΔdsbA/FSC200 (triangles) or 107 CFU/mouse of ΔiglH/FSC200 (squares) mutant strains. Mice were killed at given time interval after vaccination and cytokine levels were determined in the serum using cytokine arrays. The results shown are representatives of two independent experiments.
As for infection with parental FSC200 strain, there was a distinct early induction of IL-2, IL-23 and IL-4 in liver and of IFN-γ, Il-6 and IL-17 in serum (Figs 2 and 3). Although these mice succumbing to the infection within 5 days after inoculation.
Humoral immune response in BALB/c mice after vaccination with the ΔdsbA/FSC200 and the ΔiglH/FSC200 mutant strains
As an additional correlate of in vivo protection, we measured the development of humoral adaptive immune response. Groups of BALB/c mice (n = 3) were s.c. inoculated with 107 CFU/mouse of ΔdsbA/FSC200 or 107 CFU/mouse of ΔiglH/FSC200 mutant strains. The levels of IgM, IgG1, IgG2a, IgG2b and IgA antibodies were analyzed on days 1, 3, 5, 7, 14, 21 and 28 post-vaccination and compared to antibody levels generated in uninfected mice and mice infected with the parental FSC200 strain.
Mice vaccinated with the ΔdsbA/FSC200 mutant showed an early increase of all examined antibody classes except for IgG1 (Fig. 4; Fig. S4, Supporting Information). The most pronounced difference was found in the production of IgA and IgG2a antibodies soon after ΔdsbA/FSC200 vaccination (Fig. 4; Fig. S4, Supporting Information). It was striking that production of both antibody classes exerted the same kinetics with two peaks on days 5 and 21 post-vaccination (Fig. 4; Fig. S4, Supporting Information). Surprisingly, the antibody levels detected after the ΔiglH/FSC200 mutant vaccination did not rise over the cut off levels of uninfected mice except for IgG1, where the concentration increased early at day 3 and later at day 21 post-infection (Fig. 4; Fig. S4, Supporting Information).
Figure 4.
Humoral immune responses in BALB/c mice after ΔdsbA/FSC200 and ΔiglH/FSC200 vaccination. Groups of BALB/c mice (n = 3) were s.c. inoculated either with 102 CFU/mouse of FSC200 strain (circles) or with 107 CFU/mouse of ΔdsbA/FSC200 (triangles) or 107 CFU/mouse of ΔiglH/FSC200 (squares) mutant strains. Mice were killed at given time interval after vaccination and antibody levels were determined in the serum using cytokine arrays. The results shown are representatives of two separate experiments.
Bacterial burdens in mice organs after the ΔdsbA/FSC200 mutant vaccination
The presence of mutant bacteria in mice organs without causing animal disease is an efficient stimulus for the immune system. The groups of BALB/c mice (n = 3) were infected s.c. with the ΔdsbA/FSC200 mutant using a dose of 107 CFU/mouse. Mice tissues were then collected on days 1, 3, 5, 7, 14, 21 and 28 following infection, and bacterial numbers were determined in homogenates of spleen, lungs and liver.
As shown in Fig. 5, the ΔdsbA/FSC200 mutant strain was able to spread to the spleen, liver and lungs of BALB/c mice after s.c. infection. The highest level of recoverable bacteria was found in spleen and liver, where the CFUs reached more than 105 within 5 days of infection (Fig. 5). Thereafter, the mutant bacteria recovered from spleen and liver declined throughout the remaining study period (Fig. 5). The mutant bacteria were completely eliminated from liver samples within 21 days of infection and from spleen within 28 days (Fig. 5). It is necessary to mention that bacterial burdens in spleen and liver roughly followed level of cytokines and antibodies detected in these organs. In contrast to the liver and spleen, significantly lower CFUs were detected in lung tissue early after infection and bacteria were completely eliminated within 1 week of infection (Fig. 5).
Figure 5.
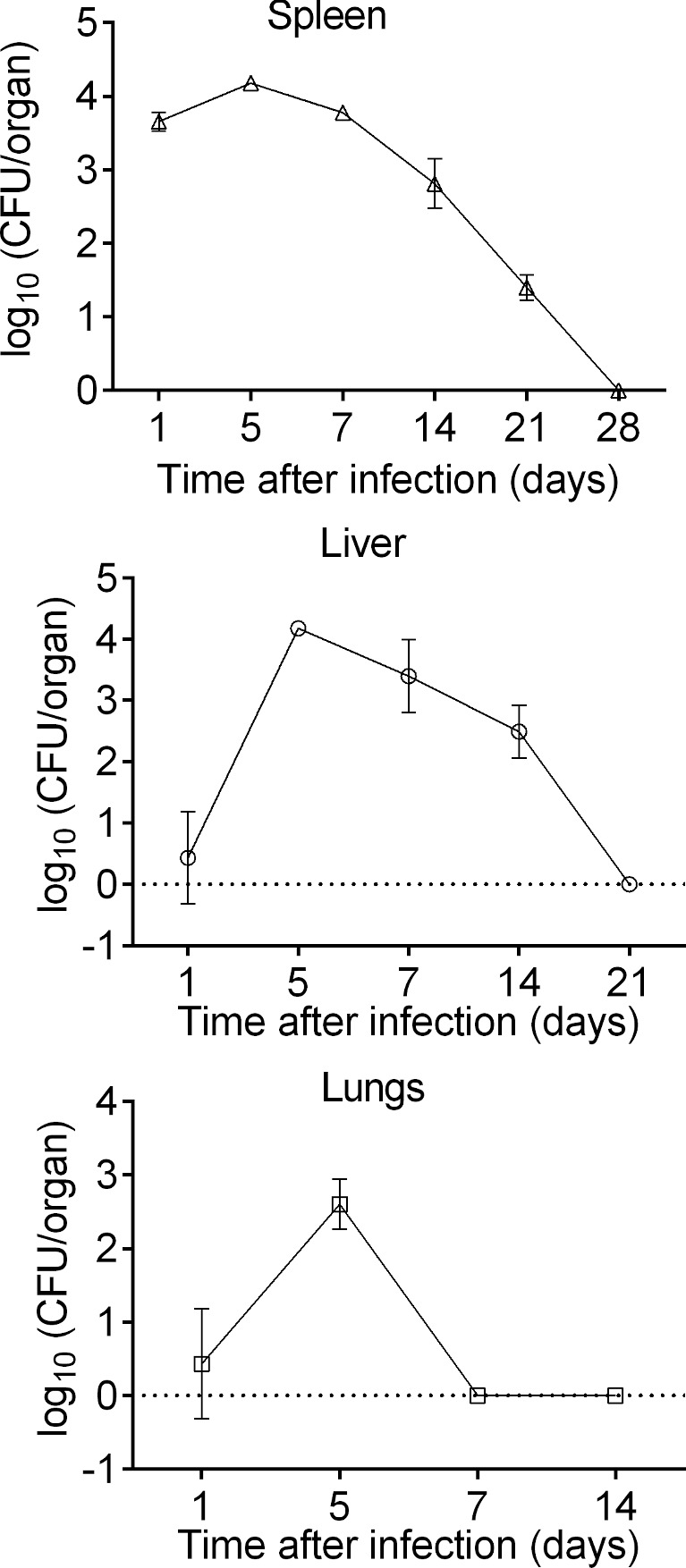
Bacterial burdens in mice organs after ΔdsbA/FSC200 mutant vaccination. BALB/c mice (n = 3 per group) were inoculated s.c. with 107 CFU/mouse of the ΔdsbA/FSC200 mutant and CFUs were determined for the lung, liver and spleen tissues at each time point indicated. Results represent means ± standard errors of CFU counts. The data are representative of three independent experiments.
Protection of the ΔdsbA/FSC200 and the ΔiglH/FSC200 mutant strains against F. tularensis SCHU S4 challenge
Based on previous data documenting protective potential of both mutants, we decided to examine their ability to protect against the challenge with highly virulent F. tularensis SCHU S4 strain. The ΔdsbA/FSC200 or the ΔiglH/FSC200 immunized mice were s.c. challenged with 100 CFU of SCHU S4 strain. Mice immunized with the ΔiglH/FSC200 mutant showed rapid signs of illness and four mice died within 6–14 days post-challenge. The remaining mice returned to health by day 21 (Table 1A). The group of mice inoculated with the ΔdsbA/FSC200 mutant survived the challenge with SCHU S4 strain without any post-infection clinical signs of tularemia (Table 1A). The control group of non-immunized animals died on days 4–5 post-challenge (Table 1A).
Table 1A.
Survival of BALB/c mice following subcutaneous immunization with the ΔdsbA/FSC200 mutant or the ΔiglH/FSC200 mutant against SCHU S4 s.c. challenge.
| Bacterial strain | s.c. dose CFU/mouse on day 0 (vaccination dose) | s.c. dose of SCHU S4 CFU/mouse on day 21 after vaccination (challenge dose) | Nr. of deaths/total |
|---|---|---|---|
| 0 (mock solution only) | 102 | 5/5 | |
| ΔdsbA/FSC200 | 10 | 102 | 4/5 |
| 102 | 102 | 4/5 | |
| 103 | 102 | 1/5 | |
| 104 | 102 | 0/5 | |
| 105 | 102 | 0/5 | |
| 107 | 102 | 0/5 | |
| ΔiglH/FSC200 | 107 | 102 | 4/5 |
BALB/c mice (n = 5) were immunized subcutaneously with the ΔdsbA/FSC200 mutant or the ΔiglH/FSC200 mutant. Animals were challenged subcutaneously 3 weeks later with virulent SCHU S4 strain. The mice were monitored daily for morbidity and mortality. The study endpoint was euthanasia when moribund or survival to 21 days following exposure.
Next, we titrated the immunization doses of the ΔdsbA/FSC200 mutant to find the lowest possible dose with protection capability. Therefore, mice were s.c. inoculated with different doses of the ΔdsbA/FSC200 mutant and after 3 weeks s.c. challenged with 100 CFU/mouse of SCHU S4 strain. We observed that the complete protection of animals against the SCHU S4 infection can be reached with doses of as low as 104 CFU/mouse (Table 1A). In addition, we tested the intranasal protection ability of the ΔdsbA/FSC200 mutant against the i.n. SCHU S4 challenge. Mice were vaccinated with different doses of the ΔdsbA/FSC200 mutant strain, where none of mice showed sickness during 28 days of observation after immunization. Immunized mice were further infected i.n. with 102 CFU/mouse of SCHU S4 strain. All mice in the groups vaccinated with 10, 102 and 103 CFU/mouse died on day 4 post-challenge. Protection was observed in the groups of animals vaccinated with 104, 105 and 106 CFU/mouse, where two of ten, three of ten and five of ten mice survived, respectively (Table 1B).
Table 1B.
Survival of BALB/c mice following intranasal immunization with the ΔdsbA/FSC200 mutant against SCHU S4 i.n. challenge.
| Bacterial strain | i.n. dose CFU/mouse on day 0 (vaccination dose) | i.n. dose of SCHU S4 CFU/mouse on day 28 after vaccination (challenge dose) | Nr. of deaths/total |
|---|---|---|---|
| 0 (mock solution only) | 102 | 10/10 | |
| ΔdsbA/FSC200 | 10 | 102 | 10/10 |
| 102 | 102 | 10/10 | |
| 103 | 102 | 10/10 | |
| 104 | 102 | 8/10 | |
| 105 | 102 | 7/10 | |
| 106 | 102 | 5/10 |
BALB/c mice (n = 10) were immunized i.n. with the ΔdsbA/FSC200 mutant, animals were challenged i.n. 28 days later with virulent SCHU S4 strain. The mice were monitored daily for morbidity and mortality. The study endpoint was euthanasia when moribund or survival to 21 days following exposure.
Our results show that the ΔdsbA/FSC200 mutant is attenuated for s.c. and i.n. infection of BALB/c mice. Moreover, the ΔdsbA/FSC200 mutant has protective ability against lethal dose of SCHU S4 strain in s.c. infection and is able to partially protect against respiratory SCHU S4 challenge.
Mapping of ΔdsbA/FSC200 post-vaccination and SCHU S4 post-challenge immunoproteome
As we have shown, the protective response of the ΔdsbA/FSC200 mutant strain is accompanied by increased levels of IgA and IgG2a antibody production. Thus, we decided to examine the profile of the antibody-recognized antigens after s.c. vaccination with the ΔdsbA/FSC200 mutant strain. Assuming that the surface-exposed and membrane-associated proteins are crucial antibody targets, detergent-enriched and pilin protein-enriched subproteomes were analyzed using a classical immunoproteomic approach with sera pooled from ΔdsbA/FSC200 vaccinated BALB/c mice. Overall, we identified 63 antigens, 22 of which had not been previously described (Table 2; Table S1, Supporting Information) (Pelletier, Raoult and La Scola 2009; Kilmury and Twine 2010; Fulton et al. 2011; Golovliov et al. 2013). It is interesting that most of them, 19 antigens in total, were found in pilin protein-enriched subproteome which seems to be a valuable source for antibody-inducing antigens. The group of novel immunoreactive antigens covered the spectrum of enzymes, as well as ribosomal and stress proteins. It is worth to mention acid phosphatase (FTT_0221), DipA (FTT_0369c), D-alanyl-D-alanine carboxypeptidase (FTT_1029), IglE (FTT_1701/FTT_1346), PdpE (FTT_1710/FTT_1355), PilA orthologs (FTT_0889c, FTT_0890c) and hypothetical proteins (FTT_0704, FTT_0903, FTT_1407c and FTT_1653). Because ΔdsbA/FSC200 vaccination helps mice to survive SCHU S4 infection, we also collected sera from SCHU S4 challenged mice to measure possible seroconversion to SCHU S4 specific antigens. In this case, we identified 71 antigens recognized by SCHU S4 post-challenge sera (Table S1, Supporting Information). Of them eight new antibody-binding antigens not found in ΔdsbA/FSC200 post-vaccination sera were discovered (Table 2). These antigens recognized uniquely by SCHU S4 post-challenge sera also formed a functionally heterogeneous group involving proteins with the known role in Francisella pathogenesis such as superoxide dismutase Fe (FTT_0068), superoxide dismutase Cu-Zn (FTT_0879) and hypothetical protein (FTT_0910).
Table 2.
Novel immunoreactive SCHU S4 antigens.
| SCHU S4 gene locus | Protein name | Detected by ΔdsbA/200 serum | Detected by ΔdsbA/200 + SCHU S4 serum | Antigen |
|---|---|---|---|---|
| FTT_0034* | NADH dehydrogenase I, D subunit | x | LP fraction | |
| FTT_0068 | Superoxide dismutase [Fe] | x | Pilin fraction | |
| FTT_0080 | Triosephosphate isomerase | x | Pilin fraction | |
| FTT_0139 | Transcription antitermination protein nusG | x | x | Pilin fraction |
| FTT_0142 | 50S ribosomal protein L10 | x | x | Pilin fraction |
| FTT_0339 | 30S ribosomal protein S8 | x | x | Pilin fraction |
| FTT_0369c | Hypothetical protein | x | x | Pilin fraction |
| FTT_0372c | AcetylCoA carboxylase beta subunit | x | x | Pilin fraction |
| FTT_0624 | ATP-dependent Clp protease subunit P | x | Pilin fraction | |
| FTT_0704 | Hypothetical protein | x | x | Pilin fraction |
| FTT_0879 | Superoxide dismutase (Cu-Zn) precursor | x | LP fraction | |
| FTT_0889c | Type IV pili fiber building block protein PilE | x | x | LP fraction |
| FTT_0890c | Type IV pili fiber building block protein PilA | x | x | LP fraction |
| FTT_0903 | Hypothetical protein | x | x | Pilin fraction LP fraction |
| FTT_0910 | Hypothetical protein | x | LP fraction | |
| FTT_1241 | Serine hydroxymethyltransferase | x | x | Pilin fraction |
| FTT_1260 | Hypothetical lipoprotein | x | LP fraction | |
| FTT_1375 | 3-Oxoacyl-(acyl-carrier-protein) reductase | x | x | Pilin fraction |
| FTT_1407c | Hypothetical membrane protein | x | x | Pilin fraction |
| FTT_1459c | NAD-dependent epimerase | x | Pilin fraction | |
| FTT_1701, FTT_1346 | Hypothetical protein | x | x | Pilin fraction |
| FTT_1710, FTT_1355 | Conserved hypothetical protein | x | x | Pilin fraction |
*Criteria for identification not fulfilled (C.I. 92% and only one peptide per protein was identified), but the protein was identified repeatedly.
DISCUSSION
So far, the development of safe and effective vaccine against tularemia is still far from realization (Pechous, McCarthy and Zahrt 2009). Recently, a hypothetical lipoprotein with high homology to the protein family of disulphide oxidoreductases DsbA was identified as a new essential virulence factor of F. tularensis. The dsbA deletion mutants in both type A and type B F. tularensis strains were constructed and exerted pronounced attenuation in mouse infection models (Qin et al. 2009; Straskova et al. 2009). Furthermore, an intranasal immunization with the ΔdsbA mutant in type A strain or subcutaneous immunization with the ΔdsbA mutant in type B strain reliably protected against the challenge with parental virulent strains (Qin et al. 2009; Straskova et al. 2009). Besides ΔdsbA/FSC200 mutant strain, our laboratory has also constructed the deletion mutant for the FPI protein encoded by iglH gene in FSC200 strain. However, compared to the ΔdsbA/FSC200 mutant, its preventive effect was dose dependent and complete protection against the wild-type parental strain was only found after administration of the highest immunization dose of 3 × 107 CFU/mouse used (Straskova et al. 2009).
In this study, we explored the features of innate and adaptive immunity induced by these two attenuated mutants with differential protective capacity. In agreement with these findings, we were able to measure an early increase of IFN-γ and IL-6 in spleen and serum in the ΔdsbA/FSC200 immunized BALB/c mice. Higher levels of IFN-γ but not IL-6 were also observed in liver samples. The strong early inflammatory response to ΔdsbA/FSC200 vaccination was further corroborated by the increased levels of IL-12 and IL-1β in spleen, liver and serum and TNF-α in spleen and serum. Furthermore, liver and serum samples exhibited an increased response of IL-2 production after ΔdsbA/FSC200 immunization. Furthermore, spleen tissue was also found to be a source of IL-23 overproduction with a maximum on day 14 post-infection. The same time interval of IL-23 upregulation was observed for serum samples. Mice deficient in IL-12p40 but not in IL-12p35 are susceptible to F. tularensis LVS infections indicating that IL-12p70 is not necessary for bacteria elimination (Elkins et al. 2002). As the IL-23 is a complex of p40 subunit with a 19-kDa protein, it is possible that only IL-23 can participate in the development of T-cell-mediated bacterial clearance. Furthermore, IL-23 is an inducer of IL-17 production. As for IL-17, we detected increased levels of this cytokine in liver tissue; however, an even more pronounced expression was found in serum with maximum at late time point (21 days post-infection). There is controversy about the role of IL-17 in protection against respiratory challenge with SCHU S4. While Skyberg et al. (2013) showed that IL-17 is inefficient for intratracheal infection of mice with SCHU S4 strain, Golovliov et al. (2013) demonstrated that increased efficacy of FSC200 clpB mutant strain in induction protective response against SCHU S4 respiratory infection is associated with the increased IL-17 pulmonary production after SCHU S4 challenge. A significant upregulation of IL-4 expression was also observed in liver and spleen samples on days 7 and 14 post-infection, respectively. Mast cells were found to be major source of IL-4 capable to restrict intramacrophage growth of F. tularensis LVS (Thathiah et al. 2011), and the production of this cytokine was dependent on mast cell TLR2 signaling (Rodriguez et al. 2012). In agreement with less efficient in vivo protection of mice against parental virulent strain, the ΔiglH/FSC200 mutant strain exerted, besides TNF-α level in serum, weak and delayed production of inflammatory cytokines compared to the ΔdsbA/FSC200. Recently, a similar study was published in which four defined gene deletion mutants of SCHU S4 were examined for their abilities to induce protective responses against dermal and respiratory challenge with SCHU S4 strain. Two of them, deletion mutants for clpB and γ-glutamyl transpeptidase exhibited the most efficient protection in both dermal and resiratory models. Likewise in our study, the protective capacity of these two mutants correlated, among others, with the increased levels of IFN γ, TNF α in serum and IFN γ, TNF α, IL-1β, IL-6, IL-17 in spleen (Ryden et al. 2013). Generally, the cytokine pattern induced by the ΔdsbA/FSC200 mutant indicates the development of a strong Th1 protective response. This finding was further supported by the array analysis of antibody induction in serum of mice immunized with the ΔdsbA/FSC200 and the ΔiglH/FSC200 mutants. We found that ΔdsbA/FSC200 vaccination induced all antibody classes; nevertheless, the most robust response concerned the IgA and IgG2a production, which had two maxima, of which the early one paralleled the traditional quick IgM antibody secretion. Very recent publication demonstrated that IgA-deficient mice exhibited enhanced susceptibility to pulmonary F. tularensis LVS infection. Additionally, these mice had significantly reduced pulmonary levels of IFN-γ and IL-12. The decline in IFN-γ amount reflects the diminished numbers of CD4+ and CD8+ T cells in the lungs (Furuya et al. 2013). In contrast to ΔdsbA/FSC200, the ΔiglH/FSC200 infected mice only upregulated IgG1 levels typical for a Th2 response. Previously, we had observed that subcutaneous vaccination of mice with ΔdsbA/FSC200 can reliably protect mice against 4 × 105 CFU of the wild-type B isolate FSC200 (Straskova et al. 2009). In this study, we looked at distribution and persistence of the ΔdsbA/FSC200 microbes in selected organs of vaccinated mice. We have shown that the bacterial burdens following s.c. infection with the mutant strain were highest in spleen followed by liver and lungs (Fig. 5). Importantly, the mutant bacteria were eliminated from all analyzed organs during the time period of 4 weeks (Fig. 5). This finding shows the ability of the host to mount the effective immune response after the ΔdsbA/FSC200 administration that even protects BALB/c mice against subcutaneous and respiratory SCHU S4 challenge (Tables 1A and 1B).
Using comparative proteome analyses, we had already showed that there are differences in protein expression between type B and type A strains (Hubalek et al. 2004; Pavkova et al. 2006). Nevertheless, the protective ability of the ΔdsbA/FSC200 mutant against type A strain indicates that some immunoreactive antigens can be shared. Data from our present immunoproteomic study confirmed that 14 antigens were recognized by both types of immune sera (Table 2). On the other hand, seroconversion to SCHU S4 unique response was reflected by production of antibodies against another eight bacterial proteins (Table 2). Among identified proteins, DipA, FTT_1407c, PilA, PdpE, FTT_0903, IglE, acid phosphatase, superoxide dismutase Fe and superoxide dismutase Cu-Zn represent potential or well-known virulence factors (Weiss et al. 2007; Melillo et al. 2009; Akimana, Al-Khodor and Abu Kwaik 2010; Forslund et al. 2010; Chong et al. 2013; Mohapatra et al. 2013; Robertson et al. 2013). It was already described that IglE, PdpE superoxide dismutase Fe and acid phosphatase can be secreted during Francisella growth (Konecna et al. 2010; Broms et al. 2012). Additionally, we observed that DipA and IglE can accumulate in a membrane of the ΔdsbA/LVS mutant (Straskova et al. 2009) and unpublished observation. It might be that these proteins are misfolded due to the loss of the DsbA protein, which has both thiol/disulfide oxidoreductase and chaperone function (Straskova et al. 2009; Schmidt et al. 2013) and are, therefore, not delivered to proper location. This situation can lead to the increased immunogenicity of these misfolded proteins.
Our results show that the ΔdsbA/FSC200 mutant is able to mount strong Th1 type immune response in vivo that is even efficient against a subcutaneous and intranasal challenge with type A Francisella SCHU S4 strain. Moreover, identification of novel immunoreactive antigens which are common or unique for the ΔdsbA/FSC200 vaccination or the SCHU S4 challenge then contributes to the list of proteins useful for a subunit vaccine design.
Supplementary Material
Acknowledgments
We are grateful to Evelyn Lawrenz for critical reading of the manuscript, to Maria Safarova for her technical assistance with mouse experiments and Jitka Zakova for help with 2D electrophoresis.
SUPPLEMENTARY DATA
FUNDING
The work was supported by Specific Research grant of Ministry of Education, Youth and Sports of Czech Republic (SV/FVZ201202), DTRA project CB3387 PA D-CZ-11-0001 and by a long-term organization development plan 1011.
Conflict of interest. None declared.
REFERENCES
- Akimana C, Al-Khodor S, Abu Kwaik Y. Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS One. 2010;5:e11025. doi: 10.1371/journal.pone.0011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balonova L, Hernychova L, Mann BF, et al. Multimethodological approach to identification of glycoproteins from the proteome of Francisella tularensis, an intracellular microorganism. J Proteome Res. 2010;9:1995–2005. doi: 10.1021/pr9011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms JE, Meyer L, Sun K, et al. Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS One. 2012;7:e50473. doi: 10.1371/journal.pone.0050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Child R, Wehrly TD, et al. Structure-function analysis of DipA, a virulence factor required for intracellular replication. PLoS One. 2013;8:e67965. doi: 10.1371/journal.pone.0067965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–73. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Elkins KL, Cooper A, Colombini SM, et al. In vivo clearance of an intracellular bacterium, Francisella tularensis LVS, is dependent on the p40 subunit of interleukin-12 (IL-12) but not on IL-12 p70. Infect Immun. 2002;70:1936–48. doi: 10.1128/IAI.70.4.1936-1948.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ME, Gregory DW, Schaffner W, et al. Tularemia: a 30-year experience with 88 cases. Medicine. 1985;64:251–69. [PubMed] [Google Scholar]
- Forslund AL, Salomonsson EN, Golovliov I, et al. The type IV pilin, PilA, is required for full virulence of Francisella tularensis subspecies tularensis. BMC Microbiol. 2010;10:227. doi: 10.1186/1471-2180-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton KM, Zhao X, Petit MD, et al. Immunoproteomic analysis of the human antibody response to natural tularemia infection with Type A or Type B strains or LVS vaccination. Int J Med Microbiol. 2011;301:591–601. doi: 10.1016/j.ijmm.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya Y, Kirimanjeswara GS, Roberts S, et al. Increased susceptibility of IgA-deficient mice to pulmonary Francisella tularensis live vaccine strain infection. Infect Immun. 2013;81:3434–41. doi: 10.1128/IAI.00408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I, Twine SM, Shen H, et al. A deltaclpB mutant of Francisella tularensis subspecies holarctica strain, FSC200, is a more effective live vaccine than F. tularensis LVS in a mouse respiratory challenge model of tularemia. PLoS One. 2013;8:e78671. doi: 10.1371/journal.pone.0078671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubalek M, Hernychova L, Brychta M, et al. Comparative proteome analysis of cellular proteins extracted from highly virulent Francisella tularensis ssp. tularensis and less virulent F. tularensis ssp. holarctica and F. tularensis ssp. mediaasiatica. Proteomics. 2004;4:3048–60. doi: 10.1002/pmic.200400939. [DOI] [PubMed] [Google Scholar]
- Johansson A, Berglund L, Sjostedt A, et al. Ciprofloxacin for treatment of tularemia. Clin Infect Dis. 2001;33:267–8. doi: 10.1086/321825. [DOI] [PubMed] [Google Scholar]
- Kilmury SL, Twine SM. The Francisella tularensis proteome and its recognition by antibodies. Front Microbiol. 2010;1:143. doi: 10.3389/fmicb.2010.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecna K, Hernychova L, Reichelova M, et al. Comparative proteomic profiling of culture filtrate proteins of less and highly virulent Francisella tularensis strains. Proteomics. 2010;10:4501–11. doi: 10.1002/pmic.201000248. [DOI] [PubMed] [Google Scholar]
- Marohn ME, Barry EM. Live attenuated tularemia vaccines: recent developments and future goals. Vaccine. 2013;31:3485–91. doi: 10.1016/j.vaccine.2013.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo AA, Mahawar M, Sellati TJ, et al. Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J Bacteriol. 2009;191:6447–56. doi: 10.1128/JB.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Rajaram MV, et al. Type A Francisella tularensis acid phosphatases contribute to pathogenesis. PLoS One. 2013;8:e56834. doi: 10.1371/journal.pone.0056834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–78. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- Pavkova I, Reichelova M, Larsson P, et al. Comparative proteome analysis of fractions enriched for membrane-associated proteins from Francisella tularensis subsp. tularensis and F. tularensis subsp. holarctica strains. J Proteome Res. 2006;5:3125–34. doi: 10.1021/pr0601887. [DOI] [PubMed] [Google Scholar]
- Pechous RD, McCarthy TR, Zahrt TC. Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol Mol Biol R. 2009;73:684–711. doi: 10.1128/MMBR.00028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier N, Raoult D, La Scola B. Specific recognition of the major capsid protein of Acanthamoeba polyphaga mimivirus by sera of patients infected by Francisella tularensis. FEMS Microbiol Lett. 2009;297:117–23. doi: 10.1111/j.1574-6968.2009.01675.x. [DOI] [PubMed] [Google Scholar]
- Qin A, Scott DW, Thompson JA, et al. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun. 2009;77:152–61. doi: 10.1128/IAI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GT, Child R, Ingle C, et al. IglE Is an outer membrane-associated lipoprotein essential for intracellular survival and murine virulence of type A Francisella tularensis. Infect Immun. 2013;81:4026–40. doi: 10.1128/IAI.00595-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AR, Yu JJ, Guentzel MN, et al. Mast cell TLR2 signaling is crucial for effective killing of Francisella tularensis. J Immunol. 2012;188:5604–11. doi: 10.4049/jimmunol.1200039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P, Eley SM, Fulop MJ, et al. The efficacy of ciprofloxacin and doxycycline against experimental tularaemia. J Antimicrob Chemoth. 1998;41:461–5. doi: 10.1093/jac/41.4.461. [DOI] [PubMed] [Google Scholar]
- Ryden P, Twine S, Shen H, et al. Correlates of protection following vaccination of mice with gene deletion mutants of Francisella tularensis subspecies tularensis strain, SCHU S4 that elicit varying degrees of immunity to systemic and respiratory challenge with wild-type bacteria. Mol Immunol. 2013;54:58–67. doi: 10.1016/j.molimm.2012.10.043. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Klimentova J, Rehulka P, et al. Francisella tularensis subsp. holarctica DsbA homologue: a thioredoxin-like protein with chaperone function. Microbiology. 2013;159:2364–74. doi: 10.1099/mic.0.070516-0. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Kida Y, Kuwano K. A dipalmitoylated lipoprotein from Mycoplasma pneumoniae activates NF-kappa B through TLR1, TLR2, and TLR6. J Immunol. 2005;175:4641–6. doi: 10.4049/jimmunol.175.7.4641. [DOI] [PubMed] [Google Scholar]
- Skyberg JA, Rollins MF, Samuel JW, et al. Interleukin-17 protects against the Francisella tularensis live vaccine strain but not against a virulent F. tularensis type A strain. Infect Immun. 2013;81:3099–105. doi: 10.1128/IAI.00203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straskova A, Cerveny L, Spidlova P, et al. Deletion of IglH in virulent Francisella tularensis subsp. holarctica FSC200 strain results in attenuation and provides protection against the challenge with the parental strain. Microbes Infect. 2012;14:177–87. doi: 10.1016/j.micinf.2011.08.017. [DOI] [PubMed] [Google Scholar]
- Straskova A, Pavkova I, Link M, et al. Proteome analysis of an attenuated Francisella tularensis dsbA mutant: identification of potential DsbA substrate proteins. J Proteome Res. 2009;8:5336–46. doi: 10.1021/pr900570b. [DOI] [PubMed] [Google Scholar]
- Tarnvik A, Berglund L. Tularaemia. Eur Respir J. 2003;21:361–73. doi: 10.1183/09031936.03.00088903. [DOI] [PubMed] [Google Scholar]
- Thathiah P, Sanapala S, Rodriguez AR, et al. Non-FcepsilonR bearing mast cells secrete sufficient interleukin-4 to control Francisella tularensis replication within macrophages. Cytokine. 2011;55:211–20. doi: 10.1016/j.cyto.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Brotcke A, Henry T, et al. In vivo negative selection screen identifies genes required for Francisella virulence. P Natl Acad Sci USA. 2007;104:6037–42. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.