Abstract
To identify immune-related genes in the larvae of white-spotted flower chafers, next-generation sequencing was conducted with an Illumina HiSeq2000, resulting in 100 million cDNA reads with sequence information from over 10 billion base pairs (bp) and >50× transcriptome coverage. A subset of 77,336 contigs was created, and ∼35,532 sequences matched entries against the NCBI nonredundant database (cutoff, e < 10−5). Statistical analysis was performed on the 35,532 contigs. For profiling of the immune response, samples were analyzed by aligning 42 base sequence tags to the de novo reference assembly, comparing levels in immunized larvae to control levels of expression. Of the differentially expressed genes, 3,440 transcripts were upregulated and 3,590 transcripts were downregulated. Many of these genes were confirmed as immune-related genes such as pattern recognition proteins, immune-related signal transduction proteins, antimicrobial peptides, and cellular response proteins, by comparison to published data.
Keywords: Protaetia brevitarsis seulensis, immune, transcription, RNA-seq, Illumina
Larvae of all kinds of beetles live on the ground, where they frequently encounter pathogenic microorganisms such as bacteria, fungi, viruses, and parasitoids. Therefore, it is conceivable that beetles have well-developed immune systems. Despite the diversity of beetle species, the immune systems of only a few beetles have been studied, including the weevil Sitophilus zeamais, the mealworm beetle Tenebrio molitor, and the burying beetle Nicrophorus vespilloides (Zou et al. 2007, Anselme et al. 2008, Vogel et al. 2011, Dobson et al. 2012, Vogel et al. 2014).
Generally, the insect immune system consists of two arms, the cellular and the humoral immune response, and is a well-organized defensive system against various pathogens despite the lack of an acquired immune response, as seen in mammals (Hoffmann 2003). Many immune-related genes are upregulated in response to microbial challenge and antimicrobial peptides (AMPs) are ultimately synthesized through Toll and Immune deficiency (Imd) pathways in insects (Lemaitre and Hoffmann 2007, Bang et al. 2012, Kwon et al. 2014b). Approximately 20, 10, and 30 genes were identified as AMP genes in Drosophila melanogaster, Anopheles gambiae, and Bombyx mori, respectively (De Gregorio 2001, Tanaka et al. 2008, Lee et al. 2013). AMPs have been characterized and isolated from many insect species. In particular, coleoptericin-like proteins (coleoptericin, acaloleptin, holotricin, and rhinocerosin), which are found in coleopterans, were characterized in Holotricia diomphalia, Zophobas atratus, Allomyrina dichotoma, Acalolepta luxuriosa, and Stitophilus oryzae (Bulet et al. 1991, Lee et al. 1994, Sagisaka et al. 2001, Imamura et al. 2009, Login et al. 2011).
By contrast, cellular immune responses are mediated by insect blood cells (hemocytes), which are activated via pattern recognition receptor (PRR)-linked signal transduction pathways and conduct cellular processes that result in pathogen killing, such as phagocytosis, encapsulation, and nodulation. Transmembrane or extracellular PRR pathways (Toll receptors, C-type lectins [CTLs], and peptidoglycan recognition proteins [PGRPs]) are activated by Spätzle in the presence of microbe-associated molecular recognition patterns (MAMPs; microbial peptidoglycan, lipopolysaccharides, β-glucans, lipoproteins, CpG dinucleotides, or flagellin) expressed by pathogens (Shelby and Popham 2012). The confusion still exists regarding hemocyte categorization because hemocyte types vary greatly depending on the insect species (Gupta 1985, Kwon et al. 2014a). Although there has been some debate on the characterization of insect blood cell types, plasmatocytes and granulocytes are considered the key players in cell-mediated immunity (Kwon et al. 2014a).
As discussed earlier, it is reasonable to assume that many genes are associated with the cellular and humoral immune response in insects. Recently, genome-wide analysis was applied for the identification of immune-related genes and study of the molecular basis of host-microorganism interactions. In particular, whole genome mRNA sequencing (RNA-seq or transcriptome sequencing) provides comprehensive insight into the immune gene repertoire of nonmodel insects, and this technology has become a powerful tool for the analysis of differential gene expression (Liu et al. 2014, Vogel et al. 2014).
This study focused on high-throughput RNA sequencing (RNA-seq) of the immune response in white-spotted flower chafers, Protaetia brevitarsis seulensis (Kolbe). This insect was very easy to maintain under laboratory conditions, and the average duration of the larval stage (the period the insect spends on the ground) is over 60 days under constant conditions. In addition, the cellular immune system in this insect is very well-developed, as we reported (Kwon et al. 2014a). Many of the components of the immune system, including the Toll pathway, the Imd pathway, and AMPs, have not been identified or studied in this insect. Therefore, we examined the differentially expressed genes (DEGs) between untreated and immunized larvae by Illumina RNA-seq digital expression profiling (de novo transcriptome assembly [TA]). More than 70,000 putative transcripts were assembled, and >30,000 were annotated to known databases. These results were validated by comparison with our previous results on humoral and cellular immune response-related genes, which revealed a comprehensive overview of gene expression changes, including additional identification of immune-related genes.
Materials and Methods
Insects, Infection, and RNA Isolation
The white-spotted flower chafers, P. brevitarsis seulensis were reared and maintained as previously described (Kwon et al. 2014a). Briefly, larvae were reared in a constant environment incubator (Sanyo Electric Biomedical, Japan) at 25 ± 1°C, 40–60% relative humidity, and a long photoperiod of 16 h light:8 h dark cycle under aseptic conditions using well-fermented sterile oakwood sawdust (Kwon et al. 2014a). The animals used in this study were always in the last larva instar. For boosting immune activity, larvae were cold-anesthetized and a finely pulled glass needle (Hematokrit-kapillaren; Hischmann Laboragerä te, GmBH & Co. KG, Germany) was shallowly inserted into the dorsal vessel; 30 µl of 2 × 103 for Escherichia coli, 4 × 104 for Saccharomyces cerevisiae in phosphate buffered saline (PBS), or sterile water (for control larvae) were injected into the hemocoel (Kwon et al. 2014a). Larva were anesthetizing by 99.5% Diethy Ether (DaeJung Chemical Metales, Korea) for 1 min at room temperature and placing them on ice, after which they were dissected in a cold 125 mM NaCl solution. We injected a total of 5 beetles per sample group (challenged and control) and 10 µg of total RNA extracted from each of challenged or control fatbodies was isolated with the SV Total RNA Isolation System kit (Promega Corp., Madison, WI). Total RNA was dissolved in sterile water, and RNA quantity was determined on a spectrophotometer (Evolution 60S; Thermo Fisher Scientific, Madison, WI). RNA integrity was checked on Agilent 2100 BioAnalyzer (Agilent Technologies, Englewood, CO). All buffers were treated with 0.1% diethylpyrocarbonate solution and autoclaved before use.
Illumina Sequencing
Total RNA pools were submitted to Seoul National University Genome Analysis Center DNA Core for Illumina NGS RNA sequencing (http://nicem.snu.ac.kr/main/). Libraries from each sample group were constructed according to the standard Illumina RNA-seq protocol (Part# 1004898 Rev. Sept 08; http://www.illumina.com) from the pooled PCR products (Shelby and Popham 2012). Briefly, polyadenylated RNA was isolated by oligo-dT hybridization and enriched poly(A) RNA of each sample was fragmented into 200–700 nt pieces with RNA Fragmentations Reagents. The fragmented RNA were converted into cDNA using random hexamer first-strand synthesis followed by second-strand cDNA synthesis (NEB, Ipswich, MA). The resulting sequencing libraries was paired-end sequenced using PE90 strategy on illumina HiSeq2000 flow cell.
De Novo Transcriptome Assembly and Differentially Expressed Genes
Contaminating adaptor sequences were filtered using fastq score and removed using CLC GENOMICS WORK-BENCH software v5.0.1 (http://www.clcbio.com). The resulting clean reads were assembled de novo using CLC GENOMICS WORK-BENCH software v5.0.1 with standard setting (Vogel et al. 2014). For functional annotation, all unigene sequences were matched with various database such as Nr, Swiss-prot, GOG, and KEGG using BLASTX, and then searched nucleotide database Nt using BLASTN, with e-value cut-off of 10−5 (Liu et al. 2014). Many of these genes were confirmed as immune-related genes such as pattern recognition proteins, immune-related signal transduction proteins, AMPs, and cellular response proteins by comparison to published data and articles. A mapping based on DEGs was performed to examine transcript level change between immune challenged and control sample. The expression of genes was calculated with fragments per kb per million fragments method (Mortazavi et al. 2008, Liu et al. 2013). P values were calculated according to the hypergeometric test described by Audic and Claverie (1997) and the calculated P values were used to identify DEGs (Liu et al. 2013). DEGs were annotated with BLAST2GO (www.blast2go.com) by querying NCBI nr using blastx with an e-value cut-off of 10−3 (Johnston and Rolff 2013). Differential expression analysis was performed using the edgeR (3.4.0) package (McCarthy et al. 2012). Gene with expression changes no <twofolds and false discovery rates < 0.001 were considered as significant DEGs (Liu et al. 2013).
Results and Discussion
An Overview of RNA-Seq Data
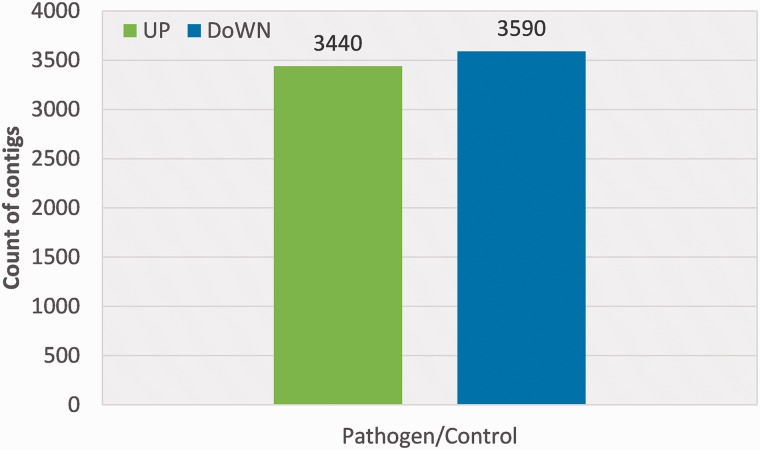
This report was mainly focused on RNA-seq expression profiling of immunized larvae of white-spotted flower chafers in response to microbial infection in vivo. An immune response was induced in these larvae by injection with bacteria and fungi, and cDNA libraries were prepared from control and immunized larvae. Analysis was carried out with an Illumina HiSeq2000 and resulted in 100 million cDNA reads with sequence information from over 10 billion base pairs (bp), and >50× transcriptome coverage. A total of 19.8 and 20.1 million raw reads were generated from challenged and control larvae libraries, respectively. After removal of adaptor sequence, ambiguous reads, and low-quality reads, challenged and control larvae libraries yielded 16.5 and 16.9 million. The de novo TA resulted in 77,336 (challenged) and 76,430 (control) contigs with a mean length, 801.5 and 766 bp, respectively. A subset of 77,336 and 76,430 contigs was created using a threshold of 50 mapped read pairs to eliminate spurious low-coverage sequences (Johnston and Rolff 2013). We identified 35,532 (challenged) and 34,230 unigenes (∼50%) matching entries against the NCBI nonredundant database (cutoff, e < 10−5). Statistical analysis was performed on these 35,532 (challenged) and 34,230 (control) contigs. However, ∼15,000 sequences did not match, indicating a large number of species-restricted transcripts, noncoding transcript segments, or chimeric sequences resulting from assembly errors and fragmented transcripts corresponding to weakly expressed genes (Beldade et al. 2008, Vogel et al. 2014). Retrotransposon and bacterial-specific sequences were detected in this assembly, and these sequences were matched with viral families and other bacterial proteins. Small amounts of these proteins may have been transcribed in these larvae or may have been detected as a result of low-level contamination by microorganisms (Shelby and Popham 2012). Expression profiling of the immune response was performed by aligning 42-base sequence tags from each treatment to the de novo reference assembly, comparing levels in immunized larvae to control levels of expression. To induce an immune response, a mixture of bacteria and yeast was injected into larvae, and 3,440 assemblies were upregulated while 3,590 assemblies were downregulated (|FC| ≥ 2) (Fig. 1).
Fig. 1.
Number of P. brevitarsis seulensis larva transcripts upregulated (green) or downregulated (blue) more than threefold by elicitation with mixture of Gram-negative bacterial and yeast.
Molecular Pattern Recognition Proteins
When pathogens invade insects, the innate immune system is activated by binding or recognizing pathogen-expressed molecules containing MAMPs (microbial peptidoglycan, lipopolysaccharides, β-glucans, lipoproteins, CpG dinucleotides, or flagellin) (Shelby and Popham 2012). These molecules are recognized by a group of proteins in insects known as PRRs, including β-1, 3-glucan recognition proteins (βGRPs), Gram-negative binding proteins (GNBPs), CTLs, scavenger receptors (SCRs), and PGRPs, which are generally classified as short (S) or long (L) forms (Liu et al. 2014, Nayduch et al. 2014). Among insect PRRs, PGRPs are highly evolutionarily conserved and enzymatically hydrolyze the bacterial cell wall in mammals. In our sequencing analysis, a number of P. brevitarsis seulensis orthologues of different PGRP types, including PGRP-1, 2, 3, LB, LC, LE, SC2, and SC3, were detected (Table 1). Among these, PGRP-1, 2, and 3 were highly upregulated in response to infection. PGRP-LB, LC, and LE were moderately upregulated, whereas PGRP-SC2 and SC3 were not significantly influenced by infection. As described in the Materials and Methods, Gram-negative bacteria and fungi were used as infection agents. Therefore, PGRP-1, 2, and 3 were specifically induced by Gram-negative bacteria or fungi, since Gram-positive bacteria were not used. Generally, the short forms of insect PGRP (PGRP-S) bind Gram-positive bacteria, activating the Toll pathway, whereas the long forms of PGRP (PGRP-L) bind Gram-negative bacteria and fungi and act via the Imd pathway (Shelby and Popham 2012). Although further confirmation is required, PGRP-SC2 and SC3 are likely activated by Gram-positive bacteria.
Table 1.
List of transcripts associated with molecular pattern recognition proteins
| Contig ID | Description | Nucleotide length (nt) | coverage (%) | Fold change | Species top blast | e-value |
|---|---|---|---|---|---|---|
| c16088_g1_i1 | PGRP-1 | 869 | 61.8 | 71.2 | H. diomphalia | 2.00E-128 |
| c16796_g1_i2 | PGRP-2 | 299 | 69.2 | 59.6 | H. diomphalia | 2.00E-40 |
| c18733_g1_i1 | PGRP-3 | 1381 | 40.6 | >100 | H. diomphalia | 3.00E-130 |
| c19802_g1_i1 | PGRP-LB | 1404 | 37.1 | 8.9 | Dr. melanogaster | 2.00E-64 |
| c24363_g1_i4 | PGRP-LC | 1917 | 51.1 | 2.3 | Dr. melanogaster | 3.00E-32 |
| c24236_g1_i1 | PGRP-LE | 2325 | 28.6 | 4.3 | Dr. melanogaster | 2.00E-57 |
| c17585_g2_i1 | βGRP | 1153 | 50.4 | 18.1 | T. molitor | 1.00E-49 |
| c18914_g1_i1 | GNBP2 | 2234 | 58.1 | 3.8 | T. castaneum | 1.00E-99 |
| c18362_g1_i2 | GNBP3 | 2107 | 62.0 | 2.7 | T. castaneum | 2.00E-120 |
| c22268_g1_i1 | SCRB | 1599 | 85.7 | 2.3 | T. castaneum | 0 |
| c122_g1_i1 | CTL | 787 | 81.9 | 2.7 | T. castaneum | 3.00E-137 |
| c18827_g1_i5 | PGRP-SC2 | 1129 | 44.3 | −1.87 | Dr. melanogaster | 4.00E-19 |
| c17295_g1_i1 | PGRP-SC3 | 1204 | 40.6 | −4.3 | An. gambiae | 2.00E-51 |
Within our de novo transcriptome, βGRP and GNBPs both were upregulated by bacterial and fungal infection. One βGRP and two GNBPs belonging to a subfamily of PRRs were also identified (Table 1). In particular, βGRP was highly expressed, as we previously reported (Bang et al. 2013). Insect βGRPs were first identified in B. mori associated with the enzyme prophenoloxidase (PPO) activation system and have since been identified in several insects such as Drosophila, Anopheles, Apis, Manduca, Spodoptera, and Tribolium (Ochiai and Ashida 1988, Kim et al. 2000, Christophides et al. 2002, Jiang et al. 2004, Evans et al. 2006, Zou et al. 2007, Bang et al. 2013, Liu et al. 2014). βGRPs have a strong affinity for β-1,3-glucan in the fungal cell wall and have been implicated in the activation of a serine protease cascade that activates a phenoloxidase cascade and AMP gene expression in insects. Although only two GNBPs were identified as moderately upregulated in our data, three GNBPs (GNBP1, GNBP2, and GNBP3) were found in Drosophila. Drosophila GNBP1 is usually upregulated by Gram-positive bacteria, and Drosophila GNBP3 is generally induced by fungal infection (Lemaitre and Hoffmann 2007).
In addition, transcripts from CTLs and SCRB were also found to be moderately upregulated in response to infection (Table 1). The SCR is specifically associated with phagocytosis by macrophages (Canton et al. 2013). We previously reported that pathogens were highly phagocytosed and killed by granulocytes in this insect (Kwon et al. 2014a). Although we do not yet have direct evidence, it seems likely that SCR expressed by granulocytes in this insect might be involved in pathogen recognition for phagocytosis.
Antimicrobial Peptides
As expected, genes encoding AMPs representing several different families (Table 2) were detected. Genes belonging to the attacin and defensin family were the most upregulated, with >100-fold higher expression in immunized larvae than in naïve ones. Attacin has a broad antibacterial range, particularly for Gram-negative bacteria, but the mode of action of attacin has not been well-studied (Bang et al. 2012). Previously, we isolated and characterized attacin from Spodoptera and demonstrated its powerful antibacterial activity. The gene encoding attacin was isolated in several insects, including Dr. melanogaster, Musca domestica, and B. mori (Sugiyama et al. 1995, Dushay et al. 2000, Geng et al. 2004). Defensin was also highly upregulated in immune-challenged larvae in our study, as previously reported by Hwang et al. (2008). Although insect defensins are known for antimicrobial effects mainly against Gram-positive bacteria, in the white-spotted flower chafer, the defensin family also seems to be active against Gram-negative bacteria and fungi (Langen et al. 2006, Vogel et al. 2014).
Table 2.
AMPs
| Contig ID | Description | Nucleotide length (nt) | Coverage (%) | Fold change | Species top blast | e-value |
|---|---|---|---|---|---|---|
| c14491_g2_i1 | Attacin family | 662 | 75.2 | >100 | Microdera dzhungarica | 1.00E-43 |
| c20401_g1_i1 | Defensin family | 1035 | 22.9 | >100 | Anomala cuprea | 1.00E-28 |
| c20145_g1_i1 | Holotricin family (Coleoptericin-like protein) | 492 | 77.4 | >100 | H. diomphalia | 7.00E-74 |
| c15432_g1_i1 | Antimicropaptide Cp1-like protein | 634 | 77.6 | >100 | C. tripartitus | 1.00E-53 |
| c17932_g1_i1 | Antimicropaptide Cp6-like protein | 488 | 78.0 | 4.4 | C. tripartitus | 2.00E-17 |
| c15466_g1_i1 | C-type Lysozme | 758 | 52.4 | 4.9 | Papilio polytes | 2.00E-37 |
| c23216_g1_i2 | Tenascin-N-like protein | 2994 | 64.5 | 17.3 | Heterocephalus glaber | 1E-07 |
| c19523_g1_i1 | Chitinase-like protein | 1996 | 71.0 | 25.9 | Holotrichia oblita | 0 |
| c6794_g1_i1 | Peritrophin | 373 | 64.3 | 2.7 | Popillia japonica | 9.00E-10 |
| c17703_g1_i1 | Mucin-like protein | 300 | 66 | 5.9 | H. oblita | 4.00E-12 |
In a separate study, we recently isolated and characterized a coleoptericin-like protein from this insect (Lee et al. 2013). Coleoptericin-like proteins (coleoptericin, acaloleptin, holotricin, and rhinocerosin) were isolated from Coleopteran such as H. diomphalia, A. dichotoma, A. luxuriosa, and S. oryzae as well as P. brevitarsis seulensis (Lee et al. 1994, Sagisaka et al. 2001, Imamura et al. 2009, Login et al. 2011). Coleoptericin-like proteins contain ∼70 amino acid residues and are glycine- and proline-rich AMPs with anti-bacteriocidal activity against Gram-positive and Gram-negative bacteria (Lee et al. 2013). Likewise, we observed that this gene was highly upregulated in immunized larvae compared with controls, and might be one of the strongest antimicrobial genes in this insect (Table 2). The antibacterial peptide PBSIP, which was previously reported by Yoon et al. (2003) in this insect, was also highly upregulated in response to immunization. In addition, two coprisin-typeAMPs that were first identified in Copris tripartitus (Hwang et al. 2009) were found to be upregulated. Previously, we reported that C (chicken)-type lysozyme from the mole cricket, Gryllotalpa orientalis, has strong antimicrobial activity against a broad range of microorganisms (Kwon et al. 2014b). In this study, C-type lysozyme was also upregulated in immunized larvae (Table 2). In addition, several putative AMPs, such as tenascin, which is involved in wound healing; peritrophin, which is thought to protect insects from invasion by microorganisms; chitinase-like protein, which is involved in protection from fungi; and mucin-like protein, which was studied in relation to pathological functions, were also detected (Du et al. 2006).
Due to their effective mechanisms of action against pathogens, AMPs are less susceptible to the development of bacterial resistance and exhibit minimal toxicity to mammalian cells (Wang et al. 2010, Lee et al. 2013). Thus, this data will provide useful information on candidate AMPs that may potentially be applied in medicine, food safety, and agriculture in place of conventional antibiotics.
Immune-Related Signal Transduction
Signal transduction pathways relating to the immune response are well-established in insects. The Toll and Imd pathways are the most well-known immune-related pathways in insects (Liu et al. 2014). The Toll pathway is usually activated by fungi and Gram-positive bacteria, while the Imd pathway is activated by the majority of Gram-negative bacteria and some Gram-positive bacteria. In this study, we identified at least 10 transcripts involved in the Toll and Imd signaling pathways (Table 3).
Table 3.
Immune-related signal transduction
| Contig ID | Description | Nucleotide length (nt) | Coverage (%) | Fold change | Species top blast | e-value |
|---|---|---|---|---|---|---|
| c7386_g1_i1 | Toll-4-like receptor (TLR4) | 1657 | 51.7 | 2.7 | T. castaneum | 1.00E-54 |
| c25207_g1_i1 | Toll-7-like receptor (TLR7) | 4335 | 89.48 | 2.3 | T. castaneum | 0 |
| c5535_g1_i1 | Toll-6-like receptor (TLR6) | 1026 | 99.71 | −2.3 | T. castaneum | 0 |
| c24168_g1_i1 | Toll-3-like receptor (TLR3) | 1099 | 45.04 | −7.0 | T. castaneum | 0.000001 |
| c23460_g1_i1 | Späetzle 3 | 1543 | 61.24 | 1.43 | T. castaneum | 4.00E-118 |
| c2924_g1_i1 | IMD | 1111 | 43.4 | 2.3 | T. castaneum | 2.00E-30 |
| c20589_g1_i1 | Relish (Rel) | 2985 | 86.33 | 32.5 | T. castaneum | 0 |
| c18352_g1_i1 | Dredd | 2192 | 65.97 | 3.8 | Drosophila pseudoobscura | 4.00E-52 |
| c22168_g1_i2 | TGF-beta activated kinase 1 (TAK) | 2042 | 74.34 | −1.0 | T. castaneum | 5.00E-180 |
| c13683_g1_i2 | Inhibitor of apoptosis protein (IAP) | 1339 | 57.80 | 13.3 | B. mori | 3.00E-61 |
In Dr. melanogaster, Toll was originally identified as a developmental gene and was later established as an innate immune gene (Anderson et al. 1985). Afterward, Toll-like receptors (TLRs) were also identified in mammals, suggesting that Toll and TLRs are evolutionarily conserved throughout insects and mammals (Rock et al. 1988). In this study, four TLRs (TLR3, 4, 6, and 7) were identified. TLR4 and 7 were moderately induced by Gram-negative bacteria and fungi, while TLR3 and 6 were unaffected. In Dr. melanogaster, there are nine TLR genes, and TLR1–4 are induced by fungal challenge (Altincicek et al. 2013). In addition, a Spätzle protein, which might be involved in the TLR signaling pathway in this insect, was detected.
The Imd pathway was originally described for its role in the humoral antibacterial response in dipterans (Nayduch et al. 2014). The Imd pathway is activated by binding between PGRP and meso-diaminopimelic acid-type peptidoglycans, which delivers a signal to the Rel protein Relish, resulting in upregulated AMP genes. There are many adaptor proteins that transduce a signal to produce AMPs, including Fas-associated death domain protein, Dredd, inhibitor of apoptosis protein 2 (IAP2), transforming growth factor β-activated kinase 1 (TAK1), transforming growth factor β-activated kinase 1/MAP3K7 binding protein 2 (Tab2), Ubc13, immune response deficient 5, Kenny, and inhibitor of nuclease factor B kinase subunits b and g (IKKb and IKKg) (Kaneko et al. 2005, Zou et al. 2007, Vogel et al. 2014). Interestingly, expression of signaling proteins associated with the Imd pathway, such as the Imd response homologues Rel, Dredd, TAK, and IAP (Table 3), was observed. Except for TAK, these genes were significantly induced by Gram-negative bacteria and fungi challenge (IAP3 [13.3-fold], Rel [32.5-fold], Dredd [3.8-fold], and Imd [2.3-fold], respectively) (Table 3). These results strongly suggest that Toll- and Imd-mediated immunity is also conserved in this beetle. A more detailed analysis will be required to verify the suggested roles of Toll- and Imd-associated components in the immune response of this insect.
Melanization, Coagulation, and Other Immune-Related Responses
Insect hemocyte defense molecules such as PPO and thioester-containing proteins are activated by invasive pathogens and are involved in opsonization of pathogens for subsequent clearance by phagocytosis (Levashina et al. 2001, Nayduch et al. 2014). PPOs are copper-containing enzymes and are synthesized in a zymogen form, which is stored in insect hemocytes. The PPO zymogen is activated by a serine protease cascade involving serine protease inhibitors such as serpin and converted to the activated form, phenoloxidase, which catalyzes the hydroxylation of monophenol to o-diphenols and the oxidation of o-diphenol to quinones. These latter compounds are active in pathogen killing, melanin synthesis, and wound healing in insects (Liu et al. 2014). Our analysis identified several serine proteases that might be involved in the PO cascade. Among these genes, the serine protease snake-like (48.6-fold), serine protease 1 (36.7-fold), and serpin B12 (63.7-fold) were highly expressed in immunized larvae (Table 4). In addition, L-3,4-dihydroxyphenylalanine (L-DOPA) decarboxylase (DDC) was highly expressed in immunized larvae and also plays a central role in the complex neuroendocrine-immune regulatory network in this insect (Zhou et al. 2011).
Table 4.
Melanization, coagulation, and other immune related gene
| Contig ID | Description | Nucleotide length (nt) | Coverage (%) | Fold change | Species top blast | -value |
|---|---|---|---|---|---|---|
| c9758_g1_i1 | Serine protease snake-like | 1582 | 66.1 | 48.6 | T. castaneum | 3.00E-61 |
| c20763_g1_i1 | Serine protease 1 | 896 | 86.7 | 36.7 | Costelytra zealandica | 5.00E-77 |
| c16255_g1_i1 | Serine protease P155 | 891 | 89.2 | 9.7 | T. castaneum | 6.00E-72 |
| c15709_g1_i1 | Serine protease inhibitors (Serpin B12) | 2453 | 60.42 | 63.7 | T. castaneum | 6.00E-157 |
| c25925_g1_i1 | Serine peptidase inhibitor (Serpin 30) | 1592 | 76.7 | 3.0 | T. molitor | 4.00E-87 |
| c20317_g1_i1 | PPO activating factor-III | 1680 | 62.5 | 6.8 | H. diomphalia | 0 |
| c21891_g1_i1 | L-DOPA DDC | 2564 | 55.34 | >100 | T. molitor | 0 |
| c15814_g1_i2 | CYP450 CYP345D2 | 1625 | 88.06 | 2.1 | T. castaneum | 2.00E-145 |
| c22699_g1_i2 | CYP450 CYP4H10 | 1607 | 87.74 | 6.1 | T. castaneum | 1.00E-133 |
| c24562_g1_i1 | CYP450 CYP4C1 | 1990 | 73.87 | 12.1 | T. castaneum | 8.00E-116 |
| c22212_g1_i4 | CYP450 CYP4C2 | 2297 | 40.62 | 2.8 | T. castaneum | 2.00E-84 |
| c22212_g1_i1 | CYP450 CYP4C3 | 2171 | 67.16 | 2.8 | T. castaneum | 4.00E-107 |
| c16765_g1_i1 | Glutathione S transferase | 833 | 74.91 | 8.8 | T. castaneum | 2.00E-59 |
| c20611_g1_i1 | HSP68a | 644 | 84.32 | 4.7 | T. castaneum | 4.00E-76 |
| c23241_g1_i1 | HSP70 | 1642 | 85.87 | 6.0 | M. dzhungarica Punctipennis | 0 |
| c24481_g1_i1 | HSP90 | 2475 | 50.42 | 7.3 | T. molitor | 0 |
| c36089_g1_i1 | Superoxide dismutase 2 | 231 | 90.91 | 2.7 | Sus scrofa | 2.00E-18 |
| c20750_g1_i1 | Apolipoprotein D | 1232 | 42.3 | −12.7 | Danaus plexippus | 9.00E-41 |
In the beetle Tribolium castaneum, stress response genes including apolipoprotein D, cytochrome P450 (CYP450), gluthathione S-transferase, and a number of heat shock proteins (HSPs) were generally induced by immunization (Altincicek et al. 2013). A similar response was observed in P. brevitarsis seulensis in our study, including CYP450s (fivefold), gluthathione S-transferase (8.8-fold), HSPs (sixfold), and superoxide dismutase (2.7-fold) (Table 4). However, apolipoprotein D was downregulated. CYP450s are among the most important xenobiotic-metabolizing enzymes in insects. Many studies demonstrated that CYP450s detoxify numerous chemicals, including plant secondary metabolites and insecticides. Although there is no direct evidence that CYP450s are associated with the insect innate immune system, CYP540 can produce reactive oxygen species (ROS) as the result of metabolizing xenobiotics, and ROS are involved in the initiation or amplification of the immune response (Namazi 2009).
In conclusion, our RNA-seq analysis revealed a comprehensive set of transcripts that are known as mediators of immune-related processes in insects. In addition, we reported the differential expression of numerous immune genes, which may help to reveal new mechanisms of the insect immune system. This study is the first whole transcriptome analysis of this beetle and included an analysis of differential gene expression in response to immune challenge. This data provides numerous candidate genes as a starting point for further studies on beetle immunity.
Acknowledgment
This research was supported by IPET (Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries) (Grant Number 314086-3).
References Cited
- Altincicek B., Elashry A., Guz N., Grundler F. A., Vilcinskas A., Dehne H. W. 2013. Next generation sequencing based transcriptome analysis of septic-injury responsive genes in the beetle Tribolium castaneum. PLoS One 8: e52004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. V., Bokla L., Nusslein-Volhard C. 1985. Establishment of dorsal-ventral polarity in the Drosophila embryo: the induction of polarity by the Toll gene product. Cell 42: 791–798. [DOI] [PubMed] [Google Scholar]
- Anselme C., Perez-Brocal V., Vallier A., Vincent-Monegat C., Charif D., Latorre A., Moya A., Heddi A. 2008. Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 6: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S., Claverie J. M. 1997. The significance of digital gene expression profiles. Genome Res. 7: 986–995. [DOI] [PubMed] [Google Scholar]
- Bang K., Park S., Yoo J. Y., Cho S. 2012. Characterization and expression of attacin, an antibacterial protein-encoding gene, from the beet armyworm, Spodoptera exigua (Hubner) (Insecta: Lepidoptera: Noctuidae). Mol. Biol. Rep. 39: 5151–5159. [DOI] [PubMed] [Google Scholar]
- Bang K., Park S., Cho S. 2013. Characterization of a beta-1,3-glucan recognition protein from the beet armyworm, Spodoptera exigua (Insecta: Lepidoptera: Noctuidae). Insect Sci. 20: 575–584. [DOI] [PubMed] [Google Scholar]
- Beldade P., McMillan W., Papanicolaou A. 2008. Butterfly genomics eclosing. Heredity 100: 150–157. [DOI] [PubMed] [Google Scholar]
- Bulet P., Cociancich S., Dimarcq J. L., Lambert J., Reichhart J. M., Hoffmann D., Hetru C., Hoffmann J. A. 1991. Insect immunity. Isolation from a coleopteran insect of a novel inducible antibacterial peptide and of new members of the insect defensin family. J. Biol. Chem. 266: 24520–24525. [PubMed] [Google Scholar]
- Canton J., Neculai D., Grinstein S. 2013. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 13: 621–634. [DOI] [PubMed] [Google Scholar]
- Christophides G. K., Zdobnov E., Barillas-Mury C., Birney E., Blandin S., Blass C., Brey P. T., Collins F. H., Danielli A., Dimopoulos G., et al. 2002. Immunity-related genes and gene families in Anopheles gambiae. Science 298: 159–165. [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P. T., Rubin G. M., Lemaitre B. 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U.S.A. 98: 12590–12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A. J., Johnston P. R., Vilcinskas A., Rolff J. 2012. Identification of immunological expressed sequence tags in the mealworm beetle Tenebrio molitor. J. Insect Physiol. 58: 1556–1561. [DOI] [PubMed] [Google Scholar]
- Du X. J., Wang J. X., Liu N., Zhao X. F., Li F. H., Xiang J. H. 2006. Identification and molecular characterization of a peritrophin-like protein from fleshy prawn (Fenneropenaeus chinensis). Mol. Immunol. 43: 1633–1644. [DOI] [PubMed] [Google Scholar]
- Dushay M. S., Roethele J. B., Chaverri J. M., Dulek D. E., Syed S. K., Kitami T., Eldon E. D. 2000. Two attacin antibacterial genes of Drosophila melanogaster. Gene 246: 49–57. [DOI] [PubMed] [Google Scholar]
- Evans J. D., Aronstein K., Chen Y. P., Hetru C., Imler J. L., Jiang H., Kanost M., Thompson G. J., Zou Z., Hultmark D. 2006. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 15: 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H., An C. J., Hao Y. J., Li D. S., Du R. Q. 2004. Molecular cloning and expression of Attacin from housefly (Musca domestica). Yi Chuan Xue Bao. 31: 1344–1350. [PubMed] [Google Scholar]
- Gupta A. P. 1985. Cellular elements in the hemolymph, pp. 401–451. In Kerkut G. A., Gilbert L. I. (eds.), Comprehensive insect physiology, biochemistry, and pharmacology. vol.3 Pergamon Press, New York. [Google Scholar]
- Hoffmann J. A. 2003. The immune response of Drosophila. Nature 426: 33–38. [DOI] [PubMed] [Google Scholar]
- Hwang J. S., Kim S. R., Park K. H., Nam S. H., Hong M. Y. 2008. Research articles: molecular characterization of a defensin-like peptide from larvae of a beetle, Protaetia brevitarsis. Int. J. Indust. Entomol. 17: 131–135. [Google Scholar]
- Hwang J. S., Lee J., Kim Y. J., Bang H. S., Yun E. Y., Kim S. R., Suh H. J., Kang B. R., Nam S. H., Jeon J. P., et al. 2009. Isolation and characterization of a defensin-like peptide (Coprisin) from the Dung beetle, Copris tripartitus. Int. J. Pept. 136284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M., Wada S., Ueda K., Saito A., Koizumi N., Iwahana H., Sato R. 2009. Multipeptide precursor structure of acaloleptin A isoforms, antibacterial peptides from the Udo longicorn beetle, Acalolepta luxuriosa. Dev. Comp. Immunol. 33: 1120–1127. [DOI] [PubMed] [Google Scholar]
- Jiang H., Ma C., Lu Z. Q., Kanost M. R. 2004. Beta-1,3-glucan recognition protein-2 (betaGRP-2)from Manduca sexta; an acute-phase protein that binds beta-1,3-glucan and lipoteichoic acid to aggregate fungi and bacteria and stimulate prophenoloxidase activation. Insect Biochem. Mol. Biol. 34: 89–100. [DOI] [PubMed] [Google Scholar]
- Johnston P. R., Rolff J. 2013. Immune- and wound-dependent differential gene expression in an ancient insect. Dev. Comp. Immunol. 40: 320–324. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Golenbock D., Silverman N. 2005. Peptidoglycan recognition by the Drosophila Imd pathway. J. Endotoxin Res. 11: 383–389. [DOI] [PubMed] [Google Scholar]
- Kim Y. S., Ryu J. H., Han S. J., Choi K. H., Nam K. B., Jang I. H., Lemaitre B., Brey P. T., Lee W. J. 2000. Gram-negative bacteria- binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J. Biol. Chem. 275: 32721–32727. [DOI] [PubMed] [Google Scholar]
- Kwon H., Bang K., Cho S. 2014a. Characterization of the hemocytes in Larvae of Protaetia brevitarsis seulensis: involvement of granulocyte-mediated phagocytosis. PLoS One 9: e103620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H., Bang K., Lee M., Cho S. 2014b. Molecular cloning and characterization of a lysozyme cDNA from the mole cricket Gryllotalpa orientalis (Orthoptera: Gryllotalpidae). Mol. Biol. Rep. 41: 5745–5754. [DOI] [PubMed] [Google Scholar]
- Langen G., Imani J., Altincicek B., Kieseritzky G., Kogel K. H., Vilcinskas A. 2006. Transgenic expression of gallerimycin, a novel antifungal insect defensin from the greater wax moth Galleria mellonella, confers resistance to pathogenic fungi in tobacco. Biol. Chem. 387: 549–557. [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Moon H. J., Kurata S., Kurama T., Natori S., Lee B. L. 1994. Purification and molecular cloning of cDNA for an inducible antibacterial protein of larvae of a coleopteran insect, Holotrichia diomphalia. J. Biochem. 115: 82–86. [DOI] [PubMed] [Google Scholar]
- Lee M., Bang K., Kwon H., Cho S. 2013. Enhanced antibacterial activity of an attacin-coleoptericin hybrid protein fused with a helical linker. Mol. Biol. Rep. 40: 3953–3960. [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25: 697–743. [DOI] [PubMed] [Google Scholar]
- Levashina E. A., Moita L. F., Blandin S., Vriend G., Lagueux M., Kafatos F. C. 2001. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell 104: 709–718. [DOI] [PubMed] [Google Scholar]
- Liu T., Zhu S., Tang Q., Yu Y., Tang S. 2013. Identification of drought stress-responsive transcription factors in ramie (Boehmeria nivea L. Gaud). BMC Plant Biol. 13: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shen D., Zhou F., Wang G., An C. 2014. Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis. PLoS One 9: e86436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Login F. H., Balmand S., Vallier A., Vincent-Monegat C., Vigneron A., Weiss-Gayet M., Rochat D., Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334: 362–365. [DOI] [PubMed] [Google Scholar]
- McCarthy D. J., Chen Y., Smyth G. K. 2012. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Namazi M. R. 2009. Cytochrome-P450 enzymes and autoimmunity: expansion of the relationship and introduction of free radicals as the link. J. Autoimmune Dis. 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayduch D., Lee M. B., Saski C. A. 2014. Gene discovery and differential expression analysis of humoral immune response elements in female Culicoides sonorensis (Diptera: Ceratopogonidae). Parasit. Vectors 7: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai M., Ashida M. 1988. Purification of a beta-1,3-glucan recognition protein in the prophenoloxidase activating system from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 263: 12056–12062. [PubMed] [Google Scholar]
- Rock F. L., Hardiman G., Timans J. C., Kastelein R. A., Bazan J. F. 1988. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA. 95: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagisaka A., Miyanoshita A., Ishibashi J., Yamakawa M. 2001. Purification, characterization and gene expression of a glycine and proline-rich antibacterial protein family from larvae of a beetle, Allomyrina dichotoma. Insect Mol. Biol. 10: 293–302. [DOI] [PubMed] [Google Scholar]
- Shelby K. S., Popham H. J. 2012. RNA-seq study of microbially induced hemocyte transcripts from larval Heliothis virescens (Lepidoptera: Noctuidae). Insects 3: 743–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M., Kuniyoshi H., Kotani E., Taniai K., Kadono-Okuda K., Kato Y., Yamamoto M., Shimabukuro M., Chowdhury S., Xu J. 1995. Characterization of a Bombyx mori cDNA encoding a novel member of the attacin family of insect antibacterial proteins. Insect Biochem. Mol. Biol. 25: 385–392. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Ishibashi J., Fujita K., Nakajima Y., Sagisaka A., Tomimoto K., Suzuki N., Yoshiyama M., Kaneko Y., Iwasaki T. 2008. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 38: 1087–1110. [DOI] [PubMed] [Google Scholar]
- Vogel H., Badapanda C., Vilcinskas A. 2011. Identification of immunity-related genes in the burying beetle Nicrophorus vespilloides by suppression subtractive hybridization. Insect Mol. Biol. 20: 787–800. [DOI] [PubMed] [Google Scholar]
- Vogel H., Badapanda C., Knorr E., Vilcinskas A. 2014. RNA- sequencing analysis reveals abundant developmental stage-specific and immunity-related genes in the pollen beetle Meligethes aeneus. Insect Mol. Biol. 23: 98–112. [DOI] [PubMed] [Google Scholar]
- Wang L. N., Yu B., Han G. Q., He J., Chen D. W. 2010. Design, expression and characterization of recombinant hybrid peptide Attacin-Thanatin in Escherichia coli. Mol. Biol. Rep. 37: 3495–3501. [DOI] [PubMed] [Google Scholar]
- Yoon H. S., Lee C. S., Lee S. Y., Choi C. S., Lee I. H., Yeo S. M., Kim H. R. 2003. Purification and cDNA cloning of inducible antibacterial peptides from Protaetia brevitarsis (Coleoptera). Arch. Insect Biochem. Physiol. 52: 92–103. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Yang J., Wang L., Zhang H., Gao Y., Shi X., Wang M., Kong P., Qiu L., Song L. 2011. A dopa decarboxylase modulating the immune response of scallop Chlamys farreri. PLoS One 6: e18596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Evans J. D., Lu Z., Zhao P., Williams M., Sumathipala N., Hetru C., Hultmark D., Jiang H. 2007. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 8: R177. [DOI] [PMC free article] [PubMed] [Google Scholar]