Abstract
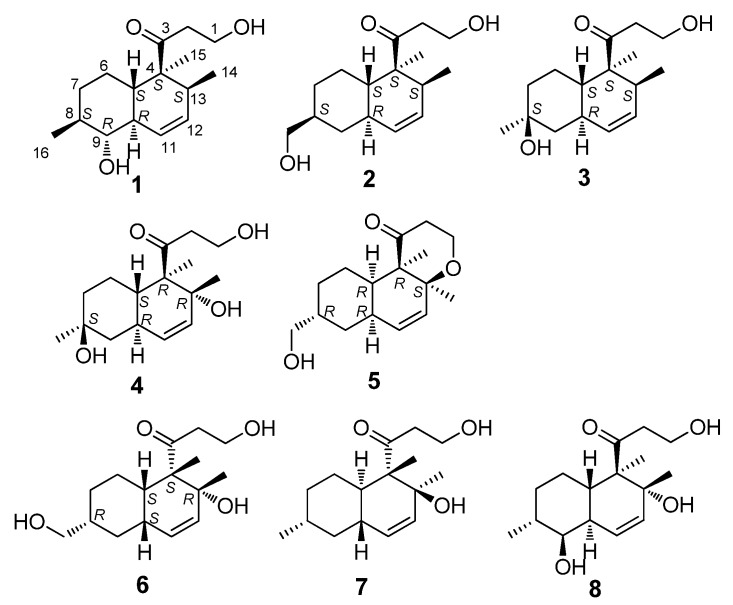
Six new compounds with polyketide decalin ring, peaurantiogriseols A–F (1–6), along with two known compounds, aspermytin A (7), 1-propanone,3-hydroxy-1-(1,2,4a,5,6,7,8,8a-octahydro-2,5-dihydroxy-1,2,6-trimethyl-1-naphthalenyl) (8), were isolated from the fermentation products of mangrove endophytic fungus Penicillium aurantiogriseum 328#. Their structures were elucidated based on their structure analysis. The absolute configurations of compounds 1 and 2 were determined by 1H NMR analysis of their Mosher esters; the absolute configurations of 3–6 were determined by using theoretical calculations of electronic circular dichroism (ECD). Compounds 1–8 showed low inhibitory activity against human aldose reductase, no activity of inducing neurite outgrowth, nor antimicrobial activity.
Keywords: mangrove endophytic fungi, Penicillium aurantiogriseum, secondary metabolites, polyketide decalin derivative
1. Introduction
Mangrove is a specialized marine ecosystem. Mangrove endophytic fungi have drawn a lot of attention for the past few years as a rich source of bioactive and novel compounds [1,2]. In the course of our exploration for the metabolites of endophytic fungi from the mangrove in the South China Sea, numerous new compounds were obtained [3,4,5]. In this study, six new compounds, peaurantiogriseols A–F (1–6), along with two known compounds, aspermytin A (7) and 1-propanone,3-hydroxy-1-(1,2,4a,5,6,7,8,8a-octahydro-2,5-dihydroxy-1,2,6-trimethyl-1-naphthalenyl) (8), were isolated from endophytic fungus Penicillium aurantiogriseum 328# from the bark of mangrove plant Hibiscus tiliaceus. Compounds 1–8 (Figure 1) had similar polyketide decalin scaffolds substituted by 3-oxopropanol side chains, and shown low inhibitory activity against 6×His-tagged recombinant human aldose reductase. Here, we report the isolation and structural elucidation of compounds 1–8 based on the studies of their NMR, EI-MS, and ECD spectra.
Figure 1.
The chemical structures of compounds 1–8.
2. Results and Discussion
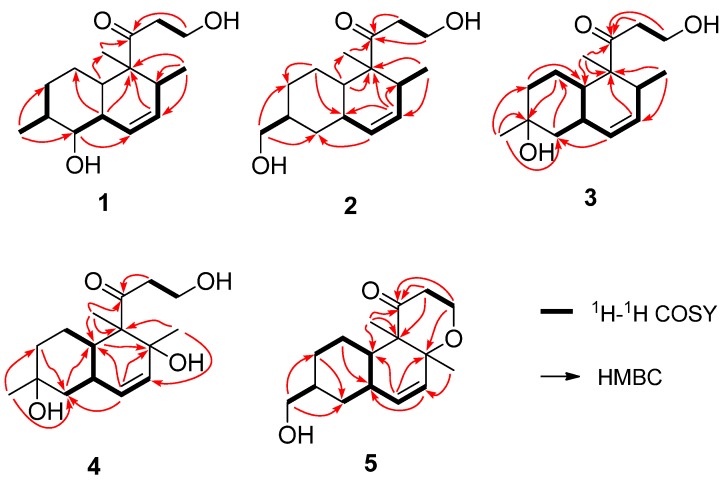
Peaurantiogriseol A (1, Figure 1) was obtained as a colorless solid and had a molecular formula of C16H26O3 as determined by HREIMS data (observed m/z 266.1878 M+, calculated 266.1876), requiring 4° of unsaturation. The 13C-NMR and DEPT spectra (Table 1) indicated the presence of a carbonyl group (δ 215.4), two olefinic carbons, four sp3 CH2 groups, five sp3 CH groups, one sp3 quaternary carbon atom, and three methyl groups. The 1H-NMR and 1H–1H COSY spectra (Table 1 and Figure 2) showed the signals of a 3-oxopropanol system (δH 3.82/2.64), and a cis double bond signals (δH 5.91 d J = 10.6 Hz; 5.58 ddd J =10.6, 4.8, 2.4 Hz). The remaining 2° of unsaturation supported a decalin segment in 1. In the HMBC spectrum (Figure 2), rich correlation data allowed us to unambiguously establish the locations of substituents on the decalin ring. A methyl singlet at δH 1.19 correlated with C-3 and C-5 respectively, which revealed that the methyl group, with the 3-oxopropanol side chain, was located at C-4 position. A methyl doublet signals at δH 0.75 (J = 8.4 Hz) correlated with C-13 and C-12, and another methyl doublet signals at δH 1.00 (J = 9.6 Hz) correlated with C-9 and C-7, revealing that the two methyl groups were located at C-8 and C-13 positions, respectively. Based on the HMBC correlations of H-11/C-9 and H-12/C-14, the cis double bond was easily assigned as C-11 and C-12. One hydroxyl group was identified at C-9 position based on the chemical shift of CH-9 (δ 2.89/79.3) and HMBC correlations.
Table 1.
1H and 13C NMR data of compounds 1–5 (400/100 MHz in CDCl3, J in Hz).
| 1 α | 2 | 3 | 4 | 5 | ||||||
| 13C | 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | 1H | |
| 1 | 58.0 t | 3.82 m | 57.9 t | 3.80 m | 58.2 t | 3.82 m | 58.4 t | 3.82 m | 61.2 t | 4.08 dd 12.0, 8.0 |
| 3.87 dt 12.0, 2.8 | ||||||||||
| 2 | 41.0 t | 2.64 ddd 18.6, 6.6, 4.2 | 41.0 t overlapped | 2.68 ddd 18.4, 6.0, 4.8 | 40.9 t | 2.66 q 4.4 | 44.1 t | 3.11 ddd 18.8, 6.4, 3.6 | 39.5 t | 2.79 ddd 14.0, 12.8, 8.0 |
| 2.63 ddd 18.6, 6.6, 4.2 | 2.61 ddd 18.4, 6.0, 4.8 | 2.67 ddd 18.8, 6.4, 3.6 | 2.19 ddd 14.0, 12.8, 8.0 | |||||||
| 3 | 215.4 s | 215.6 s | 215.8 s | 216.1 s | 212.6 s | |||||
| 4 | 52.3 s | 52.3 s | 52.4 s | 57.2 s | 57.1 s | |||||
| 5 | 45.3 d | 1.66 m | 39.0 d | 1.59 m | 38.7 d | 1.58 m | 43.3 d | 1.78 m | 43.0 d | 2.23 m |
| 6 | 26.8 t | 1.54 m | 26.7 t | 1.68 m | 23.0 t | 1.53 m | 23.1 t | 1.42 m | 25.8 t | 1.14 m |
| 0.91 m | 0.91 m | 1.26 m | 1.31 m | |||||||
| 7 | 33.5 t | 1.16 m, | 29.8 t | 1.80 m | 45.8 t | 1.71 m | 39.5 t | 1.67 m | 29.6 t | 1.83 m |
| 1.71 m | 1.08 m | 1.27 m | 1.50 m | 1.02 m | ||||||
| 8 | 41.0 d | 1.36 m | 41.0 d overlapped | 1.58 m | 70.2 s | 70.1 s | 40.8 d | 1.62 m | ||
| 9 | 79.3 d | 2.89 t 9.6 | 36.3 t | 1.84 m | 39.7 t | 1.65 m | 45.3 t | 1.74 m | 35.4 t | 1.94 m |
| 0.86 m | 1.53 m | 1.25 m | 1.03 m | |||||||
| 10 | 36.6 d | 1.67 m | 37.9 d | 1.69 m | 33.6 d | 2.13 m | 33.7 d | 2.24 tt 11.8, 2.8 | 37.4 d | 1.82 m |
| 11 | 125.0 d | 5.91 d 10.6 | 129.6 d | 5.32 d 10.0 | 129.6 d | 5.32 d 9.6 | 131.0 d | 5.34, s | 134.3 d | 5.66 dd 9.6, 1.2 |
| 12 | 130.6 d | 5.58 ddd 10.6, 4.8, 2.4 | 129.7 d | 5.45 ddd 10.0, 4.8, 2.4 | 130.0 d | 5.52 ddd 9.6, 4.8, 2.8 | 133.6 d | 5.34, s | 130.6 d | 5.52 dd 9.6, 2.8 |
| 13 | 39.5 d | 2.01 m | 39.9 d | 2.06 m | 40.0 d | 2.09 m | 74.0 s | 78.5 s | ||
| 14 | 18.6 q | 0.75 d 8.4 | 18.7 q | 0.72 d 7.2 | 18.8 q | 0.75 d 7.2 | 27.5 q | 1.13 s | 20.5 q | 1.18 s |
| 15 | 17.5 q | 1.19 s | 17.4 q | 1.17 s | 17.7 q | 1.22 s | 12.1 q | 1.33 s | 11.2 q | 0.88 s |
| 16 | 18.7 q | 1.00 d 9.6 | 68.3 t | 3.44 dd 10.8, 6.4 | 31.8 q | 1.22 s | 31.8 q | 1.25 s | 68.3 t | 3.48 m |
| 3.41 dd 10.8, 6.4 | ||||||||||
The data were recorded at α 600 MHz (1H-NMR) and 150 MHz (13C-NMR).
Figure 2.
The key 1H–1H COSY and HMBC correlations of compounds 1–5.
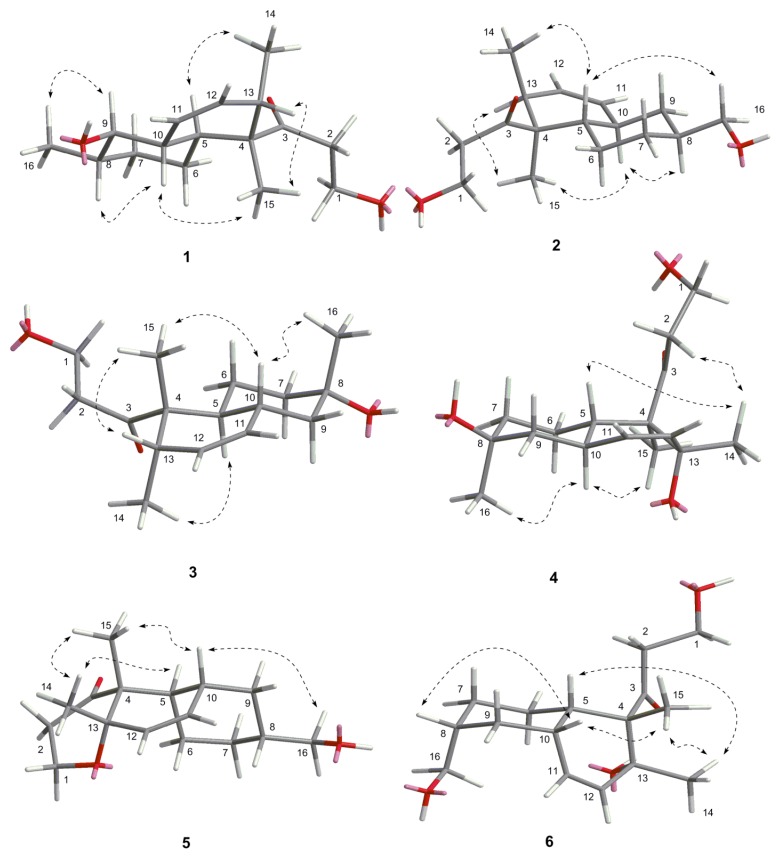
The relative stereochemistry of 1 was established by its NOESY spectrum (Figure 3). The NOE correlations of H-15/H-10, H-15/H-13 and H-14/H-5 confirmed a trans-fused decalin ring in 1. Methyl-15 and OH-9 were oriented on the same side of the decalin ring, and methyl-14 and methyl-16 were oriented on the other side based on the NOE correlations of H-8/H-10 and H-16/H-9.
Figure 3.
The key NOE correlations of compounds 1–6.
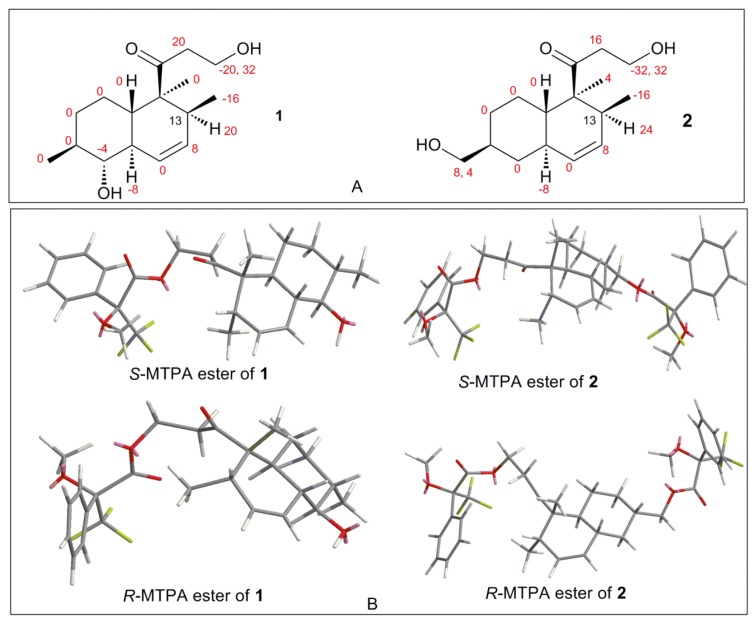
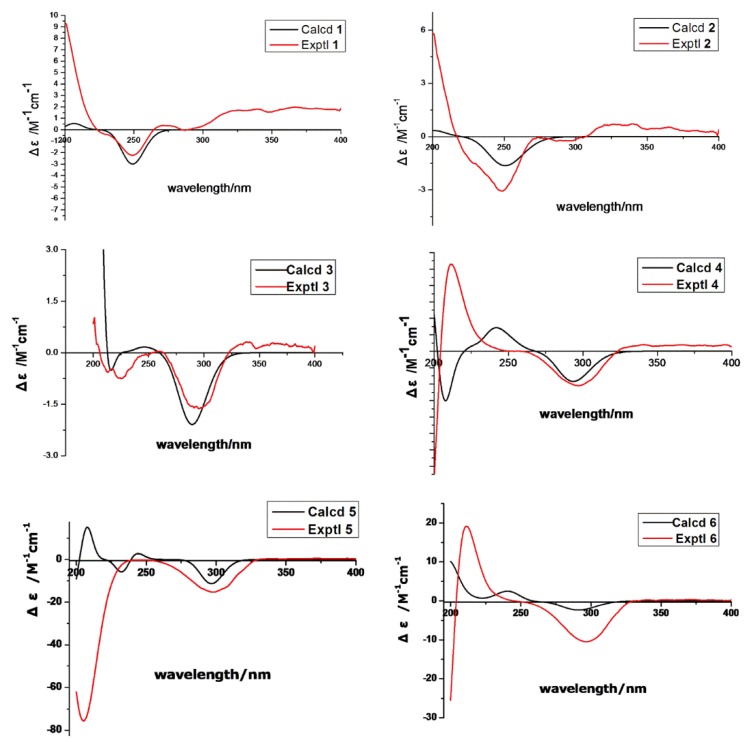
The absolute configuration of 1 was determined by comparison of the 1H-NMR spectra of corresponding (R) and (S)-Mosher esters (Figure 4; Supplementary Materials Table S2). OH-1 of compound 1 was mainly esterified by S/R-MTPA-Cl based on the larger chemical shift of H-1. There was an esterified C-1 hydroxyl group in Mosher esters of 1, which were confirmed by their 19F NMR spectra that showed 2 CF3 signals (Supplementary Materials Figure S8). The preferred conformations of Mosher esters of 1 dominating 1H NMR spectroscopic features were that the 3-oxopropanol side chains with MTPA moiety were in equatorial position, and bend to C-13 position; CH2-1–O–C=O–CF3 substructure of Mosher esters of 1 were coplanar (Figure 4). The shielding or deshielding effects of the phenyl rings of MTPA moiety on H-13 or H-14 were larger in its R-Mosher ester than that of S-Mosher ester. The absolute configuration of C-13 in 1 was deduced as S-configuration based on the positive chemical shift differences (ΔδSR) of H-13 and negative chemical shift differences (ΔδSR) of H-14 from corresponding Mosher esters [6,7]. Finally, the absolute configuration of 1 was confirmed as (4S,5S,8S,9R,10R,13S)-configuration (Figure 1) based. The absolute configuration of 1 was validated by the result that the experimental data and calculated ECD spectrum for (4S,5S,8S,9R,10R,13S)-configuration of 1 matched exactly (Figure 5).
Figure 4.
(A) Chemical shift difference values (ΔδSR = δS − δR, in Hz) of compounds 1/2 esterified by S/R-MTPA-Cl; and (B) preferred conformations of S/R-Mosher esters of 1/2.
Figure 5.
Calculated and experimental ECD spectra of 1–6.
Peaurantiogriseol B (2, Figure 1) was obtained as a white solid and had a molecular formula of C16H26O3 based on HREIMS data (observed m/z 266.1875 M+, calculated 266.1876), same as compound 1. The 1H- and 13C-NMR spectra of 2 were very similar to those of 1 (Table 1), except for the absence of one oxygenated CH-9 group signal, and the change of a doublet signal at δ 1.00/18.7 to hydroxymethyl signals at δ 3.44/3.41/68.3. These results suggested the presence on compound 2 of a hydroxymethyl group at C-16 position. The 1H-1H COSY and HMBC correlations of 2 were also similar to those of 1 (Figure 2), which confirmed that an OH group was located at C-16.
The relative stereochemistry of 2 was established by its NOESY spectrum (Figure 3). Its NOE data were very similar to those of 1. A trans-fused decalin ring in 2 was indicated by the correlations of H-15/H-13, H-15/H-10, and H-14/H-5. Methyl-15 and methyl-14 were oriented on the opposite side, and hydroxymethyl-16 and methyl-14 were oriented on the same side based on the NOE correlation of H-16/H-5.
The absolute configuration of 2 was determined by comparison of the 1H-NMR spectra of its (R) and (S)-Mosher esters like compound 1. Both alcohols (OH-1 and OH-16) in 2 were esterified based on the larger chemical shifts of H-1/H-16 (Supplementary Materials Table S2), which were confirmed based on its 19F NMR spectra (Supplementary Materials, Figure 13S); there were 3 CF3 signals in 19F NMR spectra of Mosher esters of 2. The 1H NMR spectroscopic features and preferred conformations of Mosher esters of 2 were the same as that of 1. Therefore, the absolute configuration of 2 was confirmed as (4S,5S,8S,10R,13S)-configuration (Figure 1). The absolute configuration of 2 was validated by the result that the experimental data and calculated ECD spectrum for (4S, 5S, 8S, 10R, 13S)-configuration of 2 matched exactly (Figure 5).
Peaurantiogriseol C (3, Figure 1) was obtained as a white solid and had a molecular formula of C16H26O3 based on HREIMS data (observed m/z 248.1770 [M − H2O]+, calculated for C16H24O2, 248.1771). The 1H- and 13C-NMR spectra of 3 were very similar to those of compound 2 (Table 1), except for the changes of hydroxymethyl signals at δ 3.41/68.3 to a methyl singlet at δ 1.22/31.0 in 3, and CH group signals at δ 1.58/41.0 to quaternary carbon atom signal at δ 70.2 in 3. These results suggested that compound 3 possesses a hydroxyl group at C-8 position, which were confirmed by the HMBC correlations from H-16 to C-7, and H-16 to C-9 (Figure 2). The relative stereochemistry of 3 was the same as compound 2 based of its NOESY spectrum (Figure 3). The absolute configuration of compound 3 was determined by the result that the experimental data, showing a negative Cotton effect at 291 nm, and calculated ECD spectrum for (4S,5S,8S,10R,13S)-configuration of 3 matched exactly (Figure 5).
Peaurantiogriseol D (4, Figure 1) had a molecular formula of C16H26O4 based on HREIMS data (observed m/z 282.1824 M+, calculated 282.1826), with one more oxygen atom than compound 3. The 1H-NMR, 13C-NMR, 1H–1H COSY, and HMBC correlations of 4 were very similar to those of 3 (Table 1, Figure 2), except for the change of a methyl doublet signal at δ 0.75 (d, J = 7.2 Hz) to singlet signal at δ 1.13 in 4, which suggested that compound 4 had an added OH group. The additional OH group was located at C-13 based on the chemical shift of CH-13 (δc 74.0) and HMBC correlations. The relative stereochemistry of 4 was established based on the result that the interatomic non-bonded distance of key atoms and NOESY correlation signals matched exactly in its 3D model (Table 2, Me-eea-trans conformer). The relative stereochemistry of 4 was assigned as that methyl-14 and methyl-15 were equatorial; methyl-16 was axial; and the decalin ring was trans based on the interatomic non-bonded distance of key atoms less than 4 Å. The trans-fused decalin ring of 4 was supported by the coupling constant of CH-10 signal (tt, J = 11.8, 2.8 Hz). The absolute configuration of 4 was confirmed as (4R,5S,8S,10R,13R)-conformer by the result that the experimental data, showing a negative Cotton effect at 298 nm, and calculated ECD spectrum for (4R,5S,8S,10R,13R)-conformer of 4 matched exactly (Figure 5).
Table 2.
The key NOE correlations of compound 4 and interatomic non-bonded distance of the key atoms in its main 3D conformers.
| Key Atoms | NOE Correlation | Main 3D Conformers | ||
|---|---|---|---|---|
| Me-aaa-Cis | Me-eee-Cis | Me-eea-Trans | ||
| Distance (Å) | Distance(Å) | Distance(Å) | ||
| CH3-14 | H-5 | 3.347715 | 4.256464 | 3.471439 |
| CH3-15 | H-10 | 3.608014 | 2.503497 | 1.862233 |
| CH3-16 | H-10 | 2.310649 | 5.322337 | 1.826166 |
| CH3-14 | H-2 | 5.109479 | 4.964076 | 2.470492 |
| CH-5 | H-10 | 2.265362 | 2.503497 | 3.087856 |
Me-aaa-cis conformer: methyl-14, methyl-15, and methyl-16 are axial, decalin ring is cis, hypothetically; Me-eee-cis conformer: methyl-14, methyl-15, and methyl-16 are equatorial, decalin ring is cis, hypothetically; Me-eea-trans conformer: methyl-14 and methyl-15 are equatorial, and methyl-16 is axial, decalin ring is trans, hypothetically.
Peaurantiogriseol E (5, Figure 1) had a molecular formula of C16H24O3 based on HREIMS data (observed m/z 264.1721 M+, calculated 264.1720), which was two mass units less than that of compound 2, requiring 5 degrees of unsaturation. The 1H-NMR and 13C-NMR data for 5 were similar to those of compound 2 (Table 1). The most obvious difference between 5 and 2 was that the absence of one CH group signal at δ 2.06/39.9, and the change of a doublet signal at δ 0.72 to singlet at δ 1.18/20.5 in 5. These results suggested that a pyrone moiety was formed in 5 by an O–C bond at C-13 position, which was supported by the HMBC correlations from H-1 to C-13 (Figure 2), and, from H-2 to C-4.
The relative stereochemistry of 5 was established by its NOESY spectrum (Figure 3). A cis-fused decalin ring in 5 was confirmed based on the NOE correlations of H-15/H-10, H-14/H-5, and H-15/H-14. Methyl-14, methyl-15, and hydroxymethyl-16 were oriented on the same side by the NOE correlations between H-16 and H-10. The absolute configuration of 5 was determined based on the result that the experimental ECD spectrum, showing a negative Cotton effect at 298 nm, and calculated ECD spectrum for (4R,5R,8R,10R,13S)-configuration of compound 5 matched exactly (Figure 5).
Peaurantiogriseol F (6, Figure 1) had a molecular formula of C16H26O4 based on HRESIMS data (observed m/z 283.18999 [M + H]+, calculated for C16H26O4, 283.19039 [M + H]+), with the same planar structure as that of known craterellone D based on its spectroscopic data (Supplementary Materials Table S1) [8]. The of compound 6 was +21 (c 2.2, MeOH) with opposite sign of craterellone D. The NOE correlations of H-14/H-15, H-14/H-5, H-14/H-10, H-15/H-5, and H-15/H-10 allowed us to unambiguously establish a cis-fused decalin ring in 6 different from the trans junction of craterellone D. Hydroxymethyl-16 were oriented on the opposite side of methyl-14/methyl-15 based on the NOE correlation of H-8/H-10. Compound 6 is a diastereoisomer of craterellone D. The absolute configuration of 6 was determined based on the result that the experimental ECD spectrum, showing a negative Cotton effect at 296 nm, and calculated ECD spectrum for (4S,5S,8R,10S,13R)-configuration of 6 matched exactly (Figure 5).
Compound 7 was identified as aspermytin A by comparison of its spectral data with [9]; both compound 7 and aspermytin A had the same NMR, MS and specific rotation data.
The structure of compound 8 (Figure 1) was also elucidated by its spectroscopic data (Supplementary Materials Table S1). Its was −29 (c 2.1, MeOH). Its trans-fused decalin ring was determined based on the NOE correlations of H-14/H-5 and H-15/H-10; its methyl-14 and hydroxyl-9 were oriented on the opposite side of methyl-15 and methyl-16 by the NOE correlations of H-15/H-10, H-16/H-10, and H-16/H-9. It was found that the planar structure of compound 8 was the same as that of known 1-propanone, 3-hydroxy-1-(1,2,4a,5,6,7,8,8a-octahydro-2,5-dihydroxy-1,2,6-trimethyl-1-naphthalenyl), CAS Registry Number 1235005-17-0. No reference, nor stereochemical information are presented in SciFinder.
Compounds 1−8, at a concentration of 50 mM, showed low inhibitory effect against human aldose reductase; the corresponding value of percent inhibition were 16%, 6%, 31%, 22%, 26%, 2%, 13%, and 9%. The compounds showed no activity of inducing neurite outgrowth (PC-12) [9], nor antimicrobial activity against E. coli. (ATCC 25922), Staphylococcu aureus (ATCC 25923), and Candida albicans (ATCC 60193), at a concentration of 128 μg/mL.
3. Experimental Section
3.1. General Experimental Procedures
(R)-(−)-α-Methoxy-α-trifluoromethylphenylacetyl chloride (MTPA-Cl) (99%), (S)-(+)-MTPA-Cl (99%), dimethyl sulfoxide, and pyridine-d5 (99.5%) were purchased from Sigma (St. Louis, MO, USA). 6×His-tagged recombinant human aldose reductase was presented by Xiaopeng Hu, School of Pharmaceutical Sciences, Sun Yat-sen University [10]; its substrate, β-NADPH were purchased from Sigma-Aldrich (St. Louis, MO, USA); methanol was HPLC grade; other reagents were analytical grade and commercially available; PDA medium were purchased from Beijing Land Bridge Technology Co., Ltd, Beijing, China.
Optical rotation measurements were carried out using a Bellingham-Stanley 37–440 polarimeter (Bellingham Stanley Ltd., Kent, UK). UV spectra were determined using a UV-240 spectrophotometer (Shimadzu, Tokyo, Japan). ECD spectra were measured using a Chirascan Circular Dichroism Spectrometer (Applied PhotoPhysics, Surrey, UK). IR spectra were measured on a TENSOR37 spectrometer (Bruker Optics, Ettlingen, Germany). The 1H-NMR and 13C-NMR data were acquired using a Bruker Avance 400 spectrometer at 400 MHz for 1H nuclei and 100 MHz for 13C nuclei, a Bruker Avance III 500 MHz NMR spectrometer at 470 MHz for 19F nuclei, and a Bruker Avance III 600 MHz NMR spectrometer at 600 MHz for 1H nuclei and 150 MHz for 13C nuclei (Bruker Biospin, Rheinstetten, German). TMS was used as an internal standard, and the chemical shifts (δ) were expressed in ppm or Hz. The EI mass spectra and high-resolution mass spectra were obtained using MAT95XP (ThermoFinnigan, Bremen, Germany) high resolution mass spectrometer and a LTQ-Orbitrap LC-MS (Thermo Fisher, Frankfurt, German). HPLC was performed using a 515 pump with a UV 2487 detector (Waters, Milford, MA, USA) and an Ultimate XB-C-18 column (250 mm × 10 mm, 5 μm; Welch, MD, USA). Normal pressure preparative column chromatography was carried out on RP-18 gel (25–40 μm, Daiso Inc., Osaka, Japan), silica gel (200–400 mesh, Qingdao Marine Chemical Inc., Qingdao, China), or Sephadex-LH-20 (GE Healthcare, Stockholm, Sweden) for reverse and direct phase elution modes, respectively. TLC was performed over F254 glass plates (Qingdao Marine Chemical Inc., Qingdao, China) and analyzed under UV light (254 and 366 nm).
3.2. Fungal Material
Endophytic fungus Penicillium aurantiogriseum 328# was isolated with PDA medium from the bark of Hibiscus tiliaceus collected in the Qi’ao Mangrove Nature Reserve of Guangdong Province, China and identified according to its morphological characteristics and the ITS region [11]. A voucher specimen is deposited in our laboratory at −20 °C.
3.3. Fermentation, Extraction and Isolation
Small agar slices bearing mycelia were placed in 1000 mL Erlenmeyer flasks containing rice medium (composed of 60 g rice, 80 mL distilled water, and 0.24 g sea salt); and incubated for 30 days at 28 °C. In total, 120 flasks of culture were obtained. Cultures were extracted with EtOAc. In total, 200 g crude extract was obtained by evaporation of EtOAc. The crude extract was suspended in H2O (2 L) and partitioned with n-hexane (3 L × 2) and EtOAc (3 L × 2) to give n-hexane (90 g) and EtOAc (51 g) extracts, respectively.
The EtOAc extract was subjected to silica gel column, eluted with n-hexane–EtOAc gradient (from 100:0 to 0:100) to obtain six fractions (Frs. 1–6). Fr. 2 (7.5 g) was subjected to column chromatography over silica gel, eluted with n-hexane–EtOAc (50:50) to obtain three fractions (Frs. 2.1–2.3). Frs. 2.1 and Frs. 2.2 were purified by Sephadex LH-20 (MeOH) to yield compound 1 (67.5 mg) and compound 7 (1.007 g), respectively; Fr. 2.3 was separated by HPLC (MeOH-H2O, 20:80, 2 mL/min, 254 nm) to isolate compound 5 (6 mg). Fr. 3 (4.5 g) was purified using a Sephadex LH-20 column (MeOH) and separated by HPLC (MeOH-H2O gradient from: 25:75 to 50:50, 2 mL/min, 254 nm) to yield compound 3 (40 mg) and compound 2 (45 mg). Fr. 4 (7.7g) was subjected to column chromatography over silica gel, eluted with n-hexane–EtOAc (40:69) to obtain two fractions, Fr. 5.1 and Fr. 5.2. Fr. 5.1 purified using Sephadex LH-20 (MeOH) to yield compound 6 (206 mg); and Fr. 5.2 was separated by HPLC (MeOH-H2O, 20:80, 2 mL/min, 254 nm) to yield compound 4 (4 mg) and compound 8 (27.1 mg).
3.4. Spectral Data
Peaurantiogriseol A (1): colorless solid; +73 (c 1.5, MeOH); UV (MeOH) λmax (logε) 240 (2.4) nm; ECD (MeOH) Δε249 −0.36; IR (KBr) νmax 3425, 1703 cm−1; for 1H-NMR and 13C-NMR data, see Table 1; EIMS m/z 266 (M+), 248, 217, 203, 193; HREIMS m/z 266.1878 [M+] (calculated for C16H26O3, 266.1876).
Peaurantiogriseol B (2): colorless solid; +89 (c 1.7, MeOH); UV (MeOH) λmax (logε) 240 (3.1) nm; ECD (MeOH) Δε249 −0.49; IR (KBr) νmax 3431, 1701 cm−1; for 1H-NMR and 13C-NMR data, see Table 1; EIMS m/z 266 [M+]; HREIMS m/z 266.1875 [M+] (calculated for C16H26O3, 266.1876).
Peaurantiogriseol C (3): colorless solid; +60 (c 0.5, MeOH); UV (MeOH) λmax (logε) 250 (2.1) nm; ECD (MeOH) Δε291 −0.21; IR (KBr) νmax 3434, 1701 cm−1; for 1H-NMR and 13C-NMR data, see Table 1; EIMS m/z 248 (M-H2O)+, 206, 175; HREIMS m/z 248.1770 [M − H2O]+ (calculated for C16H24O2, 248.1771).
Peaurantiogriseol D (4): white solid; +18 (c 0.4, MeOH); UV (MeOH) λmax (logε) 282 (2.6) nm; ECD (MeOH) Δε298 −0.88; IR (KBr) νmax 3434, 1688 cm−1; for 1H-NMR and 13C-NMR data, see Table 1; EIMS m/z 282 [M+]; HREIMS m/z 282.1824 [M+] (calculated for C16H26O4, 282.1826).
Peaurantiogriseol E (5): colorless solid; −153 (c 1.5, MeOH); UV (MeOH) λmax (logε) 282 (2.0) nm; ECD (MeOH) Δε298 −2.5; IR (KBr) νmax 3470, 1686 cm−1; for 1H-NMR and 13C-NMR data, see Table 1; EIMS m/z 264 [M+], 249, 231, 192; HREIMS m/z 264.1721 [M+] (calculated for C16H24O3, 264.1720).
Peaurantiogriseol F (6): white solid; +21 (c 2.2, MeOH); UV (MeOH) λmax (logε) 289 (1.9) nm; ECD (MeOH) Δε296 −1.61; for 1H-NMR and 13C-NMR data, see Supplementary Materials table S1; EIMS m/z 282 (M+), 264, 249, 203, 173; HRESIMS m/z 283.18999 [M + H]+ (calculated for C16H27O4, 283.19039 [M + H]+).
3.5. Computational Analyses
All the theoretical methods and the basis set used for optimization and spectrum calculation were recommended in previous studies [12,13]. All the theoretical calculations, including geometry optimization, frequency analysis, and ECD spectrum prediction, were carried out with the density functional theory (DFT) and time-dependent density functional theory (TDDFT) methods in the Gaussian 09 software package [14]. The geometry optimizations were performed at the B3LYP/6-31+G (d) level in the gas phase. Based on the final optimized structure, the ECD spectra were calculated at the PBE1PBE-SCRF/6-311++g (d, p) level using the PCM solvent continuum models with methanol as a solvent. The theoretical predicted ECD spectra were fitted in the SpecDis software package.
3.6. Esterification Procedure
The Mosher esters of compounds 1 and 2 were prepared by treatment of compounds 1 and 2 with corresponding (R)-(−)-α-methoxy-α-trifluoromethylphenylacetyl chloride (1.5 equivalence ratio), or (S)-(+)-α-methoxy-α-trifluoromethylphenylacetyl chloride (1.5 equivalence ratio) in pyridine-d5 under a nitrogen atmosphere. The reaction mixtures were stood at room temperature for 3.5 h. Then, 1H NMR spectra of the samples were recorded at 400 MHz.
3.7. Inhibition of Aldose Reductase
The method to examine the inhibition of aldose reductase was similar to the method used by Michael C. Van Zandt et al. [15]. Enzyme activity was measured by monitoring the rate of disappearance of NADPH at 340 nm. The reaction contents in a final volume of 300 μL were 6.6% w/v (NH4)2SO4, 33 mM NaH2PO4 (pH 6.6), 0.11 mM NADPH, 4.7 mM dl-glyceraldehyde, 0.59 μg of enzyme, 1% DMSO, and compound. Each assay was done in triplicate. Percent inhibition was calculated on the basis of enzyme activity in the presence or absence of compound.
4. Conclusions
Polyketide decalin-derived secondary metabolites are widely found in nature [16,17,18]; Abundant polyketide decalin derivatives (1–8) from endophytic fungus 328# from the mangrove in the South China Sea were reported in this work; their absolute configurations were established by theoretical calculations of electronic circular dichroism and NMR data analyses of Mosher ester derivatives. Compounds 1–8 showed low activity under our experimental conditions.
Acknowledgments
We are grateful for the financial support from the National Natural Science Foundation of China (21272286), China’s Marine Commonweal Research Project (201305017), and Department of Science and Technology of Guangdong Province (2014A020221006).
Supplementary Files
Author Contributions
Jun Wang and Lan Liu took charge of the throughout the research, structural elucidation and writing. Yanhong Ma mainly took part in the extraction and isolation. Jing Li mainly took part in the computational analyses and biological activity assay. Meixiang Huang mainly took part in the data test. Yongcheng Lin mainly checked the error about structures elucidation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hu Y., Chen J., Hu G., Yu J., Zhu X., Lin Y., Chen S., Yuan J. Statistical research on the bioactivity of new marine natural products discovered during the 28 years from 1985 to 2012. Mar. Drugs. 2015;13:202–221. doi: 10.3390/md13010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2015;32:116–211. doi: 10.1039/C4NP00144C. [DOI] [PubMed] [Google Scholar]
- 3.Song Y.X., Wang J.J., Huang H.B., Ma L., Wang J., Gu Y.C., Liu L., Lin Y.C. Four Eremophilane Sesquiterpenes from the mangrove endophytic fungus Xylaria sp. BL321. Mar. Drugs. 2012;10:340–348. doi: 10.3390/md10020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen L., Cai X., Xu F., She Z., Chan W.L., Vrijmoed L.L., Jones E.B., Lin Y. Three metabolites from the mangrove endophytic fungus Sporothrix sp. (#4335) from the South China Sea. J. Org. Chem. 2009;74:1093–1098. doi: 10.1021/jo802096q. [DOI] [PubMed] [Google Scholar]
- 5.Li J., Yang X., Lin Y., Yuan J., Lu Y., Zhu X., Li J., Li M., Lin Y., He J., et al. Meroterpenes and azaphilones from marine mangrove endophytic fungus Penicillium 303#. Fitoterapia. 2014;97:241–246. doi: 10.1016/j.fitote.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Hoye T.R., Jeffrey C.S., Shao F. Mosher ester analysis for the determination of absolute configuration of stereogenic (chiral) carbinol carbons. Nat. Protoc. 2007;2:2451–2458. doi: 10.1038/nprot.2007.354. [DOI] [PubMed] [Google Scholar]
- 7.Freire F., Seco J.M., Quiñoá E., Riguera R. Chiral 1,2-diols: The assignment of their absolute configuration by NMR made easy. Org. Lett. 2010;12:208–211. doi: 10.1021/ol9021639. [DOI] [PubMed] [Google Scholar]
- 8.Guo H., Feng T., Li Z.H., Liu J.K. Five new polyketides from the basidiomycete Craterellus odoratus. Nat. Prod. Bioprospect. 2012;2:170–173. doi: 10.1007/s13659-012-0057-5. [DOI] [Google Scholar]
- 9.Tsukamoto S., Miura S., Yamashita Y., Ohta T. Aspermytin A: A new neurotrophic polyketide isolated from a marine-derived fungus of the genus Aspergillus. Bioorg. Med. Chem. Lett. 2004;14:417–420. doi: 10.1016/j.bmcl.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 10.Zheng X., Zhang L., Zhai J., Chen Y., Luo H., Hu X. The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase. FEBS Lett. 2012;586:55–59. doi: 10.1016/j.febslet.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Cao L., Qiu Z., You J., Tan H., Zhou S. Isolation and characterization of endophytic streptomycete antagonists of fusarium wilt pathogen from surface-sterilized banana roots. FEMS Microbiol. Lett. 2005;247:147–152. doi: 10.1016/j.femsle.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., et al. Gaussian 09. Gaussian, Inc.; Wallingford, CT, USA: 2009. Revision a.02. [Google Scholar]
- 13.Bruhn T., Schaumloffel A., Hemberger Y., Bringmann G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality. 2013;25:243–249. doi: 10.1002/chir.22138. [DOI] [PubMed] [Google Scholar]
- 14.Stephens P.J., Harada N. ECD cotton effect approximated by the Gaussian curve and other methods. Chirality. 2010;22:229–233. doi: 10.1002/chir.20733. [DOI] [PubMed] [Google Scholar]
- 15.Van Zandt M.C., Jones M.L., Gunn D.E., Geraci L.S., Jones J.H., Sawicki D.R., Sredy J., Jacot J.L., Dicioccio A.T., Petrova T., et al. Discovery of 3-[(4,5,7-trifluorobenzothiazol-2-yl)methyl]indole-N-acetic acid (lidorestat) and congeners as highly potent and selective inhibitors of aldose reductase for treatment of chronic diabetic complications. J. Med. Chem. 2005;48:3141–3152. doi: 10.1021/jm0492094. [DOI] [PubMed] [Google Scholar]
- 16.Yamada T., Umebayashi Y., Kawashima M., Sugiura Y., Kikuchi T., Tanaka R. Determination of the chemical structures of tandyukisins B–D, isolated from a marine sponge-derived fungus. Mar. Drugs. 2015;13:3231–3240. doi: 10.3390/md13053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeerapong C., Phupong W., Bangrak P., Intana W., Tuchinda P. Trichoharzianol, a new antifungal from Trichoderma harzianum F031. J. Agric. Food Chem. 2015;63:3704–3708. doi: 10.1021/acs.jafc.5b01258. [DOI] [PubMed] [Google Scholar]
- 18.Nakadate S., Nozawa K., Horie H., Fujii Y., Nagai M., Hosoe T., Kawai K., Yaguchi T., Fukushima K. Eujavanicols A–C, decalin derivatives from Eupenicillium javanicum. J. Nat. Prod. 2007;70:1510–1512. doi: 10.1021/np078008a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.