Abstract
Summary: Assessing linkage disequilibrium (LD) across ancestral populations is a powerful approach for investigating population-specific genetic structure as well as functionally mapping regions of disease susceptibility. Here, we present LDlink, a web-based collection of bioinformatic modules that query single nucleotide polymorphisms (SNPs) in population groups of interest to generate haplotype tables and interactive plots. Modules are designed with an emphasis on ease of use, query flexibility, and interactive visualization of results. Phase 3 haplotype data from the 1000 Genomes Project are referenced for calculating pairwise metrics of LD, searching for proxies in high LD, and enumerating all observed haplotypes. LDlink is tailored for investigators interested in mapping common and uncommon disease susceptibility loci by focusing on output linking correlated alleles and highlighting putative functional variants.
Availability and implementation: LDlink is a free and publically available web tool which can be accessed at http://analysistools.nci.nih.gov/LDlink/.
Contact: mitchell.machiela@nih.gov
1 Motivation
Genome-wide association studies (GWAS) have identified robust genotype-phenotype associations for a range of disease phenotypes (Chanock, 2014; Welter et al., 2014). Linkage disequilibrium (LD), the non-random association of regional variants due to the low probability of meiotic recombination, has facilitated GWAS by enabling the search for markers of risk alleles based on the principle of indirect testing, namely, identification of a proxy for the underlying variant biologically responsible for the phenotype. This can be accomplished with a small fraction of known variants that ‘tag’ other highly correlated variants. Once disease susceptibility loci are identified, however, the process of choosing plausible variants to explain the observed signal requires a careful assessment of all correlated variants, based on local LD structure. Knowledge of population-specific LD structure and intuitive bioinformatic tools for interrogating LD are therefore essential for designing association studies and localizing functional variants.
Bioinformatic tools are available for assessing LD (Barrett et al., 2005; Johnson et al., 2008), but not specifically designed to winnow down perspective candidates to putative functional variants. Existing tools report standard measures of LD with different levels of functional annotation, but none report which alleles are correlated between two single nucleotide polymorphisms (SNPs) in LD. Knowing which proxy allele is correlated with a ‘risk’ allele from a GWAS associated variant is essential for functional studies, especially when plausible functional variants are not directly genotyped. While at times this relationship can be accurately inferred when correlations are high and minor allele frequencies are low, additional bioinformatic analysis is needed when correlations are modest and minor allele frequencies approach 0.5. Such analyses can be time consuming and technically difficult for researchers who lack computational training.
LDlink is a web-based LD analysis tool designed to easily query pairwise linkage disequilibrium between SNPs. The web-based modules (LDhap, LDmatrix, LDpair and LDproxy) utilize reference haplotypes from 26 different population groups in Phase 3 of the 1000 Genomes Project (1000G) (Genomes Project et al., 2012) to produce haplotype tables and interactive plots. LDlink fills in current gaps of existing LD analysis tools by integrating expanded population reference sets, updated functional annotations, and interactive output to explore possible functional variants in high LD. A thorough exploration of the variation and linkage structure that exists across populations should facilitate fine mapping disease susceptibility regions and assist researchers in characterizing functional variants based on genotype-phenotype associations with potential clinical utility.
2 Implementation
Genetic reference data for LDlink originates from the Phase 3 release of the 1000G (Genomes Project et al., 2012). The release contains over 5000 haplotypes from individuals spanning 26 ancestral population groups. Statistical phasing techniques of the genotyped data allow for the construction of extended haplotypes that are available for public download from the 1000G ftp site in VCF format. The genotyped set is complete with all individuals having called genotypes at every included locus. Sample panel files map each individual to their respective ancestral subpopulation of membership. Ancestral super-populations include African, Ad-mixed American, East Asian, European, and South Asian. LDlink is flexible to allow for any combination of super or sub-population as input based on the investigator’s interest.
The other required input for LDlink modules is reference SNP (RS) numbers of the query SNPs. An indexed SQL database of dbSNP version 142 is used to match queried RS numbers with the genomic coordinates (GRCh37) of the SNPs of interest. Only biallelic SNPs are permitted for query. SNPs with alleles other than A, C, G or T and insertions or deletions are not supported at this time. Available LDlink SNP functional annotation output includes dbSNP’s predicted functional effect of variants in coding regions and RegulomeDB scores (Boyle et al., 2012) for variants in non-coding or intergenic regions.
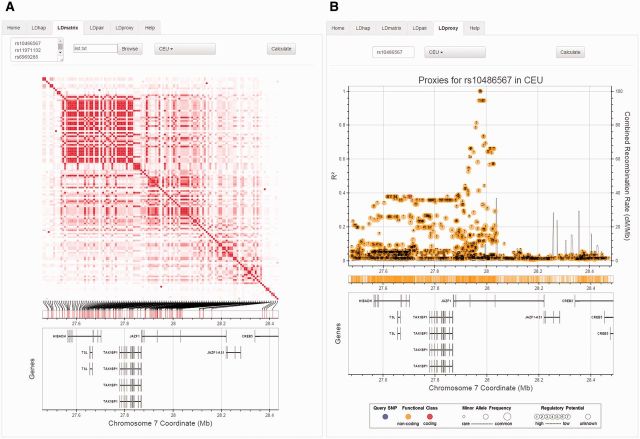
Available modules include LDhap, LDmatrix, LDpair and LDproxy (Fig. 1). LDhap calculates population-specific haplotype frequencies of all haplotypes observed for a list of query SNPs. Queried SNPs need not be contiguous and all observed 1000G haplotypes are enumerated. The LDmatrix module creates interactive heat map matrices of pairwise LD statistics from a list of SNP RS numbers and a specified population. An interactive hover tool displays LD metrics, correlated alleles, and nearby RefSeq genes. LDpair generates 2 by 2 tables of observed haplotypes for a pair of SNPs and reports haplotype and allele frequencies as well as measures of linkage disequilibrium. The LDproxy module interactively explores proxy and putatively functional SNPs for a query SNP in a selected 1000G population. Interactive plots show linkage disequilibrium over genomic distance where data point size, color and labels are used to highlight minor allele frequency and predicted function. Combined recombination rates are estimated from HapMap data. An interactive hover tool is also available to display LD metrics, correlated alleles and nearby RefSeq genes. All modules either require manual entry of RS numbers or uploading a saved list of RS numbers as well as selection of a 1000G populations of interest. Interactive output is displayed beneath the input tab and automatically updates when new input is added. An important limitation to keep in mind when using the 1000G phased genotype data is the possibility of switch rate errors, particularly when querying haplotype information on variants separated by large genomic distances.
Fig. 1.
Screen captures of LDmatrix (A) and LDproxy (B) modules
All LDlink modules are written in Python 2.7 and run on a virtual machine with UNIX operating system. Tabix version 0.2.5 is used to access phased genotypes of query SNPs from indexed VCF files (Li, 2011). The Python Bokeh package (0.8.0) is used to generate interactive plots. All web content is programed in HTML5 for cross platform compatibility.
Acknowledgements
The authors thank Sue Pan and Robert Shirley from the Center for Biomedical Informatics and Information Technology (CBIIT) for technical assistance and web development as well as the National Cancer Institute’s Laboratory of Genetic Susceptibility, Laboratory of Translational Genomics, Cancer Genomics Research Laboratory, and Division of Cancer Epidemiology and Genetics for valuable input and testing.
Funding
Support comes from the National Cancer Institute’s Intramural Research Program and the Division of Cancer Epidemiology and Genetics Informatics Tool Challenge.
Conflicts of Interest: none declared.
References
- Barrett J.C., et al. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- Boyle A.P., et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock S. (2014) Cancer biology: genome-wide association studies. In: Stewart B.W., Wild C.P. (eds), World Cancer Report 2014. International Agency for Research on Cancer, Lyon, France, pp. 193–202. [Google Scholar]
- Genomes Project et al. (2012) An integrated map of genetic variation from 1 092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.D., et al. (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics, 24, 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011) Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics, 27, 718–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D., et al. (2014) The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res., 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]