Abstract
Corticospinal motor neurons (CSMN) receive, integrate, and relay cerebral cortex's input toward spinal targets to initiate and modulate voluntary movement. CSMN degeneration is central for numerous motor neuron disorders and neurodegenerative diseases. Previously, 5 patients with mutations in the ubiquitin carboxy-terminal hydrolase-L1 (UCHL1) gene were reported to have neurodegeneration and motor neuron dysfunction with upper motor neuron involvement. To investigate the role of UCHL1 on CSMN health and stability, we used both in vivo and in vitro approaches, and took advantage of the Uchl1nm3419 (UCHL1−/−) mice, which lack all UCHL1 function. We report a unique role of UCHL1 in maintaining CSMN viability and cellular integrity. CSMN show early, selective, progressive, and profound cell loss in the absence of UCHL1. CSMN degeneration, evident even at pre-symptomatic stages by disintegration of the apical dendrite and spine loss, is mediated via increased ER stress. These findings bring a novel understanding to the basis of CSMN vulnerability, and suggest UCHL1−/− mice as a tool to study CSMN pathology.
Keywords: AAV2-mediated transduction, apical dendrite, CSMN, ER stress
Introduction
Corticospinal motor neurons (CSMN) are exceptional in their ability to receive, integrate, translate, and transmit information from the cerebral cortex to spinal cord targets (Molyneaux, Arlotta, Macklis 2007; Molyneaux Arlotta, Menezes et al. 2007; Greig et al. 2013). They are the cortical component of motor neuron circuitry that initiates and modulates very precise aspects of voluntary movement (Alstermark and Isa 2012). Execution of such complicated tasks emphasizes the importance of corticospinal motor neuron (CSMN) health and stability for proper motor neuron circuitry. Their degeneration, therefore, has significant impact on various neurodegenerative diseases in which voluntary movement is impaired, such as amyotrophic lateral sclerosis (ALS) and Parkinson's disease (PD; Novarino et al. 2014).
Identification of mutations in the ubiquitin carboxy-terminal hydrolase-L1 (UCHL1) gene of 2 patients with a PD-like phenotype (Leroy et al. 1998), and a recent clinical report describing 3 siblings with progressive and early neurodegeneration, and upper motor neuron dysfunction (Bilguvar et al. 2013) sparked interest in the potential importance of UCHL1 function in a spectrum of neurodegenerative diseases, especially those that affect voluntary movement and motor neuron circuitry.
UCHL1 is a unique deubiquitinating enzyme that has both hydrolase and ligase activities: It can remove ubiquitin from poly-ubiquitin chains (Osaka et al. 2003), and also add ubiquitin to already ubiquitinylated proteins (Liu et al. 2002). Since inhibition of UCHL1 results in a 50% reduction of free ubiquitin in vitro (Leroy et al. 1998; Cartier et al. 2009), UCHL1 was suggested to be critically important for maintaining free ubiquitin levels and for the proper function of the ubiquitin–proteasome system in neurons (Day and Thompson 2010). Abnormalities in protein homeostasis have been implicated in motor neuron diseases (Nishimura et al. 2004; Petrucelli and Dawson 2004; Johnson et al. 2010; Deng et al. 2011; Rubino et al. 2012), but the impact of a dysfunctional UCHL1 on the health and stability of motor neurons, especially upper motor neurons, remains poorly understood.
To reveal the potential link between UCHL1 function and especially the cortical component of the motor neuron circuitry, we investigated CSMN health in Uchl1nm3419 (UCHL1−/−) mice, which lack all UCHL1 function (Walters et al. 2008), and display motor function defects. Using retrograde labeling, molecular marker expression analysis, and adeno-associated virus (AAV)-mediated anterograde and retrograde transduction, we demonstrate early, selective, progressive, and profound CSMN degeneration in the absence of UCHL1 function. Other projection neurons in the motor cortex, such as callosal projection neurons (CPN), are not affected. Vulnerable CSMN display cellular abnormalities with vacuolated apical dendrites and spine loss, as early as postnatal day (P) 60, and their degeneration is linked to increased ER stress both in vivo and in vitro. Our findings begin to reveal a unique importance of UCHL1 function for CSMN health, and suggest that UCHL1−/− mice can be used as a tool to study the underlying mechanisms for CSMN dysfunction in diseases with prominent upper motor neuron involvement.
Materials and Methods
Mice
All animal procedures were approved by Northwestern University and the Massachusetts General Hospital–Harvard Medical School Animal Care and Use Committee, and conformed to the standards of the National Institutes of Health. Uchl1nm3419 (UCHL1−/−) mice carry a spontaneous 795-bp intragenic deletion that results in the removal of 24 bp of exon 6 and 771 bp of intron 6 (Fig. 1A; Walters et al. 2008). Heterozygous mice (UCHL1+/−) were viable, fertile, and bred to generate UCHL1-deficient (UCHL1−/−) mice. All mice were on a C57BL/6J background. Survival times and motor function defects were comparable between males and females with 100% penetrance. Primers used to determine genotypes are UCHL1 forward: tggacggctgtgtgtgctaatg, wild-type (WT) reverse: ctaagggaagggtcttgctcatc, mutant (Mt) reverse: gtcatctacctgaagagagccaag, yielding 668 bp WT and 334 bp Mt PCR products, respectively.
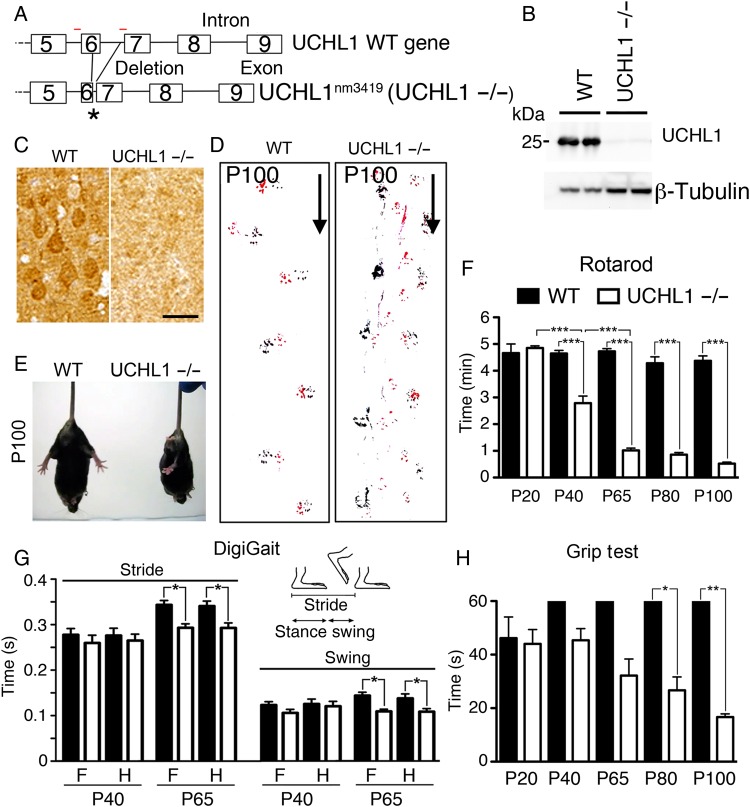
Figure 1.
UCHL1−/− mice show motor function defects. (A) Drawing of Uchl1 gene in WT and UCHL1-deficient (Uchl1nm3419; UCHL1−/−) mice. Partial deletions of exon 6 and intron 6 sequences cause a frameshift and create a de novo stop codon (asterisk). Red lines = location of PCR primers. (B) UCHL1 protein is present in the motor cortex of WT, but not in UCHL1−/−, mice by western blot analysis. β-Tubulin is used as a loading control. (C) UCHL1 expression is present in large pyramidal neurons in the motor cortex of WT, but not in UCHL1−/−, mice. (D) Directional walking patterns of WT and UCHL1−/− mice (marked by arrow) show signs of hindlimb paralysis (red: forelimb; black: hindlimb). (E) Representative screen captures of posture in WT and UCHL1−/− mice tail-hanging at P100. (F) Quantitative analysis of Rotarod test in WT and UCHL1−/− mice at P20 (n = 6 and 6), P40 (n = 14 and 10), P65 (n = 10 and 8), P80 (n = 7 and 9), and P100 (n = 7 and 9). (G) Gait parameter analysis for stride and swing in WT (n = 12) and UCHL1−/− (n = 15) mice at P40 and P65. (H) Quantitative analysis of Grip test in WT and UCHL1−/− mice at P20 (n = 6 and 6), P40 (n = 7 and 12), P65 (n = 7 and 12), P80 (n = 4 and 6), and P100 (n = 3 and 5). Bar graphs represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA with post hoc Tukey's multiple comparison test. Scale bar, 50 μm.
Postmortem Human Brain Samples
Acquisition of postmortem human tissue was carried out according to protocols approved by an institutional review board, and clinical records were available for every subject. Two control (A12–105, 52-year-old male and A12–106, 58-year-old female) cases were made available by the University of Chicago, IL, USA. A neurologist examined all human subjects and a neuropathologist reviewed autopsy tissue. Control case A12–105: 52-year-old male with a history of coronary artery disease, heart failure, morbid obesity, and implanted defibrillator who died of cardiac decompensation. Control case A12–106: 58-year-old female with pulmonary hypertension, scleroderma, and Raynaud's disease who died of cardiorespiratory failure. Brains were fixed in 20% neutral-buffered formalin for 1 week, and sections were paraffin-embedded. Areas of the primary motor cortex were retrieved; 4-μm thick serial sections were cut and used for immunocytochemical analyses.
Rotarod Analysis, DigiGait, and Grip Test
Fore and hind limbs of WT and UCHL1−/− mice were painted with red and black paints, respectively, prior to walking on white paper in an enclosed area. A rotating rod that accelerates linearly from 4 to 40 rpm (Rotarod, Ugo Basile) was used and an average time spent on the rotating rod for 3 consecutive trials was calculated for maximum 5 min. Gait analysis was performed as previously described (Gogliotti et al. 2011) using an enclosed DigiGait Imaging system (Mouse Specifics, Quincy, MA, USA). A transparent treadmill was adjusted to 17 cm/s. Swing and stride parameters were determined from a 5-s video. Time each mouse spent hanging on to an upside-down wire mesh (50 cm above a bench) was recorded in 3 consecutive trials, for Grip testing.
Generation of AAV
AAV vectors were generated by the University of Pennsylvania Vector Core facility by triple transfection of subconfluent HEK293 cells using 3 plasmids: an AAV trans-plasmid encoding AAV2 capsid, an adenovirus helper plasmid pΔF6, and an AAV cis-shuttle plasmid expressing eGFP driven by a CMV promoter (pENN.AAV.CMV.PI.eGFP .WPRE.bGH). The culture medium was collected, concentrated by tangential flow filtration, and purified by iodixanol gradient ultracentrifugation as previously described (Lock et al. 2010).
Anterograde and Retrograde Labeling and Transduction Surgeries
Surgeries were performed on a stereotaxic platform. Microinjections were performed using pulled-beveled glass micropipettes attached to a nanojector (Drummond Scientific, Broomall, PA, USA).
CSMN Retrograde Labeling and Transduction
CSMN were retrogradely labeled either by injection of fluorescent microspheres (LumaFluor, Inc., Naples, FL, USA; ∼97 nL for P2, and ∼207 nL for P38 injections), or by 0.5% Fluoro-Gold (FG; Fluorochrome LLC, Denver, CO, USA; 96 nL in 0.9% saline solution) at P30 (Ozdinler et al. 2011; Jara et al. 2012). Mice were sacrificed either at P40, P65, P80, or P100 (n = 3). To study CSMN cytoarchitecture, AAV-encoding eGFP (AAV2-eGFP; 621 nL containing 1.16 × 109 viral particles) was injected into the CST and CSMN were retrogradely transduced (n = 3) as described (Jara et al. 2012).
CSMN Anterograde Labeling
CSMN were anterogradely labeled by AAV-eGFP injection into the motor cortex. A unilateral craniotomy of approximately 5 mm2 (coordinates = +0.5 mm anterior-posterior; 1.5 mm mediolateral) was performed into the left hemisphere using a microdrill (Fine Science Tools, Foster City, CA, USA). Viral particles (1.16 × 109) were injected (10 injections, total 621 nL/mouse). Surgeries were performed at P30, and mice were sacrificed at P60 (n = 3).
CPN Retrograde Labeling
To retrogradely label CPN in the contralateral hemisphere, a craniotomy of approximately 5 mm2 (coordinates = +0.5 mm anterior-posterior; 1.5 mm mediolateral) was performed into the left hemisphere using microdrill (Fine Science Tools, Foster City, CA, USA). About 64 nL of 0.5% FG in 0.9% saline solution was injected (10 injections/mouse). Surgeries were performed at P30, and mice were sacrificed at P100 (n = 3).
Tissue Collection and Histology
Mice were deeply anesthetized with ketamine (90 mg/kg) and xylazine (10 mg/kg), and perfused with 4% PFA in PBS. The brain was removed intact from each mouse, post-fixed by 4% PFA overnight, and kept in PBS–sodium azide (0.01%) at 4 °C. Brains were sectioned (coronal; 50 μm) using a vibrating microtome (VT1000S, Leica Instruments, Nussloch, Germany). Nissl and hematoxylin–eosin (H&E) staining were performed. Optic nerves were embedded in resin and semi-thin sections were stained with toluidine blue.
Western Blot
Brains and spinal cords were isolated from WT and UCHL1−/− mice at P40 and P100. Protein lysates from primary motor cortex as well as from cervical and lumbar spinal cord were obtained, resolved by 10% SDS–PAGE, and transferred onto a PVDF membrane (Bio-Rad, Hercules, CA, USA) as previously described (Jara et al. 2007). Blots were incubated with primary antibodies overnight at 4 °C and developed by enhanced chemiluminescence (Millipore, Temecula, CA, USA) using HRP-conjugated secondary antibodies.
Immunocytochemistry and In Situ Hybridization
Immunocytochemistry was performed on every 12th coronal section of mouse brains. Antigen retrieval was performed for Ctip2, PERK, and PDI immunocytochemistry; sections were treated with 0.01 M sodium citrate, pH 9.0, at 80 °C water bath for 3 h prior to incubation with primary antibody. After PBS washes, either fluorescent-conjugated (Alexa 488, Cy3) or biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA, USA) were used.
For ER stress analysis on CSMN, PERK and PDI expression were studied on neurons that express Ctip2 (developed with alkaline phosphatase-conjugated secondary antibody and AP substrate Kit II; Vector Laboratories, Inc., Burlingame, CA, USA). Biotinylated secondary antibodies, ABC Kit (Vector Laboratories, Inc.), and NovaRED substrate (Vector Laboratories, Inc.) were used to determine PDI and PERK expression, as directed by the manufacturer.
Primary antibodies were: anti-Ctip2 (1 : 500; Abcam); anti-FG (1 : 1000; Fluorochrome, LLC); anti-GFP (1 : 1000; Invitrogen); anti-Lmo4 (1 : 500; Millipore); anti-NeuN (1 : 1000; Millipore); anti-PERK (1 : 1000; Cell Signaling Technology); anti-PDI (1 : 1000; Cell Signaling Technology); anti-ubiquitinylated proteins (1 : 1000; clone FK1; Millipore), and anti-UCHL1 (1 : 1000; ProteinTech).
For in situ hybridization, Crym and Fezf2 cRNAs were digoxigenin-labeled, and all probes were alkali hydrolyzed to an average length of 250–500 bp. Serial frozen sections (16 μm) were collected on slides, post-fixed with 4% PFA, rinsed in PBS, submerged in hybridization buffer containing a probe (100 ng/mL), and hybridized overnight at 68 °C. Sense probes were used as negative controls in all experiments and in situ hybridization was performed as previously described (Arlotta et al. 2005).
Tissue Culture
P2 motor cortices were dissected, dissociated, and cultured on glass coverslips (4.44 × 104cells per 18 mm diameter coverslip, Fisherbrand) coated with poly-l-lysine (10 mg/mL, Sigma). Neurons were cultured in serum-free medium [SFM; 0.034 mg/L BSA, 1 mM l-glutamine, 25 U/mL penicillin, 0.025 mg/mL streptomycin, 35 mM glucose, and 0.5% B27 in Neurobasal-A medium (Life Technologies)] in a humidified tissue culture incubator in the presence of 5% CO2 at 37 °C.
To induce ER stress, Tunicamycin (Sigma-Aldrich; 100 μM) or proteasome inhibitor Bortezomib (Santa Cruz Biotechnology; 100 nM) was added directly into culture medium after 1 day in vitro (DIV). Guanabenz (Sigma-Aldrich; 100 μM) was added at 1DIV to reduce ER stress. Dissociated cortical neurons were fixed prior to treatment, after 3 h and after 10 h of treatment to assess the levels of PERK and PDI and the changes in survival of CSMN and other cortical neurons. Percent survival was determined upon application of ethidium homodimer (EtHom; Invitrogen; 2 μM), the dead cell indicator, for 30 min prior to fixation (2% PFA, 15 min RT).
CSMN were identified based on their Ctip2 expression and large soma size. Ctip2/PERK and Ctip2/PDI co-expression analyses were performed in a serial staining protocol in which cells were stained with PERK or PDI antibody first, using an alkaline phosphatase-conjugated secondary antibody and AP Substrate Kit III (Vector Laboratories, Inc.), followed by Ctip2 immunocytochemistry using a biotinylated secondary antibody and ABC kit with NovaRED substrate (Vector Laboratories, Inc.). To determine percent CSMN survival, Ctip2 immunocytochemistry was performed on cultures treated with EtHom.
System Analysis
Proteins that are reported to have direct interactions with UCHL1 were determined using curated information generated from published data sets [Ingenuity Pathway Analysis (IPA); QIAGEN Comp, LA, USA] by setting stringent criterion that included only the experimental findings and excluded data generated from non-neuronal cell lines. Hits were individually subjected to confirmation and upon verification of relevance were included. The data set was used to determine the canonical pathways and the cellular networks.
Imaging, Quantification, and Statistical Analysis
Nikon SMZ1500 and Nikon Eclipse TE2000-E fluorescence microscopes equipped with Intensilight C-HGFI (Nikon, Inc.) were used. Epifluorescence images were acquired using a Digital Sight DS-Qi1MC CCD camera (Nikon, Inc.) and light images were acquired using a Ds-Fi1 camera (Nikon, Inc.). Confocal images were collected using a Zeiss 510 Meta confocal microscope (Carl Zeiss, Inc.).
For behavioral tests, including Rotarod, Gait, and Grip test, statistical differences were determined by one-way ANOVA with post hoc Tukey's multiple comparison parametric tests.
For quantification of CSMN soma diameter, progressive CSMN degeneration, percent PERK expression in Ctip2+ neurons, and percent PDI expression in Ctip2+ neurons, 2 well-defined sections spanning the motor cortex (Section 1: Bregma 0.38 mm, interaural 4.18 mm; Section 2: Bregma −0.34 mm; interaural 3.46 mm; Paxinos and Franklin 2001) were used per mouse.
Numbers and diameters of CSMN were determined in WT and UCHL1−/− mice at P40, P65, P80, and P100. CSMN numbers were counted and averaged (2 sections/per mouse; n = 3). Average CSMN diameter (at least 100 neurons/mouse; n = 3) was measured using the Elements Software (Nikon, Inc., Melville, NY, USA).
To determine the percentage of Ctip2+ neurons that express PERK and PDI, the total numbers of Ctip2+ neurons in layer V of the motor cortex (CSMN) were counted in WT and UCHL1−/− mice at P40, P65, P80, and P100 (n = 3). The Ctip2+ neurons that co-express PERK and PDI were quantified. Statistical differences between WT and UCHL1−/− mice were determined by one-way ANOVA with post hoc Tukey's multiple comparison parametric test.
The presence of total PERK and PDI in CSMN (determined by Ctip2 expression and large soma size) and other cortical neurons were determined by co-immunocytochemistry analysis in vitro using both WT and UCHL1−/− mice. Results were presented as the percent PERK and PDI expression in CSMN and other cortical neurons in the presence of compounds that either enhance (i.e. Tunicamycin and Bortezomib) or reduce (Guanabenz) ER stress. In addition, the percent survival of CSMN and other cortical neurons were quantified using EtHom exclusion. Statistical differences between compound treatments, and survival experiments in WT and UCHL1−/− mice, were determined by the unpaired t-test.
For CSMN spine density measurements, a 50-μm segment of apical dendrites (in layer II/III, in the primary apical dendrite) and basal dendrites (in layer V, separated by 30 μm from the soma) were selected and the total numbers of spines were counted (10 segments/mouse; n = 3). Statistical differences were determined by the unpaired t-test.
All statistical analyses were performed using the Prism software (version 5.0a; Graphpad Software, Inc., La Jolla, CA, USA). Statistically significant differences were determined after either one-way ANOVA with post hoc Tukey's multiple comparison tests or t-test. Statistically significant differences were considered at P < 0.05, and values were expressed as the mean ± SEM.
Results
UCHL1−/− Mice Display Motor Function Defects
To bring an insight on the potential involvement of UCHL1, especially for upper motor neuron biology, we developed a multidisciplinary approach and took advantage of Uchl1nm3419 (UCHL1−/−) mice, which lack all UCHL1 activity (Walters et al. 2008), and display prominent motor function defects (Fig. 1).
UCHL1−/− mice lack part of exon 6 and the intronic sequence between exons 6 and 7 of the Uchl1 gene due to a spontaneous intragenic deletion (Fig. 1A). This mutation results in the creation of a premature stop codon (denoted by “asterisk” in Fig. 1A) resulting in elimination of the transcript. Therefore, functional UCHL1 protein is not detected in UCHL1−/− mice brain (Fig. 1B,C; Walters et al. 2008). UCHL1−/− mice are born at the expected Mendelian ratio, and do not display any phenotypic differences or functional defects at birth and during early development. Heterozygous mice (UCHL1+/−) are viable, fertile with no signs of motor impairment at any age.
DigiGait analysis shows signs of gait impairment by P65: stride (fore limb: WT, 0.34 ± 0.01 s, n = 6; UCHL1−/−, 0.29 ± 0.01 s, n = 7; hind limb: WT, 0.34 ± 0.01 s, n = 6; UCHL1−/−, 0.29 ± 0.01 s, n = 7; P < 0.05, Fig. 1G) and swing (fore limb: WT, 0.14 ± 0.01 s, n = 6; UCHL1−/−, 0.11 ± 0.05 s, n = 7; hind limb: WT, 0.14 ± 0.01 s, n = 6; UCHL1−/−, 0.11 ± 0.01 s, n = 7; P < 0.05). Unlike WT mice, gait parameters of UCHL1−/− mice do not improve with age (Fig. 1G). Although UCHL1−/− pups are similar to their WT littermates at birth and show comparable growth, by P100 all UCHL1−/− mice display profound motor dysfunction (Fig. 1D). There was no difference at P20, but by P40 and onwards hind limb stiffness accompanied with clasping was observed when held upside-down (Fig. 1E; Supplementary Movie 1). UCHL1−/− mice spent progressively less time on the accelerating rotating rod (WT, 4.6 ± 0.1 min, n = 14; UCHL1−/−, 2.8 ± 0.3 min, n = 10; P < 0.001; Fig. 1F). In addition, UCHL1−/− mice gradually lose their ability to hold on to an inverted wire grid (WT, 60 s, n = 4; UCHL1−/−, 27 ± 5 s, n = 6; P < 0.05; Fig. 1H). Even though DigiGait results may suggest the lack of proper gait development, both Rotarod and Grip tests reveal a progressive decline in their motor function with age. These defects are observed with 100% penetrance in the UCHL1−/− mice, and the timing of the progressive motor function defect is comparable among mice and litters. However, both the optic nerve and the cerebellum appear normal, with no signs of apparent degeneration in the UCHL1−/− mice (Supplementary Fig. 1; Bilguvar et al. 2013), suggesting that not all systems are equally affected, and UCHL1−/− mice would be informative for the investigation of UCHL1’s role in motor neuron biology.
CSMN Undergo Early, Progressive, and Profound Degeneration in the UCHL1−/− Mice
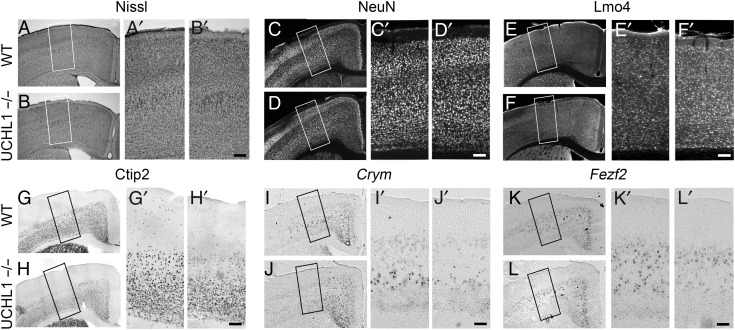
Retrograde labeling coupled with immunocytochemistry revealed high levels of UCHL1 expression particularly within CSMN in the motor cortex (Supplementary Fig. 2A–D), and initial studies reported high levels of UCHL1 in the cerebral cortex (Day and Thompson 1987, 2010). In addition, UCHL1 was primarily present in Betz cells of postmortem human brain samples (control cases, n = 2; A12–105 and A12–106; Supplementary Fig. 2E). Since CSMN have a unique function for the initiation and modulation of voluntary movement, we next investigated whether CSMN are affected in UCHL1−/− mice. WT and UCHL1−/− motor cortices were comparable with Nissl staining (Fig. 2A,B) and NeuN expression (Fig. 2C,D). Likewise, expression of Lmo4, a molecular marker for CPN (Arlotta et al. 2005), did not show any difference (Fig. 2E,F). However, the expression of Ctip2 (Fig. 2G,H), Crym (Fig. 2I,J), and Fezf2 (Fig. 2K,L), –molecular markers of CSMN in layer V of the motor cortex, and subcerebral projection neuron (SCPN) outside of motor cortex (Arlotta et al. 2005), were reduced at P100, suggesting CSMN degeneration.
Figure 2.
CSMN vulnerability in the UCHL1−/− mice. (A–L) Representative images of matching coronal sections of the motor cortex from WT and UCHL1−/− mice at P100. Nissl staining (A and B), NeuN (C and D), and Lmo4 (E and F) immunocytochemistry in the motor cortex do not show differences between WT and UCHL1−/− mice. However, expression analysis of molecular markers specific to CSMN in layer V of the motor cortex, such as Ctip2 immunocytochemistry (G and H), or Crym (I and J) and Fezf2 (K and L) in situ hybridization show selective reduction of molecular marker expression especially in large pyramidal neurons located in layer V of the motor cortex in UCHL1−/− mice. The boxed areas are enlarged in (A′–L′). Scale bars, 100 μm.
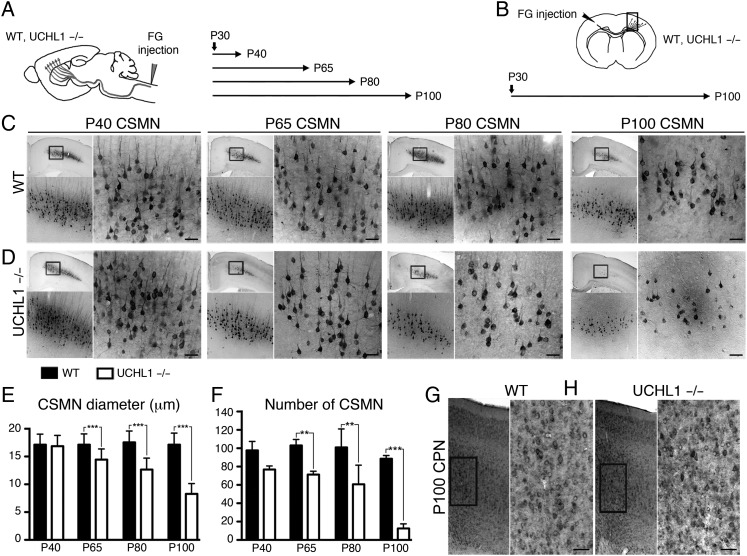
To differentiate between cellular degeneration of CSMN and a mere reduction in molecular marker expression, we performed 2 different retrograde labeling experiments (Fig. 3A,B). CSMN were labeled with FG at P30 (Fig. 3A). Their numbers and soma diameter were qualitatively (Fig. 3C,D) and quantitatively (Fig. 3E,F) analyzed at P40, P65, P80, and P100. CSMN morphology, soma diameter, and numbers were similar between WT and UCHL1−/− mice at P40 (Fig. 3C–F). However, by P65, soma diameter (WT, 17.1 ± 0.2 μm; UCHL1−/−, 14.4 ± 0.1 μm, n = 3; P < 0.001, Fig. 3E) and CSMN numbers (WT, 103 ± 3 neurons; UCHL1−/−, 71 ± 2 neurons, n = 3; P < 0.01, Fig. 3F) were significantly different. By P100, there was 88 ± 2% reduction in average CSMN numbers in the UCHL1−/− mice (Fig. 3F). The remaining CSMN had an approximately 50% reduction in soma diameter (Fig. 3E) and displayed the absence of a prominent apical dendrite (Fig. 3D). CPN were retrogradely labeled by contralateral injection of FG at P30, and analyzed at P100 to investigate whether degeneration is restricted to CSMN (Fig. 3B). CPN were comparable between WT and UCHL1−/− mice at P100 (Fig. 3G,H), suggesting that vulnerability is not common to all neurons in the motor cortex.
Figure 3.
Progressive CSMN degeneration in the UCHL1−/− mice. (A and B) Experimental design for CSMN (A) and CPN (B) retrograde labeling by FG injection in WT and UCHL1−/− mice. Surgeries were performed at P30, and mice were sacrificed at P40, P65, P80, and P100 for CSMN (A) and at P100 for CPN (B) labeling. (C and D) Representative images of matching coronal sections of WT (C) and UCHL1−/− mice (D) motor cortex, analyzed at P40, P65, P80, and P100, show progressive CSMN degeneration in UCHL1−/− mice. Boxed areas are enlarged below and to the right. (E and F) Quantitative analysis of average CSMN soma diameter (E) and average numbers of CSMN (F) show significant reduction as early as P65 in the UCHL1−/− mice (n = 3). (G and H) Retrograde labeling of CPN in WT (G) and UCHL1−/− mice (H) motor cortex at P100 do not reveal cellular degeneration (n = 3). Boxed areas are enlarged to the right. Data presented as mean ± SEM; **P < 0.01; ***P < 0.001; one-way ANOVA with post hoc Tukey's multiple comparison test. Scale bars, 50 μm.
UCHL1 Function Is Important for the Maintenance of Cytoarchitectural Stability and Protein Homeostasis
To better understand the link between the absence of UCHL1 function and CSMN vulnerability, and to determine the cellular events that would be primarily affected in the absence of UCHL1 function, a system approach was taken. The list of proteins that directly interact with UCHL1 (n = 49) were determined based on a stringent data selection paradigm using IPA. Investigation of the canonical pathways these UCHL1-interacting proteins shared in common, and the molecular networks they activate suggested involvement of UCHL1 primarily with organization of cytoskeleton (n = 21), ubiquitination of proteins (n = 12), and microtubule dynamics (n = 18), apoptosis (n = 29), degradation of proteins (n = 12), and cellular homeostasis (n = 19). Among functions and diseases, movement disorders (n = 18), motor dysfunction (n = 8), and neuromuscular disease (n = 15) were the most prominent. This protein interaction network suggested the potential importance of UCHL1 function in 2 important cellular events: Maintenance of cellular architecture (cytoarchitecture) and protein homeostasis (Supplementary Fig. 3).
Vulnerable CSMN Exhibit Spine Loss with Apical Dendrite Degeneration That Is Not Secondary to CST Defects
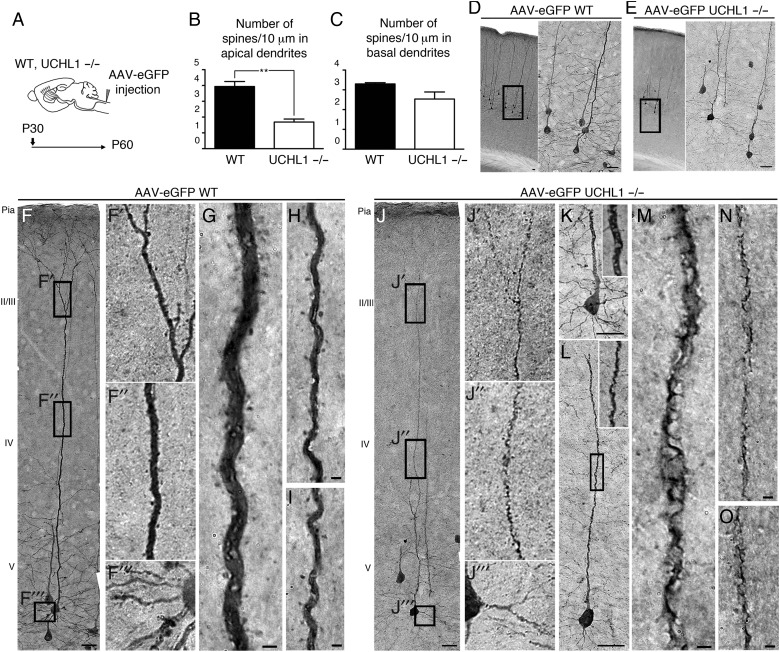
To investigate how the absence of UCHL1 function affects cytoarchitecture of CSMN at the soma, apical, and basal dendrites, CSMN were retrogradely transduced with AAV2–2 eGFP (Fig. 4A). AAV transduction did not cause pathological changes, but revealed the details of cytoarchitecture of both WT and UCHL1−/− CSMN (Fig. 4D–O). Transduced WT CSMN exhibited the morphology of healthy CSMN, with large pyramidal cell bodies, prominent apical dendrites, and spines throughout the dendrites, mostly in layers II/III (Fig. 4D,F–I; Jara et al. 2012). In striking contrast, CSMN of UCHL1−/− mice displayed massive disintegration with profound vacuolization and breakdown of cell membrane especially along the apical dendrites (Fig. 4E,J–O; UCHL1−/−, 42 ± 4%, n = 167 apical dendrites, n = 3; P < 0.01; and WT, 7%, n = 281 apical dendrites, n = 3). UCHL1−/− mice had a significant reduction in the numbers of spines throughout the apical (WT, 3.9 ± 0.3; UCHL1−/−, 1.7 ± 0.2; n = 3; P < 0.05, Fig. 4B), but not basal, dendrites (WT, 3.3 ± 0.1; UCHL1−/−, 2.5 ± 0.4; n = 3, Fig. 4C).
Figure 4.
AAV2-2-mediated retrograde CSMN transduction reveals apical dendrite degeneration and spine loss. (A) Experimental design for AAV2-2-mediated retrograde transduction of CSMN in WT and UCHL1−/− mice. Surgeries are performed at P30 and mice are sacrificed at P60. (B and C) Quantitative analysis of apical and basal dendrites reveals significant reduction in the average numbers of spines in apical (B), but not in basal (C), dendrites. (D–O) Representative images of the motor cortex of WT (D,F–I) and UCHL1−/− (E,J–O) mice. Boxed areas are enlarged to the right. WT CSMN exhibit healthy morphology with a large pyramidal cell body and prominent apical (F′ and F″, G–I) as well as basal dendrites with spines (F‴). CSMN of UCHL1−/− mice show severe cellular degeneration with spine loss throughout the vacuolated apical dendrite (J′ and J″, K–O); but, spines in basal dendrites are not significantly affected (E and J‴). Bar graphs represent mean ± SEM. **P < 0.01, unpaired t-test. Scale bars, (D–F, J) 50 μm, (G–I) and (K–O) 2 μm.
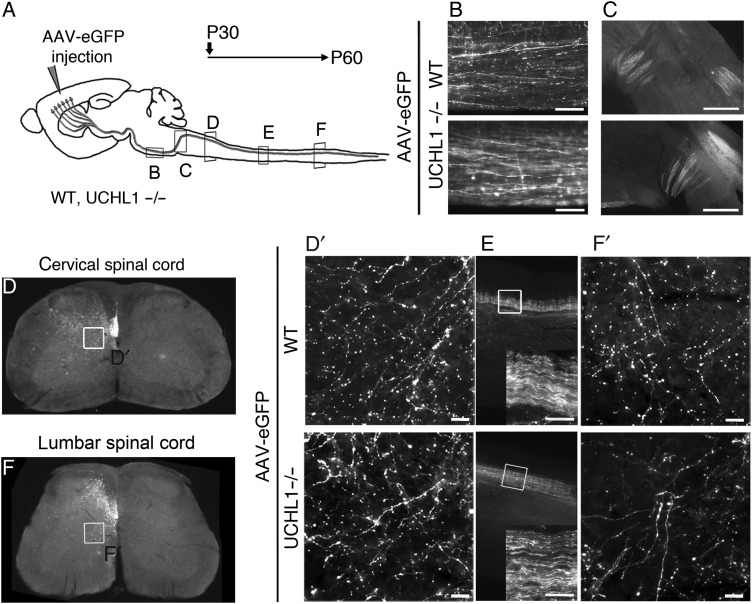
Progressive axonal degeneration is suggested as a potential mechanism for CSMN degeneration (Miura et al. 1993; Eisen and Weber 2001; Fischer et al. 2004; Morfini et al. 2013). To investigate if CST defects occur prior to apical dendrite degeneration, and to visualize CST axon fibers, AAV-eGFP was directly injected into the motor cortex (Fig. 5). Large projection neurons were transduced in the motor cortex at P30 and the axons of SCPN of WT and UCHL1−/− mice (n = 3; Fig. 5A) were examined by eGFP expression at the cervical (Fig. 5D), thoracic (Fig. 5E), and lumbar (Fig. 5F) sections of the spinal cord at P60, –when CSMN numbers and soma diameter showed a significant reduction– (Fig. 3E,F). Unilateral injection into the cerebral cortex demonstrated the presence of axons on the left side of the CST in both cervical and lumbar sections of the spinal cord, and further revealed detailed axon morphology, especially on axons that branch and project toward the spinal cord targets (boxed areas enlarged in Fig. 5D′,F′). Axon bundles were observed within the dorsal funiculus with branching toward the anterior and ventral horn (Fig. 5D,F), and they were comparable between WT and UCHL1−/− mice at all levels of the spinal cord. Subtle enlargements along the branches were observed, and in the pons area a subset of axons showed “beads on a string” appearance (UCHL1−/−, 40 ± 6%, n = 114 axons, n = 4; WT, 17 ± 7%, n = 144 axons, n = 4; P < 0.005, Fig. 5B). These results reveal the lack of massive axonal degeneration along the CST at P60, and suggest that apical dendrite defects and spine loss are not secondary to CST degeneration.
Figure 5.
AAV2-2-mediated anterograde CSMN transduction reveals absence of major corticospinal tract defects. (A) Experimental design for AAV2-2-mediated anterograde transduction of CSMN in WT and UCHL1−/− mice. Surgeries were performed at P30 and axons were visualized at the pons (B), pyramidal decussation (C), and the cervical (D), thoracic (E), and lumbar (F) sections of the spinal cord in both WT and UCHL1−/− mice at P60. Boxed areas within the cervical (D) and lumbar (F) spinal cord are enlarged in (D′) and (F′) to reveal details of axon morphology. (E) CST along the sagittal thoracic section is enlarged within insert. Scale bars, (B, D′, E, and F′) 50 μm and (C) 500 μm.
ER Stress Is Increased in CSMN of UCHL1−/− Mice
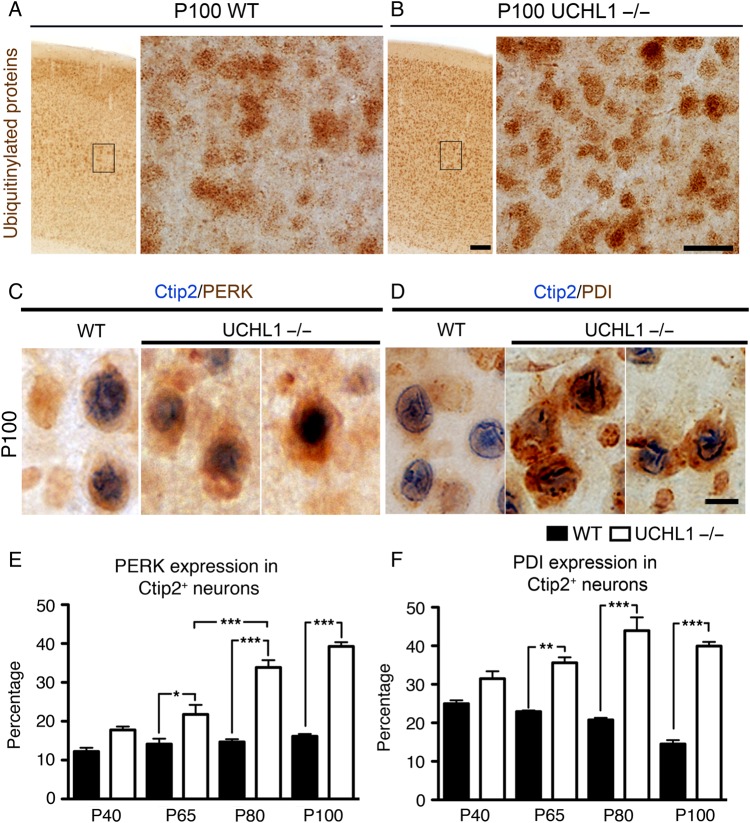
Since UCHL1 is implicated in determining the levels of free ubiquitin, and is important for maintaining protein homeostasis in the cell, we investigated ER stress in CSMN of UCHL1−/− mice. Ubiquitinylated proteins were increased in the motor cortex of UCHL1−/− mice at P100 (Fig. 6A,B). However, PDI, and other markers of ER stress, such as CHOP, BIP, and ATF4, failed to reveal differences between total lysates from motor cortices of WT and UCHL1−/− mice (Supplementary Fig. 4A,B). This could be due to selective increase in a subset of neuron population that cannot be detected when cortical homogenate is used. To investigate if ER stress is specifically increased in CSMN, 2 lines of experiments were performed. First, CSMN were retrogradely labeled with FG. The expression profile of PERK, a major ER stress sensor upregulated upon accumulation of ubiquitinylated and misfolded proteins (Walter and Ron 2011), was studied in retrogradely labeled CSMN (Supplementary Fig. 4C–I). Secondly, Ctip2 expression was used as a marker for CSMN in the motor cortex. PERK (WT, 33% ± 2; UCHL1−/−, 47% ± 4; n = 2) and PDI (WT, 30% ± 7; UCHL1−/−, 45% ± 4; n = 2) immunoreactivity were present especially in FG-labeled CSMN by P65. Even though total numbers of CSMN were decreasing, CSMN (Ctip2+, blue; Fig. 6C,D) with PERK (red; Fig. 6C) significantly increased by P65 (WT, 13.2% ± 1.7; UCHL1−/−, 21.8% ± 2,4; n = 3; P < 0.05; Fig. 6E), and continued to increase with age only in the UCHL1−/− mice. Similarly, PDI expression (red; Fig. 6D) was significantly higher in CSMN of UCHL1−/− mice by P65 (WT, 22.7% ± 0.3; UCHL1−/−, 35.6% ± 1.4; n = 3; P < 0.01; Fig. 6F) and remains significantly higher at older ages (P100 WT, 14.5% ± 1; UCHL1−/−, 39.9% ± 1; n = 3; P < 0.001; Fig. 6F). Albeit total numbers of CSMN reduce with age, the increase in the percentage of CSMN with PERK and PDI suggests a strong correlation between increased ER stress and progressive CSMN degeneration in the UCHL1−/− mice.
Figure 6.
ER stress is increased in CSMN of UCHL1−/− mice. (A and B) Ubiquitinylated proteins are present in WT mice (A), but they are increased in the motor cortex of UCHL1−/− mice (B) at P100. The boxed areas are enlarged to the right. (C and D) Double immunocytochemistry analyses of Ctip2 (blue) and PERK and PDI (red/brown) reveal the presence of ER stress in CSMN of UCHL1−/− mice. Total levels of both PERK (C) and PDI (D) are increased in Ctip2+ neurons of UCHL1−/− mice at P100. (E and F) Quantitative analyses of percent PERK (E) and percent PDI (F) expression in Ctip2+ neurons within the layer V of the motor cortex show progressively increasing ER stress in CSMN of UCHL1−/− mice, which becomes significant by P65. Bar graphs represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA with post hoc Tukey's multiple comparison test. Scale bars, (A and B) 100 μm (low magnification) and 25 μm (high magnification), and (C and D) 10 μm.
CSMN Are Vulnerable to Increased ER Stress
To investigate whether UCHL1−/− CSMN is more vulnerable to increased ER stress, dissociated cortical cultures were established from WT and UCHL1−/− mice at P2–P3. CSMN were identified among other cortical neurons by Ctip2 expression (Fig. 7A–E; red color in the nucleus) and their large soma size (Fig. 7C–E; black arrowheads).
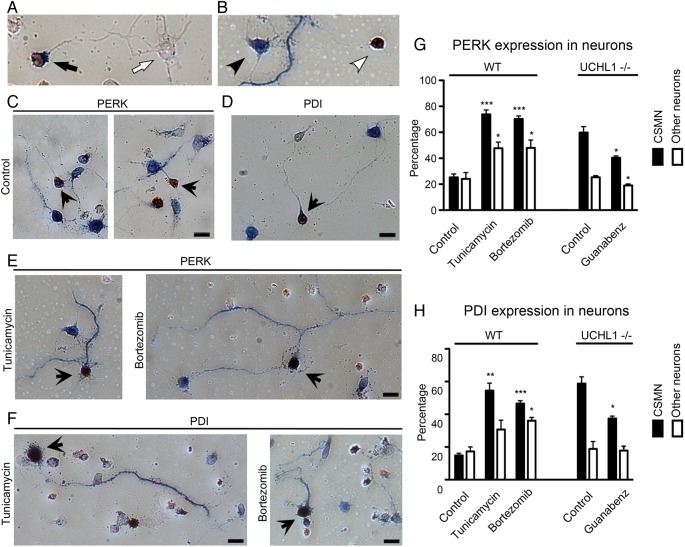
Figure 7.
Total levels of PERK and PDI are increased upon induction of ER stress and in the absence of UCHL1 function. (A and B) Ctip2+ and PERK/PDI+ CSMN (A, black arrow), Ctip2− and PERK/PDI− other neuron (A, white arrow), Ctip2− and PERK/PDI+ other neuron (B, black arrowhead), Ctip2+ and PERK/PDI− CSMN (B, white arrowhead). (C and D) In mixed cortical neurons, CSMN (brown nucleus, indicated by black arrows) and other neurons express low levels of PERK (blue; C) and PDI (blue; D). (E and F) Addition of Tunicamycin (100 μM) and Bortezomib (100 nM) increases PERK (blue; E) and PDI (blue; F) expression in distinct sets of neurons. (G) Quantitative assessment of PERK expression among CSMN and other cortical neurons that are not CSMN using dissociated cultures isolated from WT and UCHL1−/− mice, and in the presence of Tunicamycin, Bortezomib, and Guanabenz (100 μM). (H) Quantitative assessment of PDI expression among CSMN and other cortical neurons that are not CSMN using dissociated cultures isolated from WT and UCHL1−/− mice, and in the presence of Tunicamycin, Bortezomib, and Guanabenz. Bar graphs represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 pair-wise comparisons between control and drug treatments; unpaired t-test.
In the first set of experiments, dissociated mixed cultures of WT neurons were exposed to 2 different compounds that increase ER stress via different mechanisms. Tunicamycin blocks N-glycosylation of all proteins and induces ER stress (Olden et al. 1979), whereas Bortezomib induces ER stress by blocking the catalytic domain of the 26S-proteosome (Bonvini et al. 2007; Tuffy et al. 2010). In the second set of experiments, the ER stress was reduced in the dissociated mixed cortical cultures isolated from UCHL1−/− mice by the application of Guanabenz, which enhances the PERK pathway by selectively inhibiting GADD34-mediated dephosphorylation of eIF2α (100 μM; Tsaytler et al. 2011; Vaccaro et al. 2013; Wang et al. 2014). The links between the levels of ER stress, presence of total PERK, PDI, and the percent survival of CSMN in comparison with other cortical neurons were assayed based on selection criteria of PERK, PDI, and Ctip2 expression. Ctip2 expression was restricted to the nucleus (red) and both PERK and PDI were detected in the cell body (blue; Fig. 7A).
In control conditions, both CSMN and other cortical neurons expressed low levels of PERK (CSMN: 25 ± 3%, n = 237 neurons, n = 3; other neurons: 24 ± 5%, n = 134 neurons, n = 3; Fig. 7C,G) and PDI (CSMN, 15 ± 1%, n = 251 neurons, n = 3; other neurons, 17 ± 3%, n = 256, n = 3; Fig. 7D,H). Upon addition of Tunicamycin (100 μM), however, there was an overall increase in the percentage of CSMN that contained high levels of PERK (CSMN, 73 ± 3%; n = 146 neurons; n = 4 mice; other neurons, 48 ± 5%, n = 120 neurons, n = 3; P < 0.005; Fig. 7E,G) and PDI (CSMN, 54 ± 5%, n = 140 neurons, n = 3; other neurons, 31 ± 6%, n = 120 neurons, n = 3; P < 0.05; Fig. 7F,H), and significant differences between CSMN and other cortical neurons were present (Fig. 7G,H).
Addition of Bortezomib (100 nM) increased the presence of PERK and PDI in general, but the percentage of CSMN with PERK (70 ± 2%, n = 167 neurons, n = 4) was significantly higher than other cortical neurons (48 ± 6%, n = 156 neurons, n = 4, P < 0.0001; Fig. 7E,G). Similarly, there was a significant difference between the percentages of CSMN with PDI (47 ± 2%, n = 181 neurons, n = 3) and other neurons (36 ± 2%, n = 141 neurons, n = 3, P < 0.001; Fig. 7F,H). In previous studies, increases in the total levels of PERK and PDI were detected as early as 3 h (Tsaytler et al. 2011). Our current results revealed that, in the presence of increased ER stress, CSMN displayed a selective and profound increase of both PERK and PDI.
Unlike WT CSMN, CSMN of UCHL1−/− mice had intrinsically high levels of total PERK (60 ± 5%, n = 113 neurons, n = 4) and PDI (59 ± 4%, n = 117 neurons, n = 4). This could explain their immediate death upon treatment with Tunicamycin and Bortezomib, and our inability to establish reliable cultures (data not shown). However, other cortical neurons of UCHL1−/− mice had similar levels of PERK (25 ± 1%, n = 236 neurons, n = 3) and PDI (19 ± 5%, n = 245 neurons, n = 3) that are comparable with WT neurons.
Addition of Guanabenz (100 μM) significantly reduced the percentage of CSMN with PERK (40 ± 1%, n = 132 neurons, n = 2, P < 0.05) and PDI (37 ± 1%, n = 143 neurons, n = 2, P < 0.05) within 9 h, but other cortical neurons of UCHL1−/− mice were not affected (PERK; 19 ± 1%, n = 192 neurons, n = 2, P < 0.05 and PDI; 18 ± 3%, n = 180 neurons, n = 2).
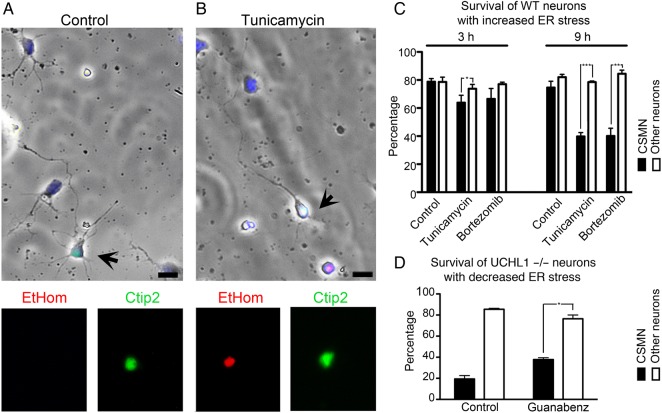
These results, together with in vivo expression analyses, suggest that CSMN show selective upregulation of ER stress in the absence of UCHL1 function. This could be one of the underlying causes of CSMN vulnerability. To test the link between increased ER stress and CSMN survival, we performed cell death assay (Fig. 8A,B). Percent survival of CSMN and other cortical neurons of WT mice were similar in control conditions (CSMN, 3 h: 79 ± 2%, n = 283 neurons; 9 h: 75 ± 4%, n = 275 neurons, n = 5; Fig. 8C). In contrast, upon induction of ER stress, more than half of CSMN were dead in culture within 9 h, whereas other cortical neurons were not affected. The percent survival of cortical neurons did not change in the presence of Tunicamycin (3 h: 74 ± 3%, n = 502 neurons; 9 h: 79 ± 1%, n = 396 neurons, n = 5) or Bortezomib (3 h: 77 ± 1%, n = 459 neurons; 9 h: 84 ± 3%, n = 469 neurons, n = 5). However, CSMN of WT mice showed rapid cellular degeneration in the presence of both Tunicamycin (3 h: 64 ± 5%, n = 308 neurons, P < 0.05; 9 h: 40 ± 6%, n = 214 neurons, n = 5, P < 0.001) and Bortezomib (3 h: 67 ± 7%, n = 335 neurons; 9 h: 40 ± 6%, n = 214 neurons, n = 5, P < 0.01). These results suggested that unlike other cortical neurons, CSMN display a unique vulnerability and undergo cellular degeneration in the presence of increased ER stress.
Figure 8.
CSMN show selective vulnerability to increased ER stress levels. (A) Mixed cortical cultures prepared from P2 cerebral cortex of WT mouse include CSMN (black arrows; Ctip2 immunofluorescence) and other cortical neurons. CSMN express Ctip2 (green) and display a healthy morphology with a pyramidal cell body, a prominent apical dendrite, and an axon. (B) Addition of Tunicamycin to the culture medium induces CSMN cell death (EtHom: red; Ctip2: green). (C) Quantitative assessment of percent survival of CSMN and other cortical neurons that are not CSMN isolated from WT mice, in the presence of Tunicamycin and Bortezomib, at 3 and 9 h, respectively. (D) Quantitative assessment of percent survival of CSMN and other cortical neurons that are not CSMN isolated from UCHL1−/− mice, in the presence of Guanabenz at 9 h. Bar graphs represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 pair-wise comparisons between control and compound treatments; unpaired t-test.
CSMN of UCHL1−/− mice displayed significantly low levels of survival in culture (WT: 85 ± 1%, n = 232 neurons; CSMN: 19 ± 3%, n = 62 neurons, n = 2, P < 0.001; Fig. 8A–C). To test whether reduction in ER stress would improve survival of UCHL1−/− CSMN, cultures were treated with Guanabenz (100 mM) for 9 h (Fig. 8D). In the presence of Guanabenz, CSMN survival significantly improved (38 ± 2%, n = 68 neurons, P < 0.05), whereas survival of other cortical neurons was not affected (76 ± 4%, n = 444 neurons, n = 2; Fig. 8D). These findings show that, in the absence of UCHL1 function, CSMN display selective cellular degeneration, and that increased ER stress is one of the cellular mechanisms responsible for CSMN vulnerability.
Discussion
UCHL1 is detected in cortical Lewy bodies and neurofibrillary tangles in patients with amyotrophic lateral sclerosis (ALS), PD, Alzheimer's disease (AD), and frontotemporal dementia (Lowe et al. 1990; Alves-Rodrigues et al. 1998; McNaught et al. 2001; Kersaitis et al. 2006; Gong and Leznik 2007). Interestingly, reduced UCHL1 levels are reported in PD, AD, and ALS patients (Choi et al. 2004; Lombardino et al. 2005; Barrachina et al. 2006; Lederer et al. 2007). Two patients with I93M mutation in the UCHL1 gene displayed PD-like symptoms associated with motor dysfunction, stiffness, and tremor (Leroy et al. 1998), and the importance of UCHL1 in the motor system was reinforced with a recent description of UCHL1GLU7ALA mutation in 3 patients displaying an early vision loss followed by progressive neurological dysfunction of the pyramidal system, cerebellum, and spinal dorsal columns, severely affecting movement. Muscle strength was grossly normal, but increased deep tendon reflexes with spasticity indicated prominent upper motor neuron degeneration (Bilguvar et al. 2013).
Mice with varied mutations in the Uchl1 gene (i.e., Uchl1gad mice; Yamazaki et al. 1988; Saigoh et al. 1999) and a targeted deletion-generated murine line, Uchl1tm1Dgen (Chen et al. 2010), have been described. They all report the lack of UCHL1 protein, but interestingly display varied phenotypes. Our goal is not to compare different lines, but to study the role of UCHL1 in motor neuron biology. Uchl1nm3419 (UCHL1−/−) mice, which lack all UCHL1 function (Walters et al. 2008), display major motor function defects without significant sensory ataxia. Unlike the Uchl1gad (Yamazaki et al. 1988; Saigoh et al. 1999) mice, the gracile nucleus is intact in the UCHL1−/− mice. Even though other lines of UCHL1 could be useful for different studies, UCHL1−/− mice offer significant advantages over other currently available lines for the investigation of motor neuron circuitry and disease. Most interestingly, UCHL1−/− mice show similarities to patients with Glu7Ala mutation (Bilguvar et al. 2013), and display early neurodegeneration, with deficits as early as P40. Pathology in the motor cortex of patients is remarkable and CSMN loss is evident by P65 with pronounced degeneration in the UCHL1−/− mice. These observations further support the choice of UCHL1−/− mice to investigate the importance of UCHL1 function for motor neuron biology. Even though CSMN vulnerability has been reported in other mouse models, such as hSOD1G93A mice (Ozdinler et al. 2011) and Prp-TDP-43A315T mice (Wegorzewska et al. 2009), UCHL1−/− mice stand out by their early, progressive, and profound CSMN degeneration that is in part due to ER stress-mediated vulnerability. Interestingly, vacuolation of the apical dendrite and cellular degeneration with spine loss is observed in both UCHL1−/− and hSOD1G93A mice, potentially a common mechanism for cellular pathology (Jara et al. 2013).
CSMN act as a “spokesperson” of the cerebral cortex for the initiation and control of voluntary movement. It is important for CSMN to receive proper input and effective modulation. Since CSMN receive much of their input from layer II/III (Anderson et al. 2010), apical dendrite degeneration, and spine loss, especially in this layer, would impair their ability to receive proper cortical input (Alstermark and Isa 2012). Thus, the message sent to the spinal targets would be cryptic and the motor neuron circuitry would be impaired.
There is a developing need for the generation and characterization of novel tools that mimic human pathology at a cellular level. Failures in clinical trials revealed that the focus has to be on the vulnerable neurons, and not on the extension of lifespan in mice (Genc and Ozdinler 2014). CSMN is of particular importance. Its degeneration is central in various neurodegenerative diseases, especially the ones that involve voluntary movement and motor function defect. Therefore, understanding the reasons behind CSMN vulnerability, and characterization of novel tools with CSMN pathology, would have implications in numerous neurodegenerative disorders, including AD and PD as well as primary lateral sclerosis, hereditary spastic paraplegia, and ALS (Novarino et al. 2014).
Our results suggest a new role for UCHL1 in promoting CSMN health and stability. In the absence of UCHL1 function, CSMN are vastly affected. UCHL1 is previously suggested to mediate cytoarchitectural plasticity by interacting with numerous cellular adhesion complexes, including paxillin, vinculin, and β-catenin (Frisan et al. 2012). Interestingly, Wnt/β-catenin complex contributes to motor neuron pathology in ALS (Yu et al. 2013). UCHL1 has also been reported on the recycling and determining the levels of NCAM (Wobst et al. 2012), one of the best-characterized components of CST. In addition, in the UCHL1-eGFP mouse, a reporter line that expresses eGFP gene under the UCHL1 promoter (Yasvoina et al. 2013), eGFP expression was restricted to CSMN in the motor cortex, further implicating an important role of UCHL1 for CSMN.
We find that CSMN are prone to cellular degeneration in the absence of UCHL1 function, and undergo early and progressive cytoarchitectural defects, especially observed with disintegration of apical dendrites. There is early upregulation of ER stress only in the CSMN of UCHL1−/− mice, and the rate of CSMN survival is proportionally related to increased ER stress. Focusing on the health of vulnerable CSMN would reveal the important information required to develop effective long-term treatment strategies for motor neuron diseases, and diseases in which voluntary movement is impaired.
Authors' Contributions
J.H.J., B.G., E.U., and P.H.O. performed experiments. All authors analyzed data and contributed to the writing of this manuscript.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This work was supported by grants from NIH-R01NS085161-01 (P.H.O.), Les Turner ALS Foundation, Wenske Foundation (P.H.O.), NUCATS (P.H.O. and M.C.B.); NIH/NINDS NS45523, NS49553, and NS41590 (J.D.M.), NIH (5T32AG020506-09 M.A.D. Postdoctoral Training; B.G.), and ALSA Safenowitz fellowship (J.H.J.). Funding to pay the Open Access publication charges for this article was provided by the Les Turner ALS Foundation.
Supplementary Material
Notes
We thank Dr Peter Pytel for the analysis of the human brain samples. D.K. Goncharoff, N.A. Khan, W.D. Weber, M.W. Tu, V. Hernandez, M.J. Standford, M. Schultz, and J. Xie helped with histochemistry and imaging. We thank UPenn Viral Vector Core, B. Goosens for help with confocal microscopy at CMRC Microscopy and Imaging Facility, and NU Behavioral Phenotyping Core Facility. Semi-thin sectioning and toluidine staining were performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. Conflict of Interest: None declared.
References
- Alstermark B, Isa T. 2012. Circuits for skilled reaching and grasping. Annu Rev Neurosci. 35:559–578. [DOI] [PubMed] [Google Scholar]
- Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME. 1998. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends Neurosci. 21:516–520. [DOI] [PubMed] [Google Scholar]
- Anderson CT, Sheets PL, Kiritani T, Shepherd GM. 2010. Sublayer-specific microcircuits of corticospinal and corticostriatal neurons in motor cortex. Nat Neurosci. 13:739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. 2005. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 45:207–221. [DOI] [PubMed] [Google Scholar]
- Barrachina M, Castano E, Dalfo E, Maes T, Buesa C, Ferrer I. 2006. Reduced ubiquitin C-terminal hydrolase-1 expression levels in dementia with Lewy bodies. Neurobiol Dis. 22:265–273. [DOI] [PubMed] [Google Scholar]
- Bilguvar K, Tyagi NK, Ozkara C, Tuysuz B, Bakircioglu M, Choi M, Delil S, Caglayan AO, Baranoski JF, Erturk O, et al. 2013. Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc Natl Acad Sci USA. 110:3489–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvini P, Zorzi E, Basso G, Rosolen A. 2007. Bortezomib-mediated 26S proteasome inhibition causes cell-cycle arrest and induces apoptosis in CD-30+ anaplastic large cell lymphoma. Leukemia. 21:838–842. [DOI] [PubMed] [Google Scholar]
- Cartier AE, Djakovic SN, Salehi A, Wilson SM, Masliah E, Patrick GN. 2009. Regulation of synaptic structure by ubiquitin C-terminal hydrolase L1. J Neurosci. 29:7857–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Sugiura Y, Myers KG, Liu Y, Lin W. 2010. Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc Natl Acad Sci USA. 107:1636–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. 2004. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem. 279:13256–13264. [DOI] [PubMed] [Google Scholar]
- Day IN, Thompson RJ. 1987. Molecular cloning of cDNA coding for human PGP 9.5 protein. A novel cytoplasmic marker for neurones and neuroendocrine cells. FEBS Lett. 210:157–160. [DOI] [PubMed] [Google Scholar]
- Day IN, Thompson RJ. 2010. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog Neurobiol. 90:327–362. [DOI] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. 2011. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 477:211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A, Weber M. 2001. The motor cortex and amyotrophic lateral sclerosis. Muscle Nerve. 24:564–573. [DOI] [PubMed] [Google Scholar]
- Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A, Khan J, Polak MA, Glass JD. 2004. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 185:232–240. [DOI] [PubMed] [Google Scholar]
- Frisan T, Coppotelli G, Dryselius R, Masucci MG. 2012. Ubiquitin C-terminal hydrolase-L1 interacts with adhesion complexes and promotes cell migration, survival, and anchorage independent growth. FASEB J. 26:5060–5070. [DOI] [PubMed] [Google Scholar]
- Genc B, Ozdinler PH. 2014. Moving forward in clinical trials for ALS: motor neurons lead the way please. Drug Discov Today. 19:441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti RG, Lutz C, Jorgensen M, Huebsch K, Koh S, Didonato CJ. 2011. Characterization of a commonly used mouse model of SMA reveals increased seizure susceptibility and heightened fear response in FVB/N mice. Neurobiol Dis. 43:142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Leznik E. 2007. The role of ubiquitin C-terminal hydrolase L1 in neurodegenerative disorders. Drug News Perspect. 20:365–370. [DOI] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. 2013. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 14:755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jara JH, Frank DD, Ozdinler PH. 2013. Could dysregulation of UPS be a common underlying mechanism for cancer and neurodegeneration? Lessons from UCHL1. Cell Biochem Biophys. 67:45–53. [DOI] [PubMed] [Google Scholar]
- Jara JH, Singh BB, Floden AM, Combs CK. 2007. Tumor necrosis factor alpha stimulates NMDA receptor activity in mouse cortical neurons resulting in ERK-dependent death. J Neurochem. 100:1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jara JH, Villa SR, Khan NA, Bohn MC, Ozdinler PH. 2012. AAV2 mediated retrograde transduction of corticospinal motor neurons reveals initial and selective apical dendrite degeneration in ALS. Neurobiol Dis. 47:174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. 2010. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 68:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersaitis C, Halliday GM, Xuereb JH, Pamphlett R, Bak TH, Hodges JR, Kril JJ. 2006. Ubiquitin-positive inclusions and progression of pathology in frontotemporal dementia and motor neurone disease identifies a group with mainly early pathology. Neuropathol Appl Neurobiol. 32:83–91. [DOI] [PubMed] [Google Scholar]
- Lederer CW, Torrisi A, Pantelidou M, Santama N, Cavallaro S. 2007. Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC Genomics. 8:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, et al. 1998. The ubiquitin pathway in Parkinson's disease. Nature. 395:451–452. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr 2002. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility. Cell. 111:209–218. [DOI] [PubMed] [Google Scholar]
- Lock M, Alvira M, Vandenberghe LH, Samanta A, Toelen J, Debyser Z, Wilson JM. 2010. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum Gene Ther. 21:1259–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardino AJ, Li XC, Hertel M, Nottebohm F. 2005. Replaceable neurons and neurodegenerative disease share depressed UCHL1 levels. Proc Natl Acad Sci USA. 102:8036–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, McDermott H, Landon M, Mayer RJ, Wilkinson KD. 1990. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J Pathol. 161:153–160. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Olanow CW, Halliwell B, Isacson O, Jenner P. 2001. Failure of the ubiquitin-proteasome system in Parkinson's disease. Nat Rev Neurosci. 2:589–594. [DOI] [PubMed] [Google Scholar]
- Miura H, Oda K, Endo C, Yamazaki K, Shibasaki H, Kikuchi T. 1993. Progressive degeneration of motor nerve terminals in GAD mutant mouse with hereditary sensory axonopathy. Neuropathol Appl Neurobiol. 19:41–51. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Macklis JD. 2007. Molecular development of corticospinal motor neuron circuitry. Novartis Found Symp. 288:3–15; discussion. 15–20, 96–18. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. 2007b. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 8:427–437. [DOI] [PubMed] [Google Scholar]
- Morfini GA, Bosco DA, Brown H, Gatto R, Kaminska A, Song Y, Molla L, Baker L, Marangoni MN, Berth S, et al. 2013. Inhibition of fast axonal transport by pathogenic SOD1 involves activation of p38 MAP kinase. PLoS ONE. 8:e65235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, et al. 2004. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 75:822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, et al. 2014. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 343:506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olden K, Pratt RM, Jaworski C, Yamada KM. 1979. Evidence for role of glycoprotein carbohydrates in membrane transport: specific inhibition by tunicamycin. Proc Natl Acad Sci USA. 76:791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, Li H, Sato Y, Nishikawa K, Sun YJ, Sakurai M, et al. 2003. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 12:1945–1958. [DOI] [PubMed] [Google Scholar]
- Ozdinler PH, Benn S, Yamamoto TH, Guzel M, Brown RH, Jr, Macklis JD. 2011. Corticospinal motor neurons and related subcerebral projection neurons undergo early and specific neurodegeneration in hSOD1G93A transgenic ALS mice. J Neurosci. 31:4166–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. 2001. The mouse brain in stereotaxic coordinates. San Diego: Academic Press. [Google Scholar]
- Petrucelli L, Dawson TM. 2004. Mechanism of neurodegenerative disease: role of the ubiquitin proteasome system. Ann Med. 36:315–320. [DOI] [PubMed] [Google Scholar]
- Rubino E, Rainero I, Chio A, Rogaeva E, Galimberti D, Fenoglio P, Grinberg Y, Isaia G, Calvo A, Gentile S, et al. 2012. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 79:1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, et al. 1999. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 23:47–51. [DOI] [PubMed] [Google Scholar]
- Tsaytler P, Harding HP, Ron D, Bertolotti A. 2011. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 332:91–94. [DOI] [PubMed] [Google Scholar]
- Tuffy LP, Concannon CG, D'Orsi B, King MA, Woods I, Huber HJ, Ward MW, Prehn JH. 2010. Characterization of Puma-dependent and Puma-independent neuronal cell death pathways following prolonged proteasomal inhibition. Mol Cell Biol. 30:5484–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro A, Patten SA, Aggad D, Julien C, Maios C, Kabashi E, Drapeau P, Parker JA. 2013. Pharmacological reduction of ER stress protects against TDP-43 neuronal toxicity in vivo. Neurobiol Dis. 55:64–75. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. [DOI] [PubMed] [Google Scholar]
- Walters BJ, Campbell SL, Chen PC, Taylor AP, Schroeder DG, Dobrunz LE, Artavanis-Tsakonas K, Ploegh HL, Wilson JA, Cox GA, et al. 2008. Differential effects of Usp14 and Uch-L1 on the ubiquitin proteasome system and synaptic activity. Mol Cell Neurosci. 39:539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Popko B, Tixier E, Roos RP. 2014. Guanabenz, which enhances the unfolded protein response, ameliorates mutant SOD1-induced amyotrophic lateral sclerosis. Neurobiol Dis. 71:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. 2009. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA. 106:18809–18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobst H, Forster S, Laurini C, Sekulla A, Dreiseidler M, Hohfeld J, Schmitz B, Diestel S. 2012. UCHL1 regulates ubiquitination and recycling of the neural cell adhesion molecule NCAM. FEBS J. 279:4398–4409. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Wakasugi N, Tomita T, Kikuchi T, Mukoyama M, Ando K. 1988. Gracile axonal dystrophy (GAD), a new neurological mutant in the mouse. Proc Soc Exp Biol Med. 187:209–215. [DOI] [PubMed] [Google Scholar]
- Yasvoina MV, Genc B, Jara JH, Sheets PL, Quinlan KA, Milosevic A, Shepherd GM, Heckman CJ, Ozdinler PH. 2013. eGFP expression under UCHL1 promoter genetically labels corticospinal motor neurons and a subpopulation of degeneration-resistant spinal motor neurons in an ALS mouse model. J Neurosci. 33:7890–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Guan Y, Wu X, Chen Y, Liu Z, Du H, Wang X. 2013. Wnt signaling is altered by spinal cord neuronal dysfunction in amyotrophic lateral sclerosis transgenic mice. Neurochem Res. 38:1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.