Abstract
Poly(ADP-ribosyl)ation is a reversible post-translational modification of proteins, characterized by the addition of poly(ADP-ribose) (PAR) to proteins by poly(ADP-ribose) polymerase (PARP), and removal of PAR by poly(ADP-ribose) glycohydrolase (PARG). Three PARPs and two PARGs have been found in Arabidopsis, but their respective roles are not fully understood. In this study, the functions of each PARP and PARG in DNA repair were analyzed based on their mutant phenotypes under genotoxic stresses. Double or triple mutant analysis revealed that PARP1 and PARP2, but not PARP3, play a similar but not critical role in DNA repair in Arabidopsis seedlings. PARG1 and PARG2 play an essential and a minor role, respectively under the same conditions. Mutation of PARG1 results in increased DNA damage level and enhanced cell death in plants after bleomycin treatment. PARG1 expression is induced primarily in root and shoot meristems by bleomycin and induction of PARG1 is dependent on ATM and ATR kinases. PARG1 also antagonistically modulates the DNA repair process by preventing the over-induction of DNA repair genes. Our study determined the contribution of each PARP and PARG member in DNA repair and indicated that PARG1 plays a critical role in this process.
In mammals such as human and mouse, a type of enzyme called poly(ADP-ribose) polymerase (PARP) can recognize and bind to the single or double strand DNA breaks in the genome and become activated1,2,3. PARPs use nicotinamide adenine dinucleotide (NAD+) as a substrate to attach the ADP-ribose moiety onto protein acceptors. The successive attachment of ADP-ribose residues produces long and branched poly(ADP-ribose) chains which are linked to glutamate, aspartate or lysine residues of the target proteins4, resulting in the poly(ADP-ribosyl)ation modification of proteins. PARPs are the primary substrates of themselves and the poly(ADP-ribosyl)ated (PARylated) PARPs recruit proteins important for DNA repair to the damaged sites, facilitating the DNA repair process1,5.
Later studies found that PARPs are also involved in other physiological processes, including chromatin remodelling, transcriptional regulation, ubiquitinylation regulation, spindle and centrosome function and stress granule formation4,6,7, in addition to DNA repair. PARPs are located in both the nucleus and cytoplasm8. The PARylated proteins can recruit PAR binding proteins, such as XRCC1, DNA ligase III, KU70, DNA-PK, ALC1, and APLF, and these proteins may also be PARylated by PARPs9,10.
So far, most of the knowledge about the cellular functions of poly(ADP-ribosyl)ation comes from animal systems. There are 17 PARP members in human and hPARP1 and hPARP2 are the most extensively studied4,11. They are localized in nucleus and involved in DNA repair. Other PARPs are mostly localized in cytoplasm and carry out functions other than DNA repair8. Among the hPARP proteins, only 6 are considered to be bona fide PARPs, including hPARP1 and hPARP2. Others are either mono(ADP-ribosyl) transferases or inactive proteins4,11.
Arabidopsis has three PARP members. All PARP enzymes have been shown to be located in nucleus12,13,14. Inhibition or silencing of PARPs improves abiotic stress tolerance, enhancing resistance to drought, high light, heat and oxidative stresses15,16,17, and perturbs innate immune responses to microbe-associated molecular patterns such as flg22 and elf1818, resulting in a compromised basal defense response13,19. Chemical inhibition of Arabidopsis PARP activity enhances plant growth and reduces anthocyanin accumulation20,21. PARP1 and PARP2 are involved in microhomology mediated end joining (MMEJ) during DNA repair process22, and a recent report indicated that PARP2 is the predominant PARP in Arabidopsis DNA damage and immune responses13. PARP3, unlike PARP1 and PARP2, lacks the conserved HYE triad important for PARP catalytic activity4,11, and is mainly expressed in developing seeds12. It is reported that PARP3 is necessary for maintaining seed viability during storage12. Whether it is involved in DNA repair during post-germination stage remains unknown.
PARGs catalyze the reverse reaction of poly(ADP-ribosyl)ation by breaking the ribose-ribose linkage in the ADP-ribose polymers23. PARGs are widely found in bacteria, filamentous fungi, animals and plants. In human, mouse and fly, a single PARG gene is found, which produces different isoforms by alternative splicing. These isoforms may exist in different subcellular locations and take part in different cellular processes24. Loss-of-function of PARG results in embryonic lethality in mouse and causes larval-stage death in Drosophila melanogaster25,26. In C. elegans, two PARG genes, PME-3 and PME-4 have been reported. They are mainly expressed in nerve cells. Silencing of each or both of them induces a hypersensitivity to ionizing radiations but has no obvious developmental effects27.
Two tandemly-arrayed PARG genes, PARG1 and PARG2, are found in Arabidopsis. They share 57% identity and 68% similarity in amino acid sequence. A recent report indicated that PARG1 is an active enzyme and PARG2 is not19. The PARG-signature motif GGL-X6-8-QEE is important for the PARG activity23,28. PARG1 contains the canonical PARG motif while PARG2 has an amino acid substitution in the motif, which is GGL-X6-8-QEE. They both have the tyrosine clasp, which is necessary for substrate binding in mammalian PARGs29. In addition, a G450R point mutation of Arabidopsis PARG1 beyond these two motifs has been shown to inactivate PARG1 and leads to enhanced immune gene expression19. The parg1 mutant in Arabidopsis is sensitive to the microbe-associated molecular pattern elf18 and to the DNA cross-linking agent MMC29, and also has reduced tolerance to drought, osmotic, and oxidative stresses30. Furthermore, PARG1 plays a role in regulating Arabidopsis circadian rhythm and in the photoperiod-dependent transition from vegetative growth to flowering31. So far no function has been assigned to PARG2.
Although the roles of PARP1 and PARP2 in DNA damage signaling have been reported, how PARPs and PARGs contribute to and coordinate this process remains elusive. DNA damage signals are mainly transduced by two sensor kinases: ATM (Ataxia telangiectasia mutated), which mediates double strand break (DSB) signaling, and ATR (ATM and Rad3-related), which responds to single strand breaks (SSB) and DNA replication stress32. These two kinases coordinately regulate most of the DNA damage responses in animals and plants. ATM phosphorylates SOG1 (suppressor of gamma response 1)33, which is the master transcription factor regulating DNA damage response in plants32,34 and is a functional counterpart of animal p53 although it has no structural similarity to p5332. Through activation of SOG1, cells undergo DNA repair, cell-cycle arrest, programmed cell death or endoreduplication32. It is interesting to understand how poly(ADP-ribosyl)ation affects the DNA damage response.
Emerging evidence shows that poly(ADP-ribosyl)ation may have a unique mode of action in plants12,13,29,31, but the detailed genetic and biochemical analyses are lacking to address the various phenotypes in different biological processes, especially the functional relevance between PARPs and PARGs in DNA repair. In this study, we compared the knock-out mutant phenotypes of all PARPs and PARGs in Arabidopsis under the same conditions. We found that PARG1 is the key enzyme mediating DNA damage response. It promotes cell survival under genotoxic stress and regulates the DNA damage response by avoiding over-induction of DNA repair genes.
Results
Phenotypic comparison of the loss-of-function mutants of all PARP and PARG genes in Arabidopsis
There are three PARP genes and two PARG genes in Arabidopsis. To reveal the critical gene(s) for DNA repair in poly(ADP-ribosyl)ation, we obtained all knock-out mutants of these genes from different sources. The T-DNA insertion sites in parp1, parp2 and parp3 mutants are listed in Supplementary Fig. S1. These mutants have been used in previous studies12,13,19,22. For PARG1 and PARG2, two mutants for each gene were used, respectively. parg1-4 and parg2-2 are mutants reported for the first time. Therefore, the T-DNA insertions in these mutants were confirmed by genomic PCR, and expression of the mutant allele was examined by reverse transcription (RT)-PCR (Supplementary Fig. S2). The results indicated that they are null mutants of PARG1 and PARG2, respectively.
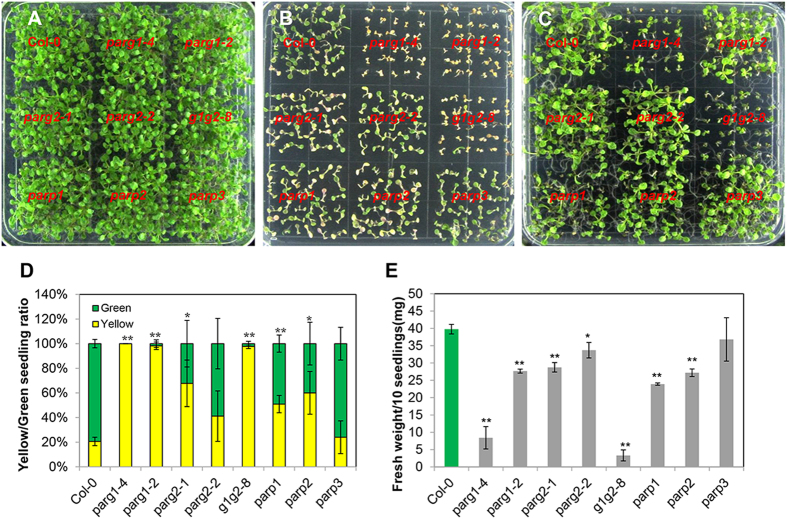
We grew these mutants on the same plate to compare their phenotypes. Two kinds of genotoxic agents, bleomycin and methyl methane sulfonate (MMS), were used in the assays, respectively. Bleomycin is a glycopeptide which mainly induces DSBs35 and MMS is a monofunctional alkylating agent that induces N-alkyl lesions and SSBs36. DSBs and SSBs can activate different DNA repair pathways37,38. DSBs are the most severe damage type since chromosomal breakage can lead to cell death32,38. On control plates, all mutants grew normal and were indistinguishable from each other (Fig. 1A). On the plate with 50 μg/ml bleomycin, parg1-2 and parg1-4 turned almost completely yellow (Fig. 1B,D), while parp1, parp2 and parg2-1 were only mildly sensitive to bleomycin compared with Col-0, and parg2-2 and parp3 responded similarly as the wild type control (Fig. 1B). MMS blocked the growth of parg1-2 and parg1-4, but did not induce yellowing (Fig. 1C), so we measured the fresh weight of each mutant and compared them with that of the Col-0 (Fig. 1E). Most of the single mutants exhibited slight sensitivity to MMS except parp3, which had no obvious phenotype, and parg1-4, which was hypersensitive to MMS. Surprisingly, another mutant allele of PARG1, parg1-2, is only slightly sensitive to MMS (Fig. 1C,E).
Figure 1. Phenotypes of the knock-out mutants of PARP and PARG genes in Arabidopsis.
(A) 1/2 MS plate. (B) 1/2 MS plate with 50 μg ml−1 bleomycin. (C) 1/2 MS plate with 100 μg ml−1 MMS. The seedlings were photographed after grown for two weeks. (D) Comparison of the yellow/green seedling numbers of each mutant grown on bleomycin plates. (E) Fresh weight per 10 seedlings of each mutant grown on MMS plates. All experiments were done for at least three times and similar results were obtained. The data were presented as means of three replicates ± SE. Significant differences (t-test) compared to Col-0 under the same conditions are indicated by asterisks: *P < 0.05; **P < 0.01.
Taken together, the results revealed that among all mutants, the parg1-4 mutant displayed the most severe phenotype while parp3 had no obvious phenotype compared to the wild type control when challenged by DNA SSB or DSB inducing agents.
Phenotypic analysis revealed the relationships between PARP and PARG family members
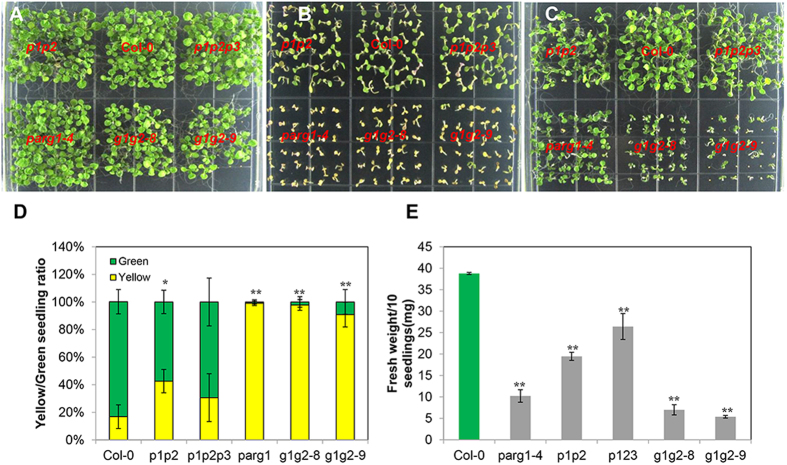
It has been reported that among the three PARPs, when PARP1 or PARP2 is absent, the expression levels of the other two PARPs are up-regulated12. We also found that when PARG1 is knocked out, the remaining PARG2 gene is up-regulated, and vice versa (Supplementary Fig. S3A). Therefore, functional redundancy might exist between the same family members. To further analyze it, we constructed parp1parp2 (p1p2) double and parp1parp2parp3 (p1p2p3) triple mutants. We were unable to generate double mutant of the closely linked PARG1 and PARG2 genes, so RNA interference was used to inhibit the expression of PARG2 in the parg1-4 mutant background (g1g2). All these mutants grew normally on 1/2 MS plate (Fig. 2A), while phenotypic comparison by bleomycin and MMS treatments revealed that the p1p2p3 triple mutant was not more sensitive than the p1p2 double mutant (Fig. 2B–E), indicating that either PARP3 does not perform the same role as PARP1 and PARP2, or is not an active enzyme. This speculation is also supported by the analysis result of the parp3 single mutant, which was not sensitive to bleomycin and MMS. Two g1g2 lines, g1g2-8 and g1g2-9, were more sensitive to bleomycin and MMS than wild type control (Fig. 2B,C). Inhibition of PARG2 expression in the parg1-4 mutant did not enhance the sensitivity of parg1-4 to bleomycin, which was already severe, but did enhance its resistance to MMS (Fig. 2B,C).
Figure 2. Phenotypic comparison of the double or triple mutants generated for the PARP or PARG gene family.
(A) 1/2 MS plate. (B) 1/2 MS plate with 50 μg ml−1 bleomycin. (C) 1/2 MS plate with 100 μg ml−1 MMS. (D) Comparison of the yellow/green seedling numbers of each mutant grown on bleomycin plates. (E) Fresh weight per 10 seedlings of each mutant grown on MMS plates. All experiments were done for at least three times and similar results were obtained. The data were presented as means of three replicates ± SE. Significant differences (t-test) compared to Col-0 under the same conditions are indicated by asterisks: *P < 0.05; **P < 0.01.
The phenotype of the g1g2 mutant was much more severe than that of the p1p2 and p1p2p3 mutants on both bleomycin and MMS plates (Fig. 2B,C), indicating a more detrimental effect of PARG mutation to DNA repair than that of PARPs mutation. Between the two PARGs, PARG1 is the major one determining plant growth and survival under bleomycin and MMS treatments. We therefore focused on the study of PARG1 and mainly used parg1-4 mutant for the following experiments due to its consistently strong phenotype on bleomycin and MMS plates. When a genomic fragment spanning from 1379 bp upstream of the translational start site to 969 bp downstream of stop codon of PARG1 was introduced into parg1-4, the bleomycin- and MMS-sensitivity phenotype of parg1-4 were rescued (Supplementary Fig. S4), demonstrating that loss of function of PARG1 caused the phenotypes. To simplify the analysis, we used bleomycin to induce DNA damages in the following experiments.
PARG1 degrades poly(ADP-ribose) in vitro
PARG is considered as the downstream enzyme of PARP, but disruption of PARG1 caused more severe phenotype than that of p1p2 double and p1p2p3 triple mutant under genotoxic stress. It raised the question whether the phenotype is directly caused by the loss of PARG activity. To answer this question, we first examined the poly(ADP-ribose) glycohydrolase activity of PARG1 in vitro using recombinant PARG1 protein. We cloned the cDNA of PARG1 and expressed PARG1 as a glutathione-S-transferase (GST) fusion protein in E. coli. The purified GST-PARG1 was used for the activity assay.
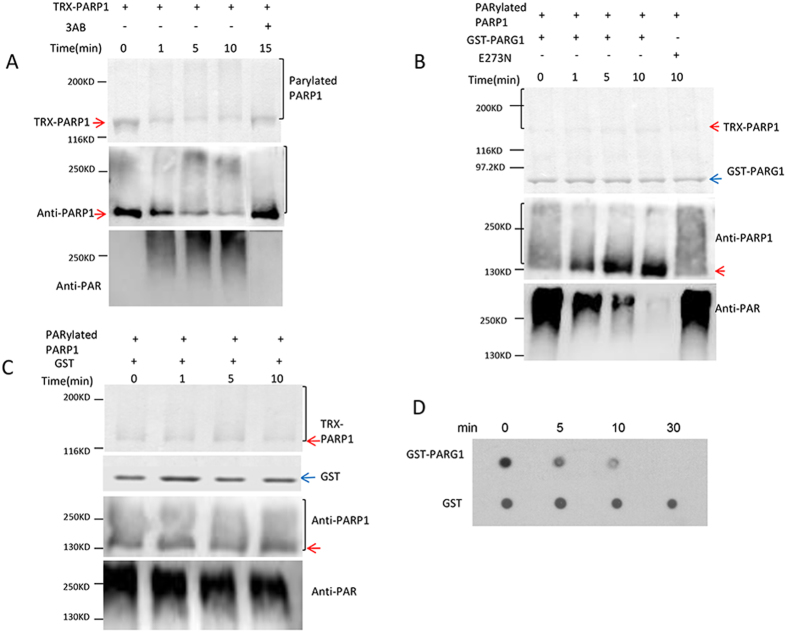
Arabidopsis PARP1 can be self-modified by adding varying numbers of ADP-riboses onto itself in the presence of broken DNA and NAD+14. The auto-PARylated protein was detected as an upwardly shifted smear on SDS-PAGE gel (Fig. 3A upper and middle panel). This auto-modified PARP1 was used as a PARylated protein substrate for the PARG1 activity assay. PAR was detected by anti-PAR antibody (Fig. 3A bottom panel). This antibody tends to recognize big ADP-ribose polymers due to their stronger immunogenicity. When the auto-modified PARP1 was incubated with the recombinant GST-PARG1 protein, the upward smear of the PARP1 disappeared and the band became concentrated again at the original size of PARP1 on the SDS-PAGE gel within one minute of adding GST-PARG1 (Fig. 3B middle panel), and PAR signal on the protein was concomitantly reduced (Fig. 3B bottom panel), indicating that PARG1 can rapidly degrade the poly(ADP-ribose) on the modified PARP1 and restore the size of the protein (Fig. 3B middle panel). For comparison, a mutant protein E273N (Glu273 → Gln) with a mutation in the conserved GGG-X6-9-QEE motif of PARG1 was generated and subjected to the same assay. The mutation completely destroyed the PAR hydrolase activity (Fig. 3B). As a control, the GST tag did not cause any mobility change of the PARylated PARP1 (Fig. 3C). When the commercial PAR was incubated with the recombinant PARG1 protein for different time periods and then detected with anti-PAR antibody, the PAR signal also disappeared gradually (Fig. 3D). These results demonstrated that PARG1 has a PAR hydrolase activity not only against PARylated proteins, but also against free PAR.
Figure 3. PARG1 has PAR-degrading activity.
(A) Generation of the PARylated PARP1 substrate. Auto-modified PARP1 protein migrated as a smeared band on SDS-PAGE gel. Anti-PARP1 and anti-PAR antibody were used for detection of the corresponding PARP1 protein and PAR on the proteins. (B) PARylated PARP1 substrate was incubated with the recombinant GST-PARG1 or mutated protein E273N for different time periods, and then subjected to SDS-PAGE analysis. Anti-PARP1 and anti-PAR antibody were used for tracking the changes of PARP1 protein and PAR on proteins. (C) PARylated PARP1 substrate was incubated with GST tag protein for different time periods, and then subjected to SDS-PAGE analysis. Anti-PARP1 and anti-PAR antibody were used for tracking the changes of PARP1 protein and PAR on proteins. (D) GST-PARG1 and GST protein were incubated with commercial PAR, and then subjected to immune-dot blot analysis by anti-PAR antibody.
Loss of PARG1 induces cell death under genotoxic stress
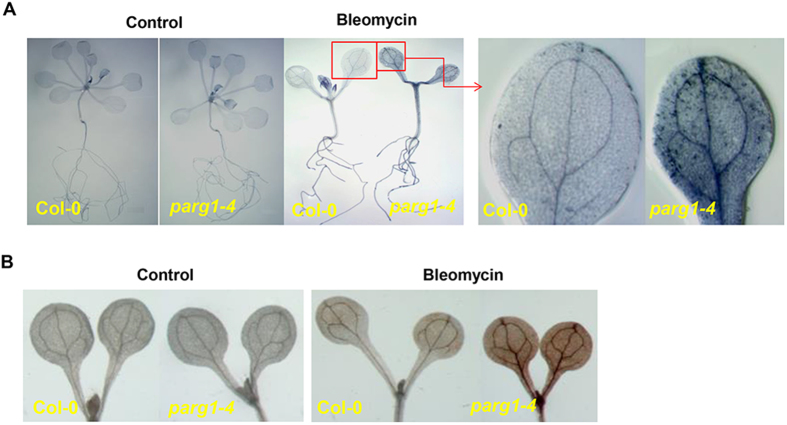
When non-lethal dose of bleomycin was included in the medium, the parg1-4 mutant grew more slowly and had fewer true leaves than wild type seedlings, reflected by a lower fresh weight of parg1 (Supplementary Fig. S5A,B). Leaf color of the parg1 mutant turned yellow gradually with the increase of bleomycin concentration (Supplementary Fig. S5C), suggesting that chlorophyll was degraded and cell death might happen. Trypan blue was used to stain the tissues for detection of cell death39. The staining results indicated that bleomycin induced more cell death in the leaves of parg1 than those of wild type (Fig. 4A). Furthermore, stronger H2O2 accumulation could be detected in the mutant cotyledons (Fig. 4B) when stained by 3, 3’-diaminobenzidine (DAB), which yields a brown precipitate in the presence of H2O240. Thus, the genotoxin treatment also led to a more oxidized status in the mutant than in wild type.
Figure 4. The parg1-4 seedling leaves show more cell death and accumulate more H2O2 than that of Col-0 seedlings under genotoxic stress.
(A) Trypan blue-stained three week-old Col-0 and parg1-4 seedlings grown on the plates without (control) or with 24 μg ml−1 bleomycin. (B) DAB stained cotyledons of one week-old Col-0 and parg1-4 seedlings grown on the plates without (control) or with 24 μg ml-1 bleomycin.
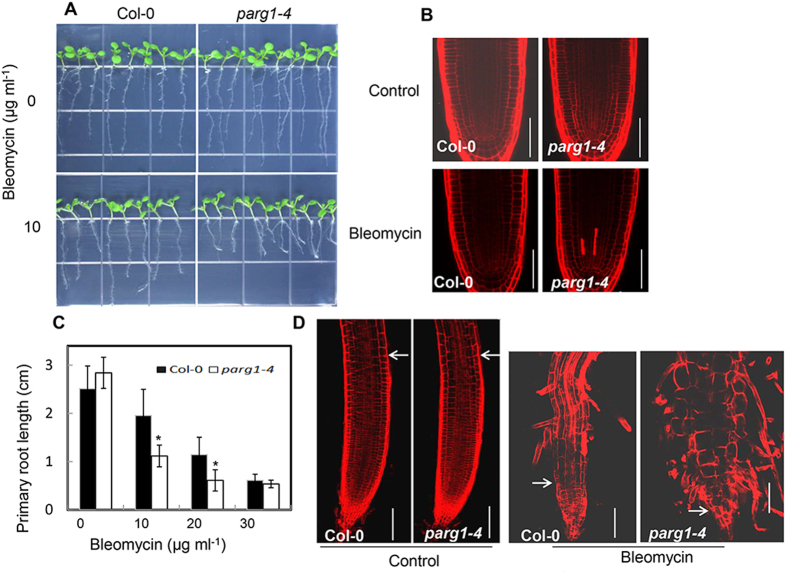
In addition, the root growth of parg1-4 was also investigated. Under normal conditions, the mutant had no significant difference from Col-0 (Fig. 5A). However, bleomycin treatment caused more cell death in the root meristem of the mutant than that of wild type, shown by propidium iodide (PI) staining (Fig. 5B). The cell death led to significant reduction of the primary root growth of the mutant compared to that of wild type (Fig. 5C). When the time period of the genotoxic stress was extended, a much shorter meristematic zone was observed at the root tip of the parg1-4 mutant (Fig. 5D), which might be due to the enhanced cell death in the parg1-4 root meristem. Meanwhile, the epidermal cells of the parg1-4 mutant were also more deformed and enlarged than that of wild type (Fig. 5D).
Figure 5. The parg1-4 mutant root is more sensitive to bleomycin than that of Col-0.
(A) Comparison of the primary root length of Col-0 and the parg1-4 mutant under normal condition and 10 μg ml−1 bleomycin treatment. (B) Propidium iodide-stained root tips of Col-0 and parg1-4 seedlings grown on plates without (control) or with 10 μg ml−1 bleomycin for 4 days. The completely reddish-stained cells indicate the dead cells. Scale bars = 75 μm. (C) Statistical analysis of the primary root lengths of Col-0 and the parg1-4 mutant grown on plates with different concentrations of bleomycin for 10 days. Significant differences (t-test) compared to Col-0 under the same conditions are indicated by asterisks: *P < 0.05; **P < 0.01. (D) Propidium iodide-stained root tips of Col-0 and parg1-4 seedlings grown on plates without (control) or with 20 μg ml−1 bleomycin for 10 days. The arrows indicate the boundary of meristematic zone at the root tip before and after bleomycin treatment. Scale bars = 100 μm.
These results indicated that disruption of PARG1 causes more cell death in both leaves and roots when the plants were treated by bleomycin.
PARG1 regulates poly(ADP-ribose) level in vivo
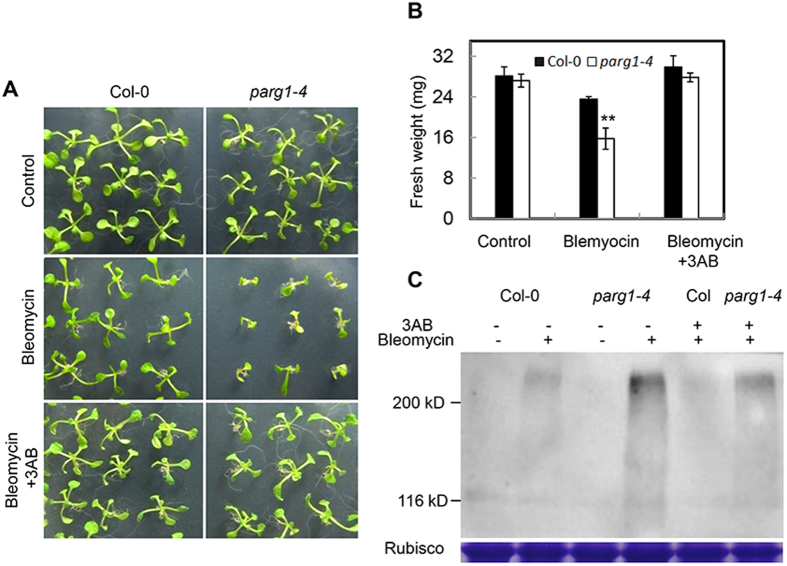
To test whether the phenotype of the parg1-4 mutant was caused by over-accumulation of poly(ADP-ribose), we added 3-AB, a cell permeable PARP inhibitor29 into the media to repress PARP activity. 3-AB reversed the phenotype of parg1-4 and restored the seedling growth of the parg1-4 mutant close to that of wild type under bleomycin treatment (Fig. 6A). In addition, the death phenotype of parg1-4 seedlings under high level of bleomycin was also rescued by 3-AB (Supplementary Fig. S6). These results suggested that PARG1 hydrolyzes PAR synthesized by PARPs in vivo and excessive PAR in the parg1-4 mutant is the reason for growth arrest and cell death under bleomycin treatment. However, parg1-4 was a little smaller than Col-0 on plates containing bleomycin and 3-AB (Fig. 6A, Supplementary Fig. S6). This may be due to incomplete suppression of PARP activity by 3-AB in vivo, or that PARG1 has other functions not affected by 3-AB.
Figure 6. The phenotype of parg1-4 is caused by the over-accumulation of PAR in vivo.
(A) The phenotype of parg1-4 can be rescued by the PARP inhibitor 3-AB. The plants were grown on 1/2 MS plate (control), or plates with 25 μg ml−1 bleomycin, and 25 μg ml−1 bleomycin plus 0.75 mM 3AB, respectively for two weeks. (B) Fresh weight per 8 seedlings of the parg1 mutant grown on the plates described in (A). Data were presented as means of three replicates ± SE. Significant differences (t-test) compared to Col-0 under the same conditions are indicated by asterisks: **P < 0.01. (C) Detection of poly(ADP-ribosyl)ated proteins in Col-0 and parg1-4 seedlings grown on plates described in (A). RuBisCo large subunit was used to indicate the protein loading amount. This experiment was repeated twice with similar results.
To further confirm the biochemical activity of PARG1 in vivo, we examined the PAR levels in parg1-4 and wild-type plants by western blot analysis using anti-PAR antibody. The results revealed that the levels of PAR were relatively low in both wild type and parg1-4 plants grown under normal conditions (Fig. 6C). When treated with bleomycin, the parg1-4 mutant accumulated more PAR than wild type, and 3-AB reduced PAR in both wild type and the parg1-4 mutant (Fig. 6C). These results once again indicated that PARG1 regulates PAR level in vivo.
PARG1 is induced primarily in mitotically active cells
Quantitative RT-PCR was performed to investigate the expression level changes of PARG1 under genotoxic stress. The transcript level of PARG1 in the seedlings subjected to a brief bleomycin treatment was examined (Supplementary Fig. S7). The results showed that PARG1 expression was induced by bleomycin. GUS staining of pPARG1::GUS transgenic line showed that PARG1 expression was initially induced in the root and shoot meristems (Supplementary Fig. S7B), then extended to other tissues.
Loss-of-PARG1 leads to the transcriptional up-regulation of DNA repair genes and increase of cellular DNA damage level
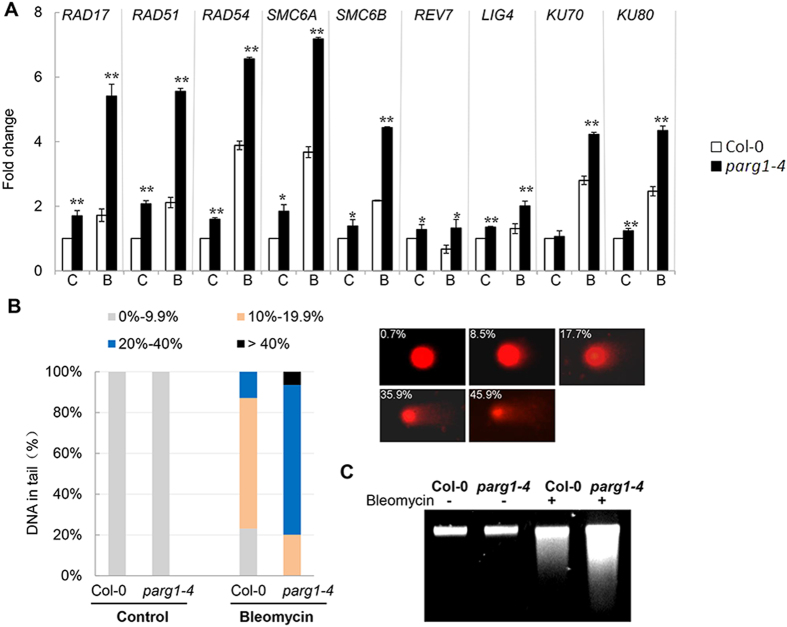
Eukaryotic organisms have two major pathways for repairing DNA double strand breaks, the homologous recombination (HR) and the non-homologous end joining (NHEJ) pathways41,42. We examined the expression levels of known genes involved in these two pathways and found that the HR pathway genes SMC6A, SMC6B43, RAD17, RAD51, RAD5444 and REV745, and the NHEJ pathway genes LIG4, KU70 and KU8046,47, were all up-regulated in the parg1-4 mutant compared to that in wild type after bleomycin treatment (Fig. 7A). These results suggested that the parg1 mutant cells might experience more DNA damage than wild type. To test this, we used the comet assay to observe DNA damages in wild type and mutant plants, where the proportion of the comet tail to the head represents the degree of DNA double-strand breaks in the nucleus48. No striking changes could be observed in the parg1-4 mutant under normal growth conditions. However, after treatment with bleomycin, the mutant showed a significantly higher level of DNA damage than wild type plants (Fig. 7B). When the genomic DNA was isolated and subjected to agarose electrophoresis, the DNA of parg1-4 migrated as a band with more smear than that of Col-0, indicating the presence of severe DNA damages in parg1-4 (Fig. 7C). Taken together, loss of function of PARG1 compromised the DNA repair process, although the DNA repair genes were still highly induced in the parg1 mutant.
Figure 7. DNA damage level is higher in the parg1-4 mutant than that in Col-0 plants under genotoxic stress.
(A) Expression level changes of genes involved in DNA double-strand break repair in roots of Col-0 and parg1-4 seedlings. The data represent the mean values of three replicates ± SD. (C) control; (B) bleomycin at 20 μg ml−1. (B) Comet assay of the DNA damage levels of Col-0 and parg1-4 seedlings grown on plates with or without (control) 20 μg ml−1 bleomycin for 10 days. The percentage of DNA in comet tails was analyzed and quantified by CASP software (http://sourceforge.net/projects/casp/) and used as an indicator of DNA damage level. 100 nuclei for each treatment were randomly selected and imaged. The bar size represents proportion of nuclei falling into the ranges of damage level indicated by different colors, and the images with percentages beside it indicate the examples of damaged nuclei. (C) DNA fragmentation assay showed that genomic DNA is more damaged in parg1-4 than in Col-0.
Induction of PARG1 gene is ATM- and ATR-dependent and PARG1 represses the transcriptional up-regulation of ATM, ATR and SOG1
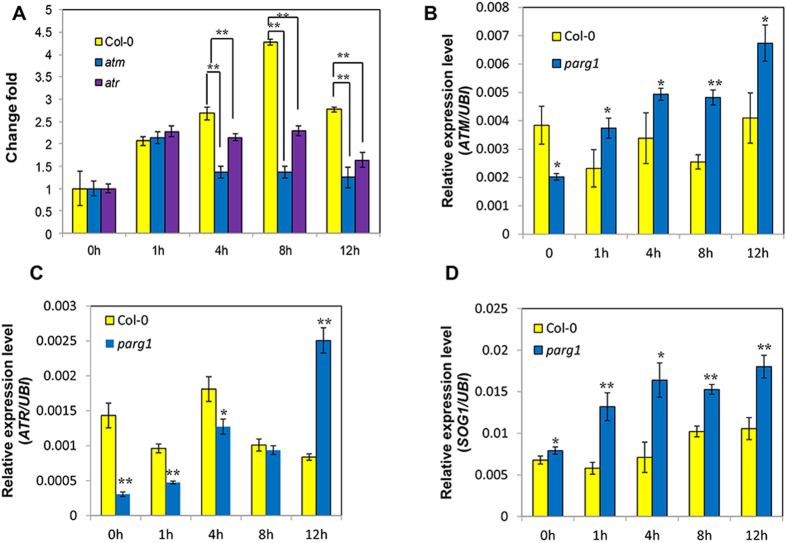
ATM and ATR are two critical kinases which transduce double and single strand break signals to DNA repair machinery, respectively32. They phosphorylate the transcription factor SOG1, which then induce the expression of DNA repair genes33,34. To understand if PARG1 participates in these two signaling pathways, we compared the expression level of PARG1 in Col-0, atm and atr mutants and found that, after bleomycin treatment, PARG1 expression levels in atm and atr mutants were obviously lower than that in wild type plants (Fig. 8A), indicating that ATM and ATR are necessary for the induction of PARG1. Surprisingly, when the transcript levels of ATM and ATR in Col-0 and parg1-4 were compared, ATM and ATR were both significantly induced in the parg1 mutant by bleomycin while not in wild type plants within the time periods we examined. The levels of ATM in the parg1 mutant at all time points were higher than those in wild type, except in the untreated plants (Fig. 8B). ATR was also induced constantly in the parg1 mutant, but the beginning level was lower than that of the wild type plants (Fig. 8C). When the expression level of the master transcription factor SOG1 was examined, it showed a similar up-regulation tendency in the parg1-4 mutant compared to Col-0 (Fig. 8D). Taken together, mutation of PARG1 caused the transcriptional up-regulation of the key factor genes ATM, ATR and SOG1 in plants, suggesting that the expression of PARG1 suppresses the induction of these genes under genotoxic stress.
Figure 8. PARG1 expression is regulated by ATM and ATR but it antagonistically represses the expression of DNA damage signaling genes.
(A) Comparison of the induction level of PARG1 in Col-0, atm and atr mutants after bleomycin treatment. The fold lines connect the columns for comparison. (B–D) Comparison of ATM, ATR and SOG1 expression levels in Col-0 and in the parg1 mutant. Seedlings grown for two weeks were treated by 50 μg ml−1 bleomycin for different time periods. The data were presented as means of three replicates ± SE. Significant differences (t-test) are indicated by asterisks: *P < 0.05; **P < 0.01.
Discussion
There is a large family of PARP genes but only one PARG gene in human and mouse25,26. The single PARG gene fulfils different functions by generating multiple isoforms24. PARG gene knock out in mouse causes embryo lethality25, indicating an essential role of PARG for embryo development. In contrast, there are two PARG genes in Arabidopsis, and the parg1 and parg2 mutants as well as parg1parg2RNAi transgenic plants all develop normally under standard growth conditions, suggesting that these genes are not critical for Arabidopsis normal development. However, under genotoxic stress, PARG1 becomes essential for plant survival. The two knock-out mutants, parg1-2 and parg1-4, are both hypersensitive to bleomycin. Loss of function of PARG1 leads to severe cell death in the mutant after bleomycin treatment, indicating a pivotal role of PARG1 in DNA DSB repair. However, only parg1-4 is highly sensitive to SSB inducing agent MMS and parg1-2 is not. This is probably due to that T-DNA in different sites interrupts the gene’s function to different extents. In parg1-4, the T-DNA is integrated between Thr311 and Gly312 residues, which are just before the Tyr313 residue in the tyrosine clasp, and the tyrosine clasp is considered important for substrate binding29. Therefore, T-DNA in the parg1-4 mutant may destroy the substrate binding site of PARG1, thus inactivates the protein. In contrast, in the parg1-2 mutant, T-DNA is integrated between Ala408 and Ser409 residues, which is far from the PARG signature motif (G262 to E274) and also the tyrosine clasp. The parg1-2 mutant may preserve residual activity of PARG1, which is enough for SSB response, but not for DSB response. As for the PARG2 gene, both mutants are slightly sensitive to MMS or bleomycin. Inhibition of PARG2 enhanced the phenotype of the parg1-4 mutant to MMS, suggesting that PARG2 does play a role, although minor, in DNA repair. A recent report indicated that PARG2 is an inactive PARG enzyme19. However, the expression data that PARG2 is induced by genotoxin (Supplementary Fig. S3B) and knock-out of PARG1 leads to a compensatory up-regulation of PARG2 expression (Supplementary Fig. S3A), together with the phenotypes of parg2 single mutant and g1g2 interference plants, all strongly support a role of PARG2 in DNA repair.
In mouse, the parp1-/-/parp2-/- double mutant, like the mouse parg mutant, is also embryo lethal49. However, the p1p2p3 triple mutant and g1g2RNAi mutant in Arabidopsis both develop normally, suggesting that poly(ADP-ribosyl)ation, different from its role in animals, does not play an essential role in Arabidopsis development under standard conditions. Under genotoxic stresses, the phenotypes of the p1p2 and p1p2p3 mutants are also weak. Although PARPs can act as detectors for DNA double or single strand breaks, there are other known DNA damage sensors, such as DSB sensor MRN (MRE11/RAD50/NBS1) complex and SSB sensors RPA protein and 9-1-1(RAD9/RAD1/HUS1) complex in plants32,44. Therefore, loss of functions of three PARPs does not cause disastrous consequence to plants. Furthermore, it is also considered that plants can bear more severe DNA damage than animals due to the immobile nature of cells and no risk for carcinogenesis32. By comparing the phenotypes of single, double and triple mutants of Arabidopsis PARP family, we conclude that PARP1 and PARP2 function in the DNA repair process at seedling stage, and PARP3 does not.
Transcriptional studies of the DNA repair genes in the parg1-4 mutant indicated that the repair pathway is activated, but not succeeded in repairing damage, resulting in the elevation of DNA damage level in the mutant. Over-accumulated DNA damage activates the cell death in the parg1 mutant, which firstly occurs in the meristems because the meristematic cells are undergoing active DNA replication, giving rise to the high sensitivity of these cells to DNA damaging agents50. PARG1 is primarily induced in root and shoot meristems, consistent with the important role of DNA repair in these cells. Besides cell death, DSBs can also induce endoreduplication in the root tips51, which is marked by an enlarged epidermal cell volume. Endoreduplication can prevent the damaged cells from propagation by undergoing DNA replication without cytokinesis. We observed a more pronounced epidermal cell size enlargement effect in the parg1-4 mutant (Fig. 5D), suggesting that the mutant also undergoes an enhanced reprogramming from mitosis to endocycle besides cell death.
Our expression data showed that PARG1 acts downstream of both ATM and ATR signaling pathways. ATM and ATR are the major kinases responsible for the activation of DNA repair pathways32,38,44,52. They regulate DNA repair process at the post-translational level by phosphorylation of the target proteins such as Chk1, Chk2 in animals and SOG1 in plants32,33. The induction of PARG1 is attenuated in both mutants, and the level is even lower in atm mutant, indicating a more pronounced effect by the absence of ATM. Interestingly, we found that the lack of PARG1 results in the expression level up-regulation of the key factors ATM, ATR and SOG1, and also other DNA damage responsive genes in plants after bleomycin treatment (Figs 7 and 8). In another word, expression of PARG1 somehow represses the induction of DNA damage responsive genes. It is recently identified in Arabidopsis that a point mutation of PARG1, which inactivates the PAR hydrolase activity, causes the overexpression of PR genes19. Therefore, PARG1 probably also regulates the amplitude of stress responses in addition to DNA damage response in plants. In animals, PARPs and PARGs can participate in stress granule formation and AGO (argonaute) proteins can be PARylated by PARPs, therefore relieves microRNA-mediated silencing of target genes53. In Arabidopsis, it remains unknown if disruption of PARG1 causes the PARylation level increase of AGOs under stress, thus alleviates the repression of stress-responsive gene by microRNA and leads to transcriptional up-regulation of these genes. Besides this, it is also possible that PAR acts as a stress signal in plants and induces a wide range of stress responses, including DNA damage response. PARG removes PAR so as to reduce the stress signaling. Poly(ADP-ribosyl)ation has been shown to be involved in biotic and abiotic responses in Arabidopsis13,15,16,19,29,30, but the molecular mechanisms are still not very clear until now.
PAR is a death signal in animal cells54, which promotes apoptosis-inducing factor (AIF) to translocate from mitochondrion to nucleus to initiate programmed cell death independent of caspase55. It is unknown whether this type of cell death exists in plants. The AIF homologue in Arabidopsis, MDAR1, is a monodehydroascorbate reductase mainly localized in peroxisome and involved in removing toxic H2O2. The MDAR genes are involved in multiple stress responses but the function in regulating cell death has not been demonstrated56,57. Our data indicated that an unknown cell death mechanism may be activated in the parg1 plants by PAR, since the parg1 mutant showed cell death phenotype after treatment with bleomycin, and a higher level of PAR is also detected in the mutant. Plants may recognize PAR as a stress signal when it is under a certain threshold but activate cell death when it exceeds the tolerance limit. A better understanding of how cell death is activated in the mutant would contribute to elucidating the molecular mechanism of cell death in plants.
Methods
Plant materials and growth conditions
All the Arabidopsis plants used in this study are of the Columbia ecotype. parp1 (GK_692A05-025067) and parp2 (SALK_140400C) were provided by de Pater Lab22. parp3 (SALK_108092C), parg1-2 (SALK_116088) and parg1-4 (SALK_012110) were ordered from ABRC (Arabidopsis Biological Resource Center, http://www.arabidopsis.org). parg2-1 (GABI_072B04) and parg2-2 (GABI_017C05) were from Nottingham Arabidopsis Seed Center (NASC, http://arabidopsis.info). The primers for identification of the new mutants parg1-4 and parg2-2 can be found in Supplementary Table S1. Plants were grown at 22 °C under long daylight conditions (12 h/8 h day/night) unless otherwise specified. For seedling phenotype observation, surface-sterilized seeds were germinated on 1/2 MS agar plates with or without bleomycin (Kayaku, Japan) and MMS (Solarbio, China), respectively.
RNA isolation and quantitative RT-PCR (qRT–PCR)
Total RNA was isolated using TRIzol Reagent (TaKaRa, Japan) and then quantified by measuring OD260. Complementary DNA (cDNA) was synthesized using PrimeScript RT-PCR Kit with gDNA eraser (Takara, Japan), and then used for qRT-PCR. PCR were performed with SyBR Premix Ex TaqTM II (TaKaRa, Japan) in a Real One Plus Real-Time PCR System (Applied Biosystem, USA), following the manufacturer’s instructions. Specific primers used can be found in Supplementary Table S1.
DNA constructs
For parg1-4 mutant complementation, a genomic DNA fragment of PARG1 was amplified using PCR (Primers are listed in Supplementary Table S2) and cloned into the Kpn I/Pst I sites of vector pZP22158.
For protein expression in E.coli, PARP1 coding sequence was cloned into pET32a vector (Novagen, USA) in frame with TRX tag using Sac I/Not I sites. PARG1 coding sequence was cloned into pGEX-4T-1 vector (GE Healthcare, UK) in frame with GST tag using EcoR I/Sal I sites (Primers are listed in Supplementary Table S2).
The RNAi construct of PARG2 was generated by amplifying the nucleotide sequence from 701 to 1140 bp downstream of the start codon of PARG2 gene, then cloning the fragment into pGEM-RNAi vector in inverted orientations to produce a hairpin fragment spanned by an intron, and finally the hairpin fragment was cloned into p35S-Fast vector58.
The construct of pPARG1::GUS was generated by amplifying 1140 bp fragment upstream of the start codon of PARG1 gene and then cloning the fragment into pAKK687 vector using Xba I and BamH I sites.
Protein expression and purification
TRX-PARP1 was produced in E.Coli Origami (DE3) (Novagen, USA) and GST-PARG1 was produced in Rosetta (DE3) (Novagen, USA). After OD600 of the culture reached 0.6, 0.3 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added into the culture and incubated at 16 °C for 20 h. Bacteria were harvested by centrifuge and then broken by ultra-sonication. The soluble proteins were used for purification. GST and GST-PARG1 were purified using the Glutathione-Agarose affinity resin (Sigma Aldrich, USA) according to the manufacturer’s instructions. TRX and TRX-PARP1 were purified using HisPur Ni-NTA resin (Thermo Scientific, USA).
Enzymatic activity assays
Preparation of auto-PARylated PARP1 substrate: Purified TRX-PARP1 and TRX proteins were dialyzed completely against 10 mM Tris·HCl pH 7.5 buffer to remove salts. 15 μg TRX-PARP1 or 3 μg TRX proteins were aliquoted into each tube containing 20 mM Tris·HCl pH 7.5, 50 mM NaCl, 7.5 mM MgCl2, 0.2 mM DTT, and 10 μM fragmented DNA. To initiate the reaction, NAD+ (Sigma Aldrich, USA) was added into each tube to a final concentration of 0.2 mM. The reactions were continued at room temperature for desired time periods and then terminated by adding 300 μl pre-chilled acetone. After centrifugation, the precipitated proteins were subjected to SDS-PAGE and visualized by Coomassie blue staining.
PARG1 activity assay on PARylated proteins: To remove contaminants from poly(ADP-ribosyl)ation reaction, the PARylated TRX-PARP1 substrate was dialyzed against pH7.0 phosphate buffer containing 1 mM 2-mercaptoethanol. Approximately 1 μg PARylated TRX-PARP1 was incubated with 0.2 μg GST or 0.5 μg GST-PARG1 for different time periods in 100 μl dialysis buffer, respectively. The reactions were terminated by the addition of 300 μl ice cold acetone. After centrifugation, the proteins in each reaction were subjected to SDS-PAGE analysis and visualized by Coomassie blue staining. Immuno-blot analysis was performed following standard protocol using anti-PAR (Abcam, UK) or anti-PARP1 antibody (Shanghai Immune Biotech, China) as the primary antibody, respectively. Enhanced chemiluminescence reagents (Thermo Scientific, USA) were used for signal detection.
PARG1 activity assay on free PAR: 0.5 μg GST-PARG1 or GST were incubated with 5 μl of 10 μM PAR (Trevigen, USA) substrate in reaction buffer (25 mM Tris pH 7.5, 150 mM NaCl) at a total volume of 20 μl for different time periods. 2 μl mixture was spotted on nitrocellulose membrane (GE Healthcare, UK). The membrane was allowed to dry at room temperature, then blocked with 1 X TBST plus 5% nonfat milk and detected with anti-PAR antibody.
Histochemistry observation
Leaf dead cell detection and H2O2 staining were carried out as described previously59. For root dead cell detection, Arabidopsis seedling roots were stained with 10 μg ml−1 propidium iodide (Sigma Aldrich, USA). Excitation and emission wavelength were set as 535 nm and 617 nm, respectively for fluorescence microscopy.
Chlorophyll determination and comet assay
The chlorophyll concentration of seedlings was measured as described60. The comet assay was performed based on the method reported before48.
DNA fragmentation assay
One week old seedlings were treated with 10 μg ml−1 bleomycin for 48 hrs. Genomic DNA was extracted using the ChargeSwitch gDNA Plant Kit (Invitrogen, USA) and resolved in agarose gel. Each lane was loaded with 100 ng DNA.
Immuno-blot analysis of PAR in plants
Arabidopsis seedlings were collected and ground in liquid nitrogen, then boiled in 1XSDS-PAGE loading buffer for 5 min and centrifuged. The supernatant was subjected to SDS-PAGE, transferred onto nitrocellulose membrane, and then detected with anti-PAR antibody. Equal loading was monitored by staining the membrane with Coomassie blue solution.
Additional Information
How to cite this article: Zhang, H. et al. Arabidopsis PARG1 is the key factor promoting cell survival among the enzymes regulating post-translational poly(ADP-ribosyl)ation. Sci. Rep. 5, 15892; doi: 10.1038/srep15892 (2015).
Supplementary Material
Acknowledgments
This work was supported by the grants to X.G. (grant no. 31070232 and 31170169) from the National Natural Science Foundation of China. We also would like to thank Prof. de Pater at Leiden University for providing parp1 and parp2 mutant seeds, and thank Dr. Ping Xu at University of East Anglia and Dr. Jeremy D. Murray in John Innes Centre for manuscript revision. Dr. Xinsheng Gu also gives helpful suggestions for the revision of the manuscript.
Footnotes
Author Contributions X.G. conceived the study and designed the experiments. H.Z., Z.G., Q.W., L.Y. and C.L. performed the experiments. Y.X. and H.M. gave very helpful suggestions to the manuscript. X.G. wrote the manuscript. All authors have read and approved the manuscript.
References
- Huber A., Bai P., de Murcia J. M. & de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst) 3, 1103–1108 (2004). [DOI] [PubMed] [Google Scholar]
- Willmitzer L. Demonstration of in vitro covalent modification of chromosomal proteins by poly(ADP) ribosylation in plant nuclei. FEBS Lett 108, 13–16 (1979). [DOI] [PubMed] [Google Scholar]
- Luo X. & Kraus W. L. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Gene Dev 26, 417–432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B. A. & Kraus W. L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Bio 13, 411–424 (2012). [DOI] [PubMed] [Google Scholar]
- Swindall A. F., Stanley J. A. & Yang E. S. PARP-1: Friend or Foe of DNA Damage and Repair in Tumorigenesis? Cancers (Basel) 5, 943–958 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisch T., Ame J. C., Dantzer F. & Schreiber V. New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem Sci 37, 381–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A., Todorova T., Ando Y. & Chang P. Poly(ADP-ribose) regulates post-transcriptional gene regulation in the cytoplasm. RNA Biol 9, 542–548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S., Chesarone-Cataldo M., Todorova T., Huang Y. H. & Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun 4, 2240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N. et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc Natl Acad Sci USA 106, 4243–4248 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J. P. et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36, 6959–6976 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K. L. Poly(ADP-ribose): An organizer of cellular architecture. J Cell Biol 205, 613–619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissel D., Losch J. & Peiter E. The nuclear protein Poly(ADP-ribose) polymerase 3 (AtPARP3) is required for seed storability in Arabidopsis thaliana. Plant Biol (Stuttg) 16, 1058–1064 (2014). [DOI] [PubMed] [Google Scholar]
- Song J. Q., Keppler B. D., Wise R. R. & Bent A. F. PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses. Plos Genet 11, 10.1371/pgen.1005200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E. et al. Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant Journal 15, 635–645 (1998). [DOI] [PubMed] [Google Scholar]
- Amor Y., Babiychuk E., Inze D. & Levine A. The involvement of poly(ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett 440, 1–7 (1998). [DOI] [PubMed] [Google Scholar]
- De Block M., Verduyn C., De Brouwer D. & Cornelissen M. Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J 41, 95–106 (2005). [DOI] [PubMed] [Google Scholar]
- Tian R., Zhang G. Y., Yan C. H. & Dai Y. R. Involvement of poly(ADP-ribose) polymerase and activation of caspase-3-like protease in heat shock-induced apoptosis in tobacco suspension cells. FEBS Lett 474, 11–15 (2000). [DOI] [PubMed] [Google Scholar]
- Ishikawa K. et al. Modulation of the Poly(ADP-ribosyl)ation Reaction via the Arabidopsis ADP-Ribose/NADH Pyrophosphohydrolase, AtNUDX7, Is Involved in the Response to Oxidative Stress. Plant Physiology 151, 741–754 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. M. et al. Protein Poly(ADP-ribosyl)ation Regulates Arabidopsis Immune Gene Expression and Defense Responses. Plos Genet 11, 10.1371/pgen.1004936 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz P. et al. Chemical PARP inhibition enhances growth of Arabidopsis and reduces anthocyanin accumulation and the activation of stress protective mechanisms. Plos One 7, 10.1371/pone.0037287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz P. et al. Poly(ADP-Ribose)Polymerase Activity Controls Plant Growth by Promoting Leaf Cell Number. Plos One 9, 10.1371/pone.0090322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q., den Dulk-Ras A., Shen H., Hooykaas P. J. & de Pater S. Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant Mol Biol 82, 339–351 (2013). [DOI] [PubMed] [Google Scholar]
- Slade D. et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477, 616–620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J. P., Hendzel M. J., Droit A. & Poirier G. G. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol 18, 145–151 (2006). [DOI] [PubMed] [Google Scholar]
- Koh D. W. et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci USA 101, 17699–17704 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai S. et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci USA 101, 82–86 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Laurent J. F., Gagnon S. N., Dequen F., Hardy I. & Desnoyers S. Altered DNA damage response in Caenorhabditis elegans with impaired poly(ADP-ribose) glycohydrolases genes expression. DNA Repair (Amst) 6, 329–343 (2007). [DOI] [PubMed] [Google Scholar]
- Kim I. K. et al. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nature structural & molecular biology 19, 653–656 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Phillips L., Briggs A. G. & Bent A. F. Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol 152, 267–280 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. J. et al. Arabidopsis poly(ADP-ribose) glycohydrolase 1 is required for drought, osmotic and oxidative stress responses. Plant Sci 180, 283–291 (2011). [DOI] [PubMed] [Google Scholar]
- Panda S., Poirier G. G. & Kay S. A. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the arabidopsis circadian oscillator. Dev Cell 3, 51–61 (2002). [DOI] [PubMed] [Google Scholar]
- Yoshiyama K. O., Sakaguchi K. & Kimura S. DNA damage response in plants: conserved and variable response compared to animals. Biology (Basel) 2, 1338–1356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K. O. et al. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO reports 14, 817–822 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K., Conklin P. A., Huefner N. D. & Britt A. B. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106, 12843–12848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Ghorai M. K., Kenney G. & Stubbe J. Mechanistic studies on bleomycin-mediated DNA damage: multiple binding modes can result in double-stranded DNA cleavage. Nucleic Acids Res 36, 3781–3790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor P. J. Interaction of chemical carcinogens with macromolecules. J Cancer Res Clin Oncol 99, 167–186 (1981). [DOI] [PubMed] [Google Scholar]
- Caldecott K. W. Single-strand break repair and genetic disease. Nat Rev Genet 9, 619–631 (2008). [DOI] [PubMed] [Google Scholar]
- Waterworth W. M., Drury G. E., Bray C. M. & West C. E. Repairing breaks in the plant genome: the importance of keeping it together. New Phytol 192, 805–822 (2011). [DOI] [PubMed] [Google Scholar]
- Strober W. Trypan Blue Exclusion Test of Cell Viability. in Current Protocols in Immunology (John Wiley & Sons, Inc., 2001). [DOI] [PubMed] [Google Scholar]
- ThordalChristensen H., Zhang Z. G., Wei Y. D. & Collinge D. B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant Journal 11, 1187–1194 (1997). [Google Scholar]
- San Filippo J., Sung P. & Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77, 229–257 (2008). [DOI] [PubMed] [Google Scholar]
- Hiom K. Coping with DNA double strand breaks. DNA Repair (Amst) 9, 1256–1263 (2010). [DOI] [PubMed] [Google Scholar]
- Watanabe K. et al. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21, 2688–2699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S. E. & Jackson S. P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25, 409–433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S. et al. Roles of arabidopsis AtREV1 and AtREV7 in translesion synthesis. Plant Physiology 138, 870–881 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A. et al. Suppression of Ku70/80 or Lig4 leads to decreased stable transformation and enhanced homologous recombination in rice. New Phytol 196, 1048–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Adachi Y., Chiba K., Oguchi K. & Takahashi H. Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant J 29, 771–781 (2002). [DOI] [PubMed] [Google Scholar]
- Olive P. L. & Banath J. P. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 1, 23–29 (2006). [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J. et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J 22, 2255–2263 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N. & Sablowski R. Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA 106, 20984–20988 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi S. et al. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc Natl Acad Sci USA 108, 10004–10009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K. M., Robertson C. E., Foreman J., Doerner P. & Britt A. B. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48, 947–961 (2006). [DOI] [PubMed] [Google Scholar]
- Leung A. K. et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42, 489–499 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi S. A. et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA 103, 18308–18313 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. W. et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297, 259–263 (2002). [DOI] [PubMed] [Google Scholar]
- Yamada T., Ichimura K., Kanekatsu M. & van Doorn W. G. Homologs of genes associated with programmed cell death in animal cells are differentially expressed during senescence of Ipomoea nil petals. Plant & cell physiology 50, 610–625 (2009). [DOI] [PubMed] [Google Scholar]
- Cande C., Cecconi F., Dessen P. & Kroemer G. Apoptosis-inducing factor (AIF): key to the conserved caspase-independent pathways of cell death? Journal of cell science 115, 4727–4734 (2002). [DOI] [PubMed] [Google Scholar]
- Ge X. et al. AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol 145, 204–215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M. E. et al. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784 (1998). [DOI] [PubMed] [Google Scholar]
- Arnon D. I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol 24, 1–15 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.