ABSTRACT
The zebrafish, Danio rerio, is a powerful model for studying bacterial colonization of the vertebrate intestine, but the genes required by commensal bacteria to colonize the zebrafish gut have not yet been interrogated on a genome-wide level. Here we apply a high-throughput transposon mutagenesis screen to Aeromonas veronii Hm21 and Vibrio sp. strain ZWU0020 during their colonization of the zebrafish intestine alone and in competition with each other, as well as in different colonization orders. We use these transposon-tagged libraries to track bacterial population sizes in different colonization regimes and to identify gene functions required during these processes. We show that intraspecific, but not interspecific, competition with a previously established bacterial population greatly reduces the ability of these two bacterial species to colonize. Further, using a simple binomial sampling model, we show that under conditions of interspecific competition, genes required for colonization cannot be identified because of the population bottleneck experienced by the second colonizer. When bacteria colonize the intestine alone or at the same time as the other species, we find shared suites of functional requirements for colonization by the two species, including a prominent role for chemotaxis and motility, regardless of the presence of another species.
IMPORTANCE
Zebrafish larvae, which are amenable to large-scale gnotobiotic studies, comprehensive sampling of their intestinal microbiota, and live imaging, are an excellent model for investigations of vertebrate intestinal colonization dynamics. We sought to develop a mutagenesis and tagging system in order to understand bacterial population dynamics and functional requirements during colonization of the larval zebrafish intestine. We explored changes in bacterial colonization dynamics and functional requirements when bacteria colonize a bacterium-free intestine, one previously colonized by their own species, or one colonized previously or simultaneously with a different species. This work provides a framework for rapid identification of colonization factors important under different colonization conditions. Furthermore, we demonstrate that when colonizing bacterial populations are very small, this approach is not accurate because random sampling of the input pool is sufficient to explain the distribution of inserts recovered from bacteria that colonized the intestines.
INTRODUCTION
Vertebrate intestinal microbial communities are complex assemblages that have important roles in the health and development of the host. The rules governing the assembly of these communities are only beginning to be understood. From a broad perspective, host-associated microbial community assembly involves both stochastic and deterministic processes (1, 2). Deterministic contributions include host-microbe interactions and within- and between-species microbial interactions. The bacterial genes responsible for such interactions can be elucidated with the use of transposon-mediated, signature-tagged mutagenesis (STM) screens, and more recently with the use of high-throughput sequencing adaptations that sequence the genomic sequence adjacent to the transposon insertion site (3–7). Stochastic factors, such as random sampling of environmental microbes, play an important role in colonization of the vertebrate intestine yet are not often addressed when interpreting the results of genetic screens aimed at uncovering bacterial genes important for colonization and competition.
The zebrafish, Danio rerio, is a model vertebrate that is increasingly being used to understand the assembly of vertebrate intestinal microbial communities (8, 9). The larval zebrafish intestine has a key advantage for the study of microbial community assembly in that it is very small for a vertebrate intestine, facilitating the interrogation of the entire gut microbiota from many individuals. Due to their high fecundity and ex utero embryonic development, large numbers of germ-free zebrafish larvae can be reared with relative ease (10). Additionally, their microbiota is dominated by gammaproteobacteria, such as Vibrio and Aeromonas species, that are readily culturable (9, 11, 12), facilitating experimental manipulations such as mono- and diassociations of germ-free zebrafish. Another strength of the model is the optical transparency of the larval zebrafish, allowing real-time imaging of intestinal colonization (13, 14). Previous work imaging defined Pseudomonas aeruginosa flagellar mutants has shown the importance of flagellum-directed motility for colonizing the zebrafish intestine (15). At present, it is unclear whether this extends to chemotaxis-directed motility, which has been shown to be important for colonization of the mouse gastrointestinal tract by Vibrio cholerae (16, 17) and Salmonella enterica serovar Typhimurium (18).
Bacterial populations colonizing intestines can undergo filtering based not only on their functional capacity for colonization or virulence but also on bottlenecks due to available space or stochastic sampling of the environment by the host. This can affect the interpretation of transposon insertional mutagenesis screens particularly when the colonizing population size is much smaller than the input library of mutants. A recent large-scale screen to identify Vibrio fischeri genes required for colonization of the small Hawaiian bobtail squid, Euprymna scolopes, acknowledged this issue and focused instead on a subset of their mutant library, which was reproducibly recovered but depleted at a defined cutoff from hundreds of colonized animals (19). Another recent Vibrio transposon insertion site sequencing study using a Vibrio cholerae rabbit intestinal infection model reported small population bottlenecks by tracking the loss of predicted neutral insertions (<5% of insertions lost) (20). Others have specifically addressed population size changes due to bottlenecks during single-pathogen infections by tracking transposon insertions (21, 22) and distinct sequence tags inserted into otherwise wild-type bacterial strains (23, 24). Particularly during intestinal colonization, population bottlenecks can also be affected by the presence of previously colonized bacteria during the phenomenon known as colonization resistance (reviewed by Lawley and Walker [25]), whereby an intestine colonized by a diverse microbiota may reduce or eliminate colonization by a pathogen. Similarly, it was recently shown that mice monoassociated with a single intestinal Bacteroides species are resistant to colonization by the same species, but not different species (26).
In the current study, we apply a high-throughput sequencing, TraDIS (transposon-directed insertion site sequencing) (6) type approach for identification of zebrafish intestinal colonization factors in an Aeromonas strain and a Vibrio strain and identify shared and distinct functional requirements in the two species. We additionally use the transposon insertion sites as tags to identify changes in the minimum colonization bottleneck size under different colonization regimes.
RESULTS
Transposon insertion site mapping in Aeromonas and Vibrio reveals colonizing population size changes and the significance of intraspecific competition in shaping zebrafish intestinal communities.
In order to identify genetic factors contributing to the colonization of the zebrafish intestinal tract, we developed a TraDIS-type system that accurately maps transposon insertion site junctions and could be used in bacteria isolated from zebrafish. The larval zebrafish intestinal microbiota contains a large proportion of Proteobacteria, with the Aeromonas and Vibrio genera being part of a core set found in zebrafish from multiple geographic locations (9, 11, 27). Therefore, we used the Tn5 transposon to mutagenize bacterial strains because it has a broad host range, it has been shown to work in Proteobacteria (28), and it has a very low insertion site bias (29, 30). Tn5 has been previously used in Aeromonas veronii Hm21 (31). This strain reverses many germ-free traits in monoassociated zebrafish (32) and has a near-complete draft genome (33); thus, we used this strain as an Aeromonas representative. For a representative Vibrio species, we have sequenced and assembled a draft genome of a zebrafish-derived Vibrio strain (ZWU0020), which exhibits motility, hemolytic activity, and resistance to polymyxin B (9). Based on gene content, including full-length 16S rRNA gene sequence, this Vibrio strain appears to be closely related to non-O1/O139 species of Vibrio cholerae and lacks genes encoding cholera toxin and toxin-coregulated pilus.
In addition to the standard method of tracking changes in the input-to-output ratio of transposon insertions, tagged bacteria such as those in our insertion libraries can be used to track changes in the relative colonization bottleneck size. Identification of the transposon-associated genomic insertion sites (TAGs) gives a genomic sequence that can be used as a marker for each unique transposon insertion in a single-species population (Fig. 1A). Although transposon libraries of greater than 105 insertions can be generated (6), libraries of such high complexity may not be completely sampled by larval zebrafish which contain approximately 105 CFU total in their intestines (32), a population which can arise from the expansion of a smaller number of founder cells (14). We therefore generated Tn5 insertion libraries in both Aeromonas and Vibrio with a complexity that is less than the total population size of the larval zebrafish intestine. In order to accurately determine the input population size and assess the reproducibility of our library preparation and read filtering methods, we sequenced three replicates of each tagged species that would serve as the inoculum. After filtering reads with low-quality mapping scores and TAGs not detected in one of the triplicate inoculum samples, 1,508 and 5,809 TAGs were detected in the input inoculums, representing insertions in 575 and 1,930 annotated genes in the Aeromonas and Vibrio genomes, respectively. We found that the relative abundance of TAGs was highly correlated between technical replicates (R2 greater than 0.99 for all six pairwise comparisons) (Fig. 1B), reflecting reproducible detection of TAGs and their relative abundances using our library preparation and sequence filtering methods.
FIG 1 .
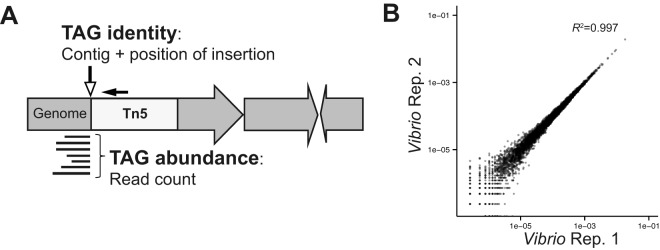
Transposon insertion site detection and reproducibility. (A) Graphical representation of a TAG in a gene and the relation of read counts to the abundance of a TAG. A TAG is defined by its insertion site whose coordinates are given by the contig and location on the contig. The small solid black arrow illustrates the position of the target for the oligonucleotide used to enrich for transposon-containing fragments, targeting inside the end of the transposon. (B) Representative plot of the correlation of a pair of input technical replicate TAG sequencing libraries after quality filtering. The axes are the detected relative abundances in each replicate (Rep.).
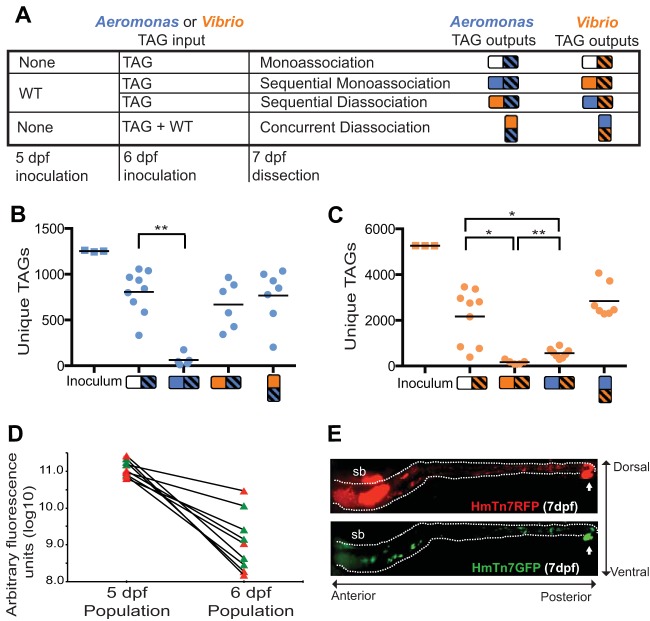
We first used these TAG populations of bacteria to track changes in a population’s colonizing bottleneck size. We generated intestinal conditions with and without other bacteria in order to explore the effect of intra- and interspecific interactions on bottleneck size. To determine the initial colonization bottleneck size of TAG libraries for each bacterial species in a monoassociation situation, zebrafish were exposed to an input TAG library of a single bacterial species at 6 days postfertilization (dpf) and then dissected after 24 h. Intestinal output libraries were then prepared for sequencing as described in Materials and Methods (Fig. 2A; see Text S1 in the supplemental material). We determined the average numbers of TAGs in the intestine after 24 h of exposure to the inoculum to be 805 and 2,167 for Aeromonas and Vibrio, respectively (Fig. 2B and C), after rarefying all samples to the minimum number of reads found within each group of samples containing the same tagged species. These numbers are below the total number of TAGs in each inoculum input (1,252 and 5,264) and below the 104 to 105 CFU per intestine previously reported to be found with Aeromonas monoassociations (14, 32). These observations suggest successful colonization by a subset of the bacteria in the population followed by expansion within the intestine. However, the abundance of TAGs is not evenly distributed in the input, so we cannot exclude the possibility that this subset of colonizing TAGs represents the full complement of bacterial cells initially colonizing the intestine
FIG 2 .
The previous state of colonization affects the ability of transposon-tagged libraries to colonize the zebrafish intestine. (A) Table showing the experimental situations where transposon-tagged libraries were exposed to zebrafish intestines at 6 days postfertilization (dpf) with different colonization states. Shapes used to represent different experimental conditions in all figures are shown, where solid squares represent WT bacterial populations and striped squares represent TAG libraries. (B and C) Abundance of unique TAGs detected among Aeromonas (B) and Vibrio (C) libraries in differentially colonized zebrafish. Each symbol represents the value for an individual zebrafish intestine, except for “inoculum,” which represents technical replicates of the same input libraries. The mean of each group of zebrafish is indicated by a short black line. Values that are significantly different are indicated by the bar and asterisk as follows: *, P < 0.01; **, P < 0.001. (D) The fluorescence intensity of two marked WT Aeromonas strains (HmTn7RFP [red] and HmTn7GFP [green]) is compared in nine fish where the second strain was given at 6 dpf to mimic the TAG experiments shown in panel B. The first marked strain allowed to colonize was swapped in four of the fish (so lines can connect red and green points or green and red points) to control for possible differences due to the incorporated fluorescent-protein-coding gene. Intensities have been scaled to reflect differences in brightness of the two colors. (E) Representative light sheet microscopy images of a zebrafish intestine colonized first by RFP-labeled bacteria (at 5 dpf) and then by GFP-labeled bacteria (at 6 dpf) and imaged at 7 dpf. This intestine had a particularly large amount of the second strain such that both can be seen well. D The intestine (inside the white dotted lines), vent (white arrows), and swim bladder (sb), which reflects fluorescence to some extent, are indicated.
We next asked whether the colonization bottleneck size changed when the same species of bacteria had already been established in the intestine. We used the same TAG input inocula to inoculate the flask media of germ-free 6 dpf zebrafish that had previously been exposed to wild-type (WT) populations of each species for 24 h (Fig. 2A). The results showed that the TAG richness of the second tagged population of bacteria was drastically reduced when the wild-type population was already present (Fig. 2B and C). This pattern was consistent for the Aeromonas and Vibrio strains assayed, suggesting that the initial population had filled the effective colonization space for each species and that competition between the WT and TAG population of the same species was occurring or that host factors were somehow limiting further colonization.
The greatly reduced ability of the second population of Aeromonas to colonize was verified independently using light sheet microscopy imaging of bacteria labeled with green fluorescent protein (GFP) (HmTn7GFP) and dTomato (HmTn7RFP [RFP stands for red fluorescent protein]) within the intestine, using the same time frame of bacterial exposure and inoculum densities used in the transposon tagging experiments. The fluorescently labeled bacterial strains harbor Tn7 insertions containing the genes for the different fluorescent proteins and antibiotic cassettes but are otherwise isogenic wild-type Aeromonas. Regardless of which labeled strain was introduced first, the second strain was present at lower levels in the intestine (Fig. 2D and E), consistent with results from the TAG experiment (Fig. 2B and C) in which bacterial populations harbored many random insertions. Imaging the bacteria in the intestine also demonstrates that the entire physical space within the intestine is not colonized by Aeromonas, suggesting that these results are not simply due to a lack of physical space for the second strain to colonize.
We also asked whether a similar pattern was observed when a different species had already established itself in the intestine. The two species may compete for the same spatial or biochemical niche or could occupy distinct niches within the intestine. The same TAG libraries were used to inoculate fish that had previously been colonized by the other wild-type species for 24 h. Interestingly, the two species showed different responses to the presence of the other species already being present. While the presence of an established Vibrio population had no significant effect on the TAG richness of the ensuing Aeromonas population relative to its monoassociation, the Vibrio TAG richness was significantly smaller when Aeromonas was already present. However, the number of Vibrio TAGs was still significantly larger than when wild-type Vibrio had been allowed to establish itself first (Fig. 2C). These results indicate that although some interspecific competition is likely occurring, intraspecific competition is more important in establishing intestinal populations for these two species.
If interspecific competition has a smaller effect on establishing intestinal populations of the two species, then equivalent access for both species to the intestine should result in similar sized colonization of the TAG populations. In agreement, when the two species were inoculated at the same time, the average TAG richness values of the tagged species were not significantly different from their respective monoassociation values (Fig. 2B and C). This result suggests that the functional requirements for colonization in monoassociation and concurrent diassociations may be similar.
A binomial sampling model of transposon mutant populations reveals that random sampling is sufficient to explain differences in transposon abundance in small populations.
In accordance with the most common use of transposon insertion libraries, we wanted to use our transposon tagging system to determine the functional requirements for colonization of the zebrafish intestine in the context of different colonization states of the host. However, we wondered whether some of the small TAG populations observed could simply be a random sampling of the available TAGs in the input. Therefore, we asked whether the distributions of TAGs in the intestines were different from the expectation based on a random sampling of the inoculum, approximated by a binomial distribution. Importantly, with our experimental setup, we can detect the presence and relative levels of all the TAGs in the input inoculum as well as the output intestinal community, due to our ability to sample the entire intestine. The binomial likelihood of the observation for each individual TAG in the intestinal output is based only on its relative level in the input inoculum (the probability of sampling that TAG), its presence in the intestine (a successful sampling of that TAG), and the total number of TAGs in the intestine (the number of observations) and does not factor in any effect of the insertion on colonization fitness. The joint log likelihood of observing the specific population of TAGs in an individual intestine is then the sum of the log likelihoods of all the TAGs in that intestine. To determine whether an individual intestinal population’s likelihood is different from what we would expect based on binomial sampling, we calculated the joint log likelihoods of 9,999 simulated populations for each intestine from a random binomial draw using the same number of TAGs and inoculum relative levels and then compared the observed log likelihood of each intestinal TAG population to the distribution of simulated communities.
To test the validity of this approach, we generated an experimental setup where we predicted that the TAG populations in the fish would be representative of subsamples of the input population. We reduced the inoculum concentration to only 100 CFU ml−1 (1,000-fold less than the other experiments) and found that some of the fish in the flask remained germ free, a situation in which we predicted that the few tagged bacteria that successfully colonized larval fish intestines did so only because of their high level in the inoculum, and thus, the higher probability of being sampled, in the input inoculum. These low-input intestinal TAG populations were not significantly different from their simulated populations of binomial draws (Table 1; see Fig. S1 in the supplemental material), confirming the utility of this approach for detecting intestinal populations that represent a simple environmental sampling.
TABLE 1 .
TAG populations significantly different from binomial draw-simulated populations
| Experimental situation | Species TAG population result significantly different from the binomial draw-simulated population resulta |
|
|---|---|---|
| Aeromonas | Vibrio | |
| Low-input monoassociation | No | NA |
| Monoassociation | Yes | Yes |
| Sequential monoassociation | No | Mixed |
| Sequential diassociation | Yes | Yes |
| Concurrent diassociation | Yes | Yes |
The Aeromonas and Vibrio TAG population results were compared to those of the binomial draw-simulated population result. The results of the comparison are shown as follows: Yes, the results were significantly different (P < 0.05); No, the results were not significantly different (P < 0.05); NA, not available; Mixed, mixed results.
Within each experimental situation, there was variation in the degree of deviation from the mean of the simulated population, but most groups revealed a consistent pattern of significance. All of the observed monoassociations and simultaneous diassociations were significantly different (P < 0.05) from their respective simulation distributions, indicating that random sampling is not sufficient to approximate them (Table 1; see Fig. S1 in the supplemental material). In contrast, nearly all the intestines from both Aeromonas and Vibrio experiments in which the same wild-type species was allowed to colonize prior to exposure to the TAG inoculum were not significantly different from the simulated random populations. Taken together, these results indicated that TAG populations from fish with the same species of bacteria already present were likely to reflect only random samples of the TAG inoculum (and therefore are not useful for identifying transposon insertions that decrease the competitive fitness of bacteria under these conditions), while monoassociated TAG populations and those with exposure to a different species experienced some selection of a subset of the TAGs in the input pool.
Functional requirements for colonization of the germ-free zebrafish intestine in monoassociation.
Having identified experimental situations that were likely to have resulted in the selection of a subset of tagged bacteria, we next asked which genetic factors in Aeromonas and Vibrio were required for colonization of the zebrafish intestine. Larval zebrafish were exposed to TAG libraries for only 24 h because we wanted to identify factors involved in colonization processes rather than long-term survival within the intestine. We first identified all differentially abundant TAGs from the two different species in monoassociation experiments across all fish in replicate flasks exposed to the same TAG inoculum for each species. After applying a multiple testing correction (34), we found that 7% (105) of the Aeromonas TAGs and 15% (851) of the Vibrio TAGs were significantly underrepresented (q < 0.05) in the intestines of monoassociated fish, representing insertions in 57 and 523 genes, respectively (see File S1 in the supplemental material). We reasoned that even over the short 24-h period since inoculation, the inoculated bacterial populations in each flask could diverge and impose different selection pressure on the set of TAGs present; therefore, we also analyzed each group of three fish from a single flask independently. This resulted in a high-confidence set of 25 genes from Aeromonas and 278 genes from Vibrio, which were consistently found to be reduced in intestines relative to the input inoculum (File S1).
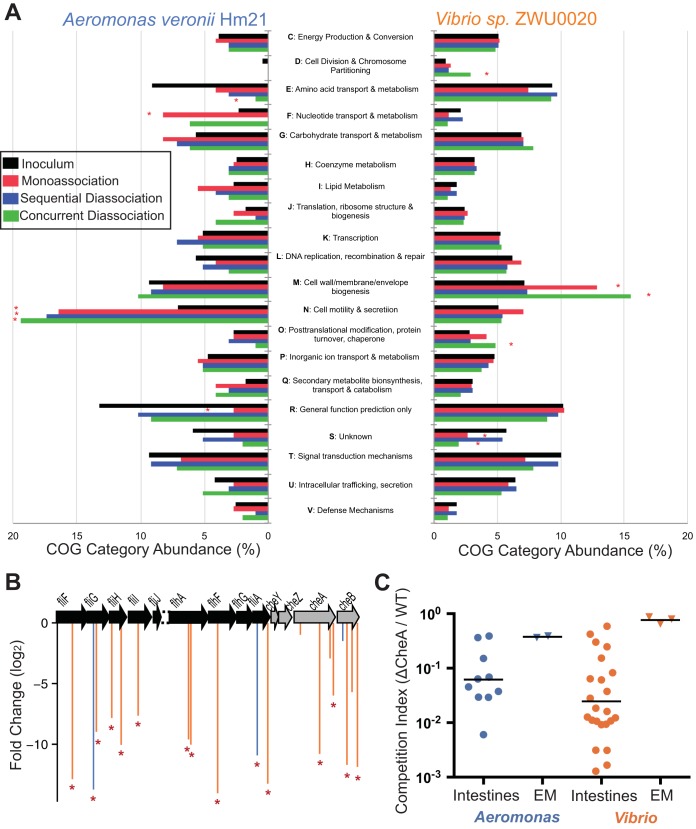
We then compared the distributions of the COG (cluster of orthologous groups) category annotations of all of the intestinally underrepresented TAGs to the distribution of categories of TAGs present in the inoculums of the two different species (Fig. 3A). Among the Vibrio TAGs, category M (cell wall/membrane/envelope biogenesis) was the only broad functional class of genes we found to be enriched among those reduced in intestines. Genes required for lipooligosaccharide synthesis are the main drivers of the observed depletion of intestinal TAGs in this category, particularly those required for the presence of O antigen, as well as genes involved in the synthesis of outer membrane proteins OmpA, OmpT, and OmpU. OmpU has been shown to be important for proper biofilm formation and resistance to bile salts during rainbow trout intestinal colonization by Vibrio anguillarum (35), illustrating the ability of these transposon libraries to recapitulate known fish colonization factors.
FIG 3 .
Key functional requirements during colonization of the zebrafish intestine. (A) COG category representation among the TAG data sets of the two species. Each COG category is shown as a percentage of all the annotated COGs within each data set. Black bars represent the input inoculum for each species. COG categories in experimental situations (monoassociations, concurrent diassociations, and sequential diassociations) that are significantly differently abundant are indicated with asterisks. (B) A cluster of motility genes with conserved synteny between Aeromonas and Vibrio. The locations of TAGs found in this region (as a percentage of each open reading frame for mapping onto the same coordinates in both species) and their log2 fold change in the intestine relative to inoculum are depicted. TAGs that were significantly depleted from intestines are indicated with asterisks. The break or dashed region indicates a lack of conserved synteny due to the presence of genes on two contigs in the Vibrio assembly. (C) The competition index (CI) of defined cheA null mutants in Vibrio and Aeromonas relative to the WT shows the reduction in the ability of this strain to colonize the germ-free zebrafish intestine. Data are from two and three replicate experiments for Aeromonas and Vibrio, respectively. EM, embryo medium.
The COG categories F (nucleotide transport and metabolism) and N (cell motility and secretion) were significantly (q < 0.05) increased in abundance among the Aeromonas TAGs that were reduced in the intestines, identifying an important role for these processes during intestinal colonization. While the cell motility and secretion COG category did not pass our threshold for significance among the Vibrio TAGs, we still observed an increase in this category in the intestinally reduced TAGs, and manual examination of the gene lists (see File S1 in the supplemental material) showed that many of the largest fold changes were in these genes. The majority of the intestinally reduced genes in this category were flagellar biosynthesis and control genes, but additional COGs in this category included genes specifically involved in chemotaxis. Many of the chemotaxis and flagellar genes have duplicates present in at least two distinct clusters in the genomes of our Aeromonas and Vibrio strains, as has also been described for Aeromonas hydrophila (36). One of these clusters with conserved synteny between the two species is shown in Fig. 3B, along with the fold change of TAGs in this region, illustrating the shared importance of motility in the two species.
As chemotaxis has previously been shown to be important for movement toward carp gut mucus by A. hydrophila (37), we were surprised by the lack of significantly depleted chemotaxis genes in the Aeromonas set and wondered whether this absence indicated that this process was less important for colonization than the presence of flagella or whether this was simply due to a low number of insertions in Aeromonas chemotaxis genes in our input transposon libraries. However, we note that two insertion sites in Aeromonas flagellar genes (fliG and fliA) upstream of the chemotaxis genes had reduced representation in monoassociated intestines (Fig. 3B) and could have polar effects on the chemotaxis genes. CheA is a key cytoplasmic signaling molecule involved in the relay of chemotactic signals from chemoreceptors to the flagellar motor. We therefore constructed an Aeromonas strain and a Vibrio strain with a marked deletion of the cheA gene that was shown to be important in the Vibrio set of TAGs (cheA2). Neither cheA mutant strain had a strong growth defect in LB medium (log phase doubling times of 33.6 min for wild-type Aeromonas and 32.9 min for the Aeromonas cheA mutant and 53.8 min for the WT Vibrio and 56.2 min for the Vibrio cheA2 mutant), but both showed a classic chemotactic defect on soft-agar swim plates (see Fig. S2 in the supplemental material). We then tested the ability of each cheA null strain to colonize the germ-free zebrafish in competition with their respective wild-type counterpart. We found that the absence of CheA had a profound effect on the ability of both species to colonize the germ-free zebrafish intestine with a >10-fold competitive disadvantage relative to the wild type (Fig. 3C). This result confirmed the importance of chemotaxis in Aeromonas and Vibrio during colonization of the intestine, but it also showed that our Aeromonas transposon insertion library may miss some critical functions due to its relatively low complexity of insertions.
Several other key functional requirements for colonization became apparent when we looked at differential abundance of individual genes instead of binning them into the broad COG functional categories. Some of these functions, such as N-acetylglucosamine (GlcNAc) metabolism, are shared between Aeromonas and Vibrio and are distributed among the COG categories M (cell wall/membrane/envelope biogenesis) and G (carbohydrate metabolism and transport). The enzyme N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25) was found to be important in the data sets of both species. This enzyme functions in the first step in the metabolism of GlcNAc resulting from import into the cytosol via a phosphotransferase system (PTS) transporter in the Vibrionaceae (38), and the subsequent enzyme in this metabolic cascade (glucosamine-6-phosphate deaminase) was also identified in the Aeromonas set of required genes. Related genes found in the Vibrio data set include two predicted chitinases and chiS, the chitin catabolic cascade sensor histidine kinase, as well as additional GlcNAc metabolism genes not found in Aeromonas and a GlcNAc-responsive transcriptional regulator (nagC).
Concurrent diassociation functional requirements are similar to monoassociation functional requirements.
Since mixed-species experiments also appeared to involve some selection from the TAG input pool in simulations, we next set out to identify the sets of intestinally depleted TAGs in those experimental situations where another species was already present or colonizing at the same time as the tagged species. We also asked to what extent the functions important for colonization in these diassociation scenarios differed from the functions important in the previously described monoassociation situations. On one extreme, if the two species have complete niche separation and do not compete at all, we would expect that the genes important for colonization would remain the same regardless of the presence of the other species before or during colonization. Alternatively, the presence of another species could result in a shift in the genes required for colonization due to multiple overlapping mechanisms of selection on the bacterial populations.
We first identified all the significantly underrepresented TAGs in the intestines in the same manner as for the monoassociation data set. We compared the COG category representations of the intestinally depleted set to the COG category representation in each species inoculum as before. At this coarse level of analysis, the concurrent diassociations had the same overrepresented COG categories as the monoassociation experiments of each species (Fig. 3A). This suggested a similar colonization process as in the monoassociation situation and is in line with the lack of changes in colonizing population sizes when looking at TAG richness in these situations. When another species was already present in the intestines, there was no significantly overrepresented COG category among the intestinally depleted TAGs, which is largely a result of the smaller number of genes that are significantly different under these conditions, failing to make significant changes in the COG category bins.
To obtain a finer view of differences in functional profiles between the TAG populations of different species colonizing different intestinal conditions, we looked at the change in the relative levels of COGs corresponding to TAGs that were depleted in the intestines in any of the experimental situations. To compare functional profiles in the two species, we first clustered the TAGs by COGs and averaged their fold changes and then trimmed this set to only those that had an ortholog in both species. The results, shown in Fig. 4A as a heatmap of hierarchical clustering of differential abundant COGs, showed that when the two species were allowed to colonize at the same time, the set of required functions was similar to those required during colonization in monoassociation experiments. Additionally, while there was an overlapping set of COGs required in all experiments (both species) and within-species experiments, some sets of genes are important only when the other species is present. The clustering of COGs from the monoassociation experiments with concurrent diassociation experiments (Fig. 4A) suggests that the two species have distinct functional preferences that drive their colonization of the zebrafish intestine. This is consistent with the results from the colonization bottleneck size analysis that suggested that interspecies competition is less important than intraspecies competition at these early time points.
FIG 4 .
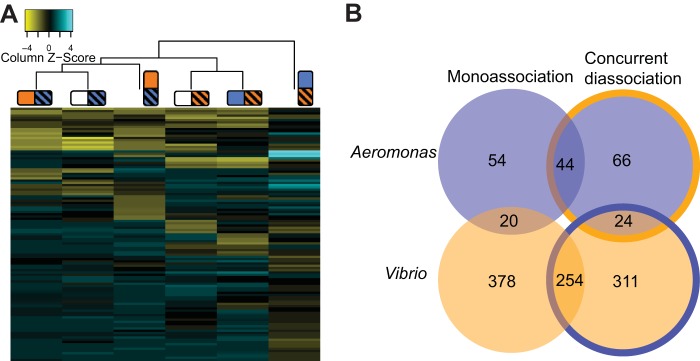
COGs important for colonization by both species show that similar functions are required in monoassociation and concurrent diassociation of the two species. (A) Heatmap and hierarchical clustering of all the COGs that were present in both species and detected as significantly depleted from intestines in any experiment. The dendrogram is based on complete linkage clustering of the distance matrix of log2 fold changes of significant, shared COGs. COGs are colored by Z-score values within an experimental group, so that negative values for COGs are less than the mean and positive values are more than the mean within that experiment. Experimental groups cluster first by tagged species but also share some of the same enriched or depleted COGs. (B) Diagram showing the degree of overlapping COGs required for colonization by each species in monoassociation and concurrent diassociations, as well as the overlap between species, which is constrained by COGs found in both genomes. Outline colors on the circles to the right represent the presence of the other species during concurrent diassociations.
Because the monoassociations and concurrent diassociations had highly similar sets of functional requirements for colonization, the overlap between these data sets in each species provides a high-confidence set of genes that are important for colonizing the germ-free zebrafish intestine. In the data set for each species, there was a large degree of overlap between the identified COGs in mono- and diassociation experiments (Fig. 4B). The overlap between important COGs in both species is constrained by the smallest set of identified genes with COG annotations in both species (in this case, the Aeromonas set) but identifies functions that are important across species. We identified a set of 14 COGs representing 14 and 18 genes in Aeromonas and Vibrio, respectively, which were important in all monoassociations and diassociations (see Table S1 in the supplemental material). These COGs represented high-confidence, shared functional requirements for colonizing the zebrafish intestine and include the N-acetylglucosamine-6-phosphate deacetylase and flagellar genes found in the monoassociation experiments.
DISCUSSION
High-throughput sequencing now allows rapid identification of large libraries of transposon insertions and the rapid generation of draft genome sequences from environmental bacterial isolates. Coupled with the large abundance of culturable bacteria from the larval zebrafish intestine, this offers great potential for the identification of shared and distinct bacterial genes and processes involved in the colonization of this model vertebrate’s intestine by many different species. In this study, we have shown the utility of transposon insertion site libraries for identification of genetic determinants of colonization of the zebrafish intestine and extended the use of these tagged populations of bacteria to address the dynamics of intestinal colonization.
In our simple two-species model, we find that monoassociated larval zebrafish are largely resistant to colonization by the same species but can be colonized by another species. However, the identity of the first species to colonize the intestine differentially affected the size of the population that could subsequently colonize the intestine, with initial Aeromonas colonization having a greater reduction on the ability of Vibrio to colonize in the subsequent 24-h period. This suggests that when there is no competition with another species, Aeromonas rapidly expands within the intestine to fill other available niches and outcompeting Vibrio for subsequent colonization, while Vibrio occupies a distinct, at least partially nonoverlapping, niche in both situations. Competition between these two species could be for spatial and/or biochemical niches within the intestine. In the future, a combination of imaging and host genetic manipulation to alter the biochemical niches should prove useful in addressing the relative influence of both restrictions during vertebrate intestinal colonization.
The clear population bottlenecks that occur during colonization of the zebrafish intestine are also likely to be factors governing colonization of much larger vertebrate intestines due to limited niche availability for particular bacterial species, and recent methods have been developed to detect and describe population bottlenecks in tagged bacterial species assayed by high-throughput sequencing methods (22, 24). Here we have shown that modeling the colonization of the intestine as a simple binomial draw from the inoculum can be used to identify experimental situations that are not notably different from a random sampling process (as in a population bottleneck) and should not be subjected to differential abundance analysis because selection is not likely to have strongly shaped the output population. The sampling model used in this study is intentionally simplistic because of our lack of knowledge of growth dynamics within the intestine, yet it is a useful first approximation for determining whether further analysis of genetic requirements is warranted. Further refinement of these models will make them more useful for modeling host colonization dynamics, as will more-refined libraries consisting of only known neutral TAGs. The creation of an archived array of transposon insertion clones, as has been done with other transposon insertion libraries (5), could allow directed subsampling of libraries and the identification of subsets of neutral insertions, which could be used to more accurately track absolute colonizing population sizes alone and in complex communities with different combinations of tagged species. However, we have shown here that such arrayed archives are not a prerequisite for the use of transposon libraries in obtaining meaningful information about population dynamics and broad functional requirements of intestinal colonization.
In this study, we confirmed the importance of motility for colonization of the zebrafish intestines by strains of Vibrio and Aeromonas, as was previously known for Pseudomonas (15). We also showed that cheA was important in establishing intestinal populations of Vibrio and Aeromonas, suggesting that it is not just the presence of flagellar motility or adhesion mediated by flagella, as has been shown for Aeromonas caviae (39), but also chemotactic behavior that is important for the colonization of zebrafish. Methyl-accepting chemotaxis proteins (MCPs) involved in transducing chemotactic signals are the second-most-abundant COG in both our Aeromonas and Vibrio genomes. Upon further inspection of transposon insertion in MCPs within the input inoculums, we detected 72 TAGs distributed among 29 of the 32 annotated Vibrio MCPs and 8 TAGs among 8 of the 41 annotated Aeromonas MCPs. Of these MCPs, only one MCP (in Vibrio) was identified as being significantly depleted from the intestines of monoassociated zebrafish. This observation is in line with the suggestion that MCPs serve redundant functions in these species or that multiple chemotactic signals are involved in colonization and an inability to sense a single signal might be complemented at the population level.
The identification of genes that were important for colonization by both species in both monoassociation and concurrent diassociation situations, such as those involved in GlcNAc metabolism and resistance to antimicrobial peptides, suggests that these genes are likely to encode components of mechanisms of general, rather than species-specific, importance during initial colonization of the zebrafish intestine. Further studies of multiple genes involved in these processes involving clean deletions followed by complementation are needed to verify the importance of these pathways during colonization of the zebrafish intestines. For example, multiple GlcNAc metabolism-related genes were identified in both species as being important for colonization, and GlcNAc derived from fish intestinal mucus might potentially be an important nutrient source for colonizing bacteria. However, as GlcNAc metabolism and modifications are important for many physiological processes in bacteria, it remains to be directly shown whether this specific sugar’s catabolism from host-derived sources is important for colonization or whether dysregulation of other bacterial pathways is the cause of the colonization defects. Notably, we also identified many hypothetical proteins and genes of general functional prediction only, showing that even in these relatively well-characterized gammaproteobacteria, a significant portion of the genome is yet to be functionally characterized with regard to colonization. The process of colonization of the fish intestine by bacteria is itself a multifaceted process involving processes external to the animal as well, such as bacterial competition and proliferation within the water, positioning within the water column, and potentially, movement toward the fish. Similarly, some mammalian intestinal pathogens must survive in an aqueous environment to be effectively transmitted and colonize a new host. A recent transposon insertional mutagenesis study looked for the genetic requirements for V. cholerae survival in rabbit hosts and a freshwater pond environment and found a more prominent role for flagellar biosynthesis and cell wall and outer membrane biogenesis genes in freshwater (20). These results highlight the notion that some of the functional requirements for colonization that we detected may have been due to defects in survival and positioning within the environment rather than direct host interactions during colonization. Future studies addressing intestinal colonization factors should consider the loss or enrichment of transposon insertions in water alone and in preconditioned fish water to obtain a more comprehensive picture of intestinal colonization by bacteria. Additionally, comparison of the colonization gene requirement profiles of these two species with other common zebrafish microbiota members will provide insight into the relative importance of functions in each species and shared processes across many diverse taxa.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
We modified a Tn5-containing pRL27 vector (28) to include an FseI restriction site inside one of the transposon ends in order to increase the annealing temperature and stringency of the targeting oligonucleotide in the modified TraDIS (transposon-directed insertion site sequencing) method (6) we used to identify transposon insertion sites for this study. The resulting pRL27F plasmid was initially electroporated into Escherichia coli EC100D-pir+ cells (Epicentre), reisolated, and electroporated into E. coli BW20767 cells (40) for conjugation with the host species. Liquid cultures of E. coli strains were grown aerobically at 37°C with shaking at 200 rpm in LB medium with 50 µg/ml kanamycin (Kan) for maintenance and 150 µg/ml for selection. Aeromonas veronii Hm21R was isolated previously (41) and provided by Joerg Graf. Vibrio sp. strain ZWU0020 was isolated from adult zebrafish intestinal contents and provided by John F. Rawls. A spontaneous rifampin (Rif)-resistant strain was isolated from a wild-type (WT) culture of Vibrio by plating on 100 µg/ml Rif. Liquid cultures of Aeromonas veronii Hm21 and Vibrio sp. strain ZWU0020 were grown aerobically at 30°C with shaking at 200 rpm in LB medium with 100 µg/ml rifampin.
Targeted deletion of cheA in Aeromonas and Vibrio strains.
Targeted deletion of cheA in Aeromonas was accomplished by allelic exchange. A targeting construct consisting of a kanamycin resistance cassette with 5′ and 3′ flanking homologous regions of the cheA gene was assembled by overlap extension and cloned into the pDMS197 (42) suicide plasmid, which contains a tetracycline resistance gene and the sacB1 counterselection marker. The resulting allelic exchange plasmid was transformed into the donor E. coli strain BW20767 for conjugation with Aeromonas. Conjugations were incubated at 37°C on LB agar for ~5 h before being picked and plated onto LB plates containing rifampin and 10% sucrose to select for Aeromonas mutants that had undergone complete allelic exchange. Verification of tetracycline sensitivity and sequencing of the targeted locus were done to confirm Aeromonas ΔcheA mutants. Similar methods were used in the engineering of the Vibrio ΔcheA2 mutant derivative with slight modification. The targeting construct containing a kanamycin resistance gene flanked by >500 bp of sequence homologous to the 5′ and 3′ regions flanking cheA2 was cloned into the alternative suicide vector pGP704sac28 (43). This vector carries a gene that encodes resistance to ampicillin along with the sacB gene, which allowed more-efficient screening of merodiploids for complete allelic exchange events. Chemotaxis defects were verified for the mutants of both strains by stabbing 1 µl of an overnight culture into soft-agar swim plates (TSA [tryptic soy agar] with 0.2% agar) (see Fig. S2 in the supplemental material) and incubating Aeromonas at 30°C for 5 h and Vibrio at room temperature overnight.
Derivation, inoculation, and sequencing of transposon insertion site libraries.
The E. coli BW20767 strain harboring the Tn5-containing pRL27F plasmid was conjugated in multiple replicates with the host strain (Vibrio or Aeromonas) overnight at 30°C by spotting mixtures of the two strains on LB agar. Conjugated bacteria were picked with sterile loops and resuspended in liquid LB medium, and multiple dilutions were plated on LB agar with Kan (150 µg/ml) and Rif (100 µg/ml). Control conjugations of only one strain (host or donor) were included to ensure that antibiotic selection was effective. Selective plates were incubated overnight at 30°C, and multiple plates with good separation of colonies were scraped and resuspended in 3 ml of LB broth. The mix of transposon-tagged bacteria was then split into three 50-ml cultures in order to reduce stochastic loss of transposon-associated genomic insertion sites (TAGs) and grown with Kan and Rif. These cultures represented an initial negative selection against insertions in essential genes. The replicate cultures were combined, and a portion was used to inoculate germ-free fish and extract DNA for sequencing libraries of the input inoculum. Fifty embryos from a single wild-type (WT) parental (strain AB/Tü) cross were derived germ free as previously described (10) and raised in 50 ml of sterile embryo medium in tissue culture flasks. Multiple fish among three replicate flasks were analyzed for monoassociation conditions, while multiple fish from a single flask were analyzed per each diassociation condition. All bacterial inoculations were given at a density of 105 CFU ml−1 in the surrounding fish water. Intestines were dissected and homogenized in 200 µl sterile embryo medium with a pellet pestle motor (catalog no. 749540-0000; Kimble-Chase) and pestle (see Text S1 in the supplemental material) at 7 days postfertilization (dpf) and briefly cultured to an optical density at 600 nm (OD600) of less than 1.0 in 1 ml of LB broth with Kan in order to increase the ratio of bacterial to zebrafish DNA used as the template in subsequent PCRs. Cultures were then pelleted, and genomic DNA was extracted using Qiagen DNeasy columns and the protocol for Gram-negative bacteria. Details on the preparation of Illumina sequencing libraries from genomic DNA for identification of transposon insertion sites are provided in Text S1, and the oligonucleotide sequences used are provided in Table S2.
Sequence read filtering, mapping, and identification of differentially abundant TAGs.
Raw Illumina reads were first quality filtered using the fastx_toolkit by end trimming reads at a quality score of less than 25 and initially discarding reads shorter than 60 bases, then reads with an overall average quality of less than 20 over 90% of their bases were discarded. Quality sequences were then filtered based on the presence of the target sequence and outside end (OE) sequence from the transposon to verify that the reads came from a sequence adjacent to a Tn5 insertion site. After trimming of transposon sequences, filtering out adapter sequence contamination and sequences shorter than 20 bases, the remaining genomic sequence of each read was mapped to the draft genomes using Bowtie 2 (44). For additional quality control, we filtered out alignments that had poor (<10) mapping quality scores because we found that at this cutoff, >96% of these alignments were derived from less than 10 reads, and we suspected that they may have been misalignments due to the draft status of our genome assemblies and sequencing error. The majority of alignments were of very high quality despite the draft status of the genomes (see Fig. S3 in the supplemental material). Alignment files were imported into an SQL database and queried to group reads from the same TAG and retrieve read counts. A unique identifier for each TAG was then assigned, and tables of abundances in each sample were imported into R for further analysis. For identification of differentially abundant TAGs, we used the R Bioconductor package edgeR (45), and estimates of common dispersion within each intestinal output and inoculum input sample comparison to identify significant changes in their relative levels (after Benjamini-Hochberg multiple testing correction).
TAG richness of populations, binomial sampling simulations, and statistical analysis.
Tables of counts of TAGs by sample are identical in structure to a community matrix of species by sites. We made use of the R package Vegan to rarefy each experiment’s table of counts to the minimum number of TAGs detected in each set of experiments and then determine the total TAG richness in each sample. Binomial sampling simulations were carried out within R using rarefied tables and the binomial distribution set of functions to generate binomial probabilities and random deviates for simulated distributions as described above. To test the significance of differences in COG category representation shown in Fig. 3A, we employed a binomial test using the relative abundance of each COG category annotation in the input inoculum TAG set as the probability of detecting a given COG category and then applied the Benjamini-Hochberg multiple hypothesis testing correction.
Light sheet microscopy.
Light sheet microscopy methods, fluorescent bacterial strains, and fluorescent intensity measurements were used as previously described (13). Briefly, germ-free fish were inoculated with 105 CFU/ml of a given labeled strain at 5 dpf, followed by inoculation with the second labeled strain at 6 dpf. Fish were rinsed in sterile media and anesthetized prior to mounting and imaging in low-melting-point agarose beginning 24 h after inoculation with the second strain.
SUPPLEMENTAL MATERIAL
Identified differentially abundant genes (q < 0.05) in each experimental situation with log fold change, q value, and functional category annotations. Genes that were detected as being significantly different in all three replicate flasks when analyzed independently in monoassociation experiments are indicated by yellow highlighting. Download
Detailed protocol of the modified TraDIS method used for preparation of transposon insertion site sequencing libraries Download
(A) Plot of P values from comparison of individual TAG intestinal populations and binomial sampling-simulated population. The dashed gray line indicates a P value of 0.05. The shapes on the x axis correspond to each experimental situation as outlined in the legend to Fig. 2A. “Low-input” denotes the Aeromonas TAG input population that was inoculated at a density of 3 orders of magnitude smaller than the others. (B) The joint log likelihood of observed “low-input” intestinal samples (red X’s) are shown against their distribution of binomial draw-simulated TAG populations. Tukey boxplots show the distribution of joint log likelihoods of 9,999 simulated TAG populations for each sample. Download
Soft-agar swim plates verify chemotaxis defects in cheA targeted deletions. (A) Aeromonas (B) Vibrio sp. strain ZWU0020. Download
Graph of the average mapping quality score for each raw TAG (after sequence quality filtering) versus their read number. The TAGs are colored by species. Download
COGs of shared importance between Vibrio and Aeromonas in monoassociation and concurrent diassociation.
Oligonucleotides used in this study.
ACKNOWLEDGMENTS
We thank Joerg Graf and Lindsey Bomar for providing Aeromonas strains and assembling the Aeromonas contigs.
Research reported in this publication was supported by NSF IGERT fellowship DGE 9972830 (to W.Z.S.) and the National Institute of General Medical Sciences of the National Institutes of Health under grants R01GM095385 and P50GM098911.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Citation Stephens WZ, Wiles TJ, Martinez ES, Jemielita M, Burns AR, Parthasarathy R, Bohannan BJM, Guillemin K. 2015. Identification of population bottlenecks and colonization factors during assembly of bacterial communities within the zebrafish intestine. mBio 6(6):e01163-15. doi:10.1128/mBio.01163-15.
Contributor Information
Andrew Goodman, Yale University.
Richard Losick, Harvard University.
REFERENCES
- 1.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJM. 21 August 2015. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J doi: 10.1038/ismej.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, Schmidt TM. 2015. Application of a neutral community model to assess structuring of the human lung microbiome. mBio 6:e02284-14. doi: 10.1128/mBio.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hensel M, Shea J, Gleeson C, Jones M, Dalton E, Holden D. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400–403. [DOI] [PubMed] [Google Scholar]
- 4.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A 106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langridge GC, Phan M-D, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong S, Stephens WZ, Burns AR, Stagaman K, David LA, Bohannan BJM, Guillemin K, Rawls JF. 2015. Ontogenetic differences in dietary fat influence microbiota assembly in the zebrafish gut. mBio 6(5):e00687-15. doi: 10.1128/mBio.00687-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens WZ, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJM. 4 September 2015. The composition of the zebrafish intestinal microbial community varies across development. ISME J doi: 10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milligan-Myhre K, Charette JR, Phennicie RT, Stephens WZ, Rawls JF, Guillemin K, Kim CH. 2011. Study of host-microbe interactions in zebrafish. Methods Cell Biol 105:87–116. doi: 10.1016/B978-0-12-381320-6.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. 2011. Evidence for a core gut microbiota in the zebrafish. ISME J 5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantas L, Sørby JRT, Aleström P, Sørum H. 2012. Culturable gut microbiota diversity in zebrafish. Zebrafish 9:26–37. doi: 10.1089/zeb.2011.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taormina MJ, Jemielita M, Stephens WZ, Burns AR, Troll JV, Parthasarathy R, Guillemin K. 2012. Investigating bacterial-animal symbioses with light sheet microscopy. Biol Bull 223:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jemielita M, Taormina MJ, Burns AR, Hampton JS, Rolig AS, Guillemin K, Parthasarathy R. 2014. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio 5:e01751-14. doi: 10.1128/mBio.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. 2007. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc Natl Acad Sci U S A 104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler SM, Camilli A. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci U S A 101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millet YA, Alvarez D, Ringgaard S, von Andrian UH, Davis BM, Waldor MK. 2014. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog 10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt W-D.. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol 10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- 19.Brooks JF, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, Whistler C, Goodman AL, Mandel MJ. 2014. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A 111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. 2013. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog 9:1–11. doi: 10.1371/journal.ppat.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troy EB, Lin T, Gao L, Lazinski DW, Camilli A, Norris SJ, Hu LT. 2013. Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infect Immun 81:2347–2357. doi: 10.1128/IAI.00266-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim CH, Voedisch S, Wahl B, Rouf SF, Geffers R, Rhen M, Pabst O. 2014. Independent bottlenecks characterize colonization of systemic compartments and gut lymphoid tissue by Salmonella. PLoS Pathog 10:e1004270. doi: 10.1371/journal.ppat.1004270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abel S, zur Wiesch PA, Chang H-H, Davis BM, Lipsitch M, Waldor MK. 2015. STAMP: sequence tag-based analysis of microbial population dynamics. Nat Methods 12:223–226. doi: 10.1038/nmeth.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawley TD, Walker AW. 2013. Intestinal colonization resistance. Immunology 138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2013. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 29.Canals R, Xia X-Q, Fronick C, Clifton SW, Ahmer BM, Andrews-Polymenis HL, Porwollik S, McClelland M. 2012. High-throughput comparison of gene fitness among related bacteria. BMC Genomics 13:212. doi: 10.1186/1471-2164-13-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reznikoff WS. 2008. Transposon Tn5. Annu Rev Genet 42:269–286. doi: 10.1146/annurev.genet.42.110807.091656. [DOI] [PubMed] [Google Scholar]
- 31.Silver AC, Rabinowitz NM, Küffer S, Graf J. 2007. Identification of Aeromonas veronii genes required for colonization of the medicinal leech, Hirudo verbana. J Bacteriol 189:6763–6772. doi: 10.1128/JB.00685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. 2006. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol 297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Bomar L, Stephens WZ, Nelson MC, Velle K, Guillemin K, Graf J. 2013. Draft genome sequence of Aeromonas veronii Hm21, a symbiotic isolate from the medicinal leech digestive tract. Genome Announc 1:e00800-13. doi: 10.1128/genomeA.00800-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
- 35.Wang S-Y, Lauritz J, Jass J, Milton DL. 2003. Role for the major outer-membrane protein from Vibrio anguillarum in bile resistance and biofilm formation. Microbiology 149:1061–1071. doi: 10.1099/mic.0.26032-0. [DOI] [PubMed] [Google Scholar]
- 36.Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol 188:8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Marel M, Schroers V, Neuhaus H, Steinhagen D. 2008. Chemotaxis towards, adhesion to, and growth in carp gut mucus of two Aeromonas hydrophila strains with different pathogenicity for common carp, Cyprinus carpio L. J Fish Dis 31:321–330. doi: 10.1111/j.1365-2761.2008.00902.x. [DOI] [PubMed] [Google Scholar]
- 38.Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol 74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirov SM, Castrisios M, Shaw JG. 2004. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect Immun 72:1939–1945. doi: 10.1128/IAI.72.4.1939-1945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metcalf WW, Jiang W, Daniels LL, Kim S-K, Haldimann A, Wanner BL. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 41.Graf J. 1999. Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect Immun 67:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 43.Lim B, Beyhan S, Meir J, Yildiz FH. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol Microbiol 60:331–348. doi: 10.1111/j.1365-2958.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- 44.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identified differentially abundant genes (q < 0.05) in each experimental situation with log fold change, q value, and functional category annotations. Genes that were detected as being significantly different in all three replicate flasks when analyzed independently in monoassociation experiments are indicated by yellow highlighting. Download
Detailed protocol of the modified TraDIS method used for preparation of transposon insertion site sequencing libraries Download
(A) Plot of P values from comparison of individual TAG intestinal populations and binomial sampling-simulated population. The dashed gray line indicates a P value of 0.05. The shapes on the x axis correspond to each experimental situation as outlined in the legend to Fig. 2A. “Low-input” denotes the Aeromonas TAG input population that was inoculated at a density of 3 orders of magnitude smaller than the others. (B) The joint log likelihood of observed “low-input” intestinal samples (red X’s) are shown against their distribution of binomial draw-simulated TAG populations. Tukey boxplots show the distribution of joint log likelihoods of 9,999 simulated TAG populations for each sample. Download
Soft-agar swim plates verify chemotaxis defects in cheA targeted deletions. (A) Aeromonas (B) Vibrio sp. strain ZWU0020. Download
Graph of the average mapping quality score for each raw TAG (after sequence quality filtering) versus their read number. The TAGs are colored by species. Download
COGs of shared importance between Vibrio and Aeromonas in monoassociation and concurrent diassociation.
Oligonucleotides used in this study.