ABSTRACT
As diet is considered the major regulator of the gut ecosystem, the overall objective of this work was to demonstrate that a detailed knowledge of the phytochemical composition of food could add to our understanding of observed changes in functionality and activity of the gut microbiota. We used metatranscriptomic data from a human dietary intervention study to develop a network that consists of >400 compounds present in the administered plant-based diet linked to 609 microbial targets in the gut. Approximately 20% of the targeted bacterial proteins showed significant changes in their gene expression levels, while functional and topology analyses revealed that proteins in metabolic networks with high centrality are the most “vulnerable” targets. This global view and the mechanistic understanding of the associations between microbial gene expression and dietary molecules could be regarded as a promising methodological approach for targeting specific bacterial proteins that impact human health.
IMPORTANCE
It is a general belief that microbiome-derived drugs and therapies will come to the market in coming years, either in the form of molecules that mimic a beneficial interaction between bacteria and host or molecules that disturb a harmful interaction or proteins that can modify the microbiome or bacterial species to change the balance of “good” and “bad” bacteria in the gut microbiome. However, among the numerous factors, what has proven the most influential for modulating the microbial composition of the gut is diet. In line with this, we demonstrate here that a systematic analysis of the interactions between the small molecules present in our diet and the gut bacterial proteome holds great potential for designing dietary interventions to improve human health.
INTRODUCTION
A substantial amount of research has established that the composition and metabolism of human microbiota play crucial roles in human health. Microbial colonization of the gastrointestinal tract varies widely, with the large intestine having not only the highest density of microbes in terms of bacterial cells per gram but also the most metabolically active microbial community (1). Firmicutes and Bacteroidetes are the most abundant phyla, but Actinobacteria, Proteobacteria, and Verrucomicrobia are also regularly found in healthy adults (2). Diet clearly has a major impact on variation in the gut microbiota composition, which is easily detected in fecal samples even after only a few days after a change in diet (3, 4). The fermentative but also (anaerobic) respiratory bacterial metabolism of dietary components produces an extraordinary chemical diversity in the large intestine with protective (e.g., short-chain fatty acids [SCFAs]) or detrimental (e.g., hydrogen sulfite or bile acids) effects on disease development (5–7). The most sensitive species in response to a change in diet belong to Firmicutes and Actinobacteria; however, more associations between species abundance and diets have been confirmed, e.g., high meat intake with an increase in Bacteroidetes, more fiber with a high proportion of Prevotella spp., etc.
Dietary phytochemicals are usually small molecules with high structural diversity, often part of the plant’s secondary metabolism, that when they reach the intestinal tract can cause selective stress or stimulus to the resident microbiota (8–10). For example, a high intake of cocoa-derived flavonoids increases the abundance of bifidobacteria and lactobacilli and reduces the level of plasma triacylglycerol (11). However, phytochemicals are also transformed by the gut microbial communities to other metabolites with altered bioactivity. For example, conversion of polyphenolic molecules from the microbiome has been shown to inhibit the tumor necrosis factor, NF-κB, and other inflammatory mediators (12–14). Furthermore, bacterial enzymatic activities, such as β-glucuronidase that converts glucuronides to their respective aglycons, are responsible for the retention time of phytochemicals in the human body. It is generally considered that the Gram-positive bacteria are relatively freely permeable to small molecules (15), while those molecules below a certain molecular mass, around 700 Da, can enter into Gram-negative bacteria through porin proteins (16).
In a previous work, we applied text mining and naive Bayes classification in 21 million abstracts from MEDLINE, and we identified 23,137 phytochemicals that are present in plant-based diets (17). Several of these phytochemicals have made it all the way to pharmacy shelves or have served as lead structures for drug development, demonstrating their important contribution to host homeostasis. We also systematically searched for and found associations between 2,768 edible plants and 1,613 human disease phenotypes and characterized the association as being positive or negative. We developed a web-based database, which to the best of our knowledge is the most complete source that links phytochemicals, diet, and disease, and we were able to provide a molecular-level mode of action on how specific diets could prevent disease development (18) or affect drug pharmacodynamics and pharmacokinetics (19). As a step further, we used this extensive source of information to generate a comprehensive picture of the interaction profile of 158 edible plants, which are positively associated with reduction in colon cancer, with a predefined candidate colon cancer target space consisting of ~1,900 human proteins (20). We uncovered the key components in colon cancer that are targeted synergistically by phytochemicals, and we identified statistically significant and highly correlated protein networks that could be perturbed by dietary habits. However, in that study, we completely ignored interactions of the phytochemicals with the gut microbiome and alterations in gut composition and activity.
The gut microbial ecosystem lies at the interface of the host and environment and is considered the third major component regulating host health and the onset of disease; therefore, developing ways to manipulate the gut microbiota represents a promising therapeutic avenue. Trying to identify the “good” and “bad” bacteria in the intestinal microbiota in order to promote the growth of healthy bacterial species and/or eliminate the others has been recognized as one strategy (21); however, not everyone is convinced that single species are to be blamed for disease development. Instead, or maybe in addition, to species-centric studies, targeting specific genes could be a more realistic approach, especially since our knowledge of harmful bacterium-specific metabolic products has grown substantially (22). In the present work, we developed the computational framework for studying possible interactions between the highly diverse phytochemical space of our diet and gut microbial proteins. By coupling metatranscriptomics, chemoinformatics, and network biology, we created a molecular-level map of diet-bacterium interactions showcasing that individual dietary components may contribute to the observed gene expression activities. To the best of our knowledge, this is the first systematic approach for a mechanistic understanding on how phytochemicals are linked to the intestinal microbiota, and it may assist in designing diets with potential therapeutic benefit.
RESULTS
Interactome analysis of bacterial genes and dietary components.
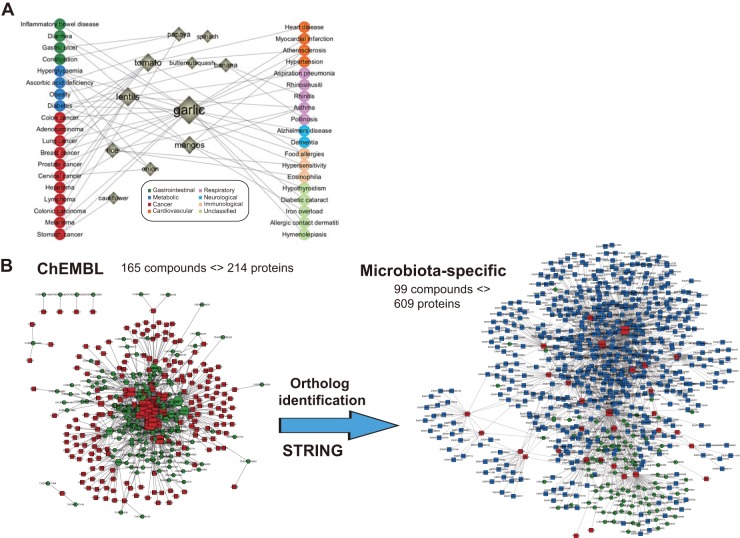
We adopted the metatranscriptomic data from a recently (2014) published study (4) that aimed to investigate whether the human gut microbiota can rapidly respond to dietary interventions in a diet-specific manner. Briefly, the original study had two diet arms, a plant-based diet and an animal-based diet; fecal samples were collected from nine individuals for baseline levels and after 2 days on each experimental diet, and transcriptome sequencing (RNA-Seq) analysis of the bacterial community in the gut was performed. In order to utilize the huge diversity of the phytochemical space and link the molecular composition of the diet with chemoinformatics and network-based analysis, we focused on only the plant-based diet in this study to achieve a molecular-level investigation of the microbial response to diet. The plant-based diet consisted of a mixture of edible plants, most of which belong to fruits and vegetables. To obtain an overall picture of the health-related effects of the administered plant-based diet, we used the plant/food names as a query in the NutriChem database (18), and we retrieved data on associated disease phenotypes. In total, 14 foods in this diet arm (see Table S1A in the supplemental material) were found in NutriChem and were used to generate a food-disease network (Fig. 1A). Specifically, for these 14 foods, there is experimental evidence for association with 38 different diseases that fall into 20 disease categories, which were further classified into 8 general disease classes, including metabolic disease, cancer, cardiovascular disease, etc. (Table S1C). Among the 14 foods, garlic was associated with the highest number of diseases (20). Notably, a considerable number of these diseases, including colon cancer, obesity, and inflammatory bowel disease, have previously been linked to the composition and diversity of the gut microbiota (23, 24).
FIG 1 .
Molecular-level view of the plant-based diet. (A) Food-disease association network for the 14 foods in the plant-based diet of this study. The general disease classes were colored differently as illustrated by the legend. The size of the food nodes reflects the number of disease connections for each food. (B) Gut microbiota-specific protein target space of the food phytochemicals. (Left) Human direct targets (red rectangles) of the food phytochemicals (green hexagons) were retrieved from the ChEMBL database. (Right) The microbiota-specific direct targets (red rectangles) and their protein-protein interaction (PPI) partners (blue rectangles) were derived after incorporating orthologous relationships with human and STRING protein-protein interaction data.
Subsequently, the chemical composition of these 14 foods was retrieved from the NutriChem database (see Table S1B in the supplemental material) and 437 unique food compounds (also called phytochemicals throughout this study) were obtained for further analysis. Of the 437 phytochemicals, 415 (95%) have molecular masses below 700 Da and thus are expected to enter even Gram-negative bacteria. There are approximately 40 compounds per food on average; among these foods, peas have the richest phytochemical profile (85 phytochemicals), while carrots contain the lowest number of compounds (4 phytochemicals). Tomatoes and rice have “the most novel chemical space,” which denotes the highest number of compounds present exclusively in one particular food. To investigate the potential bioactivity of these 437 phytochemicals, we compared them with 1,536 FDA-approved drugs in terms of chemical and physical properties. We observed that most of these food compounds cluster closely or overlap with many FDA-approved drugs indicated for metabolic diseases (see Fig. S1 in the supplemental material). There was also clear overlapping with a subset of the FDA-approved drugs that target human proteins with bacterial orthologs in the gut; thus, they (i.e., the FDA-approved drugs and subsequently their chemically similar food components) could potentially alter the activity of the microbial metabolic network (Fig. S2). Some representative food compounds, such as methoxsalen (present in papaya), quercetin (garlic, mangos, tomato, cauliflower), aminolevulinic acid (peas, tomato), trifluoperazine (garlic), which can be found in DrugBank either as experimental or FDA-approved drugs, are highlighted in Fig. S2.
To hone in on the food-microbiome interactions at the molecular level, we employed compound bioactivity data to uncover the potential targets of the compounds within the 14 foods. For that purpose, we used the ChEMBL database (25), which contains experimentally verified interactions between small molecules and proteins. According to the ChEMBL database, 165 of these 437 phytochemicals interacted with 214 proteins. The phytochemicals targeting the highest number of proteins were quercetin (CHEMBL50), apigenin (CHEMBL28), ellagic acid (CHEMBL6246), and luteolin (CHEMBL151) (listed in decreasing order) (Fig. 1B). Of the 214 protein targets that interact with those food compounds, 31 proteins were found to have bacterial orthologs in the gut (see Materials and Methods for details). To evaluate the credibility of interactions between microbial proteins and compounds generated by ortholog inference, we compared the distribution of similarities of the human-microbe orthologs defined in this study, with that of human-microbe protein pairs targeted by the same compounds (experimentally tested, 1,088 in total) retrieved from the ChEMBL database. No significant difference (P > 0.05 by the Wilcoxon rank sum test) was observed for bit score, identity, or E value between these two groups of protein pairs. In addition to this global comparison, we investigated the similarities of host-microbe pairs used here at the binding site level. It was observed that their respective binding pockets also possessed medium or high similarities, which were not lower than the similarities of human-microbe protein pairs that have been experimentally tested (based on the ChEMBL database) to share the same ligands (data not shown). This provided additional evidence for the feasibility of inferring bacterial targets based on homology to host targets. Quercetin, luteolin, and ellagic acid were found to possess the most bacterial protein targets, as well as kaempferol (CHEMBL150), resveratrol (CHEMBL165), apigenin, and tricetin (CHEMBL247484) (in decreasing order) (Fig. 1B). Quercetin and kaempferol are widespread in the plant-based diet of our study (cauliflower, mangos, onion, and peas), while other phytochemicals are present in only one or two foods. When incorporating the first-degree protein-protein interaction (PPI) partners of the 31 direct bacterial targets, we significantly expanded the phytochemical target space, generating a gut microbiota-specific compound-target interaction network (Fig. 1B). Among the 31 direct targets, pyruvate kinase (UniProt accession no. P14618) and glyceraldehyde-3-phosphate dehydrogenase (UniProt accession no. P04406) have the most PPI partners. In summary, 99 compounds present in the 14 foods of the plant-based diet were linked to 609 gut-microbial targets, which included 31 direct targets plus 578 protein-protein interaction partners.
Gene expression analysis of phytochemical targets: functional implications and topological characteristics.
To determine to what extent the gut microbial protein targets of the food compounds were significantly influenced by dietary intake, we performed a differential expression (DE) analysis using the RNA-Seq data before and after administration of the plant-based diet. Compared to the study of David et al. (4), the DE analysis on the RNA-Seq data, both in the preprocessing steps of the raw data and the statistical analysis, were modified to achieve higher accuracy (see Materials and Methods for more details). We found that 115 microbial targets (7 direct targets and 108 PPI targets), approximately 20% of the whole target space (31 direct targets and 578 PPI targets, respectively), altered gene expression significantly (false discovery rate [FDR] P value < 0.20 by Wilcoxon signed-rank test). This percentage of expression-changing targets induced by dietary phytochemicals is higher than the 8% induced by drugs (26). This may be due to the fact that phytochemicals could target multiple proteins, i.e., more promiscuous, as well as the possibility of multiple phytochemicals targeting the same protein. These 115 DE targets were not found to be significantly differentially expressed when comparing the animal-based diet (4) against its baseline (FDR P value > 0.20 by Wilcox rank sum test). Notably, the targets of food compounds had a significantly higher proportion of DE genes than the nontargeted gut-bacterial proteins (20.7% versus 14.8%; P = 0.0016 by Fisher’s exact test). All the phytochemicals targeting these 115 proteins are below 700 Da, and therefore were expected to enter the bacterial cell. On the basis of the response of the gut microbiome and in order to move beyond the restricted food set used in this study, we also attempted to identify other foods that would potentially induce the equivalent or similar effect. This endeavor was bridged by the commonalities in the phytochemical composition between the original 14 foods used in this study and the 1,772 plant-based foods available in the NutriChem database. Using as input the set of the 59 phytochemicals present in these 14 foods with DE targets, we searched the NutriChem database for alternative foods that show the highest overlap in the phytochemical space. More than 550 foods were retrieved, while some representative examples in this top 20 food list (see Table S4 in the supplemental material) include Ginkgo biloba (ginkgo; 41 shared compounds), sea buckthorn (Hippophae; 33 shared compounds), and Lonicera japonica (honeysuckle; 28 shared compounds).
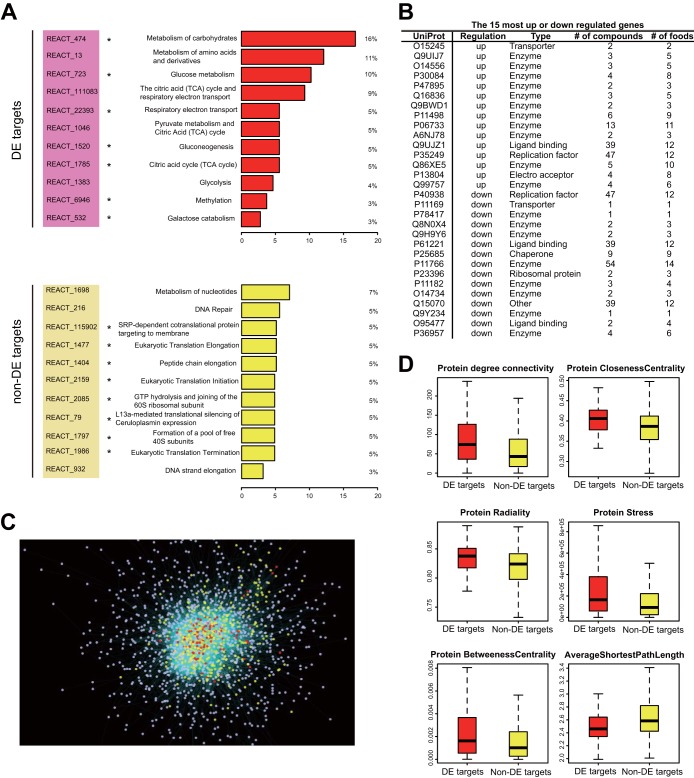
Looking into the biological processes mostly affected by the phytochemical-protein interactions, we found that the 115 DE targets were significantly enriched in 11 pathways (FDR P value < 0.05 by Fisher’s exact test) (Fig. 2A), which provided functional insights into the gut microbial targets. All of these enriched pathways are related to metabolism, such as glucose metabolism, tricarboxylic acid (TCA) cycle (also known as the citric acid cycle), and metabolism of amino acids and derivatives. In contrast, the non-DE targets displayed a very different profile of significantly enriched pathways, exemplified by DNA repair and translation-related processes (Fig. 2A). To inspect this discrepancy in a more statistical manner, we compared the pathway differences between DE and non-DE targets using a pathway overrepresentation analysis. We focused on the top 12 significantly enriched pathways for both groups as shown in Fig. 2A. Seven out of the 12 pathways for the DE targets (as indicated by asterisks in Fig. 2A) showed significant overrepresentation compared directly with the non-DE group (P < 0.05 by Fisher’s exact test); moreover, all seven pathways were metabolic processes. Conversely, all the overrepresented pathways in the non-DE group relative to the DE group were not closely related to metabolism. This suggests that the phytochemicals present in plant-based diet mainly exert their effect on gut microbiome by altering microbial metabolic activities. Taking the 15 most up- or downregulated genes (fold change) as representatives, we further zoomed in the reactions those DE targets are involved in. The solute carrier family 22 member 1 (organic cation transporter 1, UniProt accession no. Q15245) and GTP:AMP phosphotransferase AK3 (UniProt accession no. Q9UIJ7) were the two most upregulated genes, whereas replication factor C subunit 3 (UniProt accession no. P40938) and glucose transporter 3 (UniProt accession no. P11169) were the most downregulated genes in the list. The results (Fig. 2B; see Table S3 in the supplemental material) also show that the majority of the phytochemical targets with the highest fold change in gene expression activity participate in metabolic enzymatic reactions, such as the acetyl coenzyme A (acetyl-CoA) acetyltransferase (UniProt accession no. Q9BWD1) and aldehyde dehydrogenase (UniProt accession no. P47895). In the list with the most up- or downregulated genes, there are also several genes with “auxiliary” functions such as ATP binding (UniProt accession no. P61221), potentially playing a regulatory role in response to dietary intake. Phosphopyruvate hydratase (UniProt accession no. P06733) and alcohol dehydrogenase (UniProt accession no. P11766) are the enzymes interacting with the highest number of compounds within up- or downregulated gene lists, respectively. However, what also caught our attention in this list is that neither the number of compounds nor number of foods targeting gut-bacterial proteins can be used as an indicator for the observed fold changes in gene expression activity level.
FIG 2 .
Functional and topological analyses of the differentially expressed (DE) and non-DE phytochemical targets. (A) Significantly enriched pathways for DE targets and non-DE targets (only the top 11 pathways are shown for non-DE targets). The asterisks indicate the pathways that were significantly overrepresented compared to the other group in a direct comparison. (B) List of the 15 most (fold change) up- or downregulated genes (UniProt identifier [ID] or accession no. and type are also shown) targeted by phytochemicals directly or through PPI. The number of compounds targeting these genes and the number of foods containing these compounds were listed as well. (C) Complete microbiota-specific protein-protein interaction network. The DE phytochemical targets (DE targets tend to be located in more central positions within the network) are shown in red, while the non-DE ones are shown in yellow. The nodes in gray indicate proteins that are not phytochemical targets (nontargets). For clarity, not all network edges were shown here. (D) The boxplots of six network topological properties that were significantly different between DE (red) and non-DE targets (yellow) are shown. The y axes of boxplots show the calculated values of corresponding topological properties.
Next, we investigated the differences between DE and non-DE phytochemical targets, both from a chemical and biological perspective, in an attempt to understand why some of the food targets will show a significant difference in gene expression levels while others will not. First, the chemical features of the compounds were used to examine whether the DE and non-DE proteins could be distinguished. From the principal component analysis (PCA) based on the chemical descriptors of the 99 compounds, no clear separation can be observed between compounds targeting DE proteins and non-DE proteins (data not shown). In addition, the strength of the compound-protein interactions (based on bioactivity data retrieved from the ChEMBL database) was compared between DE and non-DE target groups using a set of compounds that target proteins from both groups. As discussed above, the compounds targeting both DE and non-DE proteins did not show significantly different binding affinities toward the two target groups (P > 0.05 by Student’s t test). These results suggest that the intrinsic biological properties of the target proteins, rather than the chemical characteristics of the compounds or the strength of interactions between compounds and proteins, are probably associated with the variation in the community gene expression under mediation by phytochemicals. This is also consistent with our previous observation that the targeted genes with the highest fold change activity are not necessarily the ones targeted by the highest number of compounds (Fig. 2B).
Therefore, the topological properties of the target proteins in the orthologous PPI network were further investigated (Fig. 2C). For the whole orthologous PPI network, the power-law distribution of degree connectivity was observed, consistent with the common feature of many biological networks (27) (see Fig. S3A in the supplemental material). Compared to nontarget proteins, the phytochemical targets were mainly positioned in the central area, possessing higher connectivity (Fig. 2C and Fig. S3B). Within these targets, the majority of the network centrality measures, including degree and closeness centrality, were found to be significantly different between DE and non-DE targets (P < 0.01 by Wilcoxon rank sum test). More specifically, the DE targets of the food compounds tend to be located in a more “central” position within the PPI network than the non-DE ones (Fig. 2C and D), as reflected by the higher centrality measurements (as well as the lower average shortest path length) of the DE group. The higher closeness centrality of the DE targets indicates that the information spread is generally faster from a given DE protein to all other connected nodes in the network. The DE targets also exhibited higher betweenness centrality, which measures the amount of control a node has on the information spread between other nodes. This implies that compared to non-DE targets, DE targets are more likely to be the “bridges” between clusters in the network. We also found that more than 80% of the interactions within the PPI network are between proteins found in the same genome. With the reduced PPI network (filtering out interspecies connections), the results and conclusions derived from the network topological analysis discussed above still stand.
Taxonomic contribution to gene expression variations of the phytochemical targets.
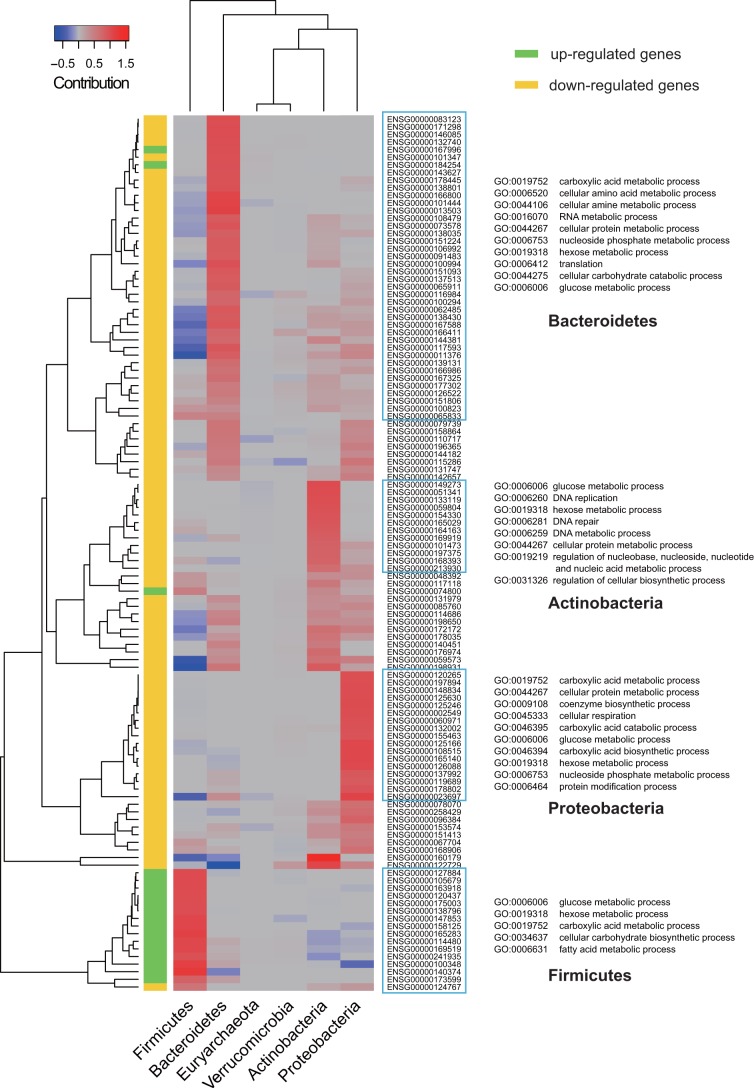
Since the phytochemicals present in the plant-based diet could potentially contribute to a gene expression response of the gut microbiota, the follow-up question was which microbe or groups of microbes were mostly influenced or contributing most to the variations in gene expression at the community level for each of the gene categories targeted by the food compounds. We calculated the overall expression change of each phylum for the 115 DE targets, and subsequently the contribution of each phylum in the whole microbial community (Fig. 3). The phylum Bacteroidetes was found to possess the “broadest responding spectrum,” i.e., it contributed highly to the differential expression of more targeted genes than other phyla. Within the group of target genes to which Bacteroidetes contributed predominantly (denoted by the bright blue rectangle in Fig. 3), it was noticed that most genes were downregulated at the community level. Gene Ontology (GO) annotation indicated that genes from this group mainly participate in the carboxylic acid metabolic process (GO:0019752), cellular amino acid metabolic process (GO:0006520), as well as RNA and protein metabolic processes. Firmicutes, although without the widest spectrum, was the phylum responsible for differential expression of most upregulated target genes, which are mainly involved in biological processes such as glucose (GO:0006006), hexose (GO:0019318), carboxylic acid (GO:0019752), and fatty acid (GO:0006631) metabolic processes. In addition to the predominant and specific contribution of Firmicutes toward upregulated genes, it also negatively contributed (i.e., opposite direction of expression change compared to that of the whole community) to a cluster of downregulated genes, which are shown as the distinct blue blocks in Fig. 3. However, this effect was overridden by the opposite effect from Bacteroidetes, leading ultimately to a community-level downregulation. The group of target genes to which Actinobacteria predominantly contributed displays a distinct profile with several regulatory processes, which are not present in the groups of other phyla. All the target genes in the groups for both Actinobacteria and Proteobacteria had decreased expression at the community level.
FIG 3 .
Phylum contribution toward the community-level gene expression variations of the differentially expressed (DE) targets. The heatmap illustrates the individual contribution of each phylum present in gut microbiota for each DE phytochemical target. The 115 DE targets were further separated into two classes, upregulated genes and downregulated genes. The red blocks represent a positive contribution of the phylum toward community-level expression variation (the direction of change for that phylum in one particular target gene was in agreement with that of the whole microbiota community, regardless of up- or downregulation). The blue blocks indicate that the expression change of these genes for that phylum were in the direction opposite that of the community (i.e., negative contribution). For each of the four main phyla, a cluster of genes was identified (enclosed by a rectangle, with the phylum name was displayed on the right), indicating the dominating effect of that phylum on gene expression variations. Gene Ontology annotations (biological processes) for the four gene clusters were also provided.
We further investigated how various microorganisms responded and contributed to variations in gene expression at the species level. As can be seen from Fig. S4 in the supplemental material, the responses were phylogenetically diverse, even within the same phylum. While a group of species in one particular phylum may be the major contributors to community-level expression change of targeted genes, the other members in that phylum may contribute very little. Taking Firmicutes as an example, 50% of the species in this phylum did not show noticeable contribution to the variation in expression of the 115 DE targets. On the other hand, the majority of those upregulated DE targets primarily stem from a few particular species, including Eubacterium rectale, Faecalibacterium prausnitzii, Roseburia hominis, Roseburia intestinalis, Coprococcus, and Ruminococcus bromii. The first three species are the main butyrate-producing bacteria in the human gut (28, 29). With a role in anti-inflammatory process, their reduced populations have been reported previously as being correlated with the development of diseases such as Crohn’s disease (30) and ulcerative colitis (31). Similar patterns were also observed in the other three main phyla. Two single species, Bifidobacterium longum from Actinobacteria and Escherichia coli from Proteobacteria, dominated the corresponding phylum contribution toward community-level expression variations. Furthermore, these two species together were mainly responsible for the community-level downregulation of most target genes.
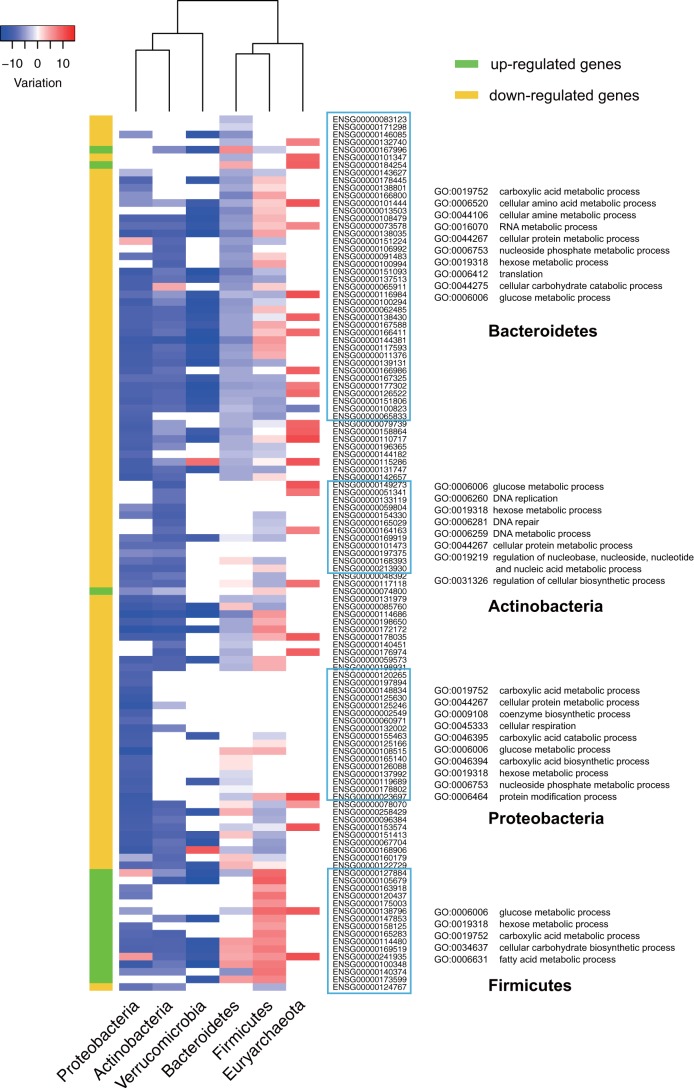
The analyses discussed above provided information about the role of each phylum within the whole gut microbial community. In order to understand better the variation of genuine transcriptional activity for each phytochemical target within each phylum, we further normalized the gene expression levels by the phylum-level abundance and compared this normalized transcriptional activity after being on the diet with that of the baseline period. This further elucidated whether the higher gene expression level was derived from a high transcription level or simply dominant phylum abundance. As shown in Fig. 4, the phyla Proteobacteria, Actinobacteria, and Verrucomicrobia had decreased transcription activities for almost all DE targets, while Firmicutes and Euryarchaeota were the two dominant phyla having higher transcription levels. For most of the upregulated DE targets, Firmicutes displayed the highest increase of transcription activity, which was in concordance with its predominant contribution to the community differential expression of upregulated genes mentioned above. Interestingly, the phylum Euryarchaeota, which consists mainly of methanogens, had increased transcription activities for most target genes that it expressed. This unique pattern was “hidden” in the contribution analysis due to its low abundance in the microbial community.
FIG 4 .
Variation of normalized transcriptional activity of DE targets in microbial phyla. The 115 DE targets were divided into two classes, upregulated genes and downregulated genes. The red blocks represent increased transcriptional level of the gene by the particular microbial phylum, while blue blocks indicate decreased transcription level. For clarity, logE transformation was performed on the values measuring the variation of transcriptional activities (when the value was negative, the absolute value was used for log transformation first and then turned into a negative value again). The four clusters of genes enclosed by rectangles were the ones from Fig. 3.
Designing dietary interventions: the case study of SCFA metabolism.
Short-chain fatty acid (SCFA) metabolism has been extensively cited as being associated with human health and disease pathology, playing a pivotal role in regulating the biological processes in the human gut and colon (32, 33). The levels of SCFAs in the gut, especially butyrate and propionate, have been connected with the development of different diseases ranging from metabolic and inflammatory diseases to cancers (34, 35). Positive correlations were also reported between the levels of certain SCFAs and particular microbes within the phylum of Firmicutes (Roseburia sp., E. rectale, and F. prausnitzii) (4). Since altering the levels of SCFAs could have potentially remarkable health benefits, we used our computational framework here for predicting diets that could potentially alter the activity of the SCFA-related pathways.
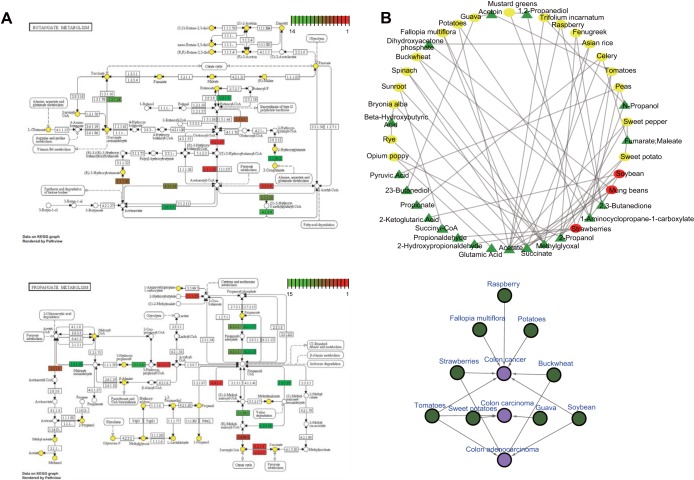
The butyrate metabolism (Kyoto Encyclopedia of Genes and Genomes [KEGG] ko00650) consists of 63 reactions (in terms of KEGG reaction identifiers [IDs]) involving 41 metabolites, while the propionate metabolism (KEGG ko00640) is composed of 64 reactions involving 44 metabolites (Fig. 5A). In order to identify foods that could alter the activity of butyrate or propionate metabolism, two approaches were adopted here: in NutriChem and ChEMBL databases, (i) we searched for food phytochemicals that have been experimentally tested to interact with proteins involved in those two SCFA metabolic pathways; (ii) we searched for foods containing phytochemicals that are essentially metabolites involved in butyrate and propionate metabolism; thus, they are expected to interact with the metabolic proteins. Although there were no phytochemicals identified in the ChEMBL database that target SCFA metabolism-related proteins, the latter approach yielded a set of 98 foods containing 31 phytochemicals (corresponding to 18 and 19 metabolites from butyrate and propionate metabolism, respectively) (Fig. 5A). Of these 98 foods, 22 contain at least two phytochemicals involved in butyrate/propionate metabolism with strawberry, mung bean, and soybean possessing the most (Fig. 5B). Interestingly, 8 of the foods used in the original study of David et al. (4)—where the authors detected significantly higher levels of SCFAs in the plant-based diet than in the animal-based diet—were part of the list containing the 98 foods (and 5 of them were part of the subgroup of the 22 foods), supporting our notion that the detailed food composition could help us partly explain the observed changes in the gut microbial function and activity.
FIG 5 .
Potential dietary interventions targeting the short-chain fatty acid (SCFA) metabolism. (A) The KEGG butyrate (top) (ko00650) and propionate (bottom) (ko00640) metabolic pathways are shown. For both pathways, the metabolites that were also found in 98 plant-based foods present in the NutriChem database were highlighted in yellow. The proteins in the two metabolic pathways were ranked in decreasing order of vulnerability and colored from red (most susceptible to change in gene expression when targeted) to green (least susceptible to change in gene expression). (B) Subgroup of the 98 plant-based foods. (Top) The 22 foods (red or yellow ellipses) containing at least two phytochemicals/metabolites involved in butyrate and/or propionate metabolism and their associated metabolites (green triangle) are shown. The network is visualized in “degree sorted” style, with a decreasing order of node connectivity from middle bottom toward counterclockwise direction. Strawberries, mung beans, and soybeans are the foods with the most metabolites and are shown in red. (Bottom) Subset of the 22 foods associated with colon cancer. Three different names for “colon cancer” in Disease Ontology were treated equivalently here.
To offer additional evidence that dietary interventions containing these 22 foods could potentially trigger changes in the SCFA gut metabolism, we developed a food-disease network based on experimental studies collected in the NutriChem database. We could retrieve 150 disease phenotypes (in terms of Disease Ontology [36] IDs) for these 22 foods (see Fig. S5 in the supplemental material) with breast cancer, hepatocellular carcinoma, and diabetes appearing as the top three diseases connected to the highest number of foods (11, 11, and 10, respectively), whereas sweet peppers, tomatoes, and buckwheat have the highest number of disease links. While hepatocellular carcinoma and diabetes have also been linked to gut microbial imbalances (37, 38), we focus here on a subset of the food-disease network, which is related to colon cancer, a disease highly associated with the levels of SCFAs (39). We found that 9 out of the 22 foods have been associated with colon cancer (Fig. 5B); interestingly, these 9 foods were significantly overrepresented in colon cancer than any other food-disease associations in the NutriChem database (P < 0.001 by Fisher’s exact test). Most of the food-disease associations present in the literature (and subsequently in the NutriChem database) rely on feeding experiments of human or animal models and monitoring the progress/development of a disease. The methodology presented here, which relies on the known phytochemical composition of diet and the potential interactions with the gut-specific bacterial genes, could serve as a route for understanding mechanistically the observed phenotypic responses.
Furthermore, we identified the most vulnerable protein targets of the butyrate and propionate metabolic pathways using their topological features (based on the comparisons between DE and non-DE proteins described above). The proteins in those two metabolic pathways were first ranked according to six significant topological features independently (degree, closeness centrality, betweenness centrality, radiality, stress, and average shortest path length). Then, these six ranked lists were aggregated into a combined ranked list in the order of decreasing centrality (or vulnerability) (see Table S5 in the supplemental material), providing information on which protein targets are more likely to change gene expression activity (Fig. 5A). In butyrate metabolism, 3-hydroxyacyl-CoA dehydrogenase (K00022), acetyl-CoA acetyltransferase (K00626), and enoyl-CoA hydratase (K07511) were found to be the proteins that if targeted by dietary phytochemicals, significant changes in the gene expression activity should be expected. As to propionate metabolism, our topology analysis indicates succinyl-CoA synthetase (GenBank accession no. K01899), l-lactate dehydrogenase (DDBJ accession no. K00016), and methylmalonyl-CoA mutase (GenBank accession no. K01847) as the most vulnerable protein targets.
DISCUSSION
The value of the gastrointestinal community members and their interaction with the host through a variety of signaling molecules and mechanisms has become widely recognized (40). Clear evidence of this realization is the research spending on the analysis of the symbiotic relationships between humans and their indigenous microbiome, which is estimated to be more than $500 million in the last 5 years (41). Metagenomic studies, such as the MetaHit project (42) and Human Microbiome Project (43), provided the multimillion-gene potential of the intestinal microbiome, whereas metatranscriptomic studies (4, 44) provided the subsets of these genes that are expressed as a response to a given stimulus. In particular, diet is considered one of the major modulators of the intestinal microbiota and a dominant source of variation in its composition (45). However, even though occasionally measurements of specific biomarkers in the feces were performed, the full interaction spectrum between the food, its molecular components, and the microbiota was neglected.
It is worthy to note that xenobiotics, including drugs and antibiotics, which can be considered small-molecule analogs of dietary phytochemicals, have been demonstrated to alter the gene expression of active gut microbiota significantly (46). Here we provide a molecular-level analysis of the extent and nature of the gut microbiome responsiveness to diet and a computational platform for understanding mechanistically how food components could drive the activity and function of the gut ecosystem to different states. The development of a chemical-protein network between the food components of a plant-based dietary intervention and the human gut-bacterial proteome space revealed that ~20% of the microbiome targets altered significantly their gene expression activity at a community level. Our analysis also indicates that the metabolic pathways in the gut microbiota are more likely to change activity and are easier to be interrupted compared to other biological processes, when targeted by dietary phytochemicals. Furthermore, while the chemical features of the phytochemicals could not shed light on which of the bacterial targets will alter their gene expression activity, the network analysis revealed the higher centrality within the bacterial PPI network as the most prominent characteristic of the DE bacterial targets of phytochemicals. Previous studies have indicated that highly connected nodes, or “hubs,” in the scale-free cellular biological network tend to be the essential genes (47, 48), which will have less expression changes in mild or moderate conditions (49). However, this might not be observed when an external stimulus is present (as shown in this study) or in a completely different physiological condition such as disease state (50). As shown here, upon dietary stimulus, these highly connected targets are more “vulnerable” (i.e., more susceptible to gene expression variation) and would generate a stronger global effect upon perturbation. From a therapeutic perspective, these more vulnerable (metabolic) proteins with high centrality could serve as promising targets in phytochemical-based dietary interventions. Furthermore, our phylum-level investigation revealed that different phyla of bacteria in the human gut would respond differently in terms of target gene expression activity upon phytochemical perturbation. More specifically, the altered expression levels of targeted genes related to metabolic processes were predominantly attributed to different dedicated phyla. The whole microbiota community thus works coordinately to achieve the overall actions in response to dietary interventions. Thus, identifying specific-disease-associated signatures in the gut microbiota and products that alter microbial populations or block specific bacterial metabolites are expected to lead to the first generation of microbiome therapies.
Even though the role of diet in health and disease states has been recognized for years, the majority of studies in the literature either treated food as a black box or the focus was on carbohydrates, lipids, and fiber, ignoring the small-molecule space. Ignoring the systemic role of phytochemicals was attributed to their presence in small amounts in our diet; however, we should not forget that drugs are also given in small amounts, and they have a profound effect on our health. We believe that the development of NutriChem, the database linking 1,772 plant-based foods with 7,898 phytochemicals and 751 diseases, opened a window of opportunities for understanding how the molecular components of diet interact with the human body, including, as shown here, the bacterial residents of our gut.
However, our analysis is not without limitations: while NutriChem is the most complete database of diet-disease associations at this point in time, the majority of our understanding on these associations so far have originated from animal studies or even cell lines, calling for more attention and research into this area to make more accurate extrapolation to humans. It should also be noted that, despite the advances of computational methods, the lack of experimental bioactivity data of small molecules on bacterial proteins in public databases is a bottleneck for producing accurate ligand-target interaction networks. Further expansion of our knowledge of the molecular composition of each food will provide a more accurate picture of the food-microbiome interaction network and one more tool for better dietary/therapeutic strategies. Last but not least, the gene expression changes of the proteins targeted by dietary phytochemicals will not necessarily be translated into changes of the protein levels. Therefore, additional metaproteomic analysis could yield important insights into causative relationships between particular perturbations and the respective enzymes/proteins. Furthermore, and since the binding of a phytochemical to a bacterial target could disrupt the function without affecting the protein’s level, metabolomic data would allow the establishment of more-accurate linkages between dietary molecules and the gut ecosystem phenotype.
Even though the framework presented here for modulating the functionality and activity of the gut microbiota relies on targeting specific bacterial genes by food phytochemicals, it can be further extended for targeting harmful bacteria, e.g., Bilophila wadsworthia (51) and Fusobacterium nucleatum (52), which can lead to acute inflammation and potentiate intestinal tumorigenesis, respectively, based on the disturbance of species-specific essential protein networks.
MATERIALS AND METHODS
RNA-Seq data description.
The transcriptome sequencing (RNA-Seq) data originated from a recently published study (4) and were deposited in the Gene Expression Omnibus (GEO) with accession number GSE46761. In that study, the community-wide gene expression status in human gut microbiota before (4 and 1 days) and after (3 and 4 days) dietary interventions was measured. To investigate the diet-microbiome interactions at a molecular level, only the plant-based diet was used here and only the plant foods for which molecular compositions are available. Nine study volunteers (subject 1 and 3 to 10) were involved in both baseline and intervention periods of the plant-based diet and were used here. Detailed data description and more information on experimental design are available in the original publication (4).
Food-phytochemical and food-disease associations.
For the foods in the plant-based diet, their phytochemical composition and disease associations were retrieved from the NutriChem database (http://www.cbs.dtu.dk/services/NutriChem-1.0/). After redundancy removal, a collection of unique compounds formed the phytochemical space of the plant-based diet. With regard to the food-disease network, only the food-disease associations supported by more than two references were included (information from NutriChem database). The third-level Disease Ontology term was used to define the disease category. In addition to the foods present in the plant-based diet, all other links between foods, phytochemicals, and diseases were obtained through NutriChem.
Similar chemical space between phytochemicals and drugs.
The SMILES (simplified molecular input line entry system) strings of 437 food compounds were retrieved from PubChem (53), while the 1,536 FDA-approved small-molecule drugs and their associated SMILES were obtained from DrugBank (54) version 4.1. Based on these structural information, the RDKit plugin (http://www.rdkit.org) in KNIME (55) was employed for the calculation of compound molecular and physical chemical descriptors, including 1,024-bit Morgan circular fingerprint, topological polar surface area (TPSA), octanol-water partition coefficient (SlogP), molecular weight (MW), and numbers of Lipinski hydrogen bond acceptors (HBA) and donors (HBD). Afterward, a matrix of compound descriptors was constructed. Compounds from different groups (e.g., phytochemicals or drugs) were distributed along the rows, whereas the 1,024-bit molecular fingerprint and five other properties constituted the 1,029 columns. All the principal component analyses (PCAs) were performed inside R.
A subset of all the FDA-approved drugs that according to DrugBank are targeting metabolic pathways was selected; the subset was defined as “drugs with metabolic targets” in this study. Within this category, drugs whose targets have bacterial orthologs based on orthologous analysis (see below), were referred to as “drugs with metabolic targets having bacterial orthologs.”
Phytochemical-protein target bioactivity data.
Initially, the food compounds were mapped to exactly matched compounds in the ChEMBL database (25) or structurally similar ChEMBL compounds, using InChI key and Morgan circular fingerprints, respectively. Two compounds were deemed similar when they had a Tanimoto coefficient (calculated from Morgan fingerprints) higher than 0.85 and their difference in molecular weight was lower than 50 g/mol. The protein targets of the food compounds were subsequently retrieved; only interactions within the following bioactivity thresholds were kept for further analysis: for Ki, dissociation constant (Kd), 50% inhibitory concentration (IC50), and 50% effective concentration (EC50), the p_chem value (calculated as the negative of the logarithm to base 10 of the measured activity) was larger than 6; for inhibition, the measurement value was greater than 30%; for potency, the measurement value was lower than 50 µM (20). To deal with the multiple measurements of the same compound on the same protein, we calculated a frequency of “positive” measurements among all candidate measurements. This was based on the aforementioned thresholds and served as evidence of compound-protein interaction. Only chemical-protein interactions with a frequency higher than 50% were considered confident and were used to derive protein targets of phytochemicals for downstream analysis (20).
Since the ChEMBL database contains mainly experimental data for interactions of small molecules with human proteins, the developed chemical-microbial protein network was based on the orthologous relationships between the gut microbial reference genomes and the human genome. For analyzing the similarity of binding sites between human-microbe protein pairs of phytochemical targets used here, we first randomly selected one microbial protein from all orthologous proteins for each human-microbe protein pair. Then, based on the full sequences of each protein pair, COACH (56) was used to identify the stretch of amino acid sequence that forms the respective binding pocket. These sequences at the predicted binding site were further compared between human and microbial proteins. The same analysis was also performed on randomly selected human-microbe protein pairs retrieved from ChEMBL that have been experimentally tested to share the same ligands.
Depletion of noncoding sequences from raw RNA-Seq data.
To achieve accurate coding sequences (CDS) and functional annotations, as well as accurate expression level (number of reads per kilobase pair per million mappable reads [RPKM]) for reads in coding region, we removed all potential rRNA sequences using the following procedures. First, all annotated noncoding sequences from all bacterial references in NCBI were downloaded (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) and combined with Silva 16S rRNA sequences into a noncoding RNA database. Second, all the reads were mapped to this noncoding RNA database using BLASTX with parameters “1e−5 -F F.” At last, we excluded the potential noncoding RNA reads if they were mapped to this noncoding RNA database with coverage >50%.
Definition of taxonomy information for reads.
To define the taxonomy of the RNA sequences, we retrieved 2,766 bacterial reference genome sequences from the NCBI database (January 2014 version). The information of the open reading frames (ORFs), their corresponding CDS and protein sequences, as well as the taxonomy information of the strains were also retrieved from NCBI. We further mapped all the reads (excluding potential noncoding RNA) using the BWA (57) version 0.7.4-r385 to these reference genomes. For each species with a relative abundance of more than 0.5% in at least one sample, the strain with the highest number of total mappable reads from all samples was selected as the representative strain for this species. Using these procedures, we identified 54 bacterial reference genomes (see Table S2 in the supplemental material). Based on the alignment with highly identical reads (both identity and coverage of the read are more than 95%), SAMtools (58) was employed to manipulate the BWA alignments. For each coding gene, per gene coverage was calculated using BEDTools (59). The relative abundances of genes in each sample were estimated using RPKM (number of reads per kilobase pair per million mappable reads) using in-house scripts.
Definition of the ortholog proteins between human and bacteria.
We first downloaded all human genes from Ensembl (60) (Ensembl Genome assembly GRCH37 release 75). We selected the longest transcript for each gene locus as the representative transcript using in-house python scripts. The ortholog relationships between each bacteria gene set from 54 reference genomes and human genes was defined using Inparanoid (61) with default parameters. The ortholog pairs between the human genome and 54 bacterial genomes were 411 on average. We also carried out the ortholog definition using reciprocal best-hit blast and found that >95% of the ortholog relationships defined are consistent between two methods.
Functional analysis of metatranscriptomic data.
Thirty-eight samples from nine subjects were kept for downstream analysis. For each subject, the two different days (if present) within the same period (baseline or dietary intervention) were merged into one sample. The reads in preprocessed merged samples were then mapped to those 54 reference genomes with BLASTN (e < 1e−05, coverage >70). The genes from the reference genomes were further annotated with human orthologous genes as described above. The expression of each phytochemical target was quantified by aggregation of all orthologous microbial genes and then normalized using RPKM. Afterward, Wilcoxon’s signed-rank test was employed to determine whether a gene displayed significantly differential expression (DE) after dietary intervention compared with the baseline status. The false discovery rate (62) (FDR) method was used for correction of multiple hypothesis testing. Pathway mapping and enrichment analysis for DE and non-DE targets were conducted with Reactome (63) version 49. A direct comparison of pathways in which DE and non-DE targets are involved was conducted with an overrepresentation analysis performed in R.
Ortholog-based PPI network and analysis.
Since the proteins within biological systems rarely act in isolation, we also included the protein-protein interaction (PPI) data that originated from STRING (64) v9.05. Only PPIs with a score higher than 400 (representing a medium-confidence interaction as defined by the STRING authors) and for pairs for which both human proteins have bacterial orthologs (as defined above) were retrieved for construction of the microbiota-specific PPI network. The PPI network, as well as all other networks displayed in this study, was visualized in Cytoscape (65) v3.2. The NetworkAnalyzer (66) plugin inside Cytoscape was employed for topological analysis, which calculated eight topological features: degree connectivity, clustering coefficient, topological coefficient, closeness centrality, radiality, stress, betweenness centrality, and average shortest path length.
Differential taxonomic contribution to the whole-community response.
The expression change of each gene present in each phylum was calculated by subtracting the mean expression level before the diet from the mean expression level after the diet. Then, for each gene, the phylum contribution was calculated by dividing the corresponding expression change by the overall community-level gene expression change. A positive value thus indicates a positive contribution toward, or consistency with the direction of, the community-level change, regardless of up- or downregulation. In contrast, a negative value means the opposite direction of change between that particular phylum and the microbiota community on the particular gene. For each of the four main phyla, a group of genes which represented an (nearly) exclusive positive contribution of the corresponding taxon toward community-level expression change of these genes was identified. The four groups of genes were functionally annotated with Gene Ontology terms (biological processes, third level) using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (67).
To measure the species-level contribution, a more stringent threshold for BLASTN mapping was used for the gene expression calculation (e < 1e−05, identical ratio >95%, where the “identical ratio” was defined as the product of identity and alignment length, which was then divided by read length). The remaining steps were analogous to the phylum-level analysis.
For each microbial phylum, the genuine transcriptional activity of each DE target gene was calculated by dividing its mean expression level (among subjects before or after the diet) by the corresponding phylum taxonomic abundance. This represented the transcriptional activity normalized by the phylum-level abundance. Then, subtraction of this normalized activity before and after the diet gave the variation of transcriptional activity of each gene within each microbial phylum. A positive value represents increased transcription level, while a negative value indicates decreased transcription level. The phylum-level abundance data (average among subjects before or after the diet) were retrieved from MG-RAST (68).
SCFA-oriented dietary interventions.
The proteins participating in butyrate (KEGG ko00650) and propionate metabolism (KEGG ko00640) were first ranked by individual topological measurements and then aggregated with the R package RankAggreg using default settings (69) to provide an overall indication of vulnerability. Last, the pathway maps with metabolites/phytochemicals and proteins highlighted were generated with R Bioconductor package Pathview (70).
SUPPLEMENTAL MATERIAL
Drug-like chemical space of plant-based foods. Principal component analysis was employed for comparison between phytochemicals in 14 foods (pink) and all FDA-approved drugs (green). FDA-approved drugs with metabolic targets were also highlighted in gold. Download
PCA highlighting the chemical closeness between food phytochemicals and FDA-approved drugs with metabolic targets (and FDA drugs with targets having bacterial orthologs). Phytochemicals in 14 foods are shown as black dots, while FDA-approved drugs with metabolic targets and metabolic targets having bacterial orthologs are shown as red dots and blue triangles, respectively. Download
Network topological analyses of phytochemical targets. (A) Power-law distribution of degree connectivity for orthologous PPI network. The fitting parameters of degree connectivity distribution were as follows: a correlation of 0.922 and R-squared value of 0.851. (B) To illustrate the difference between phytochemical targets and all bacterial orthologous proteins, several approaches, including mean connectivity, boxplot of connectivity, density distribution, and histogram, were adopted here. Download
Species contribution toward community-level differential expression of DE targets. The microbial phyla were colored differently (indicated by the color bar near the top). The red or blue blocks indicate that the direction of expression change for the species was in the same direction (“positive contribution”) or opposite direction (“negative contribution”) to that of the microbial community, respectively, regardless of up- or downregulation of the corresponding genes. It was also noticed that the expression variations of upregulated target genes were dominantly attributed to several microbial species belonging to Firmicutes, as highlighted in green (rectangle and text). Download
Food-disease network for the 22 foods having at least two SCFA-related phytochemicals/metabolites. The foods (green) and diseases (dark orange) were labeled with plant taxonomy IDs and Disease Ontology (DO) IDs, respectively. The size of the node reflects the number of connections each node has. Breast cancer (DO ID 1612), hepatocellular carcinoma (DOID 684), and diabetes (DOID 9351) are the top three diseases associated with the highest number of foods (11, 11, and 10, respectively). Download
List of 14 foods used in the plant-based diet and their associated chemicals and diseases. (A) The associated NCBI taxonomy IDs were provided. (B) Phytochemical content of the 14 foods, where the exclusive phytochemicals represent the compounds that are present only in that food. (C) Disease associations for the 14 foods in the plant-based diet. These associations were retrieved from the NutriChem database. The disease names were mapped to Disease Ontology IDs, third-level Disease Ontology disease name, and were also classified into general disease classes.
List of 54 gut microbial reference genomes used in this study.
Fifteen most up- or downregulated genes from the phytochemical targets and associated reactions (if the genes are enzymes). (A) Most upregulated genes. (B) Most downregulated genes.
Top 20 plant-based foods (or edible plants) that were predicted to elicit an effect similar to that of the 14 foods involved in this study. The foods in dark green represent those foods used in the original study.
Ranked lists of protein vulnerability in butyrate or propionate metabolism. These ranks for butyrate (A) and propionate (B) were aggregated from those six network topological properties mentioned in the text of the article.
ACKNOWLEDGMENTS
G.P. and J.L. thank the HKU-SRT of Genomics for their technical support. Y.N. thanks David Westergaard for fruitful discussions and support on the chemical-protein interaction network construction.
Footnotes
Citation Ni Y, Li J, Panagiotou G. 2015. A molecular-level landscape of diet-gut microbiome interactions: toward dietary interventions targeting bacterial genes. mBio 6(6):e01263-15. doi:10.1128/mBio.01263-15.
REFERENCES
- 1.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 2.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macfarlane G, Gibson G. 1997. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine, p 269–318. In Mackie R, White B (ed), Gastrointestinal microbiology. Springer, New York, NY. [Google Scholar]
- 6.Magee EA, Richardson CJ, Hughes R, Cummings JH. 2000. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr 72:1488–1494. [DOI] [PubMed] [Google Scholar]
- 7.Barrasa JI, Olmo N, Lizarbe MA, Turnay J. 2013. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol In Vitro 27:964–977. doi: 10.1016/j.tiv.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 8.Williamson G, Clifford MN. 2010. Colonic metabolites of berry polyphenols: the missing link to biological activity? Br J Nutr 104(Suppl 3):S48–S66. doi: 10.1017/S0007114510003946. [DOI] [PubMed] [Google Scholar]
- 9.Bialonska D, Ramnani P, Kasimsetty SG, Muntha KR, Gibson GR, Ferreira D. 2010. The influence of pomegranate by-product and punicalagins on selected groups of human intestinal microbiota. Int J Food Microbiol 140:175–182. doi: 10.1016/j.ijfoodmicro.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 10.Selma MV, Espín JC, Tomás-Barberán FA. 2009. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem 57:6485–6501. doi: 10.1021/jf902107d. [DOI] [PubMed] [Google Scholar]
- 11.Tzounis X, Rodriguez-Mateos A, Vulevic J, Gibson GR, Kwik-Uribe C, Spencer JP. 2011. Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study. Am J Clin Nutr 93:62–72. doi: 10.3945/ajcn.110.000075. [DOI] [PubMed] [Google Scholar]
- 12.Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. 2013. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem 24:1415–1422. doi: 10.1016/j.jnutbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Russell WR, Labat A, Scobbie L, Duncan SH. 2007. Availability of blueberry phenolics for microbial metabolism in the colon and the potential inflammatory implications. Mol Nutr Food Res 51:726–731. doi: 10.1002/mnfr.200700022. [DOI] [PubMed] [Google Scholar]
- 14.Larrosa M, Luceri C, Vivoli E, Pagliuca C, Lodovici M, Moneti G, Dolara P. 2009. Polyphenol metabolites from colonic microbiota exert anti-inflammatory activity on different inflammation models. Mol Nutr Food Res 53:1044–1054. doi: 10.1002/mnfr.200800446. [DOI] [PubMed] [Google Scholar]
- 15.Lambert PA. 2002. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J Appl Microbiol 92(Suppl):46S–54S. doi: 10.1046/j.1365-2672.92.5s1.7.x. [DOI] [PubMed] [Google Scholar]
- 16.Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol Res 49:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen K, Panagiotou G, Kouskoumvekaki I. 2014. Integrated text mining and chemoinformatics analysis associates diet to health benefit at molecular level. PLoS Comput Biol 10:e1003432. doi: 10.1371/journal.pcbi.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen K, Panagiotou G, Kouskoumvekaki I. 2015. NutriChem: a systems chemical biology resource to explore the medicinal value of plant-based foods. Nucleic Acids Res 43:D940–D945. doi: 10.1093/nar/gku724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen K, Ni Y, Panagiotou G, Kouskoumvekaki I. 2015. Developing a molecular roadmap of drug-food interactions. PLoS Comput Biol 11:e1004048. doi: 10.1371/journal.pcbi.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westergaard D, Li J, Jensen K, Kouskoumvekaki I, Panagiotou G. 2014. Exploring mechanisms of diet-colon cancer associations through candidate molecular interaction networks. BMC Genomics 15:380. doi: 10.1186/1471-2164-15-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 22.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. 2014. Bile acids and the gut microbiome. Curr Opin Gastroenterol 30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. 2012. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iskar M, Campillos M, Kuhn M, Jensen LJ, van Noort V, Bork P. 2010. Drug-induced regulation of target expression. PLoS Comput Biol 6:e100925. doi: 10.1371/journal.pcbi.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barabási A, Oltvai ZN. 2004. Network biology: understanding the cell’s functional organization. Nat Rev Genet 5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 28.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. 2007. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis P, Flint HJ. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 30.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. 2006. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vermeiren J, Van den Abbeele P, Laukens D, Vigsnaes LK, De Vos M, Boon N, Van de Wiele T. 2012. Decreased colonization of fecal Clostridium coccoides/Eubacterium rectale species from ulcerative colitis patients in an in vitro dynamic gut model with mucin environment. FEMS Microbiol Ecol 79:685–696. doi: 10.1111/j.1574-6941.2011.01252.x. [DOI] [PubMed] [Google Scholar]
- 32.Scheppach W. 1994. Effects of short chain fatty acids on gut morphology and function. Gut 35:S35–S38. doi: 10.1136/gut.35.1_Suppl.S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. The role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 35.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. 2011. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schriml LM, Arze C, Nadendla S, Chang Y-W, Mazaitis M, Felix V, Feng G, Kibbe WA. 2012. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res 40:D940–D946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, Sartor RB, Rabadan R, Schwabe RF. 2012. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, et al.. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 39.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. 2006. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 40.Holmes E, Li JV, Marchesi JR, Nicholson JK. 2012. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab 16:559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Reardon S. 2014. Microbiome therapy gains market traction. Nature 509:269–270. doi: 10.1038/509269a. [DOI] [PubMed] [Google Scholar]
- 42.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, et al.. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.NIH HMP Working Group, Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH Human Microbiome Project. Genome Res 19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faith JJ, McNulty NP, Rey FE, Gordon JI. 2011. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maurice CF, Haiser HJ, Turnbaugh PJ. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang YC, Lin CC, Chang JY, Mori H, Juan HF, Huang HC. 2009. Predicting essential genes based on network and sequence analysis. Mol Biosyst 5:1672–1678. doi: 10.1039/b900611g. [DOI] [PubMed] [Google Scholar]
- 48.Jeong H, Mason SP, Barabási AL, Oltvai ZN. 2001. Lethality and centrality in protein networks. Nature 411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 49.Zhou L, Ma X, Sun F. 2008. The effects of protein interactions, gene essentiality and regulatory regions on expression variation. BMC Syst Biol 2:54. doi: 10.1186/1752-0509-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ideker T, Sharan R. 2008. Protein networks in disease. Genome Res 18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, Vipperla K, Naidoo V, Mtshali L, Tims S, Puylaert PG, DeLany J, Krasinskas A, Benefiel AC, Kaseb HO, Newton K, Nicholson JK, de Vos WM, Gaskins HR, Zoetendal EG. 2015. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 6:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolton EE, Wang Y, Thiessen PA, Bryant SH. 2008. Chapter 12. PubChem: integrated platform of small molecules and biological activities, p 217–241. In Ralph AW, David CS (ed), Annual reports in computational chemistry, vol 4 Elsevier, Oxford, United Kingdom. [Google Scholar]
- 54.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, Tang A, Gabriel G, Ly C, Adamjee S, Dame ZT, Han B, Zhou Y, Wishart DS. 2014. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berthold M, Cebron N, Dill F, Gabriel T, Kötter T, Meinl T, Ohl P, Sieb C, Thiel K, Wiswedel B.. 2008. KNIME: the Konstanz information miner, p 319–326. In Preisach C, Burkhardt H, Schmidt-Thieme L, Decker R (ed), Data analysis, machine learning and applications. Springer, Berlin, Germany. doi: 10.1007/978-3-540-78246-9_38. [DOI] [Google Scholar]
- 56.Yang J, Roy A, Zhang Y. 2013. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. BioInformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kahari AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. 2015. Ensembl 2015. Nucleic Acids Res 43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Remm M, Storm CEV, Sonnhammer ELL. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Method 57:289–300. [Google Scholar]
- 63.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P. 2014. The Reactome pathway knowledgebase. Nucleic Acids Res 42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. 2008. Computing topological parameters of biological networks. Bioinformatics 24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 67.Huang DW, Sherman BT, Lempicki RA. 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 68.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pihur V, Datta S, Datta S. 2009. RankAggreg, an R package for weighted rank aggregation. BMC Bioinformatics 10:62. doi: 10.1186/1471-2105-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo W, Brouwer C. 2013. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Drug-like chemical space of plant-based foods. Principal component analysis was employed for comparison between phytochemicals in 14 foods (pink) and all FDA-approved drugs (green). FDA-approved drugs with metabolic targets were also highlighted in gold. Download
PCA highlighting the chemical closeness between food phytochemicals and FDA-approved drugs with metabolic targets (and FDA drugs with targets having bacterial orthologs). Phytochemicals in 14 foods are shown as black dots, while FDA-approved drugs with metabolic targets and metabolic targets having bacterial orthologs are shown as red dots and blue triangles, respectively. Download
Network topological analyses of phytochemical targets. (A) Power-law distribution of degree connectivity for orthologous PPI network. The fitting parameters of degree connectivity distribution were as follows: a correlation of 0.922 and R-squared value of 0.851. (B) To illustrate the difference between phytochemical targets and all bacterial orthologous proteins, several approaches, including mean connectivity, boxplot of connectivity, density distribution, and histogram, were adopted here. Download
Species contribution toward community-level differential expression of DE targets. The microbial phyla were colored differently (indicated by the color bar near the top). The red or blue blocks indicate that the direction of expression change for the species was in the same direction (“positive contribution”) or opposite direction (“negative contribution”) to that of the microbial community, respectively, regardless of up- or downregulation of the corresponding genes. It was also noticed that the expression variations of upregulated target genes were dominantly attributed to several microbial species belonging to Firmicutes, as highlighted in green (rectangle and text). Download
Food-disease network for the 22 foods having at least two SCFA-related phytochemicals/metabolites. The foods (green) and diseases (dark orange) were labeled with plant taxonomy IDs and Disease Ontology (DO) IDs, respectively. The size of the node reflects the number of connections each node has. Breast cancer (DO ID 1612), hepatocellular carcinoma (DOID 684), and diabetes (DOID 9351) are the top three diseases associated with the highest number of foods (11, 11, and 10, respectively). Download
List of 14 foods used in the plant-based diet and their associated chemicals and diseases. (A) The associated NCBI taxonomy IDs were provided. (B) Phytochemical content of the 14 foods, where the exclusive phytochemicals represent the compounds that are present only in that food. (C) Disease associations for the 14 foods in the plant-based diet. These associations were retrieved from the NutriChem database. The disease names were mapped to Disease Ontology IDs, third-level Disease Ontology disease name, and were also classified into general disease classes.
List of 54 gut microbial reference genomes used in this study.
Fifteen most up- or downregulated genes from the phytochemical targets and associated reactions (if the genes are enzymes). (A) Most upregulated genes. (B) Most downregulated genes.
Top 20 plant-based foods (or edible plants) that were predicted to elicit an effect similar to that of the 14 foods involved in this study. The foods in dark green represent those foods used in the original study.
Ranked lists of protein vulnerability in butyrate or propionate metabolism. These ranks for butyrate (A) and propionate (B) were aggregated from those six network topological properties mentioned in the text of the article.