ABSTRACT
Staphylococcus aureus capsule is an important virulence factor that is regulated by a large number of regulators. Capsule genes are expressed from a major promoter upstream of the cap operon. A 10-bp inverted repeat (IR) located 13 bp upstream of the −35 region of the promoter was previously shown to affect capsule gene transcription. However, little is known about transcriptional activation of the cap promoter. To search for potential proteins which directly interact with the cap promoter region (Pcap), we directly analyzed the proteins interacting with the Pcap DNA fragment from shifted gel bands identified by electrophoretic mobility shift assay. One of these regulators, RbsR, was further characterized and found to positively regulate cap gene expression by specifically binding to the cap promoter region. Footprinting analyses showed that RbsR protected a DNA region encompassing the 10-bp IR. Our results further showed that rbsR was directly controlled by SigB and that RbsR was a repressor of the rbsUDK operon, involved in ribose uptake and phosphorylation. The repression of rbsUDK by RbsR could be derepressed by d-ribose. However, d-ribose did not affect RbsR activation of capsule.
IMPORTANCE Staphylococcus aureus is an important human pathogen which produces a large number of virulence factors. We have been using capsule as a model virulence factor to study virulence regulation. Although many capsule regulators have been identified, the mechanism of regulation of most of these regulators is unknown. We show here that RbsR activates capsule by direct promoter binding and that SigB is required for the expression of rbsR. These results define a new pathway wherein SigB activates capsule through RbsR. Our results further demonstrate that RbsR inhibits the rbs operon involved in ribose utilization, thereby providing an example of coregulation of metabolism and virulence in S. aureus. Thus, this study further advances our understanding of staphylococcal virulence regulation.
INTRODUCTION
Staphylococcus aureus produces a large number of virulence factors that endow the organism with the ability to cause a wide range of diseases in humans and animals. The expression of virulence genes is controlled by an equally impressive number of regulators forming a complex regulatory network (1–3). Although the regulation of virulence genes has been the subject of extensive studies recently, our knowledge of the virulence regulatory network in S. aureus is still fragmented. To further understand virulence regulation, we have been studying the S. aureus virulence regulatory network by employing capsule as a model virulence factor (4–7). Capsule is an antiphagocytic virulence factor, and the majority of S. aureus strains produce either type 5 or type 8 capsule (8, 9). Sixteen cap genes, which are organized as a long operon, are required for the biosynthesis of either type of capsule (10). The genetic loci for the type 5 and type 8 capsules (cap5 and cap8) are allelic, with the four genes in the middle of the operon being type specific (11). Because of this allelic organization in the chromosome, the expression of cap5 and cap8 genes is subject to similar transcriptional regulation. To date, a large number of regulators affecting cap gene transcription, some of which are non-DNA-binding factors, have been identified and/or characterized, and they include MgrA, AgrADBC, ArlRS, SaeRS, CodY, KdpDE, SigB, SpoVG, ClpC, ClpP, SbcDC, RpiRC, CcpA, Rot, CcpE, and AirSR (4–6, 12–20).
Staphylococcal capsules are involved in immune evasion, but they can also mask cell surface components, such as adhesins, that are important for pathogenesis (21, 22). Thus, the production of capsule must be controlled properly depending on the conditions of the environment in which S. aureus resides. The surprisingly large number of regulators involved in capsule regulation further suggests that capsule is highly regulated and that the capsule regulatory network is very complex. Although many DNA-binding regulators affecting cap gene transcription have been identified, interestingly, only one cis element, a 10-bp inverted repeat (IR) located 13 bp upstream of the −35 region of the cap promoter (Pcap), has been identified to be critical for transcription of the cap genes and for capsule production (23). Among all the transcriptional regulators identified, five (CodY, KdpE, SpoVG, CcpE, and AirR) have been shown to bind directly to the Pcap region (15, 16, 19, 20, 24). However, the 10-bp IR has not been implicated in the binding of these regulators. In this study, we aimed to identify new potential Pcap-binding regulators to further understand capsule regulation. We identified 6 additional proteins that could potentially bind to Pcap in vitro to affect capsule production. We chose to focus on RbsR and showed that it is a DNA-binding regulator that directly binds to the 10-bp IR and the flanking sequences. We further demonstrated that rbsR expression is under the direct control of the alternative sigma factor SigB. In addition, we confirmed that RbsR is a repressor of the downstream rbsUDK operon.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Nebraska transposon mutants (30) were obtained through the Network of Antimicrobial Resistance in S. aureus (NARSA) program. Competent S. aureus RN4220 was used as the recipient for electroporation by the procedure of Kraemer and Iandolo (31). Phage 52A were used for plasmid and chromosomal DNA transduction between S. aureus strains. Escherichia coli strain XL1-Blue was used for plasmid construction and maintenance. S. aureus strains were cultivated with tryptic soy broth (TSB) or tryptic soy agar (TSA) (Difco Laboratory, Detroit, MI) unless indicated otherwise. E. coli was grown in Luria-Bertani broth or agar unless specified otherwise. Antibiotics were added to the culture medium when necessary, at final concentrations of 10 μg/ml for chloramphenicol, 10 μg/ml for erythromycin, and 100 μg/ml for ampicillin.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference/source |
|---|---|---|
| Strains | ||
| S. aureus strains | ||
| RN4220 | Restriction-negative laboratory strain | J. Iandolo |
| Newman | Wild-type CP5 strain | T. Foster |
| CYL6401 | Strain Becker with 4-bp mutation in 10-bp IR | 23 |
| CYL11391 | Newman ΔclpC::cat ΔsaeR ΔcodY::ermC | 6 |
| CYL11481 | Newman saeSP18L | 6 |
| CYL12847 | CYL11481 ΔNWMN_2027 | This study |
| CYL12834 | CYL11481 rbsR::bursa | This study |
| NE324 | USA300 FPR3757 recX::bursa | NARSA |
| NE425 | USA300 FPR3757 rbsR::bursa | NARSA |
| NE567 | USA300 FPR3757 sarZ::bursa | NARSA |
| NE1445 | USA300 FPR3757 xdrA::bursa | NARSA |
| NE1781 | USA300 FPR3757 NWMN_1391::bursa | NARSA |
| GP266 | RN4220 rsbU+ sigB1(Am) Tcr | 25 |
| CYL13113 | CYL11481 rsbU+ sigB1(Am) Tcr | This study |
| E. coli strains | ||
| XL1-Blue | Host strain | Stratagene |
| CYL3967 | Rosetta2(DE3)(pLysS) | Novagen |
| CYL4242 | Rosetta2(DE3)(pLysS)(pML4237) | This study |
| Plasmids | ||
| pGEM-T Easy | Cloning vector | Promega |
| pJB38 | Vector for allelic replacement | 26 |
| pML100 | Shuttle vector | 27 |
| pML4233 | pML100 with rbsR | This study |
| pET28a(+) | Expression vector | Novagen |
| pML4237 | pET28a(+) with rbsR | This study |
| pSB40N | Promoter probe plasmid | 28 |
| pAC7 | Expression vector with PBAD promoter | 28 |
| pAC7-sigB | pAC7 with sigB | 29 |
| pML4261 | pSB40N with 155-bp rbsR promoter (PrbsR1) | This study |
| pML4262 | pSB40N with 760-bp rbsR promoter (PrbsR2) | This study |
| pCL3169 | pGEM-T Easy with 614-bp Pcap fragment | This study |
| pAM3176 | pLL35 with Pcap::blaZ | 4 |
Plasmid and strain construction.
Primers used for plasmid and strain construction are listed in Table 2. To construct a deletion mutant of NWMN_2027 (CYL12847), DNA fragments flanking the gene were amplified by using primer pairs NM2027-1/NM2027-2 and NM2027-3/NM2027-4 and cloned in tandem into plasmid pJB38, followed by allele replacement as described previously (26). The mutation was confirmed by PCR. The transposon mutants CYL12833 (recX::bursa), CYL12837 (xdrA::bursa), CYL12834 (rbsR::bursa), CYL12835 (sarZ::bursa), and CYL12838 (NWMN1391::bursa) were constructed by phage transduction of the transposon insertions from the respective Nebraska transposon mutants to CYL11481 and then verified by PCR.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| cp8gs3 | CCATTATTTACCTCCCTTAAAAATTTTC |
| cp8gs4 | AACGATATGTAATATGTAAATAC |
| cp8gs5 | ACATATCGTTTAAACAATTAATTACTTT |
| cp8gs6 | CTACTTTAGAGTATAATTATTTTTAATTTC |
| cp8bla.f | CTGCAGAGCTCGCATTTGAAGATCA |
| cp8bla.r | GGATCCCTTAGTTTGATTCACTAAA |
| FAM-FP3 | VIC-CCATTATTTACCTCCCTTAAAAATTTTC |
| FAM-FP6 | FAM-CTACTTTAGAGTATAATTATTTTTAATTTC |
| NM2027-1 | GATGATATCGTCGACGAGCTCCTGATAGAATTGAAGCAGGCACATA |
| NM2027-2 | ACGTTGATCTGTTAAATCGAGCGGCCGCTTCTTCGAGATACGGACATACTTCCATC |
| NM2027-3 | TATGTCCGTATCTCGAAGAAGCGGCCGCTCGATTTAACAGATCAACGTACTGCTAA |
| NM2027-4 | GATGATATCGTCGACTAATTAATCCAGATACACCGATTGCTTC |
| rbsR-2 | GATGATATCGAATTCATGATGAGTATATTTCGGAAGATACGTAG |
| rbsR-3 | GATGATATCAAGCTTTAACTGTATGATTAATTACACAATAAAGA |
| rbsR-4 | AGCCATATGGCTAGCATGAAAAAAGTGTCAATTAAAGATGTTGCTA |
| rbsR-5 | CTCGAATTCGGATCCTTAGTTTGAAAGATGATAGCCAGTTGTTG |
| rbsR-6 | TCTCAGCACTGATGCAAAGTATTCATGAC |
| rbsR-7 | ATGCCAATAGCGAGTTCATCGTTAATAG |
| rbsR-8 | GATTCTAGAGGATCCGACAAAATGGCCATTTTCAAATATCAC |
| rbsR-9 | GATTCTAGAGGATCCTATTCTCTCGTCTCAACCTTAATCGTATACTTCAG |
| rbsR-10 | GATGGGCCCCTCGAGTCTTTATTGTGTAATTAATCATACAGTTATATAC |
| rbsR-11 | CAATCACATAGTTCAATATACATCATTTC |
| rbsR-12 | ACTGATACACCAGCTTCTCTAGCAACATC |
| rbsR-13 | CTTCAACGATTACAGCACTTAGATAAATC |
| rbsR-14 | CATGATTTGTTTTTAGTAAAACGTTTTACCAGTGCCATC |
| rbsU-1 | CATCAAATTATCGGTGCTACTGTAGGTACGTTAATC |
| rbsU-2 | AATGACTGCAACAATTACCATACCCATTGCTTG |
| rpi-1 | CTCAAATGGCGCAACTAATTAAAGAACGTGGTTAC |
| rpi-2 | CATATCTAAGAAGTATCCTGTCTCAAACACAC |
| prs-1 | CGTGCTTCTGCAGCAACAATCAATATTGTAG |
| prs-2 | GATAAAACAGGGTGTGTACAACAAGCATATAC |
| rbsR-T1 | CTAAACGTTCTGAAACAGCATGTACGTTTTTTATC |
| rbsR-T2 | CATAATGATAGTCGTTTTTCCGCAAC |
| rbsR-T3 | GTAACTGATACACCAGCTTCTCTAGCAAC |
For complementation of rbsR mutations, pML4233 carrying the S. aureus Newman rbsR gene under the control of Pxyl/tetO was constructed by cloning a 1,070-bp PCR fragment, amplified using primer pair rbsR-3/rbsR-2, into the HindIII and EcoRI sites of pML100. To express the recombinant His6-RbsR protein in E. coli, plasmid pML4237 was constructed by cloning the rbsR gene of S. aureus Newman, amplified with primer pair rbsR-4/rbsR-5, into the NheI and BamHI sites of pET-28a(+) (Novagen, Madison, WI). For two-plasmid sigB-dependent promoter assays, plasmids pML4261 and pML4262 were constructed by cloning a 153-bp fragment (amplified using primer pair rbsR-9/rbsR-10) and a 756-bp fragment (amplified using primer pair rbsR-8/rbsR-10) of the rbsR promoter region, respectively, into the promoter probe plasmid pSB40N, at the BamHI and XhoI sites. Plasmid pCL3169, which was used for footprinting analyses, was constructed by inserting a 624-bp Pcap fragment, amplified with primers cp8bla.f and cp8bla.r, into pGEM-T Easy (Promega, Madison, WI) by T/A cloning. All plasmid constructs were verified by restriction mapping and sequencing of the inserts.
Fractionation of DNA-binding proteins on heparin-agarose.
An overnight culture (200 ml) of S. aureus Newman clpC saeR codY (CYL11391) was pelleted and washed with cold saline, suspended in 10 ml TS buffer (10 mM Tris-Cl, pH 7.6, 150 mM NaCl) with a protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN), and sonicated briefly to dislodge cell aggregates. Cells were lysed with 0.1-mm zirconia-silica beads (BioSpec Products, Bartlesville, OK) in a Fast Prep homogenizer (MP Biomedicals, Solon, OH), using six 40-s pulses at speed 6, with 5-min intervals on ice between pulses. Cell lysates were collected, clarified by centrifugation at 18,000 × g for 20 min at 4°C, and applied to a 1-ml heparin-agarose column (Sigma-Aldrich, St. Louis, MO) to enrich the DNA-binding proteins. Heparin-agarose affinity column chromatography was carried out as described by Trubetskoy et al. (32). The fractions of DNA-binding proteins that eluted from the heparin-agarose column were analyzed by 12% SDS-PAGE.
EMSA and proteomic analysis.
A 156-bp DNA fragment containing the cap promoter region was generated by PCR amplification from S. aureus Newman chromosomal DNA by using oligonucleotide primers cp8gs6 and cp8gs3. The DNA fragment was labeled with digoxigenin (Dig)-dUTP by using a Dig gel shift kit (Roche Applied Science, Indianapolis, IN). Electrophoretic mobility shift assay (EMSA) was performed as described previously (27). To prepare a mutant Pcap fragment (Pcapmt), primers cp8gs6 and cp8gs3 were used for PCR amplification of chromosomal DNA from S. aureus strain CYL6401 (which has a 4-bp mutation within the 10-bp IR in Pcap) (23). To prepare truncated Pcap fragments for competition experiments, primer pairs cp8gs5/cp8gs6 and cp8gs3/gp8gs4 were used for PCR amplification of a 92-bp fragment of Pcap upstream of the 10-bp IR (Pcap5′) and a 75-bp fragment of Pcap downstream of the 10-bp IR (Pcap3′), respectively. For proteomic analysis, a preparative EMSA gel (1.5-mm thick) was used with unlabeled DNA probes and then stained with Coomassie blue G250. The gel bands were excised and submitted for proteomic analysis by in-gel trypsin digestion followed by liquid chromatography and tandem mass spectrometry (GeLC-MS/MS) at the UAMS Proteomic Core Facility.
His6-RbsR recombinant protein expression and purification.
To express the His6-RbsR protein, pML4237 was transformed into E. coli Rosetta2(DE3)(pLysS) (Novagen). Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.05 in LB medium containing 30 μg/ml kanamycin and 34 μg/ml chloramphenicol, grown at 37°C until an OD600 of about 1, and then induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C for protein expression. Bacterial cells were harvested by centrifugation and stored at −80°C until use. The cells were thawed on ice and lysed with SoluLyse (Amsbio, Lack Forest, CA), and the His6-RbsR protein was purified using a His-Bind kit (Novagen) according to the manufacturer's instructions. The buffer of the eluted protein was then changed to 20 mM HEPES buffer, pH 7.9, containing 300 mM KCl, 1 mM EDTA, 2 mM dithiothreitol (DTT), and 25% glycerol, using a Zeba Spin desalting column (Thermo Scientific, Waltham, MA) according to the manufacturer's instructions.
Northern blot analysis.
Total RNAs were isolated as described previously (27). For Northern blotting, the 517-bp rbsR-specific, 495-bp rbsU-specific, 521-bp rpi-specific, and 530-bp prs-specific DNA probes were synthesized by using PCR Dig probe synthesis kits (Roche Applied Sciences) with primer pairs rbsR-6/rbsR-7, rbsU-1/rbsU-2, rpi-1/rpi-2, and prs-1/prs-2, respectively. Denaturing RNA gel electrophoresis (1% agarose) was carried out as described by Masek et al. (33), except that the buffer was replaced with TBE (90 mM Tris-borate, 2 mM EDTA) buffer. Northern hybridization was carried out as described previously (27).
Nonradioactive DNase I footprinting.
Plasmid pCL3169, which contains a 614-bp Pcap fragment, was used as the template to synthesize a 156-bp probe by PCR, using the 6-carboxyfluorescein (FAM)-labeled primer FAM-FP6 and the VIC-labeled primer VIC-FP3, which correspond to positions −135 to +21 of Pcap with respect to the transcriptional start site of the cap operon (23). The PCR DNA fragments were purified using a NucleoSpin column (Clontech, Mountain View, CA). The procedure for DNase I footprinting was essentially as described by Zianni et al. (34). Briefly, the reaction mixture (20 μl), which consisted of 1.36 μg purified His6-RbsR, 80 ng of fluorescent dye-labeled DNA probe, 2 μg of bovine serum albumin (BSA), 0.1 μg of poly-l-lysine, and 1 μg of poly(dI-dC) in binding buffer [20 mM HEPES, pH 7.6, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% Tween 20, 30 mM KCl], was incubated at 23°C for 15 min. DNase I (0.08 U; New England BioLabs) was added to the reaction mixture, the mixture was incubated at 23°C for 4 min, and the reaction was stopped by incubation at 78°C for 10 min. The DNA fragments were purified by use of a Mini Elute PCR kit (Qiagen, Valencia, CA) and eluted in 25 μl of H2O. The experiments were repeated two times. Fifteen microliters of each purified DNA fragment, along with primers FAM-FP6 and VIC-FP3 and plasmid pCL3169, was submitted to the Ohio State University Plant-Microbe Genomic Facility for fragment analysis and sequencing using an Applied Biosystems 3730 DNA analyzer. The RbsR DNA-binding sites were determined by aligning the sizes of the fragments and sequences of the probe.
TSS determination.
The transcriptional start site (TSS) of rbsR was determined by using the adaptor- and radioactivity-free (ARF-TSS) method of Wang et al. (35). Briefly, 5 μg of total RNA isolated from S. aureus strain 11481 was used for cDNA synthesis with the 5′-end-phosphorylated primer rbsR-T1, using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNAs were then degraded using 0.25 N NaOH and neutralized with 0.25 N HCl. cDNAs were purified with a Mini Elute PCR kit (Qiagen). The 5′-phosphoryl-terminated single-stranded cDNA was then covalently joined to the 3′ end (TSS) of the cDNA by using T4 RNA ligase (Epicentre, Madison, WI). Circularized cDNAs were amplified by inverse PCR using the divergent primers rbsR-T2 and rbsR-T3. The PCR products were cloned into pGEM-T Easy (Promega) and sequenced. A QuantiTect reverse transcription kit (Qiagen) was used for reverse transcription-PCR (RT-PCR) to estimate the ends of the rbsR transcript according to the manufacturer's instructions, using primer pair rbsR-11/rbsR-12 for the 5′ end and primer pair rbsR-13/rbsR-14 for the 3′ end. PCR DNA products were analyzed using 2% agarose gel electrophoresis with TBE buffer.
Other methods.
To create a two-plasmid system for the SigB-dependent promoter assay, plasmid pML4261 or pML4262 containing the PrbsR-lacZ fusion was transformed into E. coli XL1-Blue containing pAC7 or pAC7-sigB, and clones were selected on LBACX-ARA plates as described by Homerova et al. (29). For capsule immunoblotting, capsules were prepared as described previously (6), using TSB without glucose (TSB-0G). Serially diluted samples (1.5 μl each) were applied directly to a nitrocellulose membrane by using a pipette. Membranes were treated with a specific anticapsule antibody and detected as described previously (6). BlaZ (β-lactamase) assays for Pcap::blaZ fusions were performed with cultures grown in TSB-0G according to a previously described procedure (36). Data for the promoter fusion assays were analyzed by GraphPad Prism (San Diego, CA), using the paired Student t test.
RESULTS
Identification of Pcap-binding proteins.
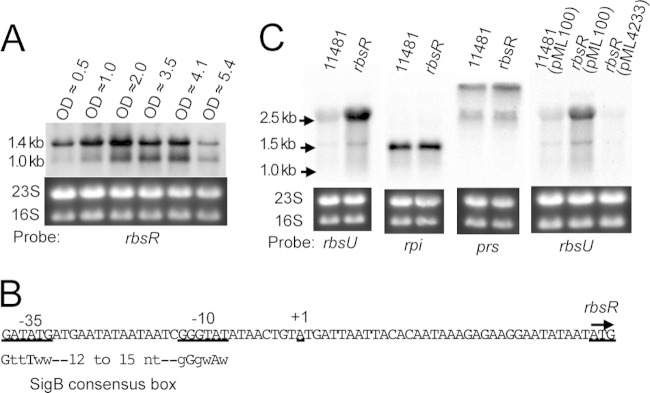
Previously, we identified CodY as a repressor of capsule by identifying proteins bound to the Pcap DNA fragment (15). Because CodY is an abundant cytoplasmic protein, we chose a codY-null strain in our experiment to avoid interference. In addition, because ClpC represses capsule through the SaeRS two-component system in strain Newman, it was possible that ClpC and SaeRS would also have a negative impact on our effort. Thus, we used a Newman clpC saeR codY triple mutant as the source to prepare cell extracts. The cell extracts were further enriched for DNA-binding proteins by using a heparin column (Fig. 1A). The enriched protein fraction was then used in EMSAs with a 156-bp digoxigenin (Dig)-labeled Pcap DNA fragment as a probe (Fig. 1B). A broad shifted band which was absent in the negative control was identified (Fig. 1C), suggesting the presence of putative DNA-binding proteins other than CodY. To identify these putative DNA-binding regulators, EMSAs were repeated using the same 156-bp Pcap DNA fragment, but without Dig labeling. Both the ethidium bromide- and Coomassie blue-stained gels revealed a shifted band just below the loading wells (Fig. 1D), at a position similar to that of the band identified by the Dig-labeled probe, suggesting that the shifted bands contained proteins (as identified by the Coomassie blue stain) interacting with the Pcap DNA fragment (as identified by the ethidium bromide stain). We also included experiments using a mutant Pcap DNA fragment, containing a 4-bp substitution within the 10-bp IR (Pcapmt) (Fig. 1B), that was amplified with the same primers. A similar shifted band was also found with the Pcapmt fragment, indicating the presence of 10-bp IR-independent DNA-binding proteins in the shifted band (Fig. 1D). The regions containing the shifted bands identified by Pcap and Pcapmt and the corresponding region in the negative-control lane (with no DNA) from the Coomassie blue-stained gels (Fig. 1D, right panel) were excised and subjected to protein analysis by GeLC-MS/MS. By comparing the spectral counts, we identified 9 proteins that were present in the EMSA using Pcap or Pcapmt but were absent or had much reduced spectral counts in the negative control (data not shown). To test the effects of these proteins on capsule, mutants of the corresponding genes were constructed either by phage transduction from NARSA Nebraska transposon mutants into strain CYL11481 or by allelic replacement in strain CYL11481. We found 6 mutants that had an effect on capsule (Fig. 2A). Among these putative capsule regulators, only XdrA was shown previously to affect cap genes, in a gene profiling study (37). In the present study, we chose to focus on NWMN_0205, which has been annotated RbsR based on homology to a repressor controlling the rbs operon involved in ribose utilization in other bacteria (38–40).
FIG 1.
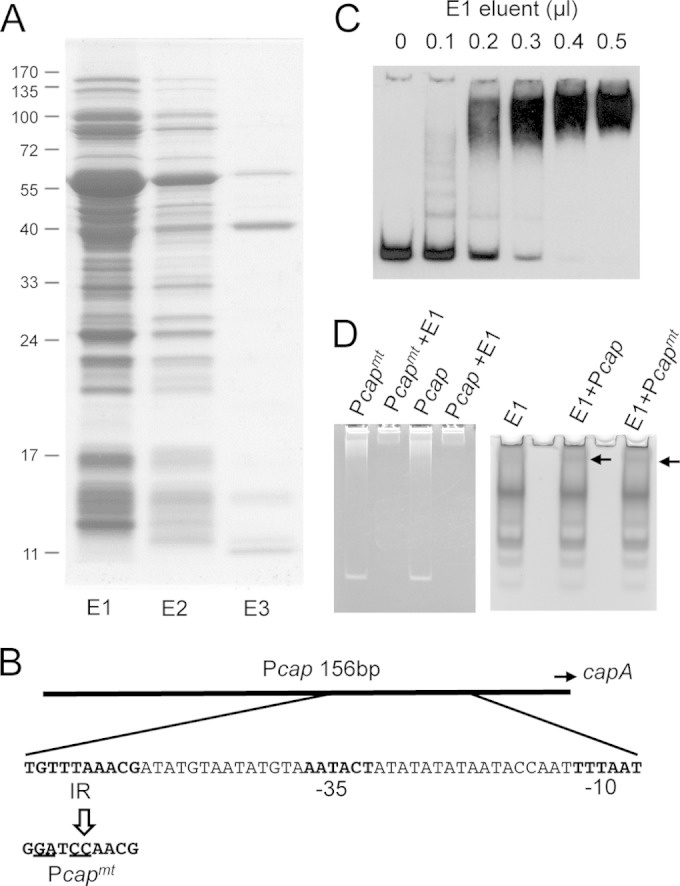
Identification of Pcap DNA-binding proteins. (A) SDS-PAGE analysis of heparin-agarose column-fractionated proteins. Proteins (10 μl) of each fraction were subjected to 12% SDS-PAGE and stained with Coomassie blue G250. E1, E2, and E3 refer to eluents of three consecutive fractionations. E1 and E2 were eluted with buffer containing 0.5 M KCl. E3 was eluted with buffer containing 1.0 M KCl. (B) cap promoter region, with the 10-bp IR and the −35 and −10 sequences shown in bold. The Pcapmt DNA sequence containing the 4-bp substitution (23) is also shown. (C) EMSA using the Dig-labeled 156-bp Pcap fragment and increasing amounts of the E1 fraction, as indicated at the top. (D) EMSA using Pcap DNA or Pcapmt DNA (∼50 ng) and 4 μl of the E1 fraction. Gels were stained with ethidium bromide (left) or Coomassie blue G250 (right). Arrows indicate shifted bands.
FIG 2.
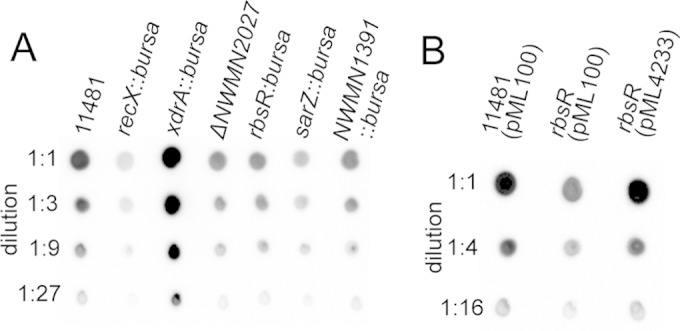
Immunoblotting of capsule. (A) Various mutants derived from the Newman P18L strain (CYL11481) were grown in TSB-0G for 4 h for capsule isolation. (B) Complementation of the rbsR mutant (CYL12834) with pML4233 (pML100-rbsR) for restoration of the capsule phenotype. Capsules were isolated from cultures grown in TSB-0G in the presence of 2.5 μg/ml chloramphenicol for 2 h and then induced with 200 ng/ml of ATc for 2 h.
RbsR activates capsule by binding to the cap promoter.
The results described above (Fig. 2A) suggest that RbsR is a putative activator of capsule production. To confirm this, we cloned the rbsR gene from strain Newman into pML100 under the control of the Pxyl/tetO promoter (pML4233). As shown in Fig. 2B, the capsule phenotype of the rbsR mutant (CYL12834) was restored to the wild type when the strain was complemented with pML4233 in the presence of anhydrotetracycline (ATc). Because there was no other gene present in the cloned fragment, the results confirmed that RbsR is an activator of capsule.
RbsR is a putative transcriptional regulator containing a helix-turn-helix DNA-binding motif, suggesting that it regulates its target genes by direct DNA binding. To determine whether RbsR binds to the cap promoter region, a His6-RbsR fusion protein was expressed in E. coli and purified by use of a His-Bind resin affinity column. The purified protein was used in EMSAs with the Dig-labeled 156-bp Pcap DNA fragment as a probe. As shown in Fig. 3A, the Pcap fragment could readily be upshifted by His6-RbsR. The shifted band could be competed away with a cold Pcap DNA fragment, suggesting that the binding was specific. The binding dissociation constant (Kd) was then determined to be ∼4.9 nM (Fig. 3B). To determine whether the 10-bp IR upstream of the −35 region of the cap promoter is required for binding, we used the 4-bp mutant Pcapmt fragment for cold competition in EMSA. The results (Fig. 3A) showed that the majority of the shifted band remained unchanged. In addition, the shifted band was not outcompeted by using a 92-bp DNA fragment upstream (Pcap5′) of the 10-bp IR or a 75-bp downstream DNA fragment (Pcap3′) (Fig. 3A). Thus, these cold competition results suggest that the 10-bp IR sequence is important for RbsR binding. To further localize the RbsR binding site, we performed a fluorescence-based footprinting experiment. The results in Fig. 4 show that, on the sense strand, RbsR protected a 46-nucleotide (nt) region that centers on the 10-bp IR sequence, which also includes the nearby downstream −35 region of the promoter. On the antisense strand, it protected a 16-nt region that centers on the −35 region of the promoter. Although the binding site is much larger than the 10-bp IR sequence, the footprinting results are consistent with those of the EMSAs. In addition, the results showing that RbsR also protected the −35 region suggest that RbsR may interact directly with RNA polymerase to activate cap mRNA transcription.
FIG 3.
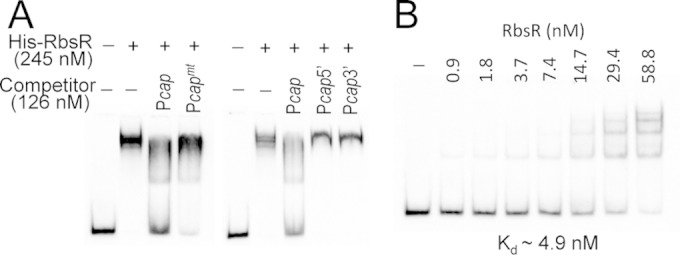
EMSAs of Pcap with the His-RbsR protein. (A) The shifted band could be competed away effectively by the cold Pcap fragment but not by the Pcapmt fragment, containing a 4-bp mutation; the Pcap5′ fragment, containing the 5′ half of the 10-bp IR with upstream sequence; or the Pcap3′ fragment, containing the 3′ half of the 10-bp IR with downstream sequence. (B) A Kd value of ∼4.9 nM was determined by using increasing amounts of His-RbsR and a constant amount of labeled Pcap fragment (0.63 nM).
FIG 4.
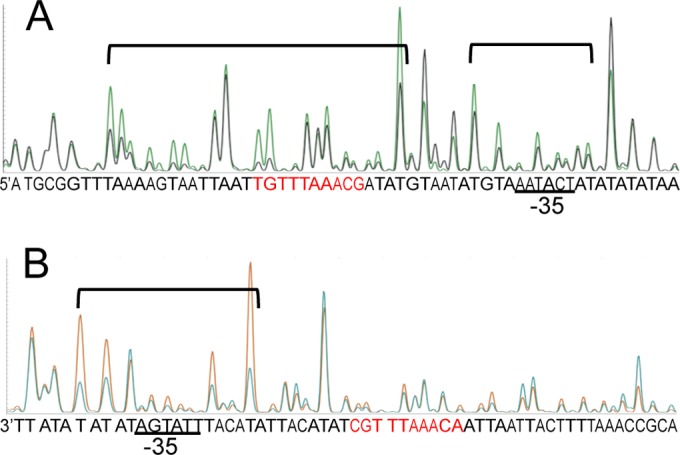
DNase I footprinting analysis of the 5′-FAM-labeled sense strand (A) and the 5′-VIC-labeled antisense strand (B) of the Pcap probe. A reduction in intensity of DNase I-digested fragments in the presence of 1.9 μM RbsR (black peaks in panel A and blue peaks in panel B) compared to that in its absence (green peaks in panel A and orange peaks in panel B) indicates protection. Protected regions are indicated by brackets. Sequences in red indicate the 10-bp IR required for Pcap activation. The results suggest that RbsR binds to the region encompassing the 10-bp IR and the −35 region of the promoter.
Characterization of rbsR transcription.
To characterize the transcription of rbsR in different growth phases, Northern analyses were performed at different time points. As shown in Fig. 5A, two transcripts, of ∼1.0 kb and ∼1.4 kb, were identified; both could carry the full-length (999 bp) rbsR gene. These results suggest that rbsR is a monocistronic gene. The larger, ∼1.4-kb transcript increased gradually but most prominently in the late exponential growth phase, at an OD600 of about 2.0 (mid-log phase), whereas the ∼1.0-kb transcript increased to the highest level at OD600s of about 3.5 and 4.1 (early stationary growth phase), suggesting that rbsR transcription is growth phase dependent. These results suggest that rbsR is transcribed from two promoters or transcribed from one promoter with two different 3′ ends. To test these possibilities, we employed the ARF-TSS method (35) to map the 5′ end of the rbsR transcript. We sequenced 8 clones, and all had the same TSS, at the A residue 35 nt upstream of the rbsR ATG start site (Fig. 5B), suggesting that there is a single rbsR promoter. To confirm this prediction, we performed RT-PCR. We were able to amplify a fragment extending from just inside the 3′ end of rbsR to about 200 bp downstream from the stop codon with the primer pair rbsR-13/rbs-14 but were unable to amplify a fragment extending from the 5′ end and encompassing about 300 bp upstream of the start codon by using the primer pair rbsR-11/rbs-12 (results not shown). Thus, taking the results of TSS mapping, RT-PCR, and Northern blotting (Fig. 5A and B) together, we suggest that rbsR is transcribed from one promoter, but with two 3′ ends. The two transcription ends could be due to transcription being terminated at two terminators or to processing of the longer transcript. However, a strong intrinsic terminator has been identified 36 bp downstream of the rbsR gene (41). Based on the start site and the predicted terminator, we estimated the rbsR transcript to be ∼1.1 kb, which matches the size of the shorter transcript that we identified by Northern blotting. This suggests that the two transcripts likely resulted from two different terminators rather than from processing of the longer transcript.
FIG 5.
Transcription analyses. Total RNA (5 μg) from each sample used for Northern blotting was denatured in formamide, applied to TBE-agarose gels, and hybridized with the Dig-labeled specific probe indicated below each blot. (A) Expression of rbsR in CYL11481 cultures grown to different OD600s, as indicated. (B) Map of TSS and predicted promoter of rbsR. The SigB binding consensus is shown below, with capital letters denoting highly conserved nucleotides and lowercase letters denoting poorly conserved nucleotides (w = A or T). (C) Effects of RbsR on rbsU, rpi, and prs transcription. Total RNAs were prepared from 4-h cultures (OD600, ∼4.0) of S. aureus CYL11481 or the rbsR mutant (CYL12834) and hybridized with specific probes, as indicated. The expression of rbsU was also assessed by complementation of rbsR by using pML100-rbsR (pML4233) in the presence of ATc.
RbsR represses transcription of the rbsUDK operon.
The operonic rbsUDK gene cluster is predicted to encode proteins involved in ribose uptake and phosphorylation. RbsR has been annotated as the repressor of the rbsUDK operon, based on its homology with RbsR repressors in other bacteria (38–40). An in silico analysis also predicted that it binds to a site just upstream of the rbsU gene (42). To test whether RbsR affects rbsUDK expression, we employed Northern hybridization using an internal fragment of the rbsU gene as a probe. The results (Fig. 5C) showed that a pronounced increase in the ∼2.5-kb band was detected for the rbsR mutant compared to the wild-type strain (Fig. 5C), indicating that RbsR is a repressor of rbsUDK. The mutant phenotype could readily be complemented with a DNA fragment carrying the wild-type rbsR gene. The size of the rbsU-specific band also indicates that rbsUDK is transcribed as an operon. In many bacteria, ribose is rapidly phosphorylated upon uptake. The resulting ribose-5-phosphate could be converted by Prs to 5-phospho-ribose-1-diphosphate, a precursor for purine, pyrimidine, and histidine synthesis, or converted by Rpi to ribulose-5-phosphate in one of the steps of the pentose phosphate cycle. Our Northern blotting results, however, showed that RbsR did not affect the expression of either the prs or rpi gene (Fig. 5C).
SigB directly activates RbsR.
Although many regulators have been found to affect capsule production, most of them are likely to regulate capsule indirectly. We therefore speculate that RbsR could serve as a downstream regulator of one or more of these upstream regulators. To test this possibility, the expression of rbsR in various mutants, including agr, mgrA, clpC, codY, saeRS, arlRS, sigB, and sbcDC mutants, was tested by Northern blotting. Among the proteins examined, only SigB had an apparent effect on rbsR expression (Fig. 6A). Indeed, rbsR was previously shown to be upregulated by SigB in a microarray transcriptional profiling study, and an imperfect SigB box upstream of the rbsR gene has been identified (Fig. 5B) (43), suggesting that SigB may bind to the rbsR promoter directly. To confirm that SigB directly affects rbsR transcription, we employed a two-plasmid system as described by Homerova et al. (29). As shown in Fig. 6B, we found that the rbsR promoter was activated in E. coli only when S. aureus SigB was also expressed, confirming that SigB is required for rbsR activation, most likely by direct promoter binding. SigB has been shown to activate capsule through SpoVG or ArlR (14, 44). Neither SpoVG nor ArlR affected rbsR transcription (not shown), indicating that the SigB-RbsR pathway affecting capsule production is independent of the SpoVG or ArlRS pathway.
FIG 6.
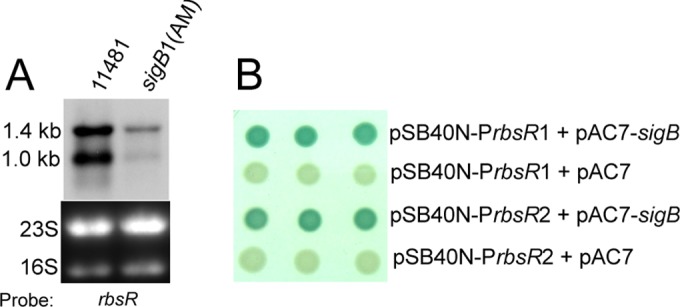
Effect of SigB on rbsR. (A) Northern blot analysis. Total RNAs were prepared from 4-h cultures (OD600, ∼4.0) of S. aureus CYL11481 or the sigB1(Am) mutant (CYL13113; contains an amber mutation in sigB) and hybridized with an rbsR-specific probe. (B) E. coli bacterial two-plasmid system. A promoter region of rbsR, PrbsR1 (153 bp) or PrbsR2 (756 bp), was cloned into the promoter probe plasmid pSB40N (giving pML4261 or pML4262, respectively) and transformed into E. coli XL1-Blue containing pAC7-sigB or pAC7. Clones were selected on LBACX-ARA plates. Positive transformants (containing pAC7-sigB) were blue, whereas negative controls (containing the pAC7 vector) were colorless. The longer PrbsR2 region was used in the study to include potential unknown upstream promoters.
The repression of rbsUDK by RbsR, but not the activation of cap, is affected by ribose.
Ribose has been shown to be an inducer controlling RbsR regulatory function in E. coli but not in Bacillus subtilis (38, 45). To determine whether ribose affects RbsR regulatory function in S. aureus, we performed Northern blotting to determine the effect of RbsR on the expression of the rbsU gene in the presence or absence of d-ribose, using TSB-0G (i.e., TSB without glucose) as the basal medium. As shown in Fig. 7A, we found that the rbsU gene was derepressed by RbsR in the presence of ribose, suggesting that ribose is an inducer that relieves the repression of the rbsUDK operon by RbsR. Likewise, because Pcap is activated by RbsR, we also tested whether RbsR activation of capsule is affected by ribose. To this end, we employed a Pcap-blaZ fusion plasmid and compared the BlaZ activities in the wild-type strain and the rbsR mutant in the presence and absence of d-ribose in the TSB-0G growth medium. As shown in Fig. 7B, we found that the Pcap promoter activity decreased in the rbsR mutants in both the presence and absence of d-ribose but that there was no significant difference between the absence and presence of d-ribose in either the wild-type strain or the rbsR mutant, suggesting that d-ribose in the medium does not affect RbsR activation of the cap operon.
FIG 7.
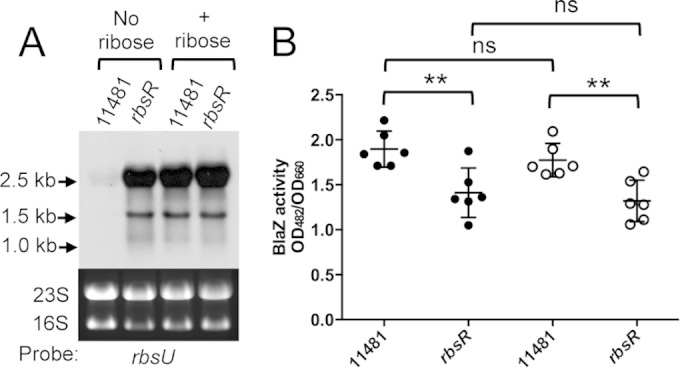
Effect of d-ribose on RbsR regulatory function as assayed in TSB-0G basal medium. (A) Expression of rbsU as assayed by Northern analyses. Total RNAs were prepared from 4-h cultures (OD600, ∼3.2) of S. aureus CYL11481 or the rbsR mutant (CYL12834) in the absence or presence of d-ribose and hybridized with an rbsU-specific probe. (B) cap promoter activities measured by BlaZ reporter assays. Four-hour cultures of CYL11481 or the rbsR mutant containing the Pcap-blaZ fusion plasmid (pAM3176) were assayed for BlaZ activity in the absence (solid circles) or presence (open circles) of d-ribose. ns, not significant; **, P < 0.01.
DISCUSSION
S. aureus can infect almost any human or animal tissues and can survive outside the host for a long time. It is therefore not surprising that the organism needs to have a large number of regulators to properly regulate various factors required for adapting to different environments. Using capsule as a target virulence factor, we and others have identified more than a dozen regulators involved in capsule gene regulation. In the present study, we attempted to identify transcriptional regulators capable of binding to the promoter region of the cap operon. By using an approach that directly analyzes the proteins bound to Pcap DNA, we found six putative DNA-binding regulators. One of these newly identified regulators, RbsR, was further characterized and shown to bind specifically to the cap promoter region. Although the other five putative regulators have not been characterized fully, our finding is rather surprising, as we did not expect to find that many potential regulators capable of binding to the cap promoter. Adding to the previously known capsule regulators, the remarkable number of regulators devoted to controlling single virulence factors further points to the extreme complexity of the virulence regulatory network in S. aureus.
RbsR is a LacI family repressor that has not been characterized previously for S. aureus. It shares 29% to 39% amino acid identity with the RbsR proteins from Lactobacillus sakei, E. coli, and B. subtilis (38, 40, 45), which have been shown to repress the rbs operon involved in ribose uptake and phosphorylation. Our results presented here demonstrate that S. aureus RbsR is also a repressor of the rbsUDK operon, predicted to be involved in ribose transport and phosphorylation. Ribose can serve as a source for energy, via the pentose phosphate pathway (PPP), and for nucleotide synthesis in many bacteria. In solution, ribose exists as α- and β-pyranose forms, with the latter being the predominant form. In S. aureus, ribose is likely imported via RbsU and converted by RbsD to the α form, which can then be recognized by RbsK and converted to ribose-5-phosphate, which either becomes part of the PPP or serves as the precursor for nucleotide synthesis. The RbsU transporter is used by S. aureus and L. sakei for ribose uptake, whereas a tricomponent ATP-binding cassette transporter composed of RbsABC is operated in E. coli and B. subtilis. In this study, we used Northern analyses (Fig. 7A) to show that the repression of the rbsUDK operon by RbsR could be derepressed by ribose in S. aureus. These results suggest that ribose or one of its derivatives may be an inducer that interacts directly with RbsR to derepress the negative regulation. This regulation is similar to the ribose induction of rbsDACBK repression by RbsR in E. coli (38). However, in B. subtilis, repression of the rbsKDACB operon by RbsR does not respond to ribose in the growth medium (45).
In this study, we discovered that RbsR was not only a repressor of the rbs operon but also an activator of the cap genes. RbsR had previously been thought to regulate genes only in the rbs operon. However, Shimada et al. (46) recently showed that E. coli RbsR also binds to the promoters of a set of genes resulting in repression and activation of the de novo and salvage purine nucleotide synthesis pathways, respectively. In Corynebacterium glutamicum, RbsR affects only the rbs genes, but in association with a coregulator, RbsR can also affect genes involved in the utilization of uridine (47). Since ribose is a direct source of ribose-5-phosphate, which is a key intermediate for synthesizing nucleotides (via phosphoribosylpyrophosphate), it is not surprising that the genes involved in nucleotide metabolism are also regulated by RbsR in these bacteria. However, our finding that the cap operon is also a direct target of RbsR in S. aureus is rather unusual, as the capsule biosynthetic pathway and the ribose utilization pathway are not closely linked.
Although RbsR regulates both the rbs and cap operons, we found that ribose had no effect on the RbsR activation of cap gene expression. These results suggest that the mechanism involved in rbsUDK repression by RbsR is different from that involved in the activation of the cap operon. The consensus RbsR binding site in the promoter region of the rbs operon in the Bacillus/Clostridium group of bacteria, including S. aureus, has been defined by a comparative approach (41). The predicted consensus RbsR box upstream of the rbsUDK operon in S. aureus bears no resemblance to the RbsR binding site in the cap promoter region as defined by footprinting in that study. The 10-bp IR sequence is also not found within a 1,000-bp region upstream of the rbsU open reading frame. These findings further corroborate that RbsR may regulate the rbs and cap genes by different mechanisms.
SigB is a stress response sigma factor that also controls the expression of a number of virulence factors. SigB has been shown to independently activate capsule through SpoVG and ArlR (44). Recently, SpoVG was shown to bind a 28-bp region that is 41 bp further upstream of the 10-bp IR (24). However, we reported earlier that deletion of a sequence further upstream of the 10-bp IR had no detectable effect on cap gene expression (23). Because there is no direct evidence that the 28-bp region of the SpoVG binding site is involved in cap gene transcription, based on our previous results (23), we speculate that SpoVG may affect capsule indirectly rather than by binding at this region. In addition to SpoVG and ArlR, through which SigB can regulate capsule, in this study we add a third circuit of regulation, through RbsR. Recently, SigB was also found to negatively regulate capsule by activating RsaA, a small RNA that inhibits MgrA translation, thereby reducing capsule production (48). Thus, at least four independent pathways are now known to be involved in SigB regulation of capsule (Fig. 8). The multiple pathways by which capsule can be regulated by SigB suggest that this regulation has a high degree of complexity, which will require additional studies to understand the biological significance of the regulation.
FIG 8.
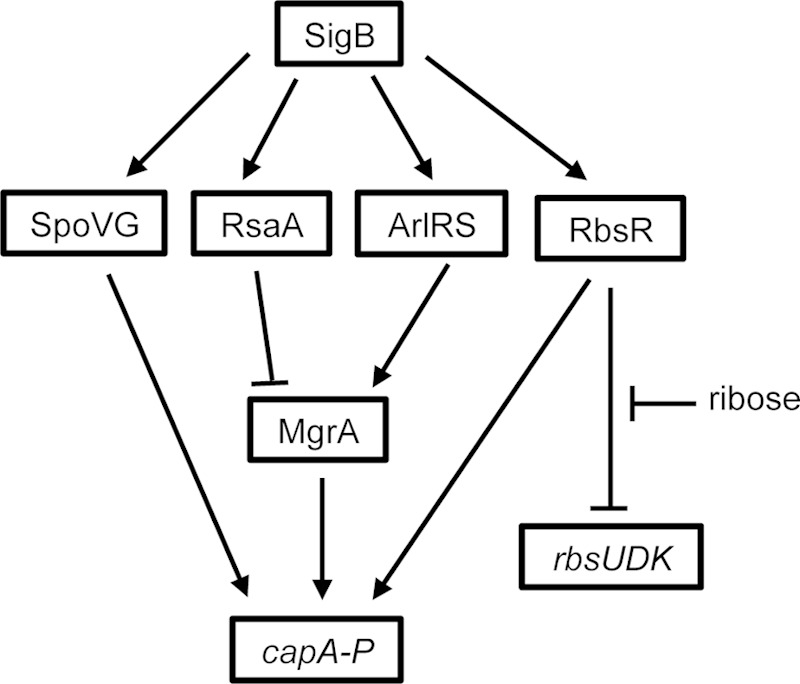
Proposed regulatory circuits for RbsR. RbsR is activated by SigB and in turn represses the rbs operon and activates the cap operon by directly binding at the promoter. Repression of the rbs operon by RbsR is relieved by d-ribose. Arrows indicate activation, and block arrows indicate repression.
There are ample examples of coregulation of metabolism and virulence in S. aureus (49). Our finding that RbsR is involved in ribose utilization as well as capsule production suggests that RbsR could also be an important regulator linking metabolism and virulence regulation. Ribose is present at ∼0.1 mM in human blood (50, 51) and in various amounts in other tissues (52). It is likely that the availability of ribose in the tissues affects S. aureus pathogenesis by promoting bacterial growth. However, our finding that repression of the rbs operon by RbsR, but not activation of capsule, is controlled by ribose suggests that ribose is not likely an effector linking the two cellular processes. On the other hand, because RbsR is highly regulated by SigB, whose activity is affected by certain in vitro and in vivo stress conditions (53–55), stress signals that modulate SigB activity are likely to be important effectors for controlling the quantity of RbsR, thereby affecting ribose uptake and capsule production. However, determining which signals are involved and how transduction of these signals through SigB affects the expression of RbsR requires further in-depth studies.
ACKNOWLEDGMENTS
This work was supported by grant AI113766 from the National Institute of Allergy and Infectious Diseases (NIAID). Nebraska transposon mutants were obtained through the Network of Antimicrobial Resistance in S. aureus (NARSA) program, which is supported by the NIAID/NIH. We also acknowledge the UAMS sequencing core and proteomic core, supported in part by National Institutes of Health grants P20GM103420, P30GM103450, and P20GM103625.
We thank M. Bischoff for providing plasmids pSB40N, pAC7, and pAC7-sigB and S. aureus GP266. We thank Ravi Gupta, Yi-Chun Pang, and Justin Graham for critical readings of the manuscript.
REFERENCES
- 1.Junecko J, Zielinska AK, Mrak LN, Ryan DC, Graham JW, Smeltzer M, Lee CY. 2012. Transcribing virulence in Staphylococcus aureus. World J Clin Infect Dis 2:63–76. doi: 10.5495/wjcid.v2.i4.63. [DOI] [Google Scholar]
- 2.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 3.Bronner S, Monteil H, Prévost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev 28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Luong TT, Lee CY. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123–3131. doi: 10.1099/mic.0.29177-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Luong TT, Lee CY. 2007. The sbcDC locus mediates repression of type 5 capsule production as part of the SOS response in Staphylococcus aureus. J Bacteriol 189:7343–7350. doi: 10.1128/JB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luong TT, Sau K, Roux C, Sau S, Dunman PM, Lee CY. 2011. Staphylococcus aureus ClpC divergently regulates capsule via sae and codY in strain Newman but activates capsule via codY in strain UAMS-1 and in strain Newman with repaired saeS. J Bacteriol 193:686–694. doi: 10.1128/JB.00987-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham JW, Lei MG, Lee CY. 2013. Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J Bacteriol 195:4506–4516. doi: 10.1128/JB.00758-13 (Erratum, 196:2324, 2014.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Riordan K, Lee JC. 2004. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CY, Lee JC. 2006. Staphylococcal capsules, p 456–463. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-positive pathogens, 2nd ed American Society for Microbiology, Washington, DC. [Google Scholar]
- 10.Sau S, Sun J, Lee CY. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol 179:1614–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sau S, Bhasin N, Wann ER, Lee JC, Foster TJ, Lee CY. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 12.Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bächi B, Bischoff M. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother 50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michel A, Agerer F, Hauck CR, Herrmann M, Ullrich J, Hacker J, Ohlsen K. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J Bacteriol 188:5783–5796. doi: 10.1128/JB.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier S, Goerke C, Wolz C, Seidl K, Homerova D, Schulthess B, Kormanec J, Berger-Bächi B, Bischoff M. 2007. σB and the σB-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus. Infect Immun 75:4562–4571. doi: 10.1128/IAI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192:2861–2877. doi: 10.1128/JB.00220-10 (Erratum, 192:4258.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L, Xue T, Shang F, Sun H, Sun B. 2010. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect Immun 78:3506–3515. doi: 10.1128/IAI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadykov MR, Mattes TA, Luong TT, Zhu Y, Day SR, Sifri CD, Lee CY, Somerville GA. 2010. Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus type 5 and 8 capsular polysaccharides. J Bacteriol 192:1459–1462. doi: 10.1128/JB.01377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mootz JM, Benson MA, Heim CE, Crosby HA, Kavanaugh JS, Dunman PM, Kielian T, Torres VJ, Horswill AR. 2015. Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol Microbiol 96:388–404. doi: 10.1111/mmi.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Y, Liu X, Chen F, Di H, Xu B, Zhou L, Deng X, Wu M, Yang CG, Lan L. 2014. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci U S A 111:E4981–E4990. doi: 10.1073/pnas.1411077111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun F, Ji Q, Jones MB, Deng X, Liang H, Frank B, Telser J, Peterson SN, Bae T, He C. 2012. AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J Am Chem Soc 134:305–314. doi: 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pöhlmann-Dietze P, Ulrich M, Kiser KB, Döring G, Lee JC, Fournier JM, Botzenhart K, Wolz C. 2000. Adherence of Staphylococcus aureus to endothelial cells: influence of capsular polysaccharide, global regulator agr, and bacterial growth phase. Infect Immun 68:4865–4871. doi: 10.1128/IAI.68.9.4865-4871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risley AL, Loughman A, Cywes-Bentley C, Foster TJ, Lee JC. 2007. Capsular polysaccharide masks clumping factor A-mediated adherence of Staphylococcus aureus to fibrinogen and platelets. J Infect Dis 196:919–927. doi: 10.1086/520932. [DOI] [PubMed] [Google Scholar]
- 23.Ouyang S, Sau S, Lee CY. 1999. Promoter analysis of the cap8 operon, involved in type 8 capsular polysaccharide production in Staphylococcus aureus. J Bacteriol 181:2492–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jutras BL, Chenail AM, Rowland CL, Carroll D, Miller MC, Bykowski T, Stevenson B. 2013. Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins. PLoS One 8:e66683. doi: 10.1371/journal.pone.0066683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bischoff M, Berger-Bächi B. 2001. Teicoplanin stress-selected mutations increasing σB activity in Staphylococcus aureus. Antimicrob Agents Chemother 45:1714–1720. doi: 10.1128/AAC.45.6.1714-1720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei MG, Cue D, Roux CM, Dunman PM, Lee CY. 2011. Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol 193:5231–5241. doi: 10.1128/JB.05454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezuchova B, Kormanec J. 2001. A two-plasmid system for identification of promoters recognized by RNA polymerase containing extracytoplasmic stress response sigma(E) in Escherichia coli. J Microbiol Methods 45:103–111. doi: 10.1016/S0167-7012(01)00237-8. [DOI] [PubMed] [Google Scholar]
- 29.Homerova D, Bischoff M, Dumolin A, Kormanec J. 2004. Optimization of a two-plasmid system for the identification of promoters recognized by RNA polymerase containing Staphylococcus aureus alternative sigma factor σB. FEMS Microbiol Lett 232:173–179. doi: 10.1016/S0378-1097(04)00063-1. [DOI] [PubMed] [Google Scholar]
- 30.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537–12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraemer GR, Iandolo JJ. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr Microbiol 21:373–376. doi: 10.1007/BF02199440. [DOI] [Google Scholar]
- 32.Trubetskoy DO, Zavalova LL, Akopov SB, Nikolaev LG. 2002. Purification of proteins specifically binding human endogenous retrovirus K long terminal repeat by affinity elution chromatography. J Chromatogr A 976:95–101. doi: 10.1016/S0021-9673(02)01236-0. [DOI] [PubMed] [Google Scholar]
- 33.Masek T, Vopalensky V, Suchomelova P, Pospisek M. 2005. Denaturing RNA electrophoresis in TAE agarose gels. Anal Biochem 336:46–50. doi: 10.1016/j.ab.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17:103–113. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Lee J, Deng Y, Tao F, Zhang LH. 2012. ARF-TSS: an alternative method for identification of transcription start site in bacteria. Biotechniques 52:1–3. doi: 10.2144/000113858. [DOI] [PubMed] [Google Scholar]
- 36.Luong T, Sau S, Gomez M, Lee JC, Lee CY. 2002. Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect Immun 70:444–450. doi: 10.1128/IAI.70.2.444-450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCallum N, Hinds J, Ender M, Berger-Bächi B, Stutzmann Meier P. 2010. Transcriptional profiling of XdrA, a new regulator of spa transcription in Staphylococcus aureus. J Bacteriol 192:5151–5164. doi: 10.1128/JB.00491-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mauzy CA, Hermodson MA. 1992. Structural and functional analyses of the repressor, RbsR, of the ribose operon of Escherichia coli. Protein Sci 1:831–842. doi: 10.1002/pro.5560010701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stentz R, Zagorec M. 1999. Ribose utilization in Lactobacillus sakei: analysis of the regulation of the rbs operon and putative involvement of a new transporter. J Mol Microbiol Biotechnol 1:165–173. [PubMed] [Google Scholar]
- 40.Woodson K, Devine KM. 1994. Analysis of a ribose transport operon from Bacillus subtilis. Microbiology 140:1829–1838. doi: 10.1099/13500872-140-8-1829. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz de los Mozos I, Vergara-Irigaray M, Segura V, Villanueva M, Bitarte N, Saramago M, Domingues S, Arraiano CM, Fechter P, Romby P, Valle J, Solano C, Lasa I, Toledo-Arana A. 2013. Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet 9:e1004001. doi: 10.1371/journal.pgen.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodionov DA, Mironov AA, Gelfand MS. 2001. Transcriptional regulation of pentose utilisation systems in the Bacillus/Clostridium group of bacteria. FEMS Microbiol Lett 205:305–314. doi: 10.1111/j.1574-6968.2001.tb10965.x. [DOI] [PubMed] [Google Scholar]
- 43.Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bächi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J Bacteriol 186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulthess B, Meier S, Homerova D, Goerke C, Wolz C, Kormanec J, Berger-Bächi B, Bischoff M. 2009. Functional characterization of the σB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob Agents Chemother 53:1832–1839. doi: 10.1128/AAC.01255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauch MA. 1995. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J Bacteriol 177:6727–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimada T, Kori A, Ishihama A. 2013. Involvement of the ribose operon repressor RbsR in regulation of purine nucleotide synthesis in Escherichia coli. FEMS Microbiol Lett 344:159–165. doi: 10.1111/1574-6968.12172. [DOI] [PubMed] [Google Scholar]
- 47.Nentwich SS, Brinkrolf K, Gaigalat L, Hüser AT, Rey DA, Mohrbach T, Marin K, Pühler A, Tauch A, Kalinowski J. 2009. Characterization of the LacI-type transcriptional repressor RbsR controlling ribose transport in Corynebacterium glutamicum ATCC 13032. Microbiology 155:150–164. doi: 10.1099/mic.0.020388-0. [DOI] [PubMed] [Google Scholar]
- 48.Romilly C, Lays C, Tomasini A, Caldelari I, Benito Y, Hammann P, Geissmann T, Boisset S, Romby Vandenesch PF. 2014. A non-coding RNA promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog 10:e1003979. doi: 10.1371/journal.ppat.1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somerville GA, Proctor RA. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol Mol Biol Rev 73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gross M, Zöllner N. 1991. Serum levels of glucose, insulin, and C-peptide during long-term d-ribose administration in man. Klin Wochenschr 69:31–36. doi: 10.1007/BF01649054. [DOI] [PubMed] [Google Scholar]
- 51.Cai Y, Liu J, Shi Y, Liang L, Mou S. 2005. Determination of several sugars in serum by high-performance anion-exchange chromatography with pulsed amperometric detection. J Chromatogr A 1085:98–103. doi: 10.1016/j.chroma.2004.11.100. [DOI] [PubMed] [Google Scholar]
- 52.Clark PM, Flores G, Evdokimov NM, McCracken MN, Chai T, Nair-Gill E, O'Mahony F, Beaven SW, Faull KF, Phelps ME, Jung ME, Witte ON. 2014. Positron emission tomography probe demonstrates a striking concentration of ribose salvage in the liver. Proc Natl Acad Sci U S A 111:E2866–E2874. doi: 10.1073/pnas.1410326111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hecker M, Pané-Farré J, Uwe V. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol 61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 54.Depke M, Burian M, Schäfer T, Bröker BM, Ohlsen K, Völker U. 2012. The alternative sigma factor B modulates virulence gene expression in a murine Staphylococcus aureus infection model but does not influence kidney gene expression pattern of the host. Int J Med Microbiol 302:33–39. doi: 10.1016/j.ijmm.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Pförtner H, Burian MS, Michalik S, Depke M, Hildebrandt P, Dhople VM, Pané-Farré J, Hecker M, Schmidt F, Völker U. 2014. Activation of the alternative sigma factor SigB of Staphylococcus aureus following internalization by epithelial cells—an in vivo proteomics perspective. Int J Med Microbiol 304:177–187. doi: 10.1016/j.ijmm.2013.11.014. [DOI] [PubMed] [Google Scholar]