Abstract
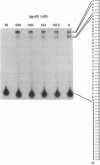
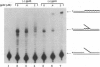
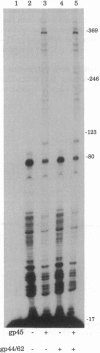
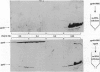
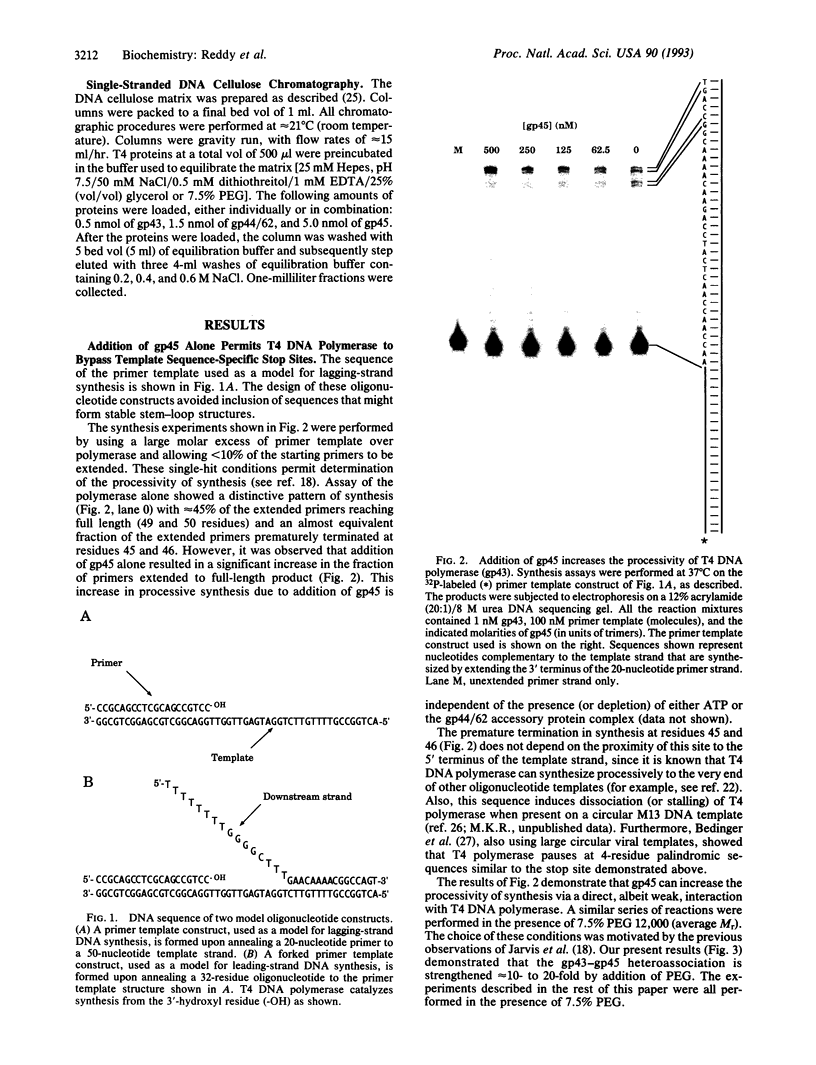
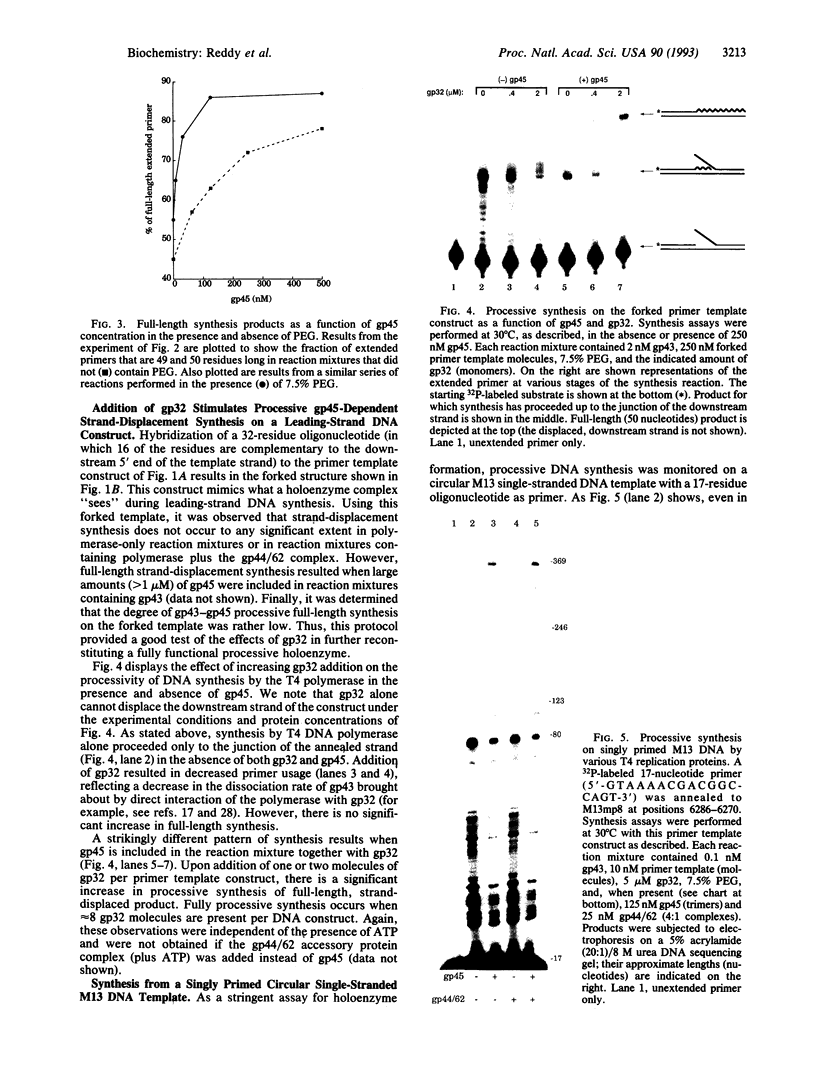
The seven-protein bacteriophage T4 DNA replication complex can be manipulated in vitro to study mechanistic aspects of the elongation phase of DNA replication. Under physiological conditions, the processivity of DNA synthesis catalyzed by the T4 polymerase (gp43) is greatly increased by the interaction of this enzyme with its accessory proteins (gp44/62 and gp45) and the T4 single-stranded DNA binding protein (gp32). The assembly of this T4 holoenzyme requires hydrolysis of ATP by the gp44/62 complex. We demonstrate here that processive T4 holoenzyme-like DNA synthesis can be obtained without hydrolysis of ATP by simply adding gp45 to the T4 DNA polymerase at extremely high concentrations, effectively bypassing the ATPase subunits (gp44/62) of the accessory protein complex. The amount of gp45 required for the gp43-gp45 heteroassociation event is reduced by addition of the macromolecular crowding agent polyethylene glycol (PEG) as well as gp32. A chromatographic strategy involving PEG has been used to demonstrate the gp43-gp45 interaction. These results suggest that gp45 is ultimately responsible for increasing the processivity of DNA synthesis via a direct and functionally significant interaction with the T4 DNA polymerase. A corollary to this notion is that the specific role of the gp44/62 complex is to catalytically link gp45 to gp43.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M. The DNA enzymology of protein machines. Cold Spring Harb Symp Quant Biol. 1984;49:1–12. doi: 10.1101/sqb.1984.049.01.003. [DOI] [PubMed] [Google Scholar]
- Bedinger P., Alberts B. M. The 3'-5' proofreading exonuclease of bacteriophage T4 DNA polymerase is stimulated by other T4 DNA replication proteins. J Biol Chem. 1983 Aug 25;258(16):9649–9656. [PubMed] [Google Scholar]
- Bedinger P., Munn M., Alberts B. M. Sequence-specific pausing during in vitro DNA replication on double-stranded DNA templates. J Biol Chem. 1989 Oct 5;264(28):16880–16886. [PubMed] [Google Scholar]
- Capson T. L., Benkovic S. J., Nossal N. G. Protein-DNA cross-linking demonstrates stepwise ATP-dependent assembly of T4 DNA polymerase and its accessory proteins on the primer-template. Cell. 1991 Apr 19;65(2):249–258. doi: 10.1016/0092-8674(91)90159-v. [DOI] [PubMed] [Google Scholar]
- Crute J. J., LaDuca R. J., Johanson K. O., McHenry C. S., Bambara R. A. Excess beta subunit can bypass the ATP requirement for highly processive synthesis by the Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 1983 Sep 25;258(18):11344–11349. [PubMed] [Google Scholar]
- Fairfield F. R., Newport J. W., Dolejsi M. K., von Hippel P. H. On the processivity of DNA replication. J Biomol Struct Dyn. 1983 Dec;1(3):715–727. doi: 10.1080/07391102.1983.10507477. [DOI] [PubMed] [Google Scholar]
- Formosa T., Burke R. L., Alberts B. M. Affinity purification of bacteriophage T4 proteins essential for DNA replication and genetic recombination. Proc Natl Acad Sci U S A. 1983 May;80(9):2442–2446. doi: 10.1073/pnas.80.9.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogol E. P., Young M. C., Kubasek W. L., Jarvis T. C., von Hippel P. H. Cryoelectron microscopic visualization of functional subassemblies of the bacteriophage T4 DNA replication complex. J Mol Biol. 1992 Mar 20;224(2):395–412. doi: 10.1016/0022-2836(92)91003-8. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Kornberg A., Alberts B. M. Stimulation of T4 bacteriophage DNA polymerase by the protein product of T4 gene 32. J Mol Biol. 1971 Nov 28;62(1):39–52. doi: 10.1016/0022-2836(71)90129-x. [DOI] [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., Hockensmith J. W., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12717–12729. [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. I. Physical characterization of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12709–12716. [PubMed] [Google Scholar]
- Jarvis T. C., Ring D. M., Daube S. S., von Hippel P. H. "Macromolecular crowding": thermodynamic consequences for protein-protein interactions within the T4 DNA replication complex. J Biol Chem. 1990 Sep 5;265(25):15160–15167. [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- LaDuca R. J., Crute J. J., McHenry C. S., Bambara R. A. The beta subunit of the Escherichia coli DNA polymerase III holoenzyme interacts functionally with the catalytic core in the absence of other subunits. J Biol Chem. 1986 Jun 5;261(16):7550–7557. [PubMed] [Google Scholar]
- Maki S., Kornberg A. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerases reconstituted from purified subunits. J Biol Chem. 1988 May 15;263(14):6561–6569. [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme. Components, structure, and mechanism of a true replicative complex. J Biol Chem. 1991 Oct 15;266(29):19127–19130. [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. DNA footprinting studies of the complex formed by the T4 DNA polymerase holoenzyme at a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20034–20044. [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. The T4 DNA polymerase accessory proteins form an ATP-dependent complex on a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20024–20033. [PubMed] [Google Scholar]
- Nossal N. G. Protein-protein interactions at a DNA replication fork: bacteriophage T4 as a model. FASEB J. 1992 Feb 1;6(3):871–878. doi: 10.1096/fasebj.6.3.1310946. [DOI] [PubMed] [Google Scholar]
- O'Donnell M. Accessory protein function in the DNA polymerase III holoenzyme from E. coli. Bioessays. 1992 Feb;14(2):105–111. doi: 10.1002/bies.950140206. [DOI] [PubMed] [Google Scholar]
- O'Donnell M., Studwell P. S. Total reconstitution of DNA polymerase III holoenzyme reveals dual accessory protein clamps. J Biol Chem. 1990 Jan 15;265(2):1179–1187. [PubMed] [Google Scholar]
- Reddy M. K., Weitzel S. E., von Hippel P. H. Processive proofreading is intrinsic to T4 DNA polymerase. J Biol Chem. 1992 Jul 15;267(20):14157–14166. [PubMed] [Google Scholar]
- Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991 Jun 15;266(17):11328–11334. [PubMed] [Google Scholar]
- Topal M. D., Sinha N. K. Products of bacteriophage T4 genes 32 and 45 improve the accuracy of DNA replication in vitro. J Biol Chem. 1983 Oct 25;258(20):12274–12279. [PubMed] [Google Scholar]
- Tsurimoto T., Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M., Nossal N. G. Bacteriophage T4 gene 44/62 and gene 45 polymerase accessory proteins stimulate hydrolysis of duplex DNA by T4 DNA polymerase. J Biol Chem. 1982 Oct 25;257(20):12435–12443. [PubMed] [Google Scholar]
- Warshaw M. M., Cantor C. R. Oligonucleotide interactions. IV. Conformational differences between deoxy- and ribodinucleoside phosphates. Biopolymers. 1970;9(9):1079–1103. doi: 10.1002/bip.1970.360090910. [DOI] [PubMed] [Google Scholar]
- Wickner S. Mechanism of DNA elongation catalyzed by Escherichia coli DNA polymerase III, dnaZ protein, and DNA elongation factors I and III. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3511–3515. doi: 10.1073/pnas.73.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. C., Reddy M. K., von Hippel P. H. Structure and function of the bacteriophage T4 DNA polymerase holoenzyme. Biochemistry. 1992 Sep 22;31(37):8675–8690. doi: 10.1021/bi00152a001. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Harrison B. Macromolecular crowding increases binding of DNA polymerase to DNA: an adaptive effect. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1871–1875. doi: 10.1073/pnas.84.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]