Abstract
Until recently, the study of mycobacterial diseases was trapped in culture-based technology that is more than a century old. The use of nucleic acid amplification is changing this, and powerful new technologies are on the horizon. Metabolomics, which is the study of sets of metabolites of both the bacteria and host, is being used to clarify mechanisms of disease, and can identify changes leading to better diagnosis, treatment, and prognostication of mycobacterial diseases. Metabolomic profiles are arrays of biochemical products of genes in their environment. These complex patterns are biomarkers that can allow a more complete understanding of cell function, dysfunction, and perturbation than genomics or proteomics. Metabolomics could herald sweeping advances in personalized medicine and clinical trial design, but the challenges in metabolomics are also great. Measured metabolite concentrations vary with the timing within a condition, the intrinsic biology, the instruments, and the sample preparation. Metabolism profoundly changes with age, sex, variations in gut microbial flora, and lifestyle. Validation of biomarkers is complicated by measurement accuracy, selectivity, linearity, reproducibility, robustness, and limits of detection. The statistical challenges include analysis, interpretation, and description of the vast amount of data generated. Despite these drawbacks, metabolomics provides great opportunity and the potential to understand and manage mycobacterial diseases.
Keywords: tuberculosis, metabolomics, nontuberculous, -omics
Tuberculosis has been a great public enemy for millennia, and nontuberculous mycobacteria are causing concern in many countries today. The study, diagnosis, and treatment of mycobacterial diseases have been based on culture-related procedures that are more than 100 years old (1). Because these organisms grow slowly, serious complications occur before diagnoses are established. As different species have different growth characteristics, complex culturing procedures require extensive time and highly specialized laboratories (2). As we enter the genomic era, however, these methods are changing. Nucleic acid amplification studies are improving the diagnosis and treatment of mycobacterial disease, with tuberculosis leading the way.
An emerging science of the “omic” era is metabolomics, the study of the set of metabolites produced in response to a disease or perturbation. The metabolites, which may arise from resident or infecting microorganisms or their hosts, have yielded important information about the pathophysiology of many conditions. Metabolites identified by proton nuclear magnetic resonance (1H-NMR) spectroscopy, gas chromatography-mass spectrometry (GC-MS), and liquid chromatography-mass spectroscopy (LC-MS) have been used to establish diagnosis and prognosis of many infectious and noninfectious diseases (3–12). Metabolomics, as with the other “omics,” is made possible only because advanced analytics are now available. In this review, we outline the potential role of metabolomics in mycobacterial disease.
Metabolomics
Metabolites are the low-molecular-weight compounds (<1 kD) resulting from life processes (metabolism). The full set of metabolites reflects the biochemical, physiological, and pathophysiological processes occurring in a life form at a particular time and appears to be unique for the various organisms or tissues sampled (13). Metabolomics is the identification and quantification of these metabolites in an unbiased analytical systems–based approach.
Metabolomics can use targeted and nontargeted approaches to investigate qualitative and quantitative endogenous metabolites. A nontargeted approach is effectively used to identify a number of unknown metabolites known as the metabolome, whereas targeted metabolomics is restricted to known metabolites, usually for specific metabolites or within a single class of compounds (14).
The metabolomics approach allows for the analysis of a variety of biological fluids, such as blood, urine, sputum, cerebral spinal fluid, and exhaled breath condensate in humans, or bacterial sources, such as culture media (15).
Metabolites are associated with all biological processes as starting, intermediate, or end products. The profile of these metabolites can provide mechanistic information about the influence of genetics, epigenetics, proteomics, and environmental factors. Metabolites change with time, and metabolomics can track the processes of disease progression and adaptation. The variations in microorganisms also can be plotted. Variation of the metabolome in different tissues may reflect how disease processes affect specific organs. Studying metabolomics with different perturbations (16, 17) can show the effects of the changes on signaling pathways. New biomarkers can be established on the basis of structurally and biochemically annotated metabolites. These biomarkers may be used for the diagnosis and prognosis of many diseases and could be useful in both basic research and clinical trials to monitor the effect of treatment (18). By profiling diseases, biomarkers can be used in drug development and to monitor the response to therapy (19). Metabolomics is a powerful tool with great potential for many areas of research in biology and medicine (19).
The great value of metabolomics is derived from the patterns of many different small molecules, which give a specific profile for different human conditions including infectious, neoplastic, cardiovascular, neurological, metabolic, and inflammatory diseases (19–21).
Metabolomics and Other “Omics”
Metabolomics is complementary to the other “omic” sciences, such as genomics, transcriptomics, and proteomics, but has less limitation because of technical and biological advantages (22). Metabolomics captures the biochemical products of the genes in their environment, and thus can provide fundamental knowledge of the biochemical networks under investigation. In this respect, metabolomics can allow for a more complete understanding of cell functions than genomics, transcriptomics, or proteomics can. This sensitive assessment of cell function at the chemical level has led to a better understanding of the cellular dysfunction caused by several disorders (23). Metabolomics has also clarified the disease development downstream of genetic or environmental changes (24) in contrast to genetics and proteomics, which give insight mainly into the predisposition for a disease.
Metabolomics can show the cellular responses to internal and external stimuli that can help identify early perturbations in cellular metabolism. The perturbations can be caused by the disorders themselves (23), by nature itself (environmental stress), or by humans (answering either research or therapeutic questions).
Analytical Platforms and Multivariate Data Analysis
Several analytical methods are used for metabolomic studies in individual as well as pooled human fluid samples. At present, NMR, GC-MS, LC-MS, and direct infusion tandem mass spectrometry (DI-MS/MS) are the most widely used techniques for the identification of large numbers of metabolites.
Each of these methods carries its own advantages, disadvantages, and predictive power. NMR, GC-MS, and LC-MS are the most common analytical tools used in metabolomics studies, and DI-MS/MS is a suitable technique for rapid diagnosis. Although GC-MS and LC-MS are more sensitive techniques with high separation efficiency, spectral resolution, mass accuracy, and resolution, NMR produces more quantifiable and reproducible data. The nondestructive NMR assay renders it more applicable to intact biomaterials (25). DI-MS/MS has high throughput and sensitivity with relatively good reproducibility (26). Each technique has the ability to detect small disturbances from stimuli, such as those caused by infectious diseases. Because of these differences, a combination of two or three analytical platforms is often needed to identify the various stages of a disease and to differentiate diseases.
Metabolomics aims to comprehensively identify and quantify a large number of metabolites from various classes. To identify the various classes of compounds, such as lipids, carbohydrates, amino acids, organic acids, sugars, sugar phosphates, biogenic amines, nucleotides, vitamins, purines, fatty acids, and steroids (27), one analytical method is not sufficient in a nontargeted approach (28, 29). Table 1 shows the most common analytical tools used (30) and Table 2 shows the strengths and weakness of these tools.
Table 1.
Analytical tools most commonly used in metabolomics studies
| Technique | Metabolites Seen | Number of Metabolites |
|---|---|---|
| NMR | • Amino acids | 50–200 |
| • Polar/nonpolar metabolites | ||
| • Sugars | ||
| • Volatile liquids | ||
| • Large metabolites | ||
| GC-MS | • Volatile/thermally stable metabolites | 100–500 |
| • Nonpolar metabolites | ||
| • Amino acids | ||
| • Medium to high lipophilicity | ||
| • Nucleosides and nucleotides | ||
| • Carbohydrates | ||
| • Esters | ||
| LC-MS | • Amino acids | 100–800 |
| • Fatty acids | ||
| • Polar metabolites | ||
| • Organic acids | ||
| • Steroids |
Definition of abbreviations: GC-MS = gas chromatography-mass spectrometry; LC-MS = liquid chromatography-mass spectroscopy; NMR = nuclear magnetic resonance.
Table 2.
Strengths and weaknesses of analytical tools used in metabolomics studies
| Technique | Strengths | Weaknesses |
|---|---|---|
| NMR | • Nondestructive technique | • Low sensitivity (only metabolites with relatively high concentration [micrograms] can be detected) |
| • Versatility for analyzing metabolites in biofluids, tissues, or in vivo | ||
| • Reproducibility and repeatability | • Overlap in peaks and high chemical degeneracy (different metabolites have resonances in the same spectral region) | |
| • More relative quantification | • Identifies mainly polar compounds | |
| • Applicable to intact biomaterial | • Usually large sample size | |
| GC-MS | • High-resolution capacity | • High molecular weight analytes |
| • High spectral resolution | • Derivatization required | |
| • Very sensitive | • Fragmentation in MS | |
| • High mass accuracy to detect compounds | • Requires technical skill | |
| • Reproducible retention time | • Extensive sample preparation steps | |
| • Highly developed compound libraries | • Poor quantification | |
| • Small sample size (50 μl) | • Possible variation due to sample preparation | |
| • High separation efficiency | • Compound degradation (high temperature) | |
| • Ideal for thermostable and volatile and nonpolar metabolites | • Problem with ionization | |
| LC-MS | • Short separation time | • Vaporization errors |
| • High resolution | • More instrumental variables than in NMR and LC-MS | |
| • Ideal for nonvolatile compounds | • High solvent consumption and lower separation power | |
| • Very sensitive (picogram quantities) | • Lower reproducibility (within and across laboratories) | |
| • Reasonable robustness | • Ionization of metabolites | |
| • Selectivity | • Selectivity | |
| • High mass accuracy to detect compounds | • Poor quantification | |
| • Simple sample preparation | • Lower reproducibility for retention time with different system | |
| • Detects a wider range of metabolites than GC-MS | • Destructive to sample | |
| • Analysis of more polar compounds without derivatization | • High instrumental cost | |
| DI-MS/MS | • High throughput | • Matrix effects |
| • Minimal sample preparation | • Lack of differentiation between isomers | |
| • Rapid analysis (1–3 min) | • Lack of accuracy of selection of ions | |
| • Good reproducibility | • Competitive ionization | |
| • Highly sensitive | ||
| • Simpler data analysis than LC-MS and GC-MS | ||
| • Considerable structural information |
Definition of abbreviations: DI-MS/MS = direct infusion tandem mass spectroscopy; GC-MS = gas chromatography-mass spectrometry; LC-MS = liquid chromatography-mass spectroscopy; NMR = nuclear magnetic resonance.
Assigning the vast array of compounds to various disease states is also a statistical challenge. The statistical methods rely on multivariate and pattern recognition approaches, which may be data driven and model driven. The products of the analyses are called “biomarker signatures” or “fingerprints.” The large data sets are analyzed by multivariate techniques, such as principal component analysis and orthogonal partial least squares discriminant analysis. Both methods are based on projection methods with the underlying assumption that the system is affected by a limited number of variables (31). Principal component analysis is essential for the visualization and subsequent removal of outliers that may prejudice further analysis. It is unsupervised, meaning no information is given when the data set is entered into a statistical program (32).
Orthogonal partial least squares, another popular technique in metabolic profiling, is frequently applied in discriminant analysis in which biological samples are classified in response to variables (32, 33). This is a supervised analysis when two cohorts take part in the analysis. It is generally used for model separation or classification.
Metabolomics and Biomarker Discovery in Mycobacterial Diseases
Metabolomics has been applied to find specific metabolic patterns for the diagnosis and prognosis of various infectious diseases (34). Studies have shown that mycobacterial exosomes could be used as biomarkers. Exosomes are small vesicles, between 40 and 200 nm, that are released from infected cells and contain mycobacterial proteins, lipoarabinomannan, and metabolites. Exosomes are being evaluated for the diagnosis of tuberculosis, and both immunogenic and nonimmunogenic exosomes are useful in vaccine studies (35).
Lipidomics and Mycobacterial Diseases
Lipidomics is the science that seeks to comprehensively identify and quantify lipid metabolites, making it a subfield of metabolomics. Mycobacterium tuberculosis has a great capacity to synthesize lipids, and its lipids are involved in many of its biological processes. They contribute to virulence and drug resistance. Despite mycobacterial lipids being extensively studied, lipidomics has uncovered many unknown lipids and changes in lipids in response to stimuli (36).
Fatty acids are the backbone of the mycobacterial cell wall lipids and contribute to many pathologic processes, such as change in immunity, virulence, antibiotic resistance, and invasiveness (37). Mass spectroscopy and NMR have been able to quantify various lipids including mycolic acids (α-alkyl, β-hydroxy long-chain fatty acids) in M. tuberculosis (38, 39). Each strain of bacteria has its own lipidome. Portevin and colleagues showed that mycolic acids have different profiles in different M. tuberculosis lineages. Mycolic acids have long been a target for antituberculous drugs, but the more recent findings may give additional clues for new medication (40).
Metabolomics and Diagnostic Biomarkers
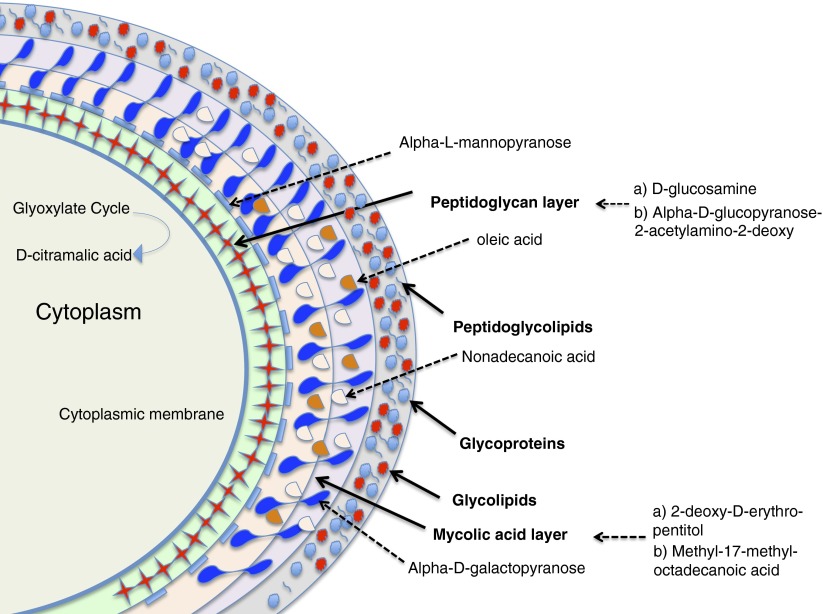
Metabolomics can be a powerful tool for the diagnosis of mycobacterial infections. du Preez and Loots used metabolomics to identify new biomarkers for tuberculosis (41). They analyzed the sputum of 34 patients with tuberculosis and 61 control subjects by two-dimensional gas chromatography time-of-flight mass spectrometry. Twenty-two metabolites (14 M. tuberculosis components and 8 host-related markers) were identified with high discriminative power. Figure 1 illustrates the potential M. tuberculosis metabolites for targeted metabolomics study adapted from the study by Schoeman and colleagues (42).
Figure 1.
Schematic illustration of the cell envelope of Mycobacterium tuberculosis and potential metabolite targets. Metabolites from the peptidoglycan layer (d-glucosamine and 2-acetylamino-2-deoxy-β-d-glucopyranose) can be used for mycobacterial infection diagnosis, and metabolites from the mycolic acid layer (2-deoxy-d-erythro-pentitol and methyl-17-methyloctadecanoic acid) can be used to differentiate mycobacterial species.
GC-MS–based lipidomics of tuberculous infected sputum samples showed that 2-acetylamino-2-deoxy-β-d-glucopyranose, α-l-mannopyranose, and d-galactose-6-deoxy could distinguish persons with tuberculosis from those without (43, 44). For these studies, ethanol homogenization was the best extraction method to differentiate the sputum of tuberculous patients from that of control subjects (42). This technique was also used to differentiate isoniazid-resistant strains of M. tuberculosis from the wild type. Isoniazid-resistant strains carried katG mutations, which produce a specific metabolomic pattern with 29 different compounds in the resistant strains. The metabolites included alkanes, alcohols, fatty acids, surfactant protein, and other compounds involved in the stress alternative energy pathway (45).
Similarly, rpoB mutations change metabolites. LC-MS–based metabolomics showed differences in the metabolic profile of rifampin-resistant M. tuberculosis: 99 molecular features are different in the rifampin-resistant strains. In addition, the studies showed the major role of rpoB mutations in the M. tuberculosis metabolism (46).
Using LC-MS–based metabolomics on plasma samples, Frediani and colleagues found 61 metabolites that were significantly different between persons with tuberculosis and their asymptomatic household contacts. These metabolites could be classified in various groups probably related to specific diet, environmental chemicals, and household microbes. This study showed eight metabolites that were specifically up-regulated by the mycobacterial infection, such as glutamate, choline derivatives, M. tuberculosis cell wall glycolipids, and lipid mediators of inflammation (47).
Nontargeted ultrahigh-pressure liquid chromatography time-of-flight mass spectroscopy (UPLC-TOF-MS) was able to identify a cohort of patients with leprosy by their bacterial indices (a gauge of bacterial burden in leprosy). Metabolomics was able to distinguish those with a bacterial index of less than 1 from those with a bacterial index greater than 4. Serum metabolomics also showed increases in metabolites, such as polyunsaturated fatty acids, eicosapentaenoic acid, and docosahexaenoic acid, in patients with high bacterial indices (48).
Feng and colleagues showed that nontargeted serum-based metabolomics using UPLC-MS was able to distinguish tuberculous patients from healthy controls and patients with other lung diseases, such as pneumonia, lung cancer, chronic obstructive disease, and bronchiectasis. They described 12 metabolites that separated tuberculous patients from control subjects. Fatty acids were the main compounds followed by amino acids, again indicating the importance of lipid metabolism in Mycobacterium infection. Palmitic acid, phosphatidylcholine, lysophosphatidylcholine, phytanic acid, and behenic acid were decreased in tuberculous patients compared with control subjects (49).
Volatile organic compounds in patient serum and urine may be biomarkers for mycobacterial infections, although the compounds differ from those found in bacterial culture (50). Taking substances from organisms or culture media or using known bacterial proteins associated with disease may not be the best strategy for discovering lipidomic candidate biomarkers because it is the profile of many metabolites that are the fingerprints of the disease or process (51, 52).
Metabolomics may be useful for distinguishing mycobacterial pathogens from nonpathogens. They could potentially distinguish persons with latent tuberculosis from those with active tuberculosis and uninfected individuals. These biomarkers could potentially also identify vaccine-induced protection against tuberculosis, which would be extremely useful for vaccine development (52).
1H-NMR–based metabolomics has been applied to diagnose M. avium subspecies paratuberculosis infection in ruminants. The metabolomic study was quicker and more sensitive at distinguishing infected from noninfected animals. The discrimination was significant regardless of infectious burden and time after infection (at 3, 6, 9, and 12 mo) (53). Although the potential is present, more work needs to be done to accurately predict the progression from latent to active mycobacterial disease, relapse after treatment (54, 55), and completion of treatment (56).
Metabolomics may help increase our understanding of the pathophysiology of nontuberculous mycobacterial diseases. These diseases are rising globally, but are less understood and have fewer treatment options that tuberculosis (57). Biomarkers, once established, may function as important tools in clinical trials (58). Although morbidity and mortality will usually remain the primary end points, biomarkers, once validated with these outcomes, may aid the analysis (compared with the current 130-yr-old gold standard of culture) and study design by allowing fewer participants to be enrolled.
Metabolomics as Predictive Biomarkers
Metabolites can be used as biomarkers of effective treatment. LC-MS–based metabolomics on urine showed specific biosignatures with treatment of tuberculosis in African patients; 45 metabolites changed in the month after treatment completion compared with baseline (59). Biomarkers could also delineate the host immune response against mycobacterial infection. The immune response differs with mycobacterial species and strains, and different inocula.
Metabolomics could clarify the interaction of genetic and environmental factors. For example, metabolomics could show if and how the composition of the diet, the occurrence of life events, and the composition of the microbiome could change the metabolomic patterns under normal conditions and in response to disease processes (60, 61).
Measurement of the number and amounts of metabolites with time provides unprecedented information for basic research of biological and pathophysiological pathways. Using isotopically labeled substrate in a targeted approach is an important area of metabolomics called “fluxomics” (62). Mass spectroscopy–based (LC and GC-MS) fluxomics can track stable isotope–labeled elements, such as 2H, 13C, and 15N, to reproducibly identify their compounds in various biological processes. This is leading to new insights in the metabolic pathways in vitro and in experimental models of infection. These methods may show the precise metabolic changes occurring as M. tuberculosis enters a dormant state, which is critical for its survival under conditions of stress, and is important for new drug development. 1H-13C-NMR–based metabolomics has been applied to determine the lethal dose of d-cycloserine and to show its effect on the metabolic pathways of peptidoglycan biosynthesis. Cycloserine inhibits several enzymes within the peptidoglycan biosynthesis pathways, particularly d-alanine–d-alanine ligase, a main enzyme of this pathway (63). These methods can be used to study the growth and replication of the organisms.
Metabolomics has shown that when mycobacteria are stressed by nitrogen limitation they produce free glucosylglycerate, by a mechanism specific for nitrogen stress but not oxidative or osmotic stress (64). These metabolic pathways help keep the mycobacteria alive by reducing the growth rate and decreasing the uptake of ammonium (64). A summary of the metabolomic profiles of various mycobacteria, using analytical tools, is shown in Table 3.
Table 3.
Summary of metabolomic profiles of various mycobacteria using analytical tools
| Analytical Tool | Main Study | Biofluids | Microorganism | Key Metabolites | Ref. No. |
|---|---|---|---|---|---|
| FT-ICR-MS | Lipidomics (Mycobacterium virulence) | Bacterial culture | M. tuberculosis | Phthiocerol dimycocerosate and sulfolipid-1 | 38 |
| 2D HSQC NMR | Lipidomics (lipid profiling and Mycobacterium virulence studies on various strains) | Bacterial culture | M. liflandii, M. tuberculosis, M. smegmatis | 11 metabolites including triacylglycerol, acylated trehaloses, mycocerosic acid* | 39 |
| MRM-based MS | Lipidomics (study on permeability barrier in resistant strain) | Mycobacterial cultures | M. tuberculosis | Mycolic acids | 40 |
| GC/GC-TOF-MS | Differentiating between TB-positive and TB-negative patients | Sputum | M. tuberculosis | 22 metabolites including d-gluconic acid lactone, glutaric acid, sebacic acid, ethane, butanal, γ-aminobutyric acid, 3,4-dihydroxybutanoic acid, and normetanephrine* | 41 |
| GC-MS | Distinguishing patients with TB from non-TB individuals | Sputum | M. tuberculosis | Tuberculosis stearic acid | 42 |
| GCxGC-TOF-MS | Metabolic profiling differentiation of drug-resistant Mycobacterium strains vs. wild-type M. tuberculosis | Mycobacterial cultures | M. tuberculosis | 23 metabolites (alkanes, alcohols, and fatty acids) including decane, hexadecane, isotridecanol, and octadecanoic acid* | 45 |
| GC-EI/MS | Metabolic profile of M. tuberculosis and differentiation of positive sputum from negative sputum | Mycobacterial cultures and sputum | M. tuberculosis | Mycocerosic acid methyl esters | 43 |
| GC/GC-TOF-MS | Metabolomics profiling to distinguish patients with TB from control subjects | Sputum and mycobacterial cultures | M. tuberculosis | 20 metabolites including nonadecanoic acid, tuberculostearic acid, myo-inositol* | 44 |
| LC-MS | Role of rpoB mutation in rifampin-resistant and -susceptible M. tuberculosis strains in metabolomics profiling differentiation | Mycobacterial cultures | M. tuberculosis | 87 features including diacylglycerol phosphocholine, hexose-N-acetylhexosamine-fucose-N-acetylhexosamine* | 46 |
| LC-MS/MS | Distinction of patients with TB from sputum-negative household contacts | Plasma | M. tuberculosis | 61 metabolites including trehalose-6-mycolate, phosphatidylinositol, and the D-series resolvins* | 47 |
| UPLC-TOF-MS | Identification of patients with leprosy with various bacterial indices | Serum | M. leprae | 48 features* | 48 |
| UPLC-MS | Differentiation of patients with TB from healthy control subjects and non-TB patients | Serum | M. tuberculosis | 12 metabolites including 3D,7D,11D-phytanic acid, behenic acid, and threoninyl-γ-glutamate* | 49 |
| GC-MS | Metabolomics profiling of M. tuberculosis | Culture, serum, urine, and EBC | M. tuberculosis | Volatile organic compounds | 50 |
| 1H-NMR | Differentiation of ruminants infected with M. avium subsp. paratuberculosis from noninfected animals | Serum | M. avium subsp. paratuberculosis | 16 metabolites including acetone, asparagine, aspartate, betaine, glycerol, isobutyrate, isoleucine, leucine, mannose, threonine, tyrosine* | 53 |
| LC-MS | Metabolic profiling of patients with TB before and after treatment | Urine | M. tuberculosis | 45 metabolites* | 59 |
| 1H-13C-NMR | Metabolic profiling for pathway analysis in multi- and extensively drug-resistant strains | Mycobacterial cultures | M. tuberculosis | d-Alanine–d-alanine ligase | 63 |
| NMR and GC-MS | Pathway analysis in nitrogen stress | Mycobacterial cultures | M. smegmatis | Glucosylglycerate | 64 |
Definition of abbreviations: 2D HSQC = two-dimensional heteronuclear single-quantum coherence; EBC = exhaled breath condensate; FT-ICR-MS = Fourier transform ion cyclotron resonance mass spectrometry; GC-EI/MS = gas chromatography-electron impact mass spectrometry; GC/GC-TOF-MS = two-dimensional gas chromatography coupled with time-of-flight mass spectrometry; LC-MS = liquid chromatography-mass spectroscopy; M. = Mycobacterium; MRM-based MS = multiple reaction monitoring–based mass spectrometry; NMR = nuclear magnetic resonance; TB = tuberculosis; UPLC-TOF-MS = ultrahigh-pressure liquid chromatography-time of flight-mass spectrometry.
For full list refer to the accompanying reference.
Precision Medicine
Precision medicine seeks to classify diseases into subgroups that in the past have been grouped together because of the lack of good discriminators. The use of metabolomics, in conjunction with genomics, pharmacogenomics, and proteomics, may allow more precise definition and treatment of the mycobacterial infections. Moreover, pharmacometabolomics is emerging as a new science aiming to understand the role of metabolites in antimycobacterial drug development and monitoring (65). Metabolites measured before and after drug administration can be used to evaluate the response to the medicine and could lead to a better understanding of the biological processes involved in effective treatment (19). Metabolomics may be able to identify susceptibility to drug toxicities and drug interactions in patients and healthy individuals (66). Snapshot measurement of hundreds of metabolites, serial analysis, and easy sampling of various body fluids are advantages that metabolomics provides for evaluating drugs in humans.
Metabolomics-based studies on animals, cell lines, and humans can be applied to characterize drug absorption, distribution, metabolism, and disposition. The specific metabolites produced during catabolism of drugs, as well as the metabolic profile changes, can be measured with sensitive analytical tools. A metabolomics approach could be used to study the success or failure of treatments in individuals. This could then determine which treatments will be effective in various individuals, based on their metabolism (67).
Challenges
Although metabolomics has great potential, it also has great challenges and limitations. Many of the studies are exploratory. By the nature of the science, there is variability in metabolite concentration caused by several sources including timing within a condition, biological variation, instrument variation, and sample preparation variation. Instrument and sample preparation variation are important sources of error and can cause both false positives and false negatives, as shown in the separation of nonsurvivors from survivors by Leichtle and colleagues (68).
Reproducibility and specificity are two major obstacles to be overcome before metabolomics can be useful in clinical studies. Impressive results found in studies of a few patients must be repeatable and specific in larger studies (68). Metabolism profoundly changes with age, sex, variations in gut microbial composition, and lifestyle (28, 69).
A great challenge for metabolomics is to describe and accurately classify the many human metabolites and their importance, using different types of instrumental and statistical analysis. Finding and validating biomarkers is a major need (51). The selection of biomarkers of interest by genomics, proteomics, or metabolomics based on performance criteria is difficult and time consuming. The validation of biomarkers is also complicated and must address important parameters such as the range of the measurements, accuracy, selectivity, linearity, reproducibility, robustness, and the limits of detection (70).
The statistical challenges include how to analyze, interpret, and describe the vast amount of data generated. Biomarkers discovered by metabolomics should be validated both internally and externally. Multivariate analyses provide prediction models creating R2 and Q2 parameters to assess variability and predictability, respectively. Cross-validation and permutation are methods for internal validation to find false-correlated and overfitted models. Q2 (goodness of prediction) is obtained by cross-validated and permutation methods in optimized potentials for liquid simulations, a statistical method of partial least squares regression.
Candidate biomarkers need to be externally validated by testing new independent samples, preferably from a new center operating with a different analytical machine and user (71). Tuberculosis biomarker signatures should be validated in different geographically and ethnically diverse populations and take into account coinfection with malaria and HIV, which affect metabolomic biosignatures (52).
Future Perspectives
Although the future of metabolomics for mycobacterial disease is great, the field is in its infancy. It should be integrated with genomics and proteomics to gain a complete picture of biologic and cellular processes. Metabolomics can be applied to the diagnosis, treatment, and prognosis of patients with mycobacterial infections (Table 4). This may be particularly important for infection with the environmental mycobacteria, about which there are more questions and less certainty than concerning infection with M. tuberculosis.
Table 4.
Potential applications of metabolomics for the diagnosis and management of mycobacterial infections
| Metabolites | Sample | Method (Ref. No.) | |
|---|---|---|---|
| Early diagnosis | d-Alanine–d-alanine ligase | Mycobacterial culture | NMR (63) |
| Mycocerosic acid methyl esters | Sputum | GC/MS (43) | |
| Latent vs. active TB | 3D,7D,11D-Phytanic acid, behenic acid, and threoninyl-γ-glutamate | Serum | UPLC-MS (49) |
| Trehalose-6-mycolate, phosphatidylinositol, and the D-series resolvins | Plasma | LC-MS/MS (47) | |
| Distinguishing pathogens from nonpathogenic mycobacteria | Triacylglycerol, acylated trehaloses, mycocerosic acid | Bacterial culture | 2D HSQC NMR (39) |
| Phthiocerol dimycocerosate and sulfolipid-1 | Bacterial culture | FT-ICR-MS (38) | |
| Treatment response | p-Aminobenzoic acid, pyridoxal/isopyridoxal, formimino-l-glutamic acid, l-α-aspartyl-l-hydroxyproline, N1,N12-diethylspermine | Urine | LC-MS (59) |
Definition of abbreviations: 2D HSQC = two-dimensional heteronuclear single-quantum coherence; FT-ICR-MS = Fourier transform ion cyclotron resonance mass spectrometry; GC/MS = gas chromatography-mass spectrometry; LC-MS/MS = liquid chromatography-tandem mass spectroscopy; NMR = nuclear magnetic resonance; TB = tuberculosis; UPLC-MS = ultrahigh-pressure liquid chromatography-mass spectrometry.
Validation studies to confirm biomarkers could be part of large multicenter studies, which may be designed for other goals. The large studies might be able to account for the heterogeneity in genetic backgrounds and environmental factors, which include age, sex, race, comorbidities, types of pathogens, and sources of infection (72). Before metabolomics can be incorporated into routine clinical practice, studies in larger and diverse groups of patients will be needed.
Conclusions
Among “omic” sciences, metabolomics is a promising approach for many different disorders, including mycobacterial disease. Its sensitivity may develop biomarkers better than other “omics,” but the field is in its infancy and the great sensitivity and enormous data produced are currently limitations as well as a source of promise. Its ability to render a complete picture of ongoing biological processes is most appealing especially when combined with the other genomic-based methods.
Footnotes
Supported by NIH grant 5 T32 HL 82547-7 (M.M.).
Author Contributions: Conception, review literature, design, and modeling for review writing of the article were done by M.M. and M.M.B. Writing the paper or substantial involvement in its revision before submission was by M.M.B., M.M., B.W.W., and D.E.S.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Mirsaeidi M, Farshidpour M, Ebrahimi G, Aliberti S, Falkinham JO., III Management of nontuberculous mycobacterial infection in the elderly. Eur J Intern Med. 2014;25:356–363. doi: 10.1016/j.ejim.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Ingen J. Diagnosis of nontuberculous mycobacterial infections. Semin Respir Crit Care Med. 2013;34:103–109. doi: 10.1055/s-0033-1333569. [DOI] [PubMed] [Google Scholar]
- 3.Ge Y, Wang TJ. Identifying novel biomarkers for cardiovascular disease risk prediction. J Intern Med. 2012;272:430–439. doi: 10.1111/j.1365-2796.2012.02589.x. [DOI] [PubMed] [Google Scholar]
- 4.Davis VW, Schiller DE, Eurich D, Sawyer MB. Urinary metabolomic signature of esophageal cancer and Barrett’s esophagus. World J Surg Oncol. 2012;10:271. doi: 10.1186/1477-7819-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farshidfar F, Weljie AM, Kopciuk K, Buie WD, Maclean A, Dixon E, Sutherland FR, Molckovsky A, Vogel HJ, Bathe OF. Serum metabolomic profile as a means to distinguish stage of colorectal cancer. Genome Med. 2012;4:42. doi: 10.1186/gm341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho WE, Xu YJ, Xu F, Cheng C, Peh HY, Tannenbaum SR, Wong WS, Ong CN. Metabolomics reveals altered metabolic pathways in experimental asthma. Am J Respir Cell Mol Biol. 2013;48:204–211. doi: 10.1165/rcmb.2012-0246OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ubhi BK, Riley JH, Shaw PA, Lomas DA, Tal-Singer R, MacNee W, Griffin JL, Connor SC. Metabolic profiling detects biomarkers of protein degradation in COPD patients. Eur Respir J. 2012;40:345–355. doi: 10.1183/09031936.00112411. [DOI] [PubMed] [Google Scholar]
- 8.Weiner J, III, Parida SK, Maertzdorf J, Black GF, Repsilber D, Telaar A, Mohney RP, Arndt-Sullivan C, Ganoza CA, Faé KC, et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS One. 2012;7:e40221. doi: 10.1371/journal.pone.0040221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mickiewicz B, Vogel HJ, Wong HR, Winston BW. Metabolomics as a novel approach for early diagnosis of pediatric septic shock and its mortality. Am J Respir Crit Care Med. 2013;187:967–976. doi: 10.1164/rccm.201209-1726OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mickiewicz B, Duggan GE, Winston BW, Doig C, Kubes P, Vogel HJ Alberta Sepsis Network. Metabolic profiling of serum samples by 1H nuclear magnetic resonance spectroscopy as a potential diagnostic approach for septic shock. Crit Care Med. 2014;42:1140–1149. doi: 10.1097/CCM.0000000000000142. [DOI] [PubMed] [Google Scholar]
- 11.Ghannoum MA, Mukherjee PK, Jurevic RJ, Retuerto M, Brown RE, Sikaroodi M, Webster-Cyriaque J, Gillevet PM. Metabolomics reveals differential levels of oral metabolites in HIV-infected patients: toward novel diagnostic targets. OMICS. 2013;17:5–15. doi: 10.1089/omi.2011.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philippeos C, Steffens FE, Meyer D. Comparative 1H NMR-based metabonomic analysis of HIV-1 sera. J Biomol NMR. 2009;44:127–137. doi: 10.1007/s10858-009-9329-8. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson JK, Lindon JC, Holmes E. “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 14.Villas-Bôas SG, Mas S, Akesson M, Smedsgaard J, Nielsen J. Mass spectrometry in metabolome analysis. Mass Spectrom Rev. 2005;24:613–646. doi: 10.1002/mas.20032. [DOI] [PubMed] [Google Scholar]
- 15.Tang J. Microbial metabolomics. Curr Genomics. 2011;12:391–403. doi: 10.2174/138920211797248619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heijne WH, Kienhuis AS, van Ommen B, Stierum RH, Groten JP. Systems toxicology: applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Expert Rev Proteomics. 2005;2:767–780. doi: 10.1586/14789450.2.5.767. [DOI] [PubMed] [Google Scholar]
- 17.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 18.Fell DA. Enzymes, metabolites and fluxes. J Exp Bot. 2005;56:267–272. doi: 10.1093/jxb/eri011. [DOI] [PubMed] [Google Scholar]
- 19.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 20.Collino S, Martin FP, Rezzi S. Clinical metabolomics paves the way towards future healthcare strategies. Br J Clin Pharmacol. 2013;75:619–629. doi: 10.1111/j.1365-2125.2012.04216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011;40:387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- 22.van Ravenzwaay B, Cunha GC, Leibold E, Looser R, Mellert W, Prokoudine A, Walk T, Wiemer J. The use of metabolomics for the discovery of new biomarkers of effect. Toxicol Lett. 2007;172:21–28. doi: 10.1016/j.toxlet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 23.Davis VW, Bathe OF, Schiller DE, Slupsky CM, Sawyer MB. Metabolomics and surgical oncology: potential role for small molecule biomarkers. J Surg Oncol. 2011;103:451–459. doi: 10.1002/jso.21831. [DOI] [PubMed] [Google Scholar]
- 24.Lindon JC, Holmes E, Nicholson JK. So what’s the deal with metabonomics? Anal Chem. 2003;75:384A–391A. doi: 10.1021/ac031386+. [DOI] [PubMed] [Google Scholar]
- 25.Banoei MM, Donnelly SJ, Mickiewicz B, Weljie A, Vogel HJ, Winston BW. Metabolomics in critical care medicine: a new approach to biomarker discovery. Clin Invest Med. 2014;37:E363–E376. doi: 10.25011/cim.v37i6.22241. [DOI] [PubMed] [Google Scholar]
- 26.Junot C, Fenaille F, Colsch B, Bécher F. High resolution mass spectrometry based techniques at the crossroads of metabolic pathways. Mass Spectrom Rev. 2014;33:471–500. doi: 10.1002/mas.21401. [DOI] [PubMed] [Google Scholar]
- 27.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, et al. The human serum metabolome. PLoS One. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 29.Lindon JC, Holmes E, Nicholson JK. Metabonomics techniques and applications to pharmaceutical research and development. Pharm Res. 2006;23:1075–1088. doi: 10.1007/s11095-006-0025-z. [DOI] [PubMed] [Google Scholar]
- 30.Johnson CH, Gonzalez FJ. Challenges and opportunities of metabolomics. J Cell Physiol. 2012;227:2975–2981. doi: 10.1002/jcp.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 32.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, Wold S. Multi- and megavariate data analysis. I. Basic principles and applications. Umeå, Sweden: Umetrics AB; 2006. [Google Scholar]
- 33.Ebbels TMD, Cavill R. Bioinformatic methods in NMR-based metabolic profiling. Prog Nucl Magn Reson Spectrosc. 2009;55:361–374. [Google Scholar]
- 34.Pacchiarotta T, Deelder AM, Mayboroda OA. Metabolomic investigations of human infections. Bioanalysis. 2012;4:919–925. doi: 10.4155/bio.12.61. [DOI] [PubMed] [Google Scholar]
- 35.Kruh-Garcia NA, Wolfe LM, Dobos KM. Deciphering the role of exosomes in tuberculosis. Tuberculosis (Edinb) 2015;95:26–30. doi: 10.1016/j.tube.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Chow ED, Cox JS. TB lipidomics—the final frontier. Chem Biol. 2011;18:1517–1518. doi: 10.1016/j.chembiol.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Layre E, Moody DB. Lipidomic profiling of model organisms and the world’s major pathogens. Biochimie. 2013;95:109–115. doi: 10.1016/j.biochi.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain M, Petzold CJ, Schelle MW, Leavell MD, Mougous JD, Bertozzi CR, Leary JA, Cox JS. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci USA. 2007;104:5133–5138. doi: 10.1073/pnas.0610634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahrous EA, Lee RB, Lee RE. A rapid approach to lipid profiling of mycobacteria using 2D HSQC NMR maps. J Lipid Res. 2008;49:455–463. doi: 10.1194/jlr.M700440-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Portevin D, Sukumar S, Coscolla M, Shui G, Li B, Guan XL, Bendt AK, Young D, Gagneux S, Wenk MR. Lipidomics and genomics of Mycobacterium tuberculosis reveal lineage-specific trends in mycolic acid biosynthesis. MicrobiologyOpen. 2014;3:823–835. doi: 10.1002/mbo3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.du Preez I, Loots DT. New sputum metabolite markers implicating adaptations of the host to Mycobacterium tuberculosis, and vice versa. Tuberculosis (Edinb) 2013;93:330–337. doi: 10.1016/j.tube.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Schoeman JC, du Preez I, Loots T. A comparison of four sputum pre-extraction preparation methods for identifying and characterising Mycobacterium tuberculosis using GCxGC-TOFMS metabolomics. J Microbiol Methods. 2012;91:301–311. doi: 10.1016/j.mimet.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Cha D, Cheng D, Liu M, Zeng Z, Hu X, Guan W. Analysis of fatty acids in sputum from patients with pulmonary tuberculosis using gas chromatography-mass spectrometry preceded by solid-phase microextraction and post-derivatization on the fiber. J Chromatogr A. 2009;1216:1450–1457. doi: 10.1016/j.chroma.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 44.O’Sullivan DM, Nicoara SC, Mutetwa R, Mungofa S, Lee OY, Minnikin DE, Bardwell MW, Corbett EL, McNerney R, Morgan GH. Detection of Mycobacterium tuberculosis in sputum by gas chromatography-mass spectrometry of methyl mycocerosates released by thermochemolysis. PLoS One. 2012;7:e32836. doi: 10.1371/journal.pone.0032836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loots T. An altered Mycobacterium tuberculosis metabolome induced by katG mutations resulting in isoniazid resistance. Antimicrob Agents Chemother. 2014;58:2144–2149. doi: 10.1128/AAC.02344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bisson GP, Mehaffy C, Broeckling C, Prenni J, Rifat D, Lun DS, Burgos M, Weissman D, Karakousis PC, Dobos K. Upregulation of the phthiocerol dimycocerosate biosynthetic pathway by rifampin-resistant, rpoB mutant Mycobacterium tuberculosis. J Bacteriol. 2012;194:6441–6452. doi: 10.1128/JB.01013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frediani JK, Jones DP, Tukvadze N, Uppal K, Sanikidze E, Kipiani M, Tran VT, Hebbar G, Walker DI, Kempker RR, et al. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PLoS One. 2014;9:e108854. doi: 10.1371/journal.pone.0108854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Mubarak R, Vander Heiden J, Broeckling CD, Balagon M, Brennan PJ, Vissa VD. Serum metabolomics reveals higher levels of polyunsaturated fatty acids in lepromatous leprosy: potential markers for susceptibility and pathogenesis. PLoS Negl Trop Dis. 2011;5:e1303. doi: 10.1371/journal.pntd.0001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng S, Du YQ, Zhang L, Zhang L, Feng RR, Liu SY. Analysis of serum metabolic profile by ultra-performance liquid chromatography-mass spectrometry for biomarkers discovery: application in a pilot study to discriminate patients with tuberculosis. Chin Med J (Engl) 2015;128:159–168. doi: 10.4103/0366-6999.149188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang NA, Janssen HG, Kolk AH. Rapid diagnosis of TB using GC-MS and chemometrics. Bioanalysis. 2013;5:3079–3097. doi: 10.4155/bio.13.288. [DOI] [PubMed] [Google Scholar]
- 51.Azuaje F. Bioinformatics and biomarker discovery. Hoboken, NJ: Wiley-Blackwell; 2010. [Google Scholar]
- 52.Ottenhoff TH, Ellner JJ, Kaufmann SH. Ten challenges for TB biomarkers. Tuberculosis (Edinb) 2012;92:S17–S20. doi: 10.1016/S1472-9792(12)70007-0. [DOI] [PubMed] [Google Scholar]
- 53.De Buck J, Shaykhutdinov R, Barkema HW, Vogel HJ. Metabolomic profiling in cattle experimentally infected with Mycobacterium avium subsp. paratuberculosis. PLoS One. 2014;9:e111872. doi: 10.1371/journal.pone.0111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sebina I, Biraro IA, Dockrell HM, Elliott AM, Cose S. Circulating B-lymphocytes as potential biomarkers of tuberculosis infection activity. PLoS One. 2014;9:e106796. doi: 10.1371/journal.pone.0106796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wallis RS, Kim P, Cole S, Hanna D, Andrade BB, Maeurer M, Schito M, Zumla A. Tuberculosis biomarkers discovery: developments, needs, and challenges. Lancet Infect Dis. 2013;13:362–372. doi: 10.1016/S1473-3099(13)70034-3. [DOI] [PubMed] [Google Scholar]
- 56.Shin JH, Yang JY, Jeon BY, Yoon YJ, Cho SN, Kang YH, Ryu H, Hwang GS. 1H NMR–based metabolomic profiling in mice infected with Mycobacterium tuberculosis. J Proteome Res. 2011;10:2238–2247. doi: 10.1021/pr101054m. [DOI] [PubMed] [Google Scholar]
- 57.Mirsaeidi M, Farshidpour M, Allen MB, Ebrahimi G, Falkinham JO. Highlight on advances in nontuberculous mycobacterial disease in North America. Biomed Res Int. 2014;2014:919474. doi: 10.1155/2014/919474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gieger C, Geistlinger L, Altmaier E, Hrabé de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahapatra S, Hess AM, Johnson JL, Eisenach KD, DeGroote MA, Gitta P, Joloba ML, Kaplan G, Walzl G, Boom WH, et al. A metabolic biosignature of early response to anti-tuberculosis treatment. BMC Infect Dis. 2014;14:53. doi: 10.1186/1471-2334-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicholson JK, Holmes E, Elliott P. The metabolome-wide association study: a new look at human disease risk factors. J Proteome Res. 2008;7:3637–3638. doi: 10.1021/pr8005099. [DOI] [PubMed] [Google Scholar]
- 61.Rezzi S, Martin FP, Kochhar S. Defining personal nutrition and metabolic health through metabonomics. Ernst Schering Found Symp Proc. 2007;4:251–264. doi: 10.1007/2789_2008_097. [DOI] [PubMed] [Google Scholar]
- 62.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem. 2007;79:7554–7559. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- 63.Halouska S, Fenton RJ, Zinniel DK, Marshall DD, Barletta RG, Powers R. Metabolomics analysis identifies d-alanine-d-alanine ligase as the primary lethal target of d-cycloserine in mycobacteria. J Proteome Res. 2014;13:1065–1076. doi: 10.1021/pr4010579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Behrends V, Williams KJ, Jenkins VA, Robertson BD, Bundy JG. Free glucosylglycerate is a novel marker of nitrogen stress in Mycobacterium smegmatis. J Proteome Res. 2012;11:3888–3896. doi: 10.1021/pr300371b. [DOI] [PubMed] [Google Scholar]
- 65.Mehta R, Jain RK, Badve S. Personalized medicine: the road ahead. Clin Breast Cancer. 2011;11:20–26. doi: 10.3816/CBC.2011.n.004. [DOI] [PubMed] [Google Scholar]
- 66.Sudhindra A, Ochoa R, Santos ES. Biomarkers, prediction, and prognosis in non-small-cell lung cancer: a platform for personalized treatment. Clin Lung Cancer. 2011;12:360–368. doi: 10.1016/j.cllc.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 67.Kumar B, Prakash A, Ruhela RK, Medhi B. Potential of metabolomics in preclinical and clinical drug development. Pharmacol Rep. 2014;66:956–963. doi: 10.1016/j.pharep.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 68.Leichtle AB, Dufour JF, Fiedler GM. Potentials and pitfalls of clinical peptidomics and metabolomics. Swiss Med Wkly. 2013;143:w13801. doi: 10.4414/smw.2013.13801. [DOI] [PubMed] [Google Scholar]
- 69.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 70.Koek MM, Jellema RH, van der Greef J, Tas AC, Hankemeier T. Quantitative metabolomics based on gas chromatography mass spectrometry: status and perspectives. Metabolomics. 2011;7:307–328. doi: 10.1007/s11306-010-0254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horvatovich PL, Bischoff R. Current technological challenges in biomarker discovery and validation. Eur J Mass Spectrom (Chichester, Eng) 2010;16:101–121. doi: 10.1255/ejms.1050. [DOI] [PubMed] [Google Scholar]
- 72.Singer M. Biomarkers in sepsis. Curr Opin Pulm Med. 2013;19:305–309. doi: 10.1097/MCP.0b013e32835f1b49. [DOI] [PMC free article] [PubMed] [Google Scholar]