Abstract
Staphylococcus aureus pathogenesis is directed by regulatory proteins and RNAs. We report the case of an RNA attenuating virulence and host uptake, possibly to sustain commensalism. A S. aureus sRNA, SprC (srn_3610), reduced virulence and bacterial loads in a mouse infection model. S. aureus deleted for sprC became more virulent and increased bacterial dissemination in colonized animals. Conversely, inducing SprC expression lowered virulence and the bacterial load. Without sprC, S. aureus phagocytosis by monocytes and macrophages was higher, whereas bacteria were internalized at lower yields when SprC expression was stimulated. Without sprC, higher internalization led to a greater number of extracellular bacteria, facilitating colonization. SprC expression decreased after phagocytosis, concurring with the facilitated growth of bacteria lacking the sRNA in the presence of an oxidant. The major staphylococcal autolysin facilitates S. aureus uptake by human phagocytes. ATL proved to be negatively regulated by SprC. The SprC domains involved in pairing with atl mRNA were analyzed. The addition of ATL reduced phagocytosis of bacteria lacking sprC with no effects on wild-type bacterial uptake, implying that SprC influences phagocytosis, at least in part, by controlling ATL. Since the control of SprC on ATL was modest, other factors must contribute to atl regulation.
INTRODUCTION
Infected hosts sense the presence of pathogens and activate immune defense mechanisms for survival. The immune system relies on antimicrobial peptides, lytic enzymes, complement activation and phagocyte recruitment. Upon interaction of bacteria with host cell receptors, the host cells protect and repair their DNA, prevent apoptosis and secrete antimicrobial peptides. Subsequently, the release of adhesins and chemo-attractants leads to phagocyte recruitment at the infection site to trigger an inflammatory response (1). Microorganisms have developed ingenious mechanisms to fight and bypass the host signaling pathways and immune responses for survival and spread into their hosts (2,3).
Staphylococcus aureus (S. aureus) is a Gram-positive bacterium that rapidly acquires antibiotic resistances, being a worldwide health problem causing nosocomial and community-associated infections. S. aureus has surface and secreted components that compromise the host immune responses, allowing bacterial escape (4,5). Staphylococcal adherence to the host cells represents a prerequisite for their uptake. S. aureus can be internalized and survive in various host cells. This represents a critical factor for persistence and also when infections recidivate (6). The ability of S. aureus to escape from the host lysosomal degradation machinery and to persist intracellularly for long time periods is an essential step during infection. If engulfed by neutrophils, the intracellular bacteria can resist reactive oxygen intermediates, nitric oxide radicals, defensins and bactericidal proteins (6). After S. aureus internalization in non-phagocytic cells, the host cells up-regulate genes involved in innate immunity, antigen presentation and cell signaling (7). Upon internalization, S. aureus also reprograms gene expression (8–10). Bacterial genes involved in capsule synthesis, oxidative stress and virulence are up-regulated following ingestion of the pathogen (7,9). After S. aureus uptake by the human lung epithelial cells, bacterial genes involved in cell division and nutrient transport are down-regulated whereas genes involved in iron scavenging and virulence are up-regulated. During infection, S. aureus generates many virulence factors whose timing and expression levels are tuned by dozens of regulatory proteins and RNAs (sRNAs). S. aureus expresses around 159 sRNAs (11,12) recently compiled into the SRD database (13) with, for the most part, no associated functions. These sRNAs are expressed from the core and variable accessory genome including Pathogenicity Islands and transposons (14). Some sRNAs either stimulate (15) or decrease (16) bacterial virulence in animal models of infection and others influence antibiotic resistance (17).
In this report, we provide evidence that the Small pathogenicity island rna C, SprC, alias srn_3610 (13), when expressed by a S. aureus strain isolated from a human infection, diminishes bacterial virulence and spread in an animal model of septicemia. An isogenic strain lacking sprC becomes more virulent and complementation of the sRNA restores a lower virulence phenotype. SprC also reduces S. aureus phagocytosis by human monocytes and macrophages. The sRNA disfavors bacterial resistance to an oxidative stress, concurrent with extinction of SprC expression after host cell uptake. The staphylococcal autolysin (ATL) functions as an adhesin/invasin and binds the heat shock cognate protein Hsc70 host cellular receptor (18). At the molecular level, SprC interacts with the atl mRNA, in turn inhibiting ATL expression through a direct pairing interaction between SprC and the atl mRNA. The SprC phagocytosis phenotype is mediated, at least in part, by ATL. Overall, SprC attenuates bacterial virulence and host cell phagocytosis. It suggests that this regulatory RNA, when expressed, may favors commensalism with the host.
MATERIALS AND METHODS
Strains, culture conditions, genetic manipulations and oxidative stress experiments
All the bacterial strains and plasmids used are listed in Supplementary Table S1. S. aureus strains were grown at 37°C in brain heart infusion broth (BHI, Oxoid). When necessary, chloramphenicol and erythromycin were added at a 10 μg/ml concentration. For the oxidative stress, overnight cultures were diluted (1/100), incubated with 5 mM hydrogen peroxide (Aldrich Chemical, St Louis, MO) and optical density was measured using the spectrophotometer Biotek Synergy 2. A chromosomal gene disruption mutant was constructed by deletion of the targeted gene and insertion of an erythromycin-resistance gene by using the temperature-sensitive vector pBT2 (19) according to (15). Double-crossover events corresponding to the desired gene disruptions were confirmed by polymerase chain reaction (PCR) and sequenced. In all the sRNA overexpression genetic constructions, the RNAs are expressed from their endogenous promoters. In pCN35ΩsprC and pCN38ΩsprC, the nucleotide sequence of sprC containing 300 upstream nucleotides was amplified from Newman genomic DNA as a 460-bp fragment possessing EcoRI and PstI restriction sites at each end. The PCR product was cloned in pCN35erm and pCN38cat. E. coli DH5α were transformed with pCN35ΩsprC and pCN38ΩsprC and selected for ampicillin resistance. The plasmids pCN35-sprC and pCN38-sprC were transferred into RN4220 and selected for erythromycin and chloramphenicol resistance respectively, then finally transferred to Newman WT and Newman ΔsprC.
S. aureus FITC labeling and microscopy
Bacteria were labeled with fluorescein isothiocyanate (FITC) for the phagocytosis assay. 50 ml of cultured cells (mid-log phase, OD600nm = 2) were harvested by centrifugation. The cell pellet was washed with phosphate-buffered saline (PBS) and re-suspended in 1 ml of carbonate/bicarbonate buffer (pH 9.5, 0.1 M Na2CO3 and 0.2 M NaHCO3). The FITC (Sigma Chemical Co., St. Louis, Mo.) was dissolved in dimethyl sulfoxide at 10 mg/ml. 100 μl of this solution was added to 500 μl of the bacterial suspension. The mixture was rotated at 180 rpm for 1 h at room temperature. Bacteria were then washed twice in PBS to remove the unbound fluorochrome and re-suspended in 500 μl of HBSS (Hanks Balanced Salt Solution). Aliquots were prepared, frozen and stored in the dark until used. Aliquots were thawed just before use. Dilutions of stock suspensions of FITC-labeled bacteria or control bacteria were plated on agar to determine the number of CFU/ml. Coverslips were mounted with Vectashield mounting medium including DAPI (Vector Laboratories, Burlingame, CA). Staining was viewed with an inverted confocal microscope (DMIRE2, Leica) equipped with a 63XNA 1.4 objective lens using the LS 3D software (Leica). Images were analyzed using the ‘MetaMorph’ (Molecular Devices, Downington, PA) and Volocity (Perkin Elmer, version 6.0.1) software.
Human macrophages production
THP1 human monocytes were obtained from ATCC (Rockville, MD) and maintained at 37°C with 5% CO2 in RPMI 1640 (Invitrogen, Carlsbad, USA) containing 10% FCS (Biowest). The THP1 cells were treated with PMA (20 ng/ml) (Sigma) for 3 days to stimulate their differentiation into macrophages. The THP1 monocytes were efficiently differentiated into adherent macrophages, as evidenced by light microscopy.
Samples preparation, flow cytometry and phagocytosis assays in the human THP1 and macrophages
Human THP1 cells (106 cells) were incubated with 107 bacteria in RPMI, 10% human serum AB (SAB) (Institut Jacques Boy, Reims, France) to stimulate opsonization, for different times, as indicated, at 37°C, 180 rpm. The extracellular bacteria were lysed by lysostaphin (10 μg/ml) for 10 min and the cells were washed with RPMI, 10% SAB and with PBS. At the indicated time points, the THP1 cells containing intracellular S. aureus were lysed for total RNA extraction. The cells were fixed in PBS / 4% paraformaldehyde and analyzed using a FC500 flow cytometer and CXP Analysis Software (Beckman Coulter, Villepinte, France). To quantify bacterial phagocytosis, the human THP1 macrophages were infected with S. aureus at a multiplicity of infection (MOI) of 1:25. Phagocytosis assays were carried out for 2 h at 37°C, in the presence of 5% CO2 in RPMI 1640 medium containing 10% human SAB. Phagocytosis was stopped by putting the plates on ice and washing the cells three times with ice-cold PBS. All the non-phagocytosed bacteria were killed by culturing the human cells for 24 h in a medium containing 50 μg/ml gentamycin (Sigma). Subsequently, the medium was removed from each well and the cells were washed three times with PBS and incubated with PBS/SDS 0.1% for 15 min to lyse the cells. The lysates were plated in various dilutions on BHI agar plates. The colony counts were performed to determine the number of intra- and extracellular bacteria. All the samples were prepared and tested in triplicate. For the inhibition of phagocytosis mediated by ATL, the macrophages were pre-incubated with ATL in a concentration range of 5 to 50 nM for 30 min at 37°C, 5% CO2 prior to the infection with a S. aureus strain lacking sprC expression (Newman ΔsprC).
Cytochalasin D treatment and intracellular bacteria counting
To study the role of the human cell cytoskeleton in S. aureus uptake and provide evidence for phagocytosis, human THP1 macrophages were incubated with 2.5 and 5 μg/ml cytochalasin D (Sigma) for 60 min at 37°C before S. aureus interaction and also during the experiment. Human THP1 macrophages were infected with S. aureus and phagocytosis was carried out for 2 h at 37°C, 5% CO2, in RPMI 1640 medium. Phagocytosis was stopped by putting the plates on ice and washing the cells three times with ice-cold PBS. The remaining non-phagocytosed bacteria were killed by adding 100 μg/ml gentamycin (Sigma) and cytochalasin D (1 to 2.5 μg/ml, Sigma) in the medium for 60 min, at 37°C, 5% CO2. Subsequently, the medium was removed from each well, the cells were washed with PBS and cold distilled water was added to lyse the cells by repeated pipetting. The lysates were plated, in various dilutions, on BHI agar plates and incubated overnight at 37°C to determine the number of intracellular bacteria.
RNA extractions, Northern blots, reverse transcription and qPCR assays
Isolated colonies were suspended in BHI and incubated at 37°C overnight. The culture was diluted 1:100 then incubated at 37°C with agitation and stopped at various phases of growth. RNA extraction was performed as described (20). The DNA probes used to detect sRNA are listed in Supplementary Table S2. Total RNA (10 μg) was separated on denaturing 6% PAGE and transferred onto a Zeta probe GT membrane (Bio-Rad) in 0.5× TBE. The membranes were hybridized with specific 32P-labeled probes in ExpressHyb solution (Clontech), washed, exposed and scanned with a PhosphorImager (Molecular Dynamics). Contaminating DNA was removed from RNA samples by treatment with DNase (DNase amplification grade, InVitrogen, Carlsbad, USA) according to the manufacturer's protocol. Two micrograms of RNA was used for first strand cDNA synthesis using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, USA) following the manufacturer's protocols. Real-time quantitative PCR was performed with the fluorescent dye SYBR Green methodology using the Real Master Mix (Eppendorf) and the Mastercycler realplex (Eppendorf). Supplementary Table S3 shows primer pairs for each transcript chosen with the Primer 3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Gene specificity of each pair of primers was checked by comparing their sequences to the GenBank database using the program BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/). Using the comparative Ct method, the amount of target sequence in unknown samples was normalized to Hu or 16S reference.
Protein isolation, mass spectrometry and immunoblots
For the preparation of the protein extracts, bacteria were grown until the exponential or stationary phases and the cells are pelleted for 10 min at 4°C (8000g). To purify the extracellular proteins, the supernatants were collected and precipitated with 10% trichloroacetic acid. The precipitates were washed with ice-cold acetone and loaded onto SDS-PAGE gels according to (21) and stained with Coomassie blue R-250. The proteins of interest were extracted from the gel, digested with trypsin and the peptides identified by MALDI MS/MS and RP-HPLC/NanoLC/ESI-MS-MS. For the total protein extractions, pellets of 2-ml cultures were washed with TE (50 mM EDTA, 50 mM Tris pH 7.5), and suspended in 0.2 ml of the same buffer containing 0.1 mg/ml lysostaphin. Following incubation at 37°C for 10 min, the samples are boiled for 5 min and analyzed by SDS-PAGE. For the immunoblots, proteins were transferred to PVDF membranes (Immobilon-P, Millipore) and incubated with polyclonal anti-autolysin antibodies (a gift from Dr Sugai, Hiroshima University, Japan, (22). Signals were visualized using a STORM 840 Phosphor-Imager (Molecular Dynamics).
Toeprints and gel retardation assays
Annealing mixtures containing 0.15 pmol of unlabeled atl154 mRNA (154 nts, including the mRNA 5′-UTR, starting with three additional Gs for transcriptional efficiency) and 1.5 pmol of labeled primer in 20 mM Tris-acetate (pH 7.5), 60 mM NH4Cl and 1 mM DTT were incubated for 1 min at 90°C and quickly chilled on ice for 1 min. Renaturation proceeded in 10 mM MgCl2 at 20°C for 15 min. The influence of SprC (a 150 nt-long synthetic transcript with 3 additional Gs at the 5′-end for transcription efficiency), SprCΔ78–89 (a 138 nt-long RNA) or SprCΔ55–67 (a 137 nt-long RNA) was assayed by adding increasing amounts of each RNA (7.5 to 30 pmol) to the annealing mixtures and incubating for 5 min at 37°C. The purified E. coli 70S ribosomes was diluted in the reaction buffer in the presence of 1 mM MgCl2, and then activated for 15 min at 37°C. For each sample, 0.3 pmol of 70S ribosomes was added, followed by 5 min incubation at 37°C and adjustment of the MgCl2 concentration to 10 mM. After 10 min at 37°C, 10 pmol of uncharged tRNAfMet were added and the samples incubated for 5 min at 37°C. The cDNAs were synthesized using three units of AMV RT (New England Biolabs, Ipswich, USA) for 15 min at 43°C. Reactions were stopped by adding 10 μl of loading buffer II (Ambion, Foster City, USA). The cDNAs were loaded and separated on a urea 8% PAGE gel. The toeprint position on atl154 mRNA was determined by DNA sequencing. The results were analyzed on a PhosphorImager. For the electrophoretic mobility shift assays, the atl154 mRNA with SprC or SprCΔ78–89, or SprCΔ55–67 RNAs were denatured in 80 mM Hepes (pH 7.5), 330 mM KCl and 4 mM MgCl2 for 2 min at 80°C, chilled on ice and refolded for 20 min at 20°C. For the binding reaction (10 μl reaction volume), the incubation was for 30 min at 30°C. 0.005 pmol of labeled atl154 mRNA was incubated with 6 to 200 pmol of unlabeled SprC for Kd determination, or with the mutants from 5 to 40 pmol. To demonstrate specificity, 250 or 500 pmol of unlabeled SprA2AS were added. The samples were supplemented with 10% glycerol (final concentration) and loaded on a native 8% polyacrylamide gel containing 5% glycerol. The electrophoresis was performed in 0.5X Tris-borate EDTA at 4°C (100V). The results were analyzed on a PhosphorImager.
Filter binding assays
0.5 pmol of labeled atl154 mRNA or SprC were incubated in 20 mM Tris-acetate (pH 7.5), 60 mM NH4Cl, 1 mM DTT for 1 min at 90°C and chilled on ice for 1 min. Renaturation was performed in 10 mM MgCl2 at 20°C for 15 min. Purified E. coli 70S ribosomes were diluted in the reaction buffer in the presence of 1 mM MgCl2 and activated for 15 min at 37°C. For the samples containing 70S ribosomes, 5 pmol of 70S ribosomes were added, followed by 5 min incubation at 37°C and the concentration of MgCl2 adjusted to 10 mM. After 10 min at 37°C, 25 pmol of uncharged tRNAfMet were added and the samples incubated for 15 min at 37°C. The final volume was set to 20 μl. For the binding assays, we used nitrocellulose membranes (AppliChem, Pure Nitrocellulose). After equilibrating the membrane with 50 μl of reaction buffer per well, the samples were loaded into the well and sucked through. We washed twice with 50 μl reaction buffer and air-dried the membrane prior exposition onto a phosphor imaging device. The results were analyzed on a PhosphorImager.
ATL protein purification
A plasmid encoding the 6XHis-fusion ATL protein was a gift from Dr C. Heilmann (Münster University, Germany). Purification of the 6XHis-fusion ATL protein from an E. coli culture containing plasmid pQatl was performed under denaturing conditions. Its production, in E. coli, was induced by supplementing the culture with 1 mM IPTG during 3 h. The cell pellets were dissolved in a lysis buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M Urea, pH 8) and centrifuged. Supernatant fluid was loaded onto an ‘AKTA purifier’ (GE Healthcare) equipped with a Ni2+ column. The washes were performed with a buffer containing 100 mM NaH2PO4, 10 mM Tris-HCl and 8 M Urea, pH 6.3. The ATL protein was eluted with a buffer containing 100 mM NaH2PO4, 10 mM Tris-HCl, 8 M Urea, pH 5.9 and then with the same elution buffer at pH 4.5. ATL was additionally purified under native conditions. The 6XHis-fusion ATL protein was dialyzed against dialysis buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8) and loaded onto an ‘AKTA purifier’ equipped with a Ni2+ column. The washes and binding were performed with a buffer containing 500 mM NaCl, 50 mM NaH2PO4, 5 mM Tris-HCl and 5 mM Imidazole, pH 8. The ATL protein was eluted with a buffer containing 500 mM NaCl, 50 mM NaH2PO4, 5 mM Tris-HCl, 250 mM Imidazole, pH 8. The fractions containing the overexpressed His-tagged ATL were pooled and dialyzed against 50 mM Tris-HCl pH 7.2. The purity of the protein was verified by 10% SDS-PAGE and its concentration estimated with a Qubit fluorometer (InVitrogen).
Animal infection experiments
Experiments were monitored in the central animal laboratory ARCHE-BIOSIT- Rennes and all protocols were approved by the ethics committee (agreement No. R-2011-PT-01). Virulence levels of strain Newman, its isogenic deletion mutant ΔsprC-pCN38 and the complemented strain ΔsprC-pCN38ΩsprC were compared using a murine intravenous sepsis model. Groups of 10 female Swiss mice, 6- to 8-weeks old (Janvier Laboratories, Le Genest Saint Isle, France) were inoculated intravenously with 200 μl of bacterial suspensions containing 5×107 S. aureus cells in 0.9% NaCl. The survival of the mice was monitored for 14 days and the statistical significance of differences between groups was evaluated using the Mantel-Haenszel test. A P-value <0.05 was considered significant. Four days after inoculation, mice were euthanized with CO2 and their kidneys excised. After photographs were taken, the organs were homogenized, diluted in 0.9% NaCl and plated on BHI agar for determination of bacterial titers, expressed as log10 CFU per pair of kidneys. The statistical significance of differences between groups was evaluated using the Mann-Whitney U test. A P-value <0.05 was considered significant. The stability of plasmid ΔsprC-pCN38ΩsprC (encoding chloramphenicol resistance) in the complemented ΔsprC mutant was assessed by plating randomly selected colonies grown from kidney homogenates on nutrient agar containing chloramphenicol.
Statistical analysis of the data
For the statistical analysis of the data, the Mann-Whitney and Wilcoxon (animal infection experiments) tests were performed, as indicated for each set of experiments (Figure legends), on at least three independent experiments, to evaluate significance. The data were expressed as mean ± standard deviations (SD).
RESULTS
SprC negatively controls the expression of the major autolysin ATL in S. aureus
In a large-scale analysis of the S. aureus transcriptome, we identified a small RNA, SprC (srn_3610, SRD database, (13)), expressed from the genome of a converting phage (14). We determined the 5′- and 3′-ends of SprC by RACE (Rapid Amplification of cDNA Ends) mapping. They were located, respectively, at positions 1918280 and 1918432 within the S. aureus Newman sequence. In strain Newman, sprC is located in the νSAβ pathogenicity island (PI) that contains several virulence factors (14). Another S. aureus sRNA, SprD, also situated within a PI, has a positive role on virulence (15). According to the location of sprC within a PI, we postulated that it could also be involved in staphylococcal pathogenesis. In order to identify molecular targets of SprC, we analyzed whether the sRNA modifies the expression of the extracellular proteins that include virulence factors, as reported (15). To achieve this goal, a sprC deletion mutant was constructed by homologous recombination in strain Newman, isolated from a human infection. Northern blots confirmed the absence of SprC expression in strain Newman-ΔsprC and the growth curves of strains Newman and Newman-ΔsprC were identical (Figure 1A). Overexpression of SprC was achieved using a ‘multicopy’ plasmid expressing SprC from its endogenous promoter, with no impact on cell growth (pCN38-ΩsprC, Figure 1B).
Figure 1.
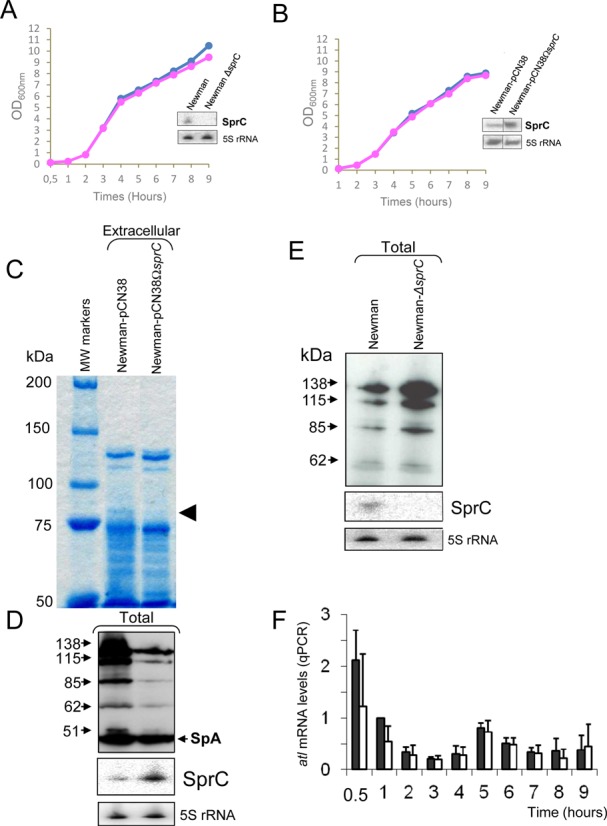
S. aureus SprC sRNA (srn_3610) lowers the expression levels of the major autolysin ATL. (A) Monitoring the bacterial growth and SprC expression levels, by Northern blots (OD600nm: 2), in S. aureus strains Newman (blue diamonds), Newman-ΔsprC (pink rectangles). (B) Monitoring the bacterial growths and SprC expression levels, by Northern blots (OD600nm: 2) of Newman-pCN38 (blue diamonds) and Newman-pCN38ΩsprC (pink rectangles). The 5S rRNAs are the loading controls. (C) Coomassie staining of SDS-PAGE of the exoproteins in strain Newman overexpressing (WT-pCN38ΩsprC), or not (WT-pCN38), SprC (OD600 = 7). The arrows point to the reduced levels of a protein when SprC is overexpressed. (D) Immunoblot analysis with anti-autolysin (anti-ATL) antibodies to monitor the levels of the ATL protein produced as five distinct translational products (23) in strain Newman overexpressing (WT-pCN38ΩsprC), or not (WT-pCN38), SprC. Reduced levels are detected for all the ATL protein products when SprC is overexpressed. Northern blot analyses of SprC expression levels in strains ‘WT-pCN38’ and ‘WT-pCN38ΩsprC’ demonstrating SprC overexpression when collecting the exoproteins. 5S rRNAs are the loading controls, ‘Spa’ corresponds to ‘Surface protein A’. (E) Lacking sprC increases S. aureus ATL expression levels. Immunoblots with anti-ATL antibodies to monitor the expression levels of ATL in S. aureus wild type strain (WT) and its isogenic strain lacking sprC (ΔsprC). Northern blot analyses of SprC expression levels in strains ‘WT’ and ‘ΔsprC’ demonstrating the absence of SprC expression in the mutant strain. 5S rRNAs are the loading controls. (F) qPCR analysis of the atl mRNA expression levels during bacterial growth in strains Newman ‘WT-pCN38’ (black bars) and ‘WT-pCN38ΩsprC’ (white bars). Using the comparative Ct method, the amount of RNAs was normalized against the expression of the hu reference gene and referred to the Newman ‘WT-pCN38’ after 1 h of bacterial growth. The data shown are the mean and the error bars indicated representing Standard Deviation (SD) are derived from three independent experiments.
In the extracellular proteins extracted from strain Newman-pCN38ΩsprC, levels of an ∼85 kD protein decreased as compared to the isogenic Newman strain (Figure 1C, arrow). Proteins from that band were eluted, tryptic digests were prepared and the fragments analyzed by MALDI-TOF mass spectrometry. Twenty-three peptides were identified (Table 1), all matching the sequence of the major autolysin ATL protein encoded by atl (23). The ATL proteins are associated with the bacterial surface by ionic or hydrophobic interactions and have enzymatic (peptidoglycan-hydrolytic) and adhesive functions. The ATL protein is detected within the cytoplasm as well as at the cell surface and extracellularly, after cleavage (22). An independent confirmation of the decrease in the autolysin levels, when SprC expression is induced, was obtained by monitoring the autolysin protein within a total protein extract by immunoblots with anti-autolysin antibodies (Figure 1D). The immunoblots detected five proteins in the cellular extracts, with apparent molecular weights of 138, 115, 85, 62 and 51 kDa (Figure 1D). According to (22), the 138 kDa bi-functional ATL protein contains both the amidase (Ami) and the glucosaminidase (GL) domains that is processed into two intermediates of 115 and 85 kDa, both of which are subsequently cleaved to produce, respectively, an N-terminal amidase domain (Ami, N-acetylmuramoyl-L-alanine amidase; 62 kDa) and a C-terminally located glucosaminidase domain (GL,endo-b-N-acetylglucosaminidase; 51 kDa, (23), that correspond to the two lower bands detected by immunoblots (Figure 1D). As additional and independent experimental evidence of the negative regulation of ATL by SprC, immunoblots performed in strain Newman ΔsprC indicated that the expression levels of the ATL protein were increased compared to the isogenic Newman strain (Figure 1E).
Table 1. Mass spectrometry identification of the bi-functional ATL autolysin protein by detecting 23 autolysin peptides.
| Observed | Mr (expected) | Mr (calculated) | Peptides |
|---|---|---|---|
| 1000.5056 | 999.4984 | 999.5349 | K.DLNVQNLGK.E |
| 1045.5361 | 1044.5288 | 1044.5564 | K.ASKQQQIDK.S |
| 1151.5647 | 1150.5574 | 1150.6023 | K.LYTVPWGTSK.Q |
| 1164.5835 | 1163.5762 | 1163.5935 | K.VTTFSASAQPR.S |
| 1213.5896 | 1212.5824 | 1212.6179 | K.LYSVPWGTYK.Q |
| 1310.7420 | 1309.7348 | 1309.7466 | K.IAQVKPNNTGIR.A |
| 1391.6556 | 1390.6483 | 1390.6292 | K.IEEDYTSYFPK.Y |
| 1447.6911 | 1446.6839 | 1446.6779 | K.FYLVQDYNSGNK.F |
| 1466.7259 | 1465.7186 | 1465.7161 | K.QVAGSVSGSGNQTFK.A |
| 1671.7965 | 1670.7892 | 1670.7801 | K.YLGGTDHADPHGYLR.S |
| 1696.8794 | 1695.8721 | 1695.8580 | K.YKPQVNSSINDYIR.K |
| 1718.9045 | 1717.8972 | 1717.8999 | K.SPVNVNQSYSIKPGTK.L |
| 1749.8965 | 1748.8892 | 1748.9131 | K.SPVNVMQTYTVKPGTK.L |
| 1765.8677 | 1764.8604 | 1764.9080 | K.SPVNVMQTYTVKPGTK.L + Oxidation (M) |
| 1794.8242 | 1793.8169 | 1793.8908 | K.QEAGAVSGTGNQTFKATK.Q |
| 1866.8675 | 1865.8602 | 1865.8445 | K.NNYQNAFVHAFVDGDR.I |
| 2001.9170 | 2000.9098 | 2000.9115 | R.SHNYSYDQLYDLINEK.Y |
| 2019.0255 | 2018.0182 | 2018.0010 | R.FINVEIVHTHDYASFAR.S |
| 2216.1376 | 2215.1304 | 2215.1273 | R.IIETAPTDYLSWGVGAVGNPR.F |
| 2298.1750 | 2297.1677 | 2297.1652 | K.NPTQNISGTQVYQDPAIVQPK.T |
| 2640.4100 | 2639.4027 | 2639.3806 | K.AYLVDTAKPTPTPTPKPSTPTTNNK.L |
| 2841.4854 | 2840.4781 | 2840.4556 | K.VAPWGTQSTTTPTTPSKPTTPSKPSTGK.L |
| 3322.5774 | 3321.5701 | 3321.5120 | K.TNTNVTNAGYSLVDDEDDNSENQINPELIK.S |
ATL regulation by SprC is not at the transcriptional level but SprC reduces ATL expression by pairing with the mRNA and preventing ribosome loading
To determine whether ATL regulation by SprC occurs at the transcriptional level, we examined the effect of SprC on the steady-state levels of atl mRNA. The atl mRNA levels were monitored by quantitative PCR (qPCR), in strains Newman WT-pCN38 and WT-pCN38ΩsprC, using hu as a reference gene. The atl mRNA expression profile was similar in both strains, elevated early, decreasing at the E phase, rising at the early S phase and decreasing thereafter (Figure 1F). During S. aureus growth, the atl mRNA expression profile is not influenced by stimulating SprC expression, indicating that SprC induction does not modify the atl mRNA steady-state level (Figure 1F). Thus, the regulation of ATL by SprC is not at the mRNA accumulation level, implying that SprC may prevent atl mRNA translation.
The above data prompted us to test for the existence of an interaction between SprC and the atl mRNA that could reduce the mRNA translation. We did electrophoretic mobility shift assays (EMSA) to analyze duplex formation between SprC and a 154 nt-long atl mRNA fragment containing its 5′ UTR sequence (34 nt plus 3Gs at the 5′-end), followed by the first 39 codons. SprC formed a specific complex with the atl151 mRNA in vitro (Figure 2A, left panel), suggesting that the atl mRNA is a direct target of SprC. The estimated Kd between the two RNAs is approximatively 35 μM, suggesting that additional ligands might be involved in the recognition process between the two RNAs in vivo. The binding between SprC and the atl mRNA was specific, since a ten-fold excess of an unrelated RNA, SprA2AS, was unable to displace SprC from a preformed ‘atl mRNA-SprC’ complex (Figure 2A, right panel). SprC secondary structure was inferred by MFold structure prediction (24) and consists of three stem-loops (H1-H3) connected by two single strands (Figure 2B). SprC sequence comparison among the currently published S. aureus genomes indicated a ∼98% sequence conservation, with two base-pair compensatory changes located within helices H1 and H3, whereas the majority of the other changes are within the predicted RNA single strands (Figure 2B).
Figure 2.
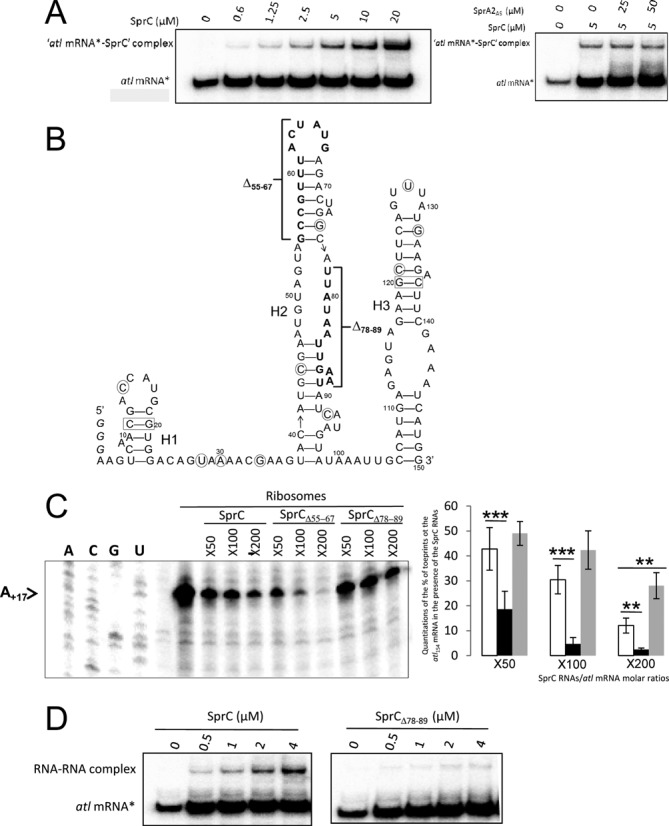
SprC (srn_3610) interacts with the atl mRNA to prevent ribosome loading; SprC secondary structure and SprC mutants enhancing or reducing atl mRNA translation initiation. (A) (left panel). Complex formation between an atl mRNA fragment and SprC. Native gel retardation assays of purified labeled atl154 mRNA with increasing amounts of purified, unlabeled SprC. (right panel) Competition assays performed with 5 to 10-fold excess of unlabeled, unrelated SprA2AS RNA. (B) SprC secondary structure prediction. A careful sprC gene sequence comparison among 49 available staphylococcal genomic sequences have evidenced two compensatory base pair changes maintaining base pairings within H1 and H3 (rectangles) and the other nucleotide changes (circles) are located, for the most part, within predicted RNA single strands. The three Gs added at SprC 5′-end for transcription are in italic. (C) (left panel) Toeprint assays on the atl154 mRNA in the presence of increasing concentrations of purified SprC, SprCΔ55–67 or SprCΔ78–89. ‘-’ indicates the absence of purified 70S ribosomes. The arrow points to the location of the experimentally-determined toeprint. U, A, G and C refer to the atl mRNA sequencing ladders. (right panel) Quantitations of the toeprints on the atl mRNA in the presence of increasing concentrations of SprC (white), SprCΔ55–67 (black) or SprCΔ78–89 (gray). Without any RNAs, the amount of toeprints was arbitrarily set to 100. The data and error bars, representing SD, indicated are derived from three to six independent experiments. Two and three asterisks corresponded to the P-values ≤0.01 and ≤0.001, respectively. (D) Native gel retardation assays of purified labeled atl154 mRNA with increasing amounts of purified, unlabeled SprC or SprCΔ78–89.
We conjectured that SprC could lower ribosome loading to the atl mRNA ribosomal binding site (RBS). We tested this hypothesis by performing toeprint analysis. A ternary initiation complex consisting of purified ribosomes, initiator tRNAfMet and the atl mRNA was formed. The ribosome blocked the elongation of reverse transcription and produced a toeprint 17 nt downstream from the AUG initiation codon of the atl mRNA (Figure 2C). SprC reduced the toeprint in a concentration-dependent manner, indicating that SprC inhibits ribosome binding onto the atl mRNA RBS. It implies that the pairing interaction between SprC with the atl mRNA negatively influences translation initiation. A possible pairing between SprC and the atl mRNA was detected between U74-U85 (SprC)/A+12-A+23 (atl mRNA). Therefore, the capacity of a SprC mutant lacking nucleotides U78-U89 (SprCΔ78–89) to influence atl mRNA translation was analyzed. As expected and compared to SprC, SprCΔ78–89 had an impaired ability to lower translation initiation onto the atl mRNA (Figure 2C). EMSA performed between SprCΔ78–89 and the atl mRNA reinforces further these results by showing that complex formation is severely impaired (Figure 2D). Another SprC mutant lacking G55-G67 (SprCΔ55–67) was constructed, because it was predicted, based on SprC secondary structure, to destabilize helix H2, in turn possibly enhancing its downregulation of atl mRNA translation. Indeed, increasing amounts of SprCΔ55–67 reduced the toeprint in a concentration-dependent manner, significantly more than SprC (Figure 2C), indicating that mutant SprCΔ55–67 has increased regulatory activity onto the atl mRNA than SprC. As internal control for the toeprint assays, the putative interaction between SprC and the purified ribosomes was challenged experimentally by filter binding assays and Supplementary Figure S1A shows that whereas the atl mRNA associates with the ribosomes, SprC does not. Moreover, EMSA performed between labeled SprC and the tRNAfMet have not detected any interaction between the two RNAs (Supplementary Figure S1B). These data indicate that SprC does not interact with either ribosomes or tRNAfMet. Overall, the atl mRNA is a direct molecular target of SprC and the sRNA likely reduces its protein expression levels by preventing translation initiation, but the interaction between the two RNAs is elaborate and will need further structural investigations.
Implementing an S. aureus internalization assay in human phagocytes
SprC lowers the expression of the major autolysin ATL. ATL has implications in cell wall turnover, cell separation, division, autolysis, and mediates S. aureus internalization by endothelial cells (18,25). We hypothesized that SprC's regulation of ATL synthesis could influence bacterial uptake by the host's professional phagocytes. To test that assumption, an assay was implemented monitoring S. aureus uptake by human phagocytes, including THP1 monocytes and macrophages. Flow cytometric internalization assays were set up by using a THP1 monocyte cell line. The S. aureus Newman cells were labeled with FITC. The human THP1 cells were incubated with the bacteria. Various ratios between the human cells and the bacteria were assayed, and a 1:10 ratio was selected. Incubations were from 5 min to 3 h, after which the extracellular bacteria were removed by lysostaphin treatment. Phagocytosis was monitored by flow cytometry. The absence of any remaining extracellular microorganisms, after the lysostaphin treatment, was verified by microscopy (Figure 3A, panels 1 and 2) and confocal microscopy showed S. aureus internalization by the human host cells (Figure 3A, panel 3). Cytochalasin D, an actin depolymerizing agent, inhibits actin-dependent S. aureus uptake by the host cells (26). Cytochalasin was used as an inhibitor of host cell S. aureus uptake, providing evidence that the number of internalized bacteria reflects host cell uptake into the intracellular compartment. Increasing amounts of cytochalasin D drastically reduced the number of S. aureus cells present in the human macrophages (Figure 3B). It indicates that the number of internalized bacteria is dependent on uptake into the intracellular compartment, demonstrating phagocytosis in our assay.
Figure 3.
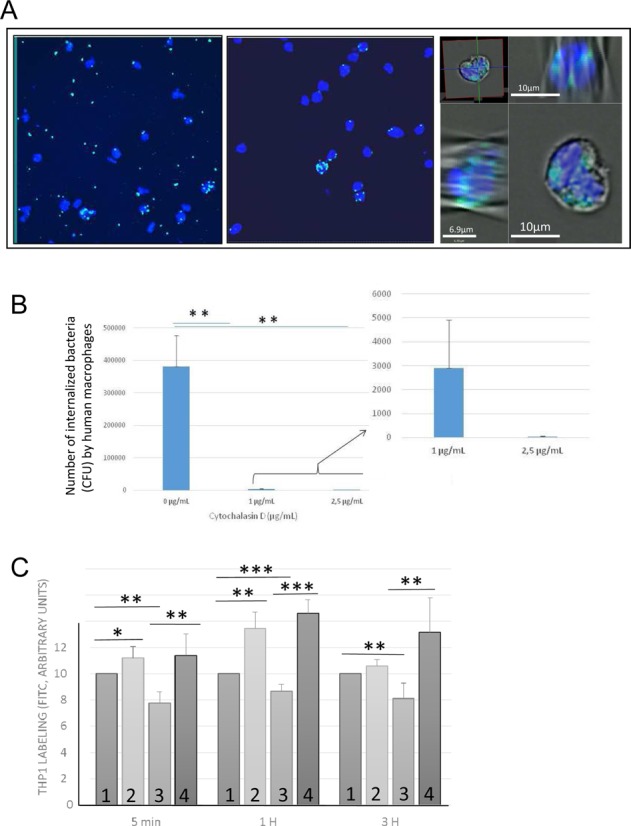
Implementing an S. aureus-host cell phagocytosis assay showing that Atl facilitates S. aureus uptake by the human phagocytes. (A) Human THP1 cells are incubated for 1 h with FITC labeled S. aureus Newman cells (MOI 1:10). Coverslips are mounted with the ‘Vectashield mounting medium’ containing DAPI. The human cell nucleus is blue (DAPI) and the S. aureus cell wall is green (FITC). The extracellular bacteria were (panel 2), or not (panel 1), lysed with lysostaphin for 10 min. After the lysostaphin treatment, microscope imaging did not detect extracellular bacteria (compare panels 1 and 2). The bacteria were also viewed by confocal microscopy (panel 3). In panel 3, the S. aureus cells are stained green (FITC), the host cells nuclei are blue (DAPI) and the host cell membranes are gray, imaged by Differential Image Contrast (DIC). In panel 3, two orthogonal sections are presented, demonstrating S. aureus internalization. (B) Staphylococcal phagocytosis by human macrophages as evidenced by Cytochalasin D treatment. Cytochalasin D-treated THP1 macrophages were infected with S. aureus (Newman) for 2 h followed by adding gentamycin for 1 h to remove the extracellular bacteria. Infected THP1 macrophages without Cytochalasin D treatment served as internal positive controls. The data shown are the mean ± SD of three independent experiments, each realized in triplicates. The data are considered highly significant for P-values ≤0.01 (**). (C) The major autolysin ATL facilitates S. aureus internalization in human professional phagocytes. Amount of labeled THP1 cells after the internalization of FITC-labeled S aureus Newman strains (1), Newman strain in the presence of pLI50atl (2), strain lacking atl expression (3) or Δatl Newman strain complemented with pLI50atl (4). FITC labeled S. aureus strains were incubated for 5 min, 1 or 3 h with the human THP1 monocytes (MOI 1:10). The cells were analyzed by flow cytometry. The data shown are the mean ± SD of three independent experiments, each realized in triplicates. The data are considered significant for P-values <0.05 (*), highly significant for P-values ≤0.01 (**) and extremely significant for P-values ≤0.001 (***).
Atl facilitates S. aureus internalization by the human phagocytes
We analyzed the capacities of a S. aureus Newman atl-deficient mutant (a gift from Prof. J. O'Gara, Dublin, Ireland), a complemented strain with a plasmid expressing the protein (27) and an isogenic Newman strain to be internalized by the human THP1 cells. The absence of atl mRNA expression in strain Newman Δatl was confirmed by qPCR, as well as its expression in the complemented strain Newman Δatl-pLI50 atl (not shown). Compared to a Newman strain, flow cytometric internalization assays indicated that the Δatl mutation reduced the ability of the Newman strain to be internalized by the THP1 host cells by ∼20%. The effect was observed early, after 5 min incubation (Figure 3C). Compared to an isogenic S. aureus strain, the internalization of Newman Δatl by the human host cells was decreased and that reduction was reversed by atl complementation (Figure 3C). These experiments indicated that the ATL protein facilitates S. aureus internalization by human professional phagocytes and that its absence reduces uptake by host cells.
SprC lowers internalization by human phagocytes and its absence increases S. aureus uptake
ATL protein facilitates S. aureus uptake by the host cells and SprC reduces Atl protein expression. To test if SprC influences S. aureus internalization by the host cells, phagocytosis of S. aureus strain Newman-ΔsprC by human monocytes was compared to an isogenic Newman strain. Host cell internalization of strain Newman ΔsprC gradually increased over time, reaching a level ∼40% higher than for strain Newman (Figure 4). SprC was overexpressed from a low copy (∼20–25 copies per cell) pCN38ΩsprC plasmid in which sprC was cloned with its endogenous promoter. Strains Newman-pCN38 and Newman-pCN38ΩsprC had similar growth curves and Northern blots confirmed SprC overexpression in the latter strain (Figure 4). qPCR quantifications indicated a 50±10 fold increase in SprC expression levels in strain Newman pCN38ΩsprC compared to the strain carrying an empty plasmid (not shown). The level of induction of SprC expression in strains with pCN38ΩsprC reduced the amount of internalized bacteria by the human phagocytes by ∼20% (Figure 4A). When the S. aureus Newman cells were transformed with a high copy (∼300–400 copies per cell) pCN35ΩsprC plasmid, SprC expression was significantly enhanced whereas bacterial growth was unaffected (Figure 4B). qPCR quantifications of SprC levels in strain Newman pCN35ΩsprC, compared to the strain with an empty plasmid, indicated a 500±50 fold increase in SprC expression (not shown). Compared to Newman pCN35, the number of Newman pCN35ΩsprC bacteria internalized by the human monocytes was reduced by ∼40% (Figure 4A). Inducing SprC expression reduces S. aureus uptake by the host cells and the effect increases with the amount of SprC produced.
Figure 4.
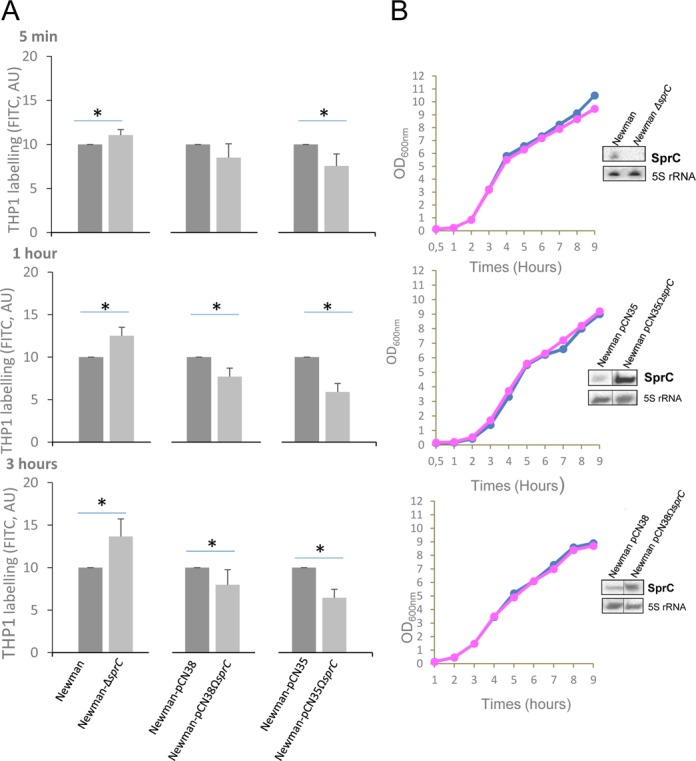
SprC reduces S. aureus internalization by human monocytes and its absence augments phagocytosis. (A) Percentage of THP1 cells labeled with FITC after S aureus internalization. FITC labeled S. aureus Newman and Newman-ΔsprC, Newman-pCN38 and Newman-pCN38ΩsprC, Newman-pCN35 and Newman-pCN35ΩsprC were incubated with the human monocytes (MOI 1:10) for the indicated times. The cells were analyzed by flow cytometry. For a direct comparison of the S. aureus strains, we set up the labeling of control strains ‘Newman’, ‘Newman-pCN35′ and ‘Newman-pCN38’ arbitrarily at ‘10’. The data presented and the error bars indicated are the mean ± SD derived from three independent experiments (p* < 0.025). (B) Lacking or stimulating SprC expression in vivo does not impact S. aureus growth. Monitoring bacterial growth and SprC expression levels, by Northern blots (OD600nm = 2), in S. aureus strains Newman pCN35 (blue) and Newman-pCN35ΩsprC (pink). The 5S rRNAs are the loading controls. The data presented are the mean ± SD of three independent experiments realized in triplicates. The data are considered significant for P-values < 0.025 (*).
The influence of SprC on S. aureus phagocytosis was also evaluated in human macrophages, whose primary roles are to ingest foreign particles and infectious microorganisms. Phagocytosis of S. aureus strain Newman-ΔsprC by PMA-induced THP1 macrophages was compared to an isogenic Newman strain. Host cell internalization of strain Newman-ΔsprC was significantly enhanced compared to strain Newman (Figure 5A). A trans-complemented Newman ΔsprC-pCN38ΩsprC strain was constructed. Northern blots confirmed the re-expression of SprC in strain Newman ΔsprC-pCN38ΩsprC, and its growth was identical to that of the strain Newman-ΔsprC-pCN38 (Figure 5C). Conversely, the phagocytosis of a trans-complemented strain (Newman ΔsprC-pCN38ΩsprC) was decreased compared to a Newman ΔsprC-pCN38 strain. Interestingly, bacterial release from the THP1 macrophages, after 4 days, was significantly increased in the absence of the sRNA, and reduced in the trans-complemented strains (Figure 5B). Therefore, these observations suggest that the bacteria lacking SprC could possess higher virulence by a higher uptake and release from the immune host cells.
Figure 5.
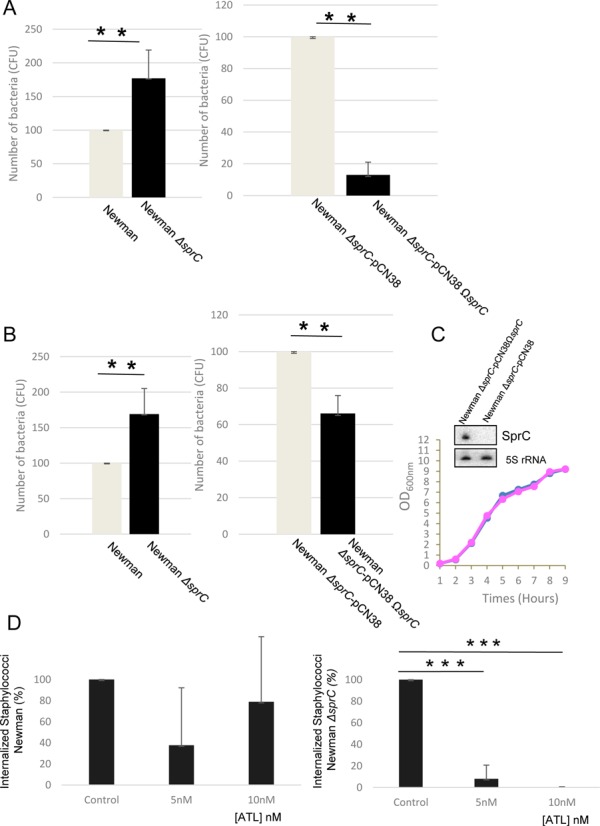
SprC reduces S. aureus internalization by human macrophages and its absence augments phagocytosis and bacterial release from the host cells. (A) Number of S. aureus cells (CFU) after internalization. Newman, Newman-ΔsprC, Newman-ΔsprC pCN38 and Newman-ΔsprC pCN38ΩsprC cells were incubated with the THP1 macrophages (MOI 1:25) for 2 h. The non-phagocytosed bacteria were killed by culturing in medium containing 50 μg/ml gentamycin for 24 h. The medium was then replaced with fresh media for up to 4 days. (B) After 4 days, colony counts correspond to the bacteria that were released by the human macrophages. The data presented are the mean ± SD of three independent experiments realized in triplicates. The data are considered highly significant for P-values ≤0.01 (**). (C) Monitoring bacterial growth and SprC expression levels, by Northern blots (OD600nm = 2), in S. aureus strains Newman ΔsprC-pCN38 (pink) and Newman ΔsprC-pCN38ΩsprC (blue). The 5S rRNAs are the loading controls. Each blot presented is from identical experiments. (D) Inhibition of S. aureus strain Newman ΔsprC phagocytosis by the THP1 macrophages by adding increasing concentrations of purified ATL protein (right panel) whereas exogenous ATL has limited, if any, effects on S. aureus strain Newman internalization (left panel).
To provide experimental evidence about a link between the ATL and SprC regarding the S. aureus internalization phenotype, we purified the ATL protein and monitored its influence on phagocytosis of S. aureus strain Newman lacking sprC, in comparison with strain Newman (attempts to construct a ΔsprC-Δatl double mutant failed). Pre-incubation of the macrophages with a purified full-length ATL protein, dose-dependently reduced internalization of S. aureus bacteria lacking SprC, but has no or limited effects on S. aureus bacteria expressing SprC (Figure 5D). This result suggests that the effect of SprC on phagocytosis is mediated, at least in part, by its regulatory action onto ATL.
SprC attenuates S. aureus virulence and its induction lowers bacterial spread
S. aureus survives and multiplies within several host cells including endothelial and epithelial cells, keratinocytes, fibroblasts and osteoclasts (28). Intracellular S. aureus uses human phagocytes, especially the neutrophils, to travel and infect distant sites (29). Bacteria lacking sprC may use the host phagocytes for dissemination and multiplication into the colonized host, in turn providing a significant advantage during infection and spread into the infected hosts. To assay that hypothesis, the implication of SprC expression was tested in a mouse systemic infection model, as performed for SprD (15). The virulence of strain Newman-ΔsprC was significantly and reproducibly enhanced, with half the animals already deceased at day 9 of infection (Figure 6A), five days earlier than what is observed with strain Newman. Compared to strain Newman-ΔsprC-pCN38, the virulence of the trans-complemented strain Newman ΔsprC-pCN38ΩsprC was partially restored (Figure 6B). After day 6 of infection, the in vivo persistence of plasmid pCN38ΩsprC in the Newman ΔsprC-pCN38ΩsprC complemented strain was verified in selected colonies obtained from mice kidney homogenates. All the selected colonies retained chloramphenicol resistance, a specific marker of pCN38. In a different intravenous infection experiment, the animals were sacrificed at day 4, the kidneys of groups of five mice inoculated with strains Newman, Newman ΔsprC, Newman ΔsprC-pCN38 and the trans-complemented Newman ΔsprC-pCN38ΩsprC strain were collected and viable bacteria counted. Kidneys of mice inoculated with the ΔsprC mutant are swollen and displayed mottled discoloration with numerous abscesses, whereas those of mice inoculated with strain Newman are homogenously red-brown with only a few abscesses (Figure 6C). Kidneys of mice infected with the SprC-complemented strains display homogenous coloration with very few abscesses (Figure 6C). Results of the macroscopic observation are confirmed, in the same experiment, by viable bacteria counts per kidney pairs for the different strains (Figure 6C). Comparing the viable bacteria number per kidney in strains Newman versus Newman-ΔsprC, and Newman-ΔsprC-pCN38 versus Newman-ΔsprC-pCN38ΩsprC, using the Mann-Whitney U test, indicates a significant P-value (<0.05). Altogether, these results indicated that SprC has a negative impact on the virulence of a S. aureus human isolate in an animal infection model. SprC is therefore a virulence attenuator, reducing bacterial spread into the colonized mice.
Figure 6.
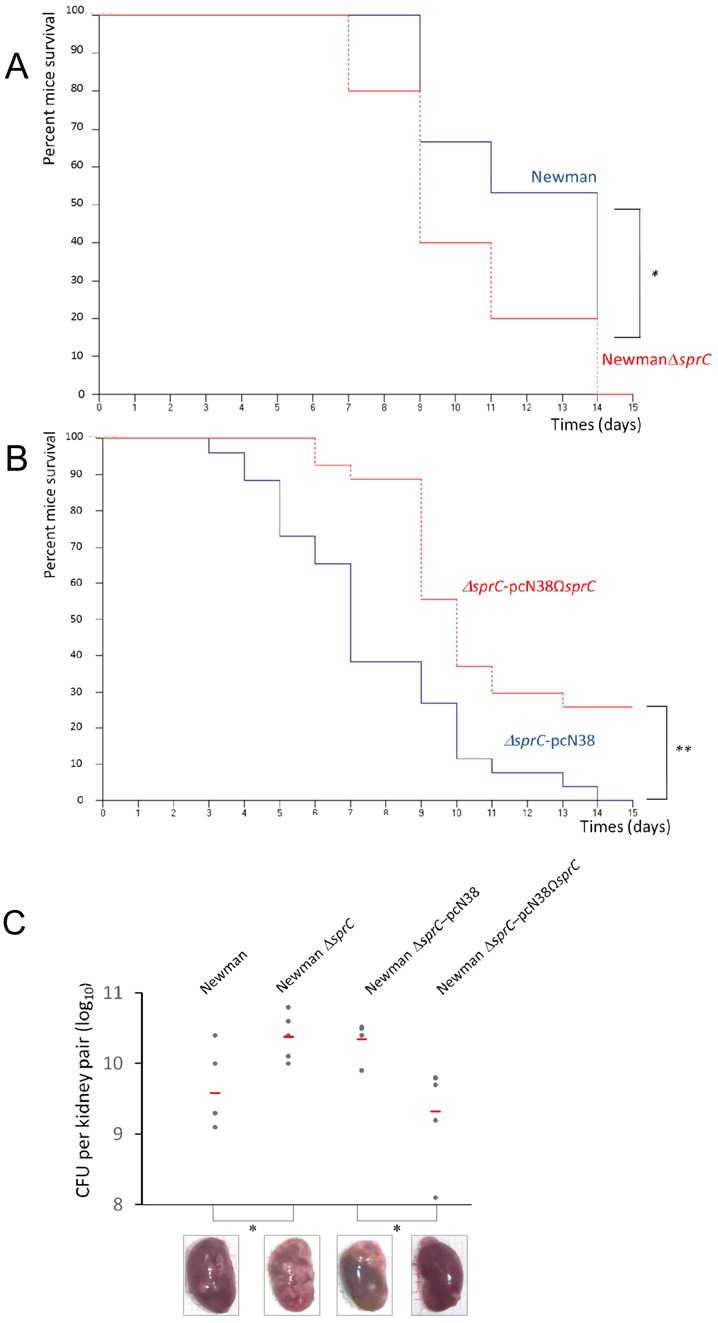
SprC attenuates the virulence and spread of a S. aureus clinical isolate on infected mice. (A) Survival probability plots (Kaplan-Meier) of mice infected either with S aureus Newman (blue, 5.107 cfu per mouse) or isogenic S. aureus Newman-ΔsprC (red; 5.107 cfu); 0.05 < P-value < 0.1. Groups of 10 ‘six to eight-weeks old’ Swiss mice were inoculated intravenously with 5.109 bacteria and monitored daily for 2 weeks. The Mantel-Haenszel test P-value is between 0.01 and 0.05. The results are representative of three independent experiments representing 30 mice per strain. (B) Survival probability plots (Kaplan-Meier) of mice infected either with S. aureus Newman-ΔsprCpCN38 (blue; 5.107 cfu) or S. aureus Newman-ΔsprCpCN38ΩsprC (red, 5.107 cfu per mouse); The Mantel-Haenszel test P-value is lower than 0.01. Results are representative of three independent experiments representing 30 mice per strain. (C) Recovery of S. aureus from the kidneys of infected mice four days after bacterial challenge. Groups of 5 mice were inoculated intravenously with the indicated strains. Each individual is indicated by a circle symbol, with mean bacterial titers as red lines. The Mann-Whitney test P-value is between 0.01 and 0.05. The red line represents the mean value of 5 mice. Bottom: Macroscopic aspects of the recovered kidneys after intravenous infection with the respective S. aureus strains.
SprC is detrimental for S. aureus resistance in the presence of a sublethal oxidative stress
Since SprC has a negative impact on virulence and host cell internalization, the sRNA might also influence S. aureus intracellular survival and persistence. In the presence of a sublethal oxidative stress triggered by hydrogen peroxide (5 mM), strain Newman ΔsprC grew better than its isogenic control strain (Figure 7A). The trans-complemented strain Newman ΔsprC-pCN38ΩsprC has a restored growth phenotype in the presence of hydrogen peroxide (Figure 7B). In the presence of hydrogen peroxide, inducing SprC expression substantially (Figure 7C) or moderately (Figure 7D) delayed S. aureus growth, with respect to their internal controls without sRNA induction. These data indicate that, when S. aureus adapts to oxidative stress, the absence of sprC is beneficial whereas inducing SprC expression is prejudicial for growth, in agreement with the experiments performed at the cellular and animal levels.
Figure 7.
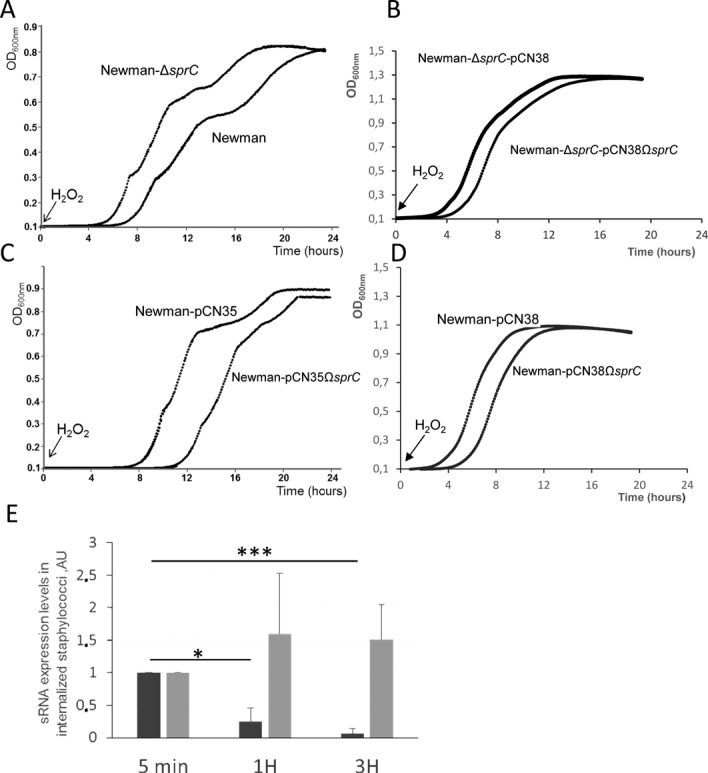
SprC reduces S. aureus resistance toward hydrogen peroxide and its expression level decreases in internalized bacteria. Growth curves of Newman, Newman-ΔsprC(A), Newman-ΔsprC-pCN38 and Newman-ΔsprC-pCN38ΩsprC (B), Newman-pCN35’, ‘Newman-pCN35ΩsprC’ strains (C), ‘Newman-pCN38’ and ‘Newman-pCN38ΩsprC’ (D), after exposure to 5 mM hydrogen peroxide (H2O2). Optical density was measured using the spectrophotometer Biotek Synergy from 0 for 24 h. Growth curves are representative of three experiments. (E), SprC expression levels decrease in bacteria internalized by human monocytes. Monitoring the expression levels of SprC (dark gray) and of tmRNA (light gray) by qPCR during phagocytosis of S. aureus wild-type strain Newman. The bacteria were labeled by FITC, incubated with the human monocytes (MOI 1:10) for the indicated periods of times and total RNAs extracted. Using the comparative Ct method, the amount of sRNAs was normalized against the expression of the hu reference gene and referred to the internalized bacteria after 5 min of phagocytosis. The data presented are the mean ± SD of three independent experiments realized in triplicates. The data are considered significant for P-values <0.05 (*) and extremely significant for P-values ≤0.001 (***).
SprC expression decreases after S. aureus host cell internalization
Since the absence of SprC facilitates S. aureus host cell internalization (Figures 4 and 5), virulence (Figure 6) and alleviates resistance against an oxidant (Figure 7), we conjectured that SprC expression might be diminished after bacterial uptake by the host cells. To test this hypothesis, total eukaryotic and prokaryotic RNAs were extracted from human THP1 cells infected with labeled S. aureus strain and deprived of extracellular bacteria Newman after 5 min, 1 h and 3 h of incubation, according to the experimental set up described above. SprC expression levels were monitored by qPCR. Within the infected host cells, SprC expression is detected early and decreases ∼4-fold after 1 h and ∼10-fold after 3 h (Figure 7E). As an internal control, the expression of another S. aureus sRNA, transfer-messenger RNA (tmRNA), responsible of ribosome rescue and trans-translation (30) was monitored during host cell phagocytosis. At the three time points after S. aureus internalization, tmRNA expression is maintained relatively stable over time (Figure 7E). The steady state levels of the hu mRNA, from 5 min to 3 h, were stable, indicating that the internalized S. aureus cells are alive and transcriptionally active. The lower expression of SprC within the ingested staphylococci could increase their resistance to oxidative killing by the immune cells, in turn promoting their dissemination into the colonized host.
DISCUSSION
We report the case of a regulatory RNA expressed from the accessory genome of a major bacterial pathogen, S. aureus, that reduces virulence and host immune cell phagocytosis. When SprC is not expressed, the bacteria are internalized more, resist better to an oxidative burst, are released further by the human phagocytes and possess increased virulence in an animal infection model. SprC expression is detrimental for bacterial spread into the colonized organisms, induces a disadvantage for intracellular entry and resistance against an oxidative environment. This may explain why its expression is turned off after host cell internalization. Another sRNA expressed from the S. aureus core genome, RsaA, is also a virulence suppressor that favors chronic infection (16). Interestingly, whereas SprC lowers both the host cell phagocytosis and the oxidative stress resistance, RsaA renders the bacteria more sensitive to opsono-phagocytosis by the host PMNs. These two sRNAs, expressed from the core (RsaA) and accessory (SprC) genome, attenuate staphylococcal virulence by distinct mechanisms. Why does S. aureus express sRNAs exerting negative influences on pathogenicity? sprC is detected in numerous S. aureus clinical strains but its expression levels has not been monitored yet and we also do not know if its negative impact on virulence can be extrapolated to many clinical isolates. Nevertheless, recent results in the lab performed on clinical strains isolated from thirty patients with bloodstream infections indicate that 30% of the strains lack sprC whereas only 20% of the strains isolated from a set of thirty asymptomatic colonized individuals lack sprC. A negative selection pressure against sprC may operate faster in the clinical strains than for those present in the asymptomatic carriers, but this result should be confirmed on a larger set of individuals.
sRNAs impacting host cell entry and invasion were reported in several bacteria. The PI-encoded IsrM sRNA is required for Salmonella invasion of epithelial cells, intracellular replication inside macrophages, virulence and colonization in mice (31). Also, the expression of the AfaD invasins that mediate entry of E. coli clinical isolates into the host cells from various tissues is controlled and reduced by the AfaR antisense sRNA (32). As for the E. coli AfaR, SprC protects the bacteria against their engulfment by the human host cells. Neutrophils and monocytes struggle to eradicate many invading pathogens, including S. aureus (33). In the Gram positive bacterium Listeria monocytogenes, several sRNAs are specifically expressed when the bacteria are within the macrophages (34), but their implications for bacterial survival are unknown. In the Gram negative Salmonella enterica serovar Typhi, two sRNAs, RfrA and RfrB, are also required for optimal bacterial intracellular replication in macrophages (35). The expression of RNAIII, the effector of the agr quorum-sensing system in S. aureus, is also modified after ingestion of S. aureus clinical strain USA300 by the human neutrophils but, in that case, its expression rapidly increases to produce α-hemolysin and, in turn, to trigger PMN lysis (36). We monitored RNAIII expression levels, by qPCR, on the total eukaryotic and prokaryotic extracted RNAs, during human THP1 infection by labeled S. aureus strain Newman. After infection, the expression of RNAIII reproducibly increases (6±1-fold) compared to the non-internalized bacteria (not shown). Therefore, in clinical strain Newman, the expression of another S. aureus sRNA is modified upon host cell uptake for survival and escape immune clearance. Is there a functional link between SprC and RNAIII? Preliminary experiments have monitored, by Northern blots, the RNAIII levels during S. aureus growth in strains lacking or overexpressing SprC. At three time points during growth, RNAIII levels were not influenced by the presence/absence of SprC (not shown). These recent examples, including our findings, indicate that once bacterial pathogens are internalized, the expression of dedicated sRNAs and various toxins can be required for their intracellular persistence and growths.
The human phagocytes are the first line of host defenses against the invading pathogens. Intracellular bacterial persistence includes ‘phagosome-lysosome’ fusion inhibition, survival inside the phagolysosomes and cytoplasm escape (37). When the bacteria survive, multiply and are released from the host cells, phagocytosis becomes an advantage for colonization and spread. Within the host phagocytes, S. aureus cells lacking SprC possess a higher ability to disseminate and spread, ultimately stimulating the overall infectious process. S. aureus phagocytosis by neutrophils induce global changes in gene expression, including proteins involved in resistance to oxidants (9). In S. aureus, the expression of many protein-encoding genes, including virulence genes, is modified in response to an oxidative stress (38,39). When S. aureus is phagocytosed, it faces and resists the host defenses and resides in phagosomes containing antimicrobial peptides, proteolytic enzymes and reactive oxygen intermediates. The aim of the reactive oxygen species secreted by the host cells is to destroy the engulfed bacteria. The increased virulence of bacteria lacking SprC could be linked to its higher survival rate in an oxidative environment such as the phagolysosome. It suggests a tight coupling between resistance against oxidants and bacterial virulence, exemplified here by SprC. Future studies should address if SprC is involved in S. aureus phagosomal escape (40).
We identified one direct molecular target of SprC whose expression is down-regulated during translation initiation, by antisense pairings, the ATL staphylococcal autolysin. Many sRNAs are multifunctional and it might also be the case of SprC, having additional regulatory functions involved in S. aureus fitness with its environment. The interaction between SprC and the atl mRNA is weak and complex, probably implying the existence of additional RNA and/or protein associates involved in the interaction. This is of noticeable interest in the S. aureus sRNA field because, until now, there are no facilitator/chaperone involved in RNA-RNA interactions characterized in this bacterium. In addition to its positive implication in host cell phagocytosis, ATL has activities in cell division, autolysis and biofilm formation (41), suggesting that SprC may also influence these bacterial functions. Preliminary experiments reported here suggest that SprC influences host phagocytosis, at least in part, by its control of ATL expression. The functional link between SprC and Atl regarding phagocytosis could therefore be established. Strain ΔsprC expresses more ATL at the bacterial surface, compared to wild-type (Figure 1E), consequently predicted to interact more efficiently with the ATL receptors located at the surface of the human phagocytes, therefore increasing bacterial phagocytosis. When the ATL protein is provided externally, it binds to and blocks some of the human phagocytes receptors, subsequently preventing bacterial uptake via these ATL receptors. Thus, the progressive decrease of internalized staphylococci lacking sprC, when the ATL is provided externally, may be rationalized by the above arguments. This is probably not observed in the case of the wild-type cells because they express lower amounts of ATL at the bacterial surface (Figure 1E) and, therefore, may be less affected by the blocking of the ATL receptors from the human phagocytes via the ‘externally’ provided ATL. The physiological associations and implications between the S. aureus phagocytosis, SprC and ATL ‘triumvirate’ are currently being addressed experimentally.
Supplementary Material
Acknowledgments
We thank Dr P. Ame-Thomas (UMR U917, Rennes University) for her advices and help in THP1 cell culture and flow cytometry and Dr S. Dutertre (UMS 3480– US018 Biosit Ibisa Plateform of Microscopy, Rennes Imaging Center MRic-Photonics) for her help with the microscopes and data collection. We also thank M1 student S. Raynaud for his help in the ATL purification and cell culture experiments and Dr M. Sassi for the SprC sequence alignments. We also thank the colleagues from the lab for their comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale pour la Recherche [ANR-09-MIEN-030–01]; Inserm; French Department of Research and Education [to B.F.]. Funding for open access charge: Inserm; [to B.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hossain H., Tchatalbachev S., Chakraborty T. Host gene expression profiling in pathogen-host interactions. Curr. Opin. Immunol. 2006;18:422–429. doi: 10.1016/j.coi.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Bestebroer J., De Haas C.J., Van Strijp J.A. How microorganisms avoid phagocyte attraction. FEMS Microbiol. Rev. 2010;34:395–414. doi: 10.1111/j.1574-6976.2009.00202.x. [DOI] [PubMed] [Google Scholar]
- 3.Shimada T., Park B.G., Wolf A.J., Brikos C., Goodridge H.S., Becker C.A., Reyes C.N., Miao E.A., Aderem A., Gotz F., et al. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe. 2010;7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zecconi A., Scali F. Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol. Lett. 2013;150:12–22. doi: 10.1016/j.imlet.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Broker B.M., Holtfreter S., Bekeredjian-Ding I. Immune control of Staphylococcus aureus - regulation and counter-regulation of the adaptive immune response. Int. J. Med. Microbiol. 2014;304:204–214. doi: 10.1016/j.ijmm.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Loffler B., Tuchscherr L., Niemann S., Peters G. Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Med. Microbiol. 2014;304:170–176. doi: 10.1016/j.ijmm.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Matussek A., Strindhall J., Stark L., Rohde M., Geffers R., Buer J., Kihlstrom E., Lindgren P.E., Lofgren S. Infection of human endothelial cells with Staphylococcus aureus induces transcription of genes encoding an innate immunity response. Scand. J. Immunol. 2005;61:536–544. doi: 10.1111/j.1365-3083.2005.01597.x. [DOI] [PubMed] [Google Scholar]
- 8.Vriesema A.J., Beekhuizen H., Hamdi M., Soufan A., Lammers A., Willekens B., Bakker O., Welten A.G., Veltrop M.H., van De Gevel J.S., et al. Altered gene expression in Staphylococcus aureus upon interaction with human endothelial cells. Infect. Immun. 2000;68:1765–1772. doi: 10.1128/iai.68.4.1765-1772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voyich J.M., Braughton K.R., Sturdevant D.E., Whitney A.R., Said-Salim B., Porcella S.F., Long R.D., Dorward D.W., Gardner D.J., Kreiswirth B.N., et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 10.Garzoni C., Francois P., Huyghe A., Couzinet S., Tapparel C., Charbonnier Y., Renzoni A., Lucchini S., Lew D.P., Vaudaux P., et al. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics. 2007;8:171. doi: 10.1186/1471-2164-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillet J., Hallier M., Felden B. Emerging functions for the Staphylococcus aureus RNome. PLoS Pathog. 2013;9:e1003767. doi: 10.1371/journal.ppat.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fechter P., Caldelari I., Lioliou E., Romby P. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS Lett. 2014;588:2523–2529. doi: 10.1016/j.febslet.2014.05.037. [DOI] [PubMed] [Google Scholar]
- 13.Sassi M., Augagneur Y., Mauro T., Ivain L., Chabelskaya S., Hallier M., Sallou O., Felden B. SRD: a Staphylococcus regulatory RNA database. RNA. 2015;21:1005–1017. doi: 10.1261/rna.049346.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichon C., Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chabelskaya S., Gaillot O., Felden B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010;6:e1000927. doi: 10.1371/journal.ppat.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romilly C., Lays C., Tomasini A., Caldelari I., Benito Y., Hammann P., Geissmann T., Boisset S., Romby P., Vandenesch F. A non-coding RNA promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog. 2014;10:e1003979. doi: 10.1371/journal.ppat.1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyraud A., Tattevin P., Chabelskaya S., Felden B. A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res. 2014;42:4892–4905. doi: 10.1093/nar/gku149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirschhausen N., Schlesier T., Schmidt M.A., Gotz F., Peters G., Heilmann C. A novel staphylococcal internalization mechanism involves the major autolysin Atl and heat shock cognate protein Hsc70 as host cell receptor. Cell. Microbiol. 2010;12:1746–1764. doi: 10.1111/j.1462-5822.2010.01506.x. [DOI] [PubMed] [Google Scholar]
- 19.Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 20.MacCallum A., Hardy S.P., Everest P.H. Campylobacter jejuni inhibits the absorptive transport functions of Caco-2 cells and disrupts cellular tight junctions. Microbiology. 2005;151:2451–2458. doi: 10.1099/mic.0.27950-0. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Komatsuzawa H., Sugai M., Nakashima S., Yamada S., Matsumoto A., Oshida T., Suginaka H. Subcellular localization of the major autolysin, ATL and its processed proteins in Staphylococcus aureus. Microbiol. Immunol. 1997;41:469–479. doi: 10.1111/j.1348-0421.1997.tb01880.x. [DOI] [PubMed] [Google Scholar]
- 23.Oshida T., Sugai M., Komatsuzawa H., Hong Y.M., Suginaka H., Tomasz A. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U.S.A. 1995;92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vollmer W., Joris B., Charlier P., Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 26.Sinha B., Francois P.P., Nusse O., Foti M., Hartford O.M., Vaudaux P., Foster T.J., Lew D.P., Herrmann M., Krause K.H. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cell. Microbiol. 1999;1:101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 27.Houston P., Rowe S.E., Pozzi C., Waters E.M., O'Gara J.P. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immunol. 2011;79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garzoni C., Kelley W.L. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 2009;17:59–65. doi: 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Thwaites G.E., Edgeworth J.D., Gkrania-Klotsas E., Kirby A., Tilley R., Torok M.E., Walker S., Wertheim H.F., Wilson P., Llewelyn M.J. Clinical management of Staphylococcus aureus bacteraemia. Lancet Infect. Dis. 2011;11:208–222. doi: 10.1016/S1473-3099(10)70285-1. [DOI] [PubMed] [Google Scholar]
- 30.Janssen B.D., Hayes C.S. The tmRNA ribosome-rescue system. Adv. Protein Chem. Struct. Biol. 2012;86:151–191. doi: 10.1016/B978-0-12-386497-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong H., Vu G.P., Bai Y., Chan E., Wu R., Yang E., Liu F., Lu S. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog. 2011;7:e1002120. doi: 10.1371/journal.ppat.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichon C., du Merle L., Lequeutre I., Le Bouguenec C. The AfaR small RNA controls expression of the AfaD-VIII invasin in pathogenic Escherichia coli strains. Nucleic Acids Res. 2013;41:5469–5482. doi: 10.1093/nar/gkt208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mraheil M.A., Billion A., Mohamed W., Mukherjee K., Kuenne C., Pischimarov J., Krawitz C., Retey J., Hartsch T., Chakraborty T., et al. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 2011;39:4235–4248. doi: 10.1093/nar/gkr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leclerc J.M., Dozois C.M., Daigle F. Role of the Salmonella enterica serovar Typhi Fur regulator and small RNAs RfrA and RfrB in iron homeostasis and interaction with host cells. Microbiology. 2013;159:591–602. doi: 10.1099/mic.0.064329-0. [DOI] [PubMed] [Google Scholar]
- 36.Pang Y.Y., Schwartz J., Thoendel M., Ackermann L.W., Horswill A.R., Nauseef W.M. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate Immunol. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urban C.F., Lourido S., Zychlinsky A. How do microbes evade neutrophil killing? Cell. Microbiol. 2006;8:1687–1696. doi: 10.1111/j.1462-5822.2006.00792.x. [DOI] [PubMed] [Google Scholar]
- 38.Chang W., Small D.A., Toghrol F., Bentley W.E. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 2006;188:1648–1659. doi: 10.1128/JB.188.4.1648-1659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee L.Y., Liang X., Hook M., Brown E.L. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb) J. Biol. Chem. 2004;279:50710–50716. doi: 10.1074/jbc.M408570200. [DOI] [PubMed] [Google Scholar]
- 40.Fraunholz M., Sinha B. Intracellular Staphylococcus aureus: live-in and let die. Front. Cell Infect. Microbiol. 2012;2:43. doi: 10.3389/fcimb.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bose J.L., Lehman M.K., Fey P.D., Bayles K.W. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One. 2012;7:e42244. doi: 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


