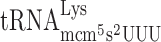Abstract
In Saccharomyces cerevisiae, 11 out of 42 tRNA species contain 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), 5-methoxycarbonylmethyluridine (mcm5U), 5-carbamoylmethyluridine (ncm5U) or 5-carbamoylmethyl-2′-O-methyluridine (ncm5Um) nucleosides in the anticodon at the wobble position (U34). Earlier we showed that mutants unable to form the side chain at position 5 (ncm5 or mcm5) or lacking sulphur at position 2 (s2) of U34 result in pleiotropic phenotypes, which are all suppressed by overexpression of hypomodified tRNAs. This observation suggests that the observed phenotypes are due to inefficient reading of cognate codons or an increased frameshifting. The latter may be caused by a ternary complex (aminoacyl-tRNA*eEF1A*GTP) with a modification deficient tRNA inefficiently being accepted to the ribosomal A-site and thereby allowing an increased peptidyl-tRNA slippage and thus a frameshift error. In this study, we have investigated the role of wobble uridine modifications in reading frame maintenance, using either the Renilla/Firefly luciferase bicistronic reporter system or a modified Ty1 frameshifting site in a HIS4A::lacZ reporter system. We here show that the presence of mcm5 and s2 side groups at wobble uridines are important for reading frame maintenance and thus the aforementioned mutant phenotypes might partly be due to frameshift errors.
INTRODUCTION
Transfer of genetic information from mRNA into proteins is the most energy consuming process in the cell and the translation machinery needs to decode mRNAs with high efficiency and fidelity (1). Even though the translational machinery transfers the information in mRNA into protein with high fidelity, errors occur at a low frequency. Missense errors are in most cases not harmful to the function of a protein, since such errors alter only one single amino acid, which will not interfere with the function or stability of the protein if they occur in non-critical positions. In contrast, processivity errors, like frameshift errors, are detrimental, since they completely change the amino acid sequence downstream of the frameshift site. Moreover, following such an error, the ribosome frequently encounters a stop codon in the new reading frame resulting in premature termination of translation. Accordingly, the frequency of frameshift errors is about 10-fold lower than the frequency of missense errors (1,2).
There are many examples where alterations in the tRNA structure, e.g. lack of a modified nucleoside, will affect the fidelity of reading frame maintenance (3,4). In bacteria, modified nucleosides of different chemical structures, present in different positions, and in different species of the tRNA all prevent frameshifts errors (5,6). In eukaryotes, both wyosin (yW) and queosine (Q) in rabbit reticulocytes as well as other modified nucleosides present in the anticodon loop of eukaryotic tRNAs are important to maintain the reading frame (7,8). Synthesis of yW in yeast tRNA occurs in several steps and whereas fully modified yW has a low frequency of frameshifting, presence of any of the various intermediates in the synthesis of yW all increase frameshifting (9). Also, lack of either cyclic N6-threonylcarbamoyladenosine (ct6A) at position 37 or pseudouridine (Ψ) at position 38 and 39 in yeast tRNA increases +1 frameshifting (10–13). Relevant for this study, the modified wobble nucleoside 5-methylaminomethyl-2-thiouridine (mnm5s2U34) present in bacterial tRNA specific for Gln, Lys and Glu, is important for proper reading frame maintenance (The wobble nucleoside is in position 34 of the tRNA and we denote such a nucleoside as N34 where N is any nucleoside.) (6,14–17). Apparently, modification status both in bacteria and in eukaryotes is important for a proper reading frame maintenance (3,4).
A peptidyl-tRNA slippage model of how tRNA modification deficiency may induce frameshifting errors is well established (3,4,6,18–24). According to this model (Figure 1) modification deficient aminoacyl-tRNAs present in a ternary complex, i.e. aminoacyl-tRNA*eEF1A*GTP (here shorten as aminoacyl-tRNA) induces frameshifts either by causing an A- or a P-site effect, or a combination thereof. Lack of modification causes a defect in the cognate aminoacyl-tRNA selection step (we denote such an error as an A-site effect by modification deficiency), allowing a ternary complex with a near cognate wild type aminoacyl-tRNA instead of a cognate aminoacyl-tRNA to be accepted in the A-site. After translocation to the P-site, the fit of the near cognate peptidyl-tRNA is not optimal why it slips one nucleotide forward (+1 frameshift) (Figure 1A). Alternatively, lack of a modified nucleoside reduces the efficiency by which a cognate aminoacyl-tRNA is accepted to the A-site, which induces a ribosomal pause allowing the wild type peptidyl-tRNA to slip forward one nucleotide (denoted an A-site effect by modification deficiency, Figure 1B). When frameshifting is caused by a P-site effect, the hypomodified tRNA is efficiently accepted to the A-site, translocates to the P-site where its fit is not optimal why it slips into an alternative reading frame due to a reduced ribosomal grip (P-site effect by modification deficiency, Figure 1C) (3,6,20,21,23). Thus, in some cases, the modification deficiency reduces the rate of selection of the aminoacyl-tRNA (A-site effect) but also lack of the modification reduces the ribosomal grip in the P-site (P-site effect). Note, in all cases explained above, the error in reading frame maintenance is due to a peptidyl-tRNA slippage.
Figure 1.
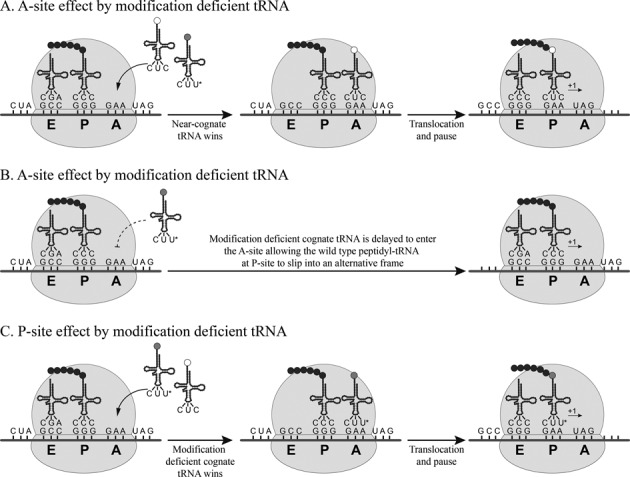
Dual-error frameshifting model. Modification deficient tRNAs can induce frameshifting by either an A- or a P-site effect, or a combination thereof. (A) Lack of wobble uridine modification reduces the efficiency of the ternary complex (aminoacyl-tRNA*eEF1A*GTP, here shorten as aminoacyl-tRNA) to be accepted to the A-site, allowing a near cognate aminoacyl-tRNA to be accepted in the A-site. After translocation to the P-site, the near cognate tRNA slips into an alternative reading frame, as it does not perfectly fit in the P-site. (B) Lack of wobble uridine modification reduces the efficiency of the cognate aminoacyl-tRNA to be accepted to the A-site, which induces a pause that allows the tRNA in the P-site to frameshift. (C) The hypomodified aminoacyl-tRNA is able to enter the A-site and translocate to the P-site where it then slips into an alternative reading frame due to a reduced ribosomal grip.
Modifications of uridines in the wobble position of tRNAs are frequent in all three domains of life. In Saccharomyces cerevisiae, there are 11 tRNA species having four related modified uridine nucleotides at wobble position (25–32). These modified nucleosides are 5-carbamoylmethyluridine (ncm5U34) present in five (26,27,32), 5-carbamoylmethyl-2′-O-methyluridine (ncm5U34m) present in one (25), 5-methoxycarbonylmethyluridine (mcm5U34) present in two (29,30) and 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U34) present in three tRNA species (Figures 2 and 3) (28,30,31).
Figure 2.
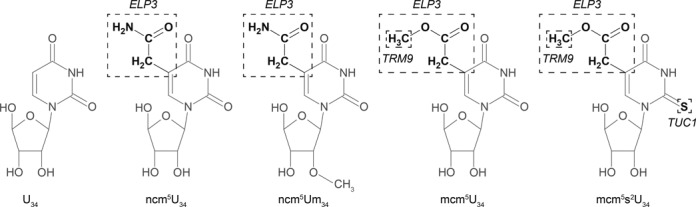
Chemical structures of uridine, 5-carbamoylmethyluridine (ncm5U), 5-carbamoylmethyl-2′-O-methyluridine (ncm5Um), 5-methoxycarbonylmethyluridine (mcm5U) and 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) nucleosides. Each dotted box indicates the side group that is removed by mutating the indicated gene.
Figure 3.
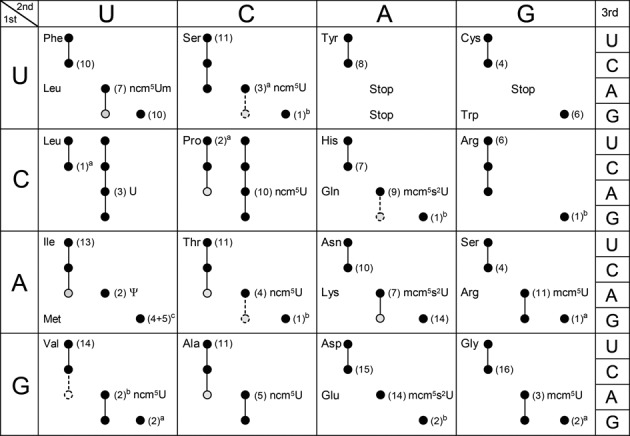
The genetic code and decoding abilities of individual tRNA species. Circles and connecting lines indicate codons read by the same tRNA isoacceptor. Gray circles connected with a dashed line indicate that the tRNA species reads the codon only when it is overexpressed. The empty dashed circle for 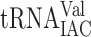 is shown only to indicate that this inosine containing tRNA species does not efficiently read the GUA codon. The nucleoside at the wobble position is given for the 13 wobble uridine containing tRNA species. Black and gray circles represent decoding abilities predicted by the wobble hypothesis, the revised wobble rules and the distribution of tRNA species. A gray circle indicates that the tRNA species is less likely to read the codon. The number of genes coding for a tRNA species is indicated next to the circle for the complementary codon. a The gene(s) encoding the tRNA is nonessential. b The gene(s) encoding the tRNA is essential. c Four genes code for
is shown only to indicate that this inosine containing tRNA species does not efficiently read the GUA codon. The nucleoside at the wobble position is given for the 13 wobble uridine containing tRNA species. Black and gray circles represent decoding abilities predicted by the wobble hypothesis, the revised wobble rules and the distribution of tRNA species. A gray circle indicates that the tRNA species is less likely to read the codon. The number of genes coding for a tRNA species is indicated next to the circle for the complementary codon. a The gene(s) encoding the tRNA is nonessential. b The gene(s) encoding the tRNA is essential. c Four genes code for 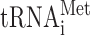 and five for
and five for 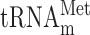 . Copyright © American Society for Microbiology, [Molecular and Cellular Biology, 28, 2008, 3301–3312 and doi:10.1128/MCB.01542–07](26).
. Copyright © American Society for Microbiology, [Molecular and Cellular Biology, 28, 2008, 3301–3312 and doi:10.1128/MCB.01542–07](26).
The first step in the synthesis of the mcm5 and ncm5 groups of the uridine modifications mentioned above requires the six-subunit Elongator complex and its seven associated proteins (Reviewed in Karlsborn et al. (33)). Mutations in any of the corresponding genes result in deficiency of these xm5-uridine modifications without affecting stability or aminoacylation of tRNA (26). These mutants also show strong pleiotropic phenotypes, such as defects in growth, transcription, chromatin remodelling, DNA repair and secretion (Reviewed in Karlsborn et al. (33)). All these phenotypes, except lack of xm5 side chains, are suppressed by overexpression of hypomodified tRNAs specific for Gln, Lys and Glu that in a wild type contains mcm5s2U34 (34,35). It was concluded that lack of this wobble nucleoside reduces the efficiency to recognize the cognate codons for these tRNAs, which is compensated by an increased concentration of the modification deficient tRNA. Thus, the many different phenotypes of Elongator mutants are due to reduced efficiency in translating some key mRNAs encoding proteins important for manifesting a correct phenotype.
In bacteria, modified wobble uridines are important to prevent +1 frameshifting (6,36). In eukaryotes, only a limited study has been done, which focused on the influence of the esterified methyl group of mcm5U34 in reading frame maintenance (37). However, no specific conclusion was made where the frameshift errors occur, since the frameshift window used was very large. Therefore, no extensive information of the role of modified wobble uridines in reading frame maintenance is available for eukaryotic tRNA. It was therefore important to investigate whether or not lack of the xm5U or mcm5s2U modifications are crucial for reading frame maintenance. Here, we show that presence of xm5- (x, any substitution) or s2 side groups at wobble uridines in yeast is pivotal in maintaining the translational reading frame.
MATERIALS AND METHODS
Strains, media and genetic procedures
The source and genotypes of yeast strains used in this study are listed in Table 1. E. coli strain used was DH5α (Bethesda Research Laboratories). Yeast transformation (38), media and genetic procedures have been described previously (39).
Table 1. Yeast strains used in this study.
| Strains | Genotype | Source |
|---|---|---|
| W303–1A | MATa leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15 | (52) |
| UMY3269 | MATa leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15 elp3::KanMX4 | (30) |
| UMY3164 | MATa leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15 tuc1::TRP1 | (48) |
| UMY3267 | MATa leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15 trm9::KanMX4 | (53) |
Plasmid constructions
Plasmid pJD375 contains a Renilla/Firefly luciferase bicistronic reporter system (40). To introduce various frameshifting windows between the luciferase genes, a BamHI-XhoI fragment from plasmid pJD375 containing the Firefly luciferase gene was cloned into corresponding sites of YCp50, generating plasmid YCp50-Firefly. Two complementary oligonucleotides carrying various frameshifting windows (see Supplementary Table S1) were annealed into the BamHI and SacI sites of YCp50-Firefly. The newly constructed plasmids were digested with restriction enzymes (BamHI and XhoI) and fragments containing the frameshifting sites linked to the Firefly luciferase gene were cloned back into the corresponding sites of pJD375 restoring the bicistronic reporter system with the frameshifting window.
Plasmids pMB38–9mer (FF and WT) contain a HIS4A::lacZ reporter cassette. In pMB38–9merFF (in-frame control construct), the lacZ gene is in 0 frame, while in pMB38–9merWT (test construct), the lacZ gene is in +1 frame (Figure 4B) (41). These plasmids were used as templates for PCR oligonucleotide directed mutagenesis to alter the Ty1 sequence (CTT-AGG-C) (Figure 4B and Supplementary Table S2).
Figure 4.
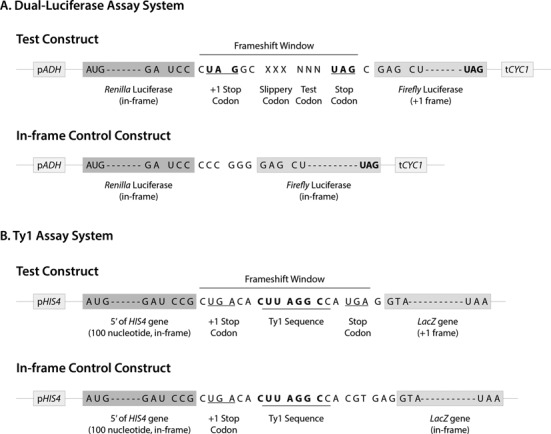
(A) Schematic drawing of the dual-luciferase assay system. Transcription of the genes encoding the Renilla- and Firefly-luciferase is under the ADH1 promoter and terminated by CYC1 terminator. Frameshift sites were cloned between the luciferase genes and expression of the Firefly luciferase gene requires +1 frameshifting. The frameshifting site is as follows: XXX-slippery site, NNN-assay codon and UAG-stop codon (all in-frame). An upstream stop codon (UAG) was placed in the +1 frame to eliminate frameshifting events occurring before the assay site. The frame of the different luciferase genes is indicated. In the in-frame control construct, Renilla- and Firefly-luciferase genes are in-frame. (B) Schematic drawing of the Ty1 assay system. Transcription from HIS4 promoter generates a transcript containing the first 100 nucleotides of the HIS4 gene in the in-frame and the lacZ gene of Escherichia coli in the +1 frame. Expression of the lacZ gene is dependent on a +1 ribosomal frameshift event taking place within Ty1 sequence. An upstream stop codon (UGA) was placed in the +1 reading frame to eliminate frameshifting events occurring before the assay site. In the in-frame control construct, the first 100 nucleotides of the HIS4 gene and lacZ gene are in-frame.
For the overexpression of the Lys-tRNA encoded by the tK(UUU)L gene, we first introduced SphI and NheI restriction sites to plasmids pMB38–9mer (FF and WT) carrying the ‘CUU-AAA-C’ sequence by PCR oligonucleotide directed mutagenesis. Oligonucleotides used were 5′-GGTGTCGGGGCGCATGCATGACCCAGTCAC-3′ and 5′-AGAGTGCACCATATGCGGTGTGAGCTAGCGCACAGATGCG-3′. The tK(UUU)L gene was amplified from strain UMY2067 by using oligonucleotides 5′ AAAAGCATGCCGGTAGAGTCTCTT-CTTGGTC-3′ and 5′ AAAAGCTAGCCGGTA-AGAGAGAAACCTCCA-3′ and cloned between SphI and NheI sites of these plasmids.
Dual-luciferase assays
Three individual transformants of each dual luciferase assay construct (biological replicates) were grown at 30°C in synthetic complete (SC)-Ura medium to an optical density at 600 nm (OD600) of 0.5. For each transformant triplicate samples (technical replicates) of 10 μl cells were collected and kept at -80°C. The luciferase assays were performed according to the instructions of Dual-Luciferase Reporter Assay System (Promega, Catalog No. E1960). The luciferase activities were determined in a white 96-well plate (Thermo Scientific, #436111) using a TECAN infinite 200 luminometer. The levels of +1 frameshifting (%) were determined by normalization of each biological test replicate with the average of the three biological replicates of the in-frame control. Each value of the biological replicates was determined by taking the median of the three technical replicates. The significant differences between wild type and mutant were determined by two-tail t-test.
β-galactosidase assays
Three transformants of each Ty1 assay construct (biological replicates) were grown in SC-Ura to OD600≈0.5 and 20 OD600-units were collected and kept at -20°C. For each transformant, β-galactosidase measurements were done three times (technical replicates). β-galactosidase activities were determined as described previously (39). Values of the biological replicates were determined by taking the median of the technical replicates. The levels of +1 frameshifting (%) were determined by normalization of each biological test replicate with the average of the three biological replicates of the in-frame control. The significant differences between wild type and mutant were determined by two-tail t-test.
RESULTS AND DISCUSSION
Assay system
To analyze the role of wobble uridine modifications ncm5U, ncm5Um, mcm5U or mcm5s2U in reading frame maintenance, we used defined yeast mutants unable to form the s2 group (tuc1Δ; also denoted as ncs6Δ), the ncm5 or mcm5 groups (elp3Δ) or the esterified methyl group (trm9Δ) of the mcm5 side chain.
The ribosomal +1 frameshifting assay system used contains a Renilla luciferase (R-luc)/ Firefly luciferase (F-luc) bicistronic reporter system (Figure 4A) (see Material and Methods) (40). This bicistronic mRNA synthesizes a two domain protein with the indicated enzymatic activities. To analyze a +1 frameshift event a sequence is introduced between these two cistrons in such a way that translation of R-luc is in the 0 frame and the F-luc is in the +1 frame (Figure 4A). To obtain F-luc activity the ribosome must shift into the +1 frame before entering the F-luc gene. The inserted sequence between the R-luc and the F-luc reporter genes consists of a slippery codon (XXX) at which the peptidyl-tRNA will slip, the codon to be assayed for A-site selection (NNN), followed by a stop codon in zero frame (UAG) (Figure 4A). To terminate all ribosomes that have accidentally slipped into the +1 frame upstream the slippery codon, a stop codon was inserted in the +1 frame just a few nucleotides upstream the slippery codon (See Figure 4). Thus, to obtain F-luc activity a +1 frameshift must occur at the +1 frameshift sequence upstream of the stop codon in the zero frame. This construct results in a very short frameshifting window between the upstream stop codon in the +1 frame and the downstream in-frame stop codon. The slippery codon is determined individually for different assay sites in order to optimize the slippage of the peptidyl tRNA at the P-site. We chose UUU, CCC or GGG codons as the slippery codons (Supplementary Figure S1 and Table 2 and Supplementary Table S1). Codon UUU is decoded by 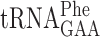 , which has the wobble nucleoside Gm34 (42), and its structure is not affected by the elp3, tuc1 or trm9 mutations. Codon CCC is read by the I34 (inosine) containing
, which has the wobble nucleoside Gm34 (42), and its structure is not affected by the elp3, tuc1 or trm9 mutations. Codon CCC is read by the I34 (inosine) containing 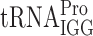 and the ncm5U34 containing
and the ncm5U34 containing 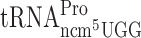 and the slippery codon GGG is read by the mcm5U34 containing
and the slippery codon GGG is read by the mcm5U34 containing 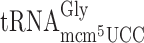 and the C34 containing
and the C34 containing 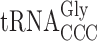 (Figure 3) (26,30,43). Note that the structures of the ncm5U containing
(Figure 3) (26,30,43). Note that the structures of the ncm5U containing 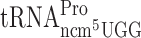 reading the slippery codon CCC and the mcm5U34 containing
reading the slippery codon CCC and the mcm5U34 containing 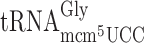 reading the slippery codon GGG are affected by the elp3 mutation and might therefore obscure the monitoring of an A-site effect at these test codons. These issues will be addressed below. As a control, we used a construct carrying the R-luc and F-luc genes in-frame. By dividing the ratio of F-luc/R-luc activities generated from the frameshifting construct with the ratio of activities from the F-luc/R-luc in-frame control, the level of frameshifting was revealed. Using these reporter systems, the level of frameshifting for specific tRNA isoacceptors was investigated in the presence or absence of s2, ncm5, mcm5 groups or the esterified methyl group of the mcm5 side chain at U34.
reading the slippery codon GGG are affected by the elp3 mutation and might therefore obscure the monitoring of an A-site effect at these test codons. These issues will be addressed below. As a control, we used a construct carrying the R-luc and F-luc genes in-frame. By dividing the ratio of F-luc/R-luc activities generated from the frameshifting construct with the ratio of activities from the F-luc/R-luc in-frame control, the level of frameshifting was revealed. Using these reporter systems, the level of frameshifting for specific tRNA isoacceptors was investigated in the presence or absence of s2, ncm5, mcm5 groups or the esterified methyl group of the mcm5 side chain at U34.
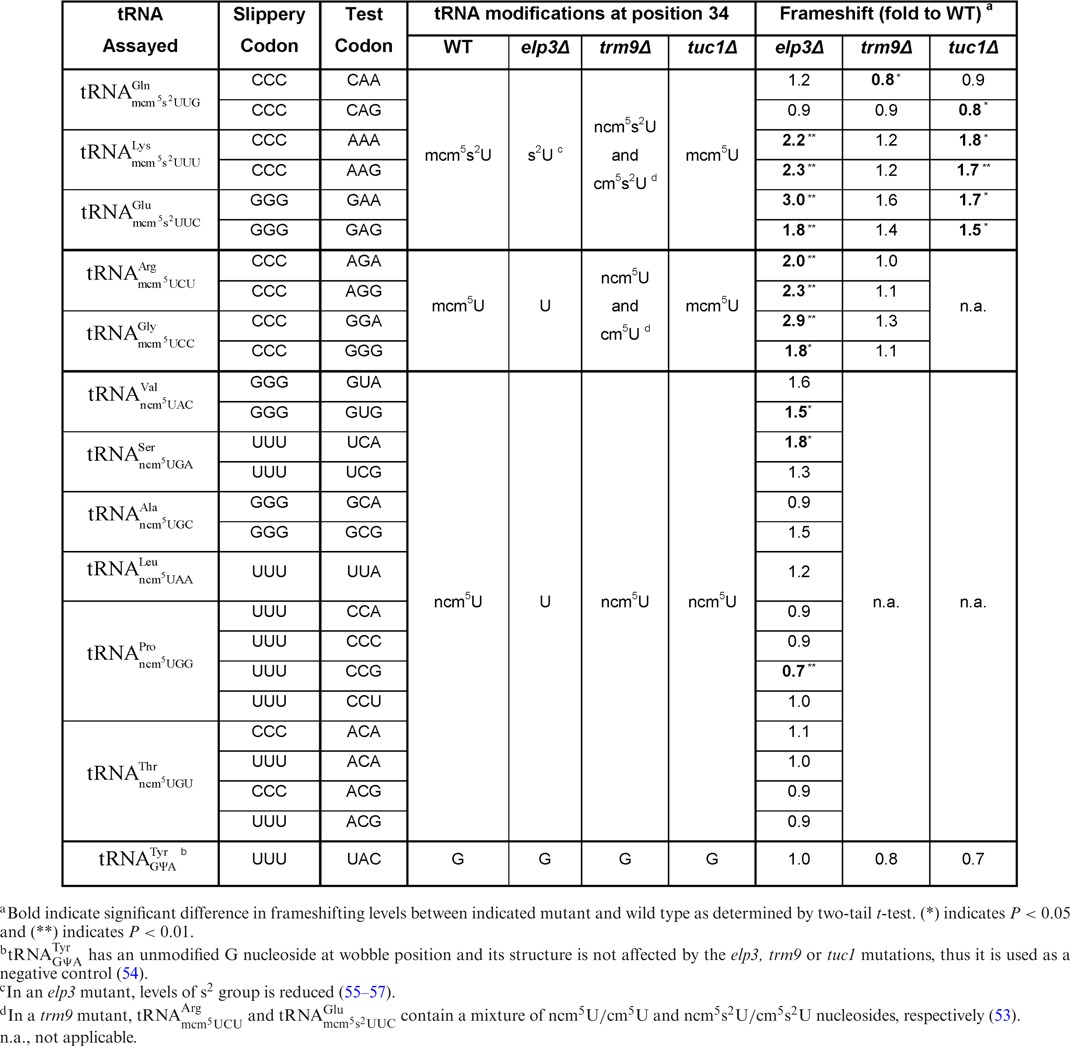
Table 2. Influence of tRNA modifications mcm5s2U34, mcm5U34, ncm5U34 and ncm5U34m on reading frame maintenance based on data using the Renilla/Firefly luciferase bicistronic reporter system.
In the bacterial system, modification deficiency of aminoacyl-tRNA in the ternary complex causes in most cases a slow entry of it to the A-site and thereby induces a peptidyl-tRNA slippage (Figure 1A and B) (6). Therefore, we suspected that in the cases below where we observed an effect on the frequency of frameshifting in the modification deficient mutants, it would primarily be due to an A-site effect, i.e. slow entry of the ternary complex containing aminoacyl-tRNA cognate to the test codon allowing a peptidyl-tRNA interacting with the slippery codon XXX to slip (Figure 1A and B). In the constructs used, all have a UAG stop codon just after the test codon NNN (i.e. the sequence is in zero frame -XXX-NNN-UAG). Translational termination in yeast is controlled by two interacting protein chain release factors, eRF1 and eRF3. Whereas eRF1 recognizes all three stop codons, binds to ribosomal A-site, and promotes hydrolysis of the P-site located peptidyl-tRNA, eRF3 stimulates the termination activity of eRF1 (Reviewed in Kisselev and Buckingham (44)). A poor eRF1 binding to the UAG in the A-site may induce slippage by the modification deficient peptidyl-tRNA from cognate NNN codon to NN-U codon. Therefore, the +1 frameshifting observed using the luciferase assay may be caused by either an A-site or a P-site effect or both. Note that, in all these cases the error occurs in the P-site (either by the tRNA reading the slippery codon or a tRNA cognate to the test codon). Although the luciferase system used by us is unable to distinguish between an A- or a P-site effect caused by modification deficiency, it is still a valuable method to address whether or not modification is important for maintaining the reading frame. To address specifically if modification deficiency induces an A- or a P-site effect, we used the Ty1 system, which is explained below.
Role of xm5U or mcm5s2U nucleosides in reading frame maintenance
In yeast, there are 11 tRNA species having mcm5s2U34, mcm5U34, ncm5U34 or ncm5U34m nucleosides at wobble position (Figures 2 and 3). The role of these modified uridines was analyzed for ribosomal +1 frameshifting using the Renilla/Firefly luciferase bicistronic reporter system described in the previous section.
In an elp3 mutant these tRNA species are missing the ncm5 and mcm5 groups at wobble position (U34) (26,45). The role of the ncm5 and mcm5 groups present in these tRNAs in reading frame maintenance was investigated in a wild type and in an elp3 mutant strain using cognate or near cognate codons as test codons. Lack of the mcm5 side chain in 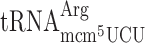 ,
, 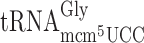 ,
, 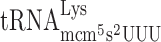 and
and 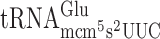 resulted in significantly higher levels of +1 frameshifting with either A-ending cognate or G-ending near cognate codons (Table 2 and Supplementary Figure S1). However, absence of the mcm5 group in
resulted in significantly higher levels of +1 frameshifting with either A-ending cognate or G-ending near cognate codons (Table 2 and Supplementary Figure S1). However, absence of the mcm5 group in 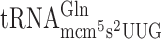 did not have any significant effect on reading frame maintenance for the Gln codons CAA or CAG (Table 2). Lack of the ncm5 group of U34 in
did not have any significant effect on reading frame maintenance for the Gln codons CAA or CAG (Table 2). Lack of the ncm5 group of U34 in 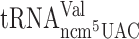 and
and 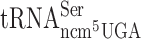 resulted in an increased level of +1 frameshifting with near cognate Val codon GUG or cognate Ser codon UCA. In contrast, absence of the ncm5 group of U34 in
resulted in an increased level of +1 frameshifting with near cognate Val codon GUG or cognate Ser codon UCA. In contrast, absence of the ncm5 group of U34 in 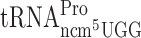 resulted in a decreased level of +1 frameshifting with near cognate Pro codon CCG. Lack of the ncm5 group of U34 of the remaining tRNAs did not cause a significant difference in levels of +1 frameshifting (Table 2 and Supplementary Figure S1). We conclude that in the xm5U and mcm5s2U tRNA isoacceptors, the mcm5 group plays a more vital role than the ncm5 group in reading frame maintenance.
resulted in a decreased level of +1 frameshifting with near cognate Pro codon CCG. Lack of the ncm5 group of U34 of the remaining tRNAs did not cause a significant difference in levels of +1 frameshifting (Table 2 and Supplementary Figure S1). We conclude that in the xm5U and mcm5s2U tRNA isoacceptors, the mcm5 group plays a more vital role than the ncm5 group in reading frame maintenance.
Codon CCC that can be read by ncm5U34 containing 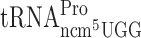 is used as a slippery codon upstream next to Gln-, Lys, Arg-, Gly- and Thr- test codons (Table 2 and Supplementary Figure S1). Thus, the ncm5U34 present in the potential peptidyl-Pro-tRNA might influence the ribosomal grip in the P-site and thus influence the slippage. To test directly the influence of the ncm5 group in Pro-tRNA in peptidyl-Pro-tRNA slippage, we used a construct -UUU-CCC-UAG- in the luciferase system. The stop codon UAG is in the zero frame just after the Pro codon CCC. Since eukaryotic release factor 1 (eRF1) acts in the A-site (Reviewed in Kisselev and Buckingham (44)) a possible +1 frameshift by Pro-tRNA lacking ncm5U would occur in the P-site. However, no significant +1 frameshifting was observed when the CCC codon was just upstream the stop codon and thus in the P-site (Table 2 and Supplementary Figure S1). We conclude that the ncm5 group in Pro-tRNA does not increase peptidyl-tRNA slippage at the slippery codon CCC.
is used as a slippery codon upstream next to Gln-, Lys, Arg-, Gly- and Thr- test codons (Table 2 and Supplementary Figure S1). Thus, the ncm5U34 present in the potential peptidyl-Pro-tRNA might influence the ribosomal grip in the P-site and thus influence the slippage. To test directly the influence of the ncm5 group in Pro-tRNA in peptidyl-Pro-tRNA slippage, we used a construct -UUU-CCC-UAG- in the luciferase system. The stop codon UAG is in the zero frame just after the Pro codon CCC. Since eukaryotic release factor 1 (eRF1) acts in the A-site (Reviewed in Kisselev and Buckingham (44)) a possible +1 frameshift by Pro-tRNA lacking ncm5U would occur in the P-site. However, no significant +1 frameshifting was observed when the CCC codon was just upstream the stop codon and thus in the P-site (Table 2 and Supplementary Figure S1). We conclude that the ncm5 group in Pro-tRNA does not increase peptidyl-tRNA slippage at the slippery codon CCC.
The slippery codon GGG is decoded by both C34 containing 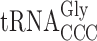 and mcm5U34 containing
and mcm5U34 containing 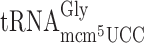 . The structure of the latter tRNA is affected by the elp3 mutation and this tRNA reads the GGG codon very inefficiently compared to the cognate
. The structure of the latter tRNA is affected by the elp3 mutation and this tRNA reads the GGG codon very inefficiently compared to the cognate 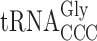 (26). Therefore, in the elp3 mutant it is not likely that
(26). Therefore, in the elp3 mutant it is not likely that 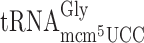 lacking the mcm5 side chain at U34 will out-compete the efficiently decoding cognate
lacking the mcm5 side chain at U34 will out-compete the efficiently decoding cognate 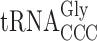 at GGG codons. Consequently, the observed +1 frameshifting for
at GGG codons. Consequently, the observed +1 frameshifting for 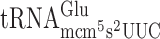 and
and 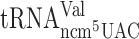 (Table 2) is most likely not caused by slippage of an unmodified peptidyl-
(Table 2) is most likely not caused by slippage of an unmodified peptidyl-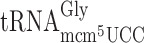 but rather a poor A-site entry by
but rather a poor A-site entry by 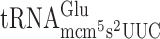 and
and 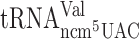 , respectively.
, respectively.
The formation of the esterified methyl group of mcm5U, which is the last step in the synthesis of the mcm5 side chain, is catalyzed by the dimeric Trm9/Trm112 protein complex (46,47).
The influence of the esterified methyl group of the mcm5U34 and mcm5s2U34 nucleoside in reading frame maintenance was investigated in a wild type and in a trm9 mutant strain using cognate or near cognate codons as test codons (Table 2). Lack of the esterified methyl group of the mcm5 side chain of U34 in 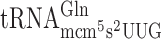 resulted in significantly decreased +1 frameshifting at the Gln codon CAA (Table 2 and Supplementary Figure S1). There were no significant differences in the levels of frameshifting between wild type and trm9 mutant in the remaining test constructs (Table 2 and Supplementary Figure S1). We conclude that presence or absence of the esterified methyl group in the mcm5 side chain only seem to play a minor role in +1 frameshifting.
resulted in significantly decreased +1 frameshifting at the Gln codon CAA (Table 2 and Supplementary Figure S1). There were no significant differences in the levels of frameshifting between wild type and trm9 mutant in the remaining test constructs (Table 2 and Supplementary Figure S1). We conclude that presence or absence of the esterified methyl group in the mcm5 side chain only seem to play a minor role in +1 frameshifting.
In a tuc1 mutant, the s2 group of mcm5s2U at the wobble position (U34) is absent in 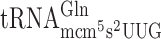 ,
, 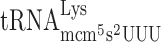 and
and 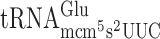 (48). The role of the s2 group present in these tRNAs in reading frame maintenance was investigated in a wild type and a tuc1 mutant strain using cognate or near cognate codons as test codons (Table 2). Absence of the s2 group in
(48). The role of the s2 group present in these tRNAs in reading frame maintenance was investigated in a wild type and a tuc1 mutant strain using cognate or near cognate codons as test codons (Table 2). Absence of the s2 group in 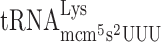 and
and 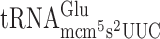 resulted in significantly higher levels of +1 frameshifting with either A-ending cognate or G-ending near cognate codons (Table 2 and Supplementary Figure S1). However, lack of the s2 group in
resulted in significantly higher levels of +1 frameshifting with either A-ending cognate or G-ending near cognate codons (Table 2 and Supplementary Figure S1). However, lack of the s2 group in 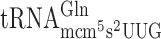 resulted in significantly decreased +1 frameshifting with the near cognate CAG (Table 2 and Supplementary Figure S1).
resulted in significantly decreased +1 frameshifting with the near cognate CAG (Table 2 and Supplementary Figure S1).
In the cases stated above, Gln- and Pro-tRNAs showed reduced levels of +1 frameshifting due to lack of esterified methyl, s2 or ncm5 groups (Table 2). This reduced level of frameshifting might be surprising but similar observations were noted earlier. In bacteria the Gln-, Lys- and Glu-tRNA contain as wobble nucleoside the mnm5s2U, which is structurally related to the mcm5s2U present in the corresponding yeast tRNAs. Lack of either the mnm5 side chain or the sulphur at position 2 reduced frameshifting similarly as noted by us for the two aforementioned cases (15,16). Although these results seems counterintuitively strange, one has to remember that the structure of the different tRNA species is optimized and in fact has evolved to have similar decoding activity, which is obtained partly due to modification of it (49). Therefore, a modification may improve the activity of one tRNA whereas it might reduce the activity of another tRNA species (See discussion of this issue in Björk and Hagervall (4)). From such considerations, one would expect that when measuring a specific activity of a tRNA, like influencing reading frame maintenance, a modification might improve or reduce the fidelity of it.
The frameshifting error occurs by peptidyl-tRNA slippage
A key feature of the peptidyl-tRNA slippage model is that the error in reading frame maintenance, induced either by an A- or a P-site effect due to modification deficient tRNA, occurs in the P-site by peptidyl-tRNA slippage. There are two ways to establish if the frameshift errors occur in the ribosomal A- or P-site. Either one determines the amino acid sequence of the frameshift peptide covering the frameshift window or by overexpressing the tRNA cognate to the A-site codon. In the latter case, if the frameshift error occurs due to an A-site effect, such overexpression would decrease the frameshift error, since it reduces the ribosomal pause and thereby reduces the ability of the peptidyl-tRNA to slip forward. We chose the latter method, since this approach is relevant for this study, as such a treatment also suppresses all the pleiotropic phenotypes induced by a mutation in, e.g. the ELP3 gene. Thus, the strong pleiotropic phenotypes observed in an elp3 mutant might be due, at least partly, to errors in reading frame maintenance of some key mRNAs.
As stated in the description of the assay system, the dual-luciferase assay system is not designed to clarify the difference between an A- or a P-site effect caused by modification deficiency, we decided to use Ty1 assay system to address this question. The expression of the TYB gene of yeast Ty retrotransposon requires a ribosomal +1 frameshift event caused by a peptidyl-tRNA slippage (41). Only a seven nucleotide sequence CUU-AGG-C is required for the +1 frameshift event to occur and thus only two tRNA species—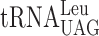 and
and 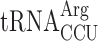 —are participating in this event. In the yeast strain used, the availability of
—are participating in this event. In the yeast strain used, the availability of 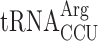 is low resulting in a low rate of ribosomal A-site selection, which induces a slippage by
is low resulting in a low rate of ribosomal A-site selection, which induces a slippage by 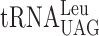 at the CUU P-site codon into the +1 frame (UU-A) (41). Therefore, we decided to use an altered version of the Ty1 +1 frameshift system to study whether or not the +1 frameshifting caused by lack of the mcm5 side chain in the R-luc-F-luc system, is due to a peptidyl-tRNA slippage. We altered the ‘CUU-AGG-C’ +1 frameshift site by changing the Arg codon (AGG) into either a Lys codon AAA decoded by
at the CUU P-site codon into the +1 frame (UU-A) (41). Therefore, we decided to use an altered version of the Ty1 +1 frameshift system to study whether or not the +1 frameshifting caused by lack of the mcm5 side chain in the R-luc-F-luc system, is due to a peptidyl-tRNA slippage. We altered the ‘CUU-AGG-C’ +1 frameshift site by changing the Arg codon (AGG) into either a Lys codon AAA decoded by 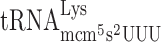 or an Arg codon AGA decoded by
or an Arg codon AGA decoded by 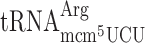 to test whether or not lack of mcm5 side chain of these tRNAs induce +1 frameshifting (Table 3). If the hypomodified tRNA is inefficiently accepted to the A-site in an elp3 mutant, the AAA (Lys) and/or AGA (Arg) test codons will act similarly as codons decoded by the low available
to test whether or not lack of mcm5 side chain of these tRNAs induce +1 frameshifting (Table 3). If the hypomodified tRNA is inefficiently accepted to the A-site in an elp3 mutant, the AAA (Lys) and/or AGA (Arg) test codons will act similarly as codons decoded by the low available 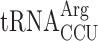 resulting in a slow entry to the A-site by the ternary complex containing the unmodified tRNA. If so, the
resulting in a slow entry to the A-site by the ternary complex containing the unmodified tRNA. If so, the 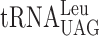 in the P-site will slip into the +1 frame (from cognate CUU to non-cognate UU-A). All alterations of the Ty1 sequence were made in the HIS4A::lacZ frameshift reporter plasmid (see Materials and Methods) (Figure 4B). The levels of frameshifting were calculated by dividing the β-galactosidase values generated from the test construct with the values from the in-frame control construct. Table 3 shows that for the ‘CUU-AAA-C’ Lys codon test construct, lack of mcm5 side group in the mcm5s2U nucleoside of
in the P-site will slip into the +1 frame (from cognate CUU to non-cognate UU-A). All alterations of the Ty1 sequence were made in the HIS4A::lacZ frameshift reporter plasmid (see Materials and Methods) (Figure 4B). The levels of frameshifting were calculated by dividing the β-galactosidase values generated from the test construct with the values from the in-frame control construct. Table 3 shows that for the ‘CUU-AAA-C’ Lys codon test construct, lack of mcm5 side group in the mcm5s2U nucleoside of 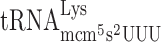 resulted in 10-fold increased +1 frameshifting in the elp3 mutant compared to wild type. In contrast, for the ‘CUU-AGA-C’ Arg codon test construct, lack of mcm5 side group in
resulted in 10-fold increased +1 frameshifting in the elp3 mutant compared to wild type. In contrast, for the ‘CUU-AGA-C’ Arg codon test construct, lack of mcm5 side group in 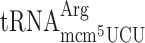 did not increase frameshifting in the elp3 mutant compared to the wild type (Table 3). Thus, similar to the results obtained by the luciferase system lack of the mcm5 group of
did not increase frameshifting in the elp3 mutant compared to the wild type (Table 3). Thus, similar to the results obtained by the luciferase system lack of the mcm5 group of 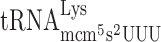 induced increased +1 frameshifting. Although we observed an increased frameshifting for mcm5 deficient
induced increased +1 frameshifting. Although we observed an increased frameshifting for mcm5 deficient 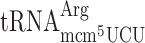 in the luciferase assay system (Table 2), this was not the case using the Ty1 assay system (Table 3).
in the luciferase assay system (Table 2), this was not the case using the Ty1 assay system (Table 3).
Table 3. Influence of mcm5 side chain on reading frame maintenance based on data using the HIS4A::Ty1::lacZ reporter system.
| tRNA decoding the codon and Its Gene Copy Number | Normalized Frameshift Rates (%) | ||||
|---|---|---|---|---|---|
| Ty1 Frameshift Site | at the P-site | at the A-site | WT | elp3Δ | Frameshift Ratioselp3Δ/WT |
| CUU-AGA-C |
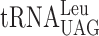 , (3 a) , (3 a) |
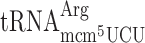 , (11 a) , (11 a) |
0.142 ± 0.066 | 0.114 ± 0.020 | 0.80 d |
| CUU-AAA-C |
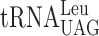 , (3 a) , (3 a) |
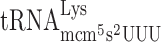 , (7 a) , (7 a) |
0.191 ± 0.092 | 1.907 ± 0.787 | 9.99c |
| CUU-AAA-C |
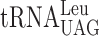 , (3 a) , (3 a) |
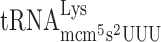 , (7 a +4 b) , (7 a +4 b) |
0.076 ± 0.013 | 0.239 ± 0.020 | 3.14c |
| AAA-AGG-C |
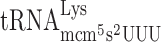 , (7 a) , (7 a) |
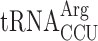 , (1 a) , (1 a) |
1.203 ± 0.558 | 0.892 ± 0.225 | 0.74 d |
| AAA-CGT-C |
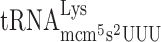 , (7 a) , (7 a) |
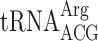 , (6 a) , (6 a) |
0.053 ± 0.021 | 0.039 ± 0.010 | 0.74 d |
| AAA-ATT-C |
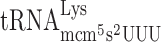 , (7 a) , (7 a) |
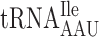 , (13 a) , (13 a) |
0.023 ± 0.008 | 0.025 ± 0.009 | 1.07 d |
aGenomic copy numbers of the tRNA genes.
cDifference in frameshifting between elp3 mutant and wild type was significant as determined by two-tail t-test (P < 0.02).
dDifference in frameshifting between elp3 mutant and wild type was not significant as determined by two-tail t-test (P > 0.05).
To analyze whether lack of the mcm5 group of 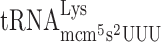 could induce +1 frameshifting due to a P-site effect by modification deficiency, we placed a Lys codon AAA instead of the CUU codon in the Ty1 assay system and varied the following codon. The concentration of a tRNA species is proportional to the number of the corresponding tRNA genes in the yeast genome (50). Accordingly, by placing different codons in the A-site, the concentration of the corresponding tRNAs in the cell reading this codon is changed and thereby the efficiency of reading the A-site codon is altered. Thus, to test for a possible P-site effect induced by a lack of the mcm5 group of mcm5s2U in Lys-tRNA we placed an Arg codon AGG read by the rare cognate
could induce +1 frameshifting due to a P-site effect by modification deficiency, we placed a Lys codon AAA instead of the CUU codon in the Ty1 assay system and varied the following codon. The concentration of a tRNA species is proportional to the number of the corresponding tRNA genes in the yeast genome (50). Accordingly, by placing different codons in the A-site, the concentration of the corresponding tRNAs in the cell reading this codon is changed and thereby the efficiency of reading the A-site codon is altered. Thus, to test for a possible P-site effect induced by a lack of the mcm5 group of mcm5s2U in Lys-tRNA we placed an Arg codon AGG read by the rare cognate 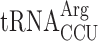 (1 genomic copy) and the near cognate
(1 genomic copy) and the near cognate 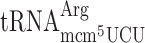 (11 genomic copies) after the Lys codon AAA. In an elp3 mutant
(11 genomic copies) after the Lys codon AAA. In an elp3 mutant 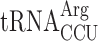 is essential, demonstrating that mcm5 group of the near cognate Arg-tRNA is required for efficient reading of the Arg codon AGG (26). Therefore, in an elp3 mutant, a situation is generated where the AGG codon is read slowly since it is read mainly by the rare cognate
is essential, demonstrating that mcm5 group of the near cognate Arg-tRNA is required for efficient reading of the Arg codon AGG (26). Therefore, in an elp3 mutant, a situation is generated where the AGG codon is read slowly since it is read mainly by the rare cognate 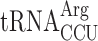 and inefficiently by the more abundant modification deficient near cognate
and inefficiently by the more abundant modification deficient near cognate 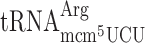 . Such a condition would allow
. Such a condition would allow 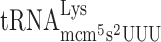 at the P-site to slip to the +1 translational frame. Furthermore, we made test constructs to increase the rate of A-site selection by introducing either an Ile codon AUU decoded by cognate
at the P-site to slip to the +1 translational frame. Furthermore, we made test constructs to increase the rate of A-site selection by introducing either an Ile codon AUU decoded by cognate 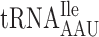 present in 13 genomic copies or an Arg codon CGU decoded by cognate
present in 13 genomic copies or an Arg codon CGU decoded by cognate 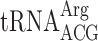 present in 6 genomic copies after Lys codon AAA (Table 3). By varying concentration of the potential A-site coding tRNAs from 1 genomic copy to 13 genomic copies, we did not observe any significant difference in the levels of +1 frameshifting between wild type and elp3 mutant (Table 3). Apparently, the possible peptidyl-
present in 6 genomic copies after Lys codon AAA (Table 3). By varying concentration of the potential A-site coding tRNAs from 1 genomic copy to 13 genomic copies, we did not observe any significant difference in the levels of +1 frameshifting between wild type and elp3 mutant (Table 3). Apparently, the possible peptidyl-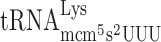 slippage is not sensitive to the rate of A-site selection suggesting that lack of mcm5s2U does not cause any P-site effect and thus an increased peptidyl-tRNA slippage.
slippage is not sensitive to the rate of A-site selection suggesting that lack of mcm5s2U does not cause any P-site effect and thus an increased peptidyl-tRNA slippage.
If the frameshifting event occurring at the modified Ty1 site ‘CUU-AAA-C’ was caused by a slow entry of the ternary complex containing the hypomodified 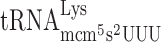 causing a peptidyl-
causing a peptidyl-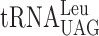 slippage to +1 translational frame, an elevated level of the hypomodified
slippage to +1 translational frame, an elevated level of the hypomodified 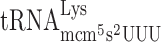 should increase the rate of A-site selection and thereby reducing +1 frameshifting (Figure 1B). We therefore cloned the tK(UUU)L gene, which encodes
should increase the rate of A-site selection and thereby reducing +1 frameshifting (Figure 1B). We therefore cloned the tK(UUU)L gene, which encodes 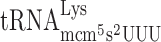 into either plasmid pMB38–9merWT (test construct, containing the CUU-AAA-C frameshift site) and pMB38–9merFF (corresponding in-frame control construct, Figure 4 and Supplementary Table S2). Thus, the plasmids harbor both the tRNA gene and the β-galactosidase gene with either a frameshift site or an in-frame control. The plasmid encoded tK(UUU)L gene results in overexpression of
into either plasmid pMB38–9merWT (test construct, containing the CUU-AAA-C frameshift site) and pMB38–9merFF (corresponding in-frame control construct, Figure 4 and Supplementary Table S2). Thus, the plasmids harbor both the tRNA gene and the β-galactosidase gene with either a frameshift site or an in-frame control. The plasmid encoded tK(UUU)L gene results in overexpression of 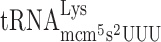 and concomitantly reduced the levels of +1 frameshifting in the elp3 mutant from 10- to 3-fold compared to wild type (Table 3). This data strongly suggest that the +1 frameshifting event at ‘CUU-AAA-C’ Lys codon test construct occurs by peptidyl-
and concomitantly reduced the levels of +1 frameshifting in the elp3 mutant from 10- to 3-fold compared to wild type (Table 3). This data strongly suggest that the +1 frameshifting event at ‘CUU-AAA-C’ Lys codon test construct occurs by peptidyl-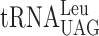 slippage due to an A-site effect caused by a slow entry of the hypomodified
slippage due to an A-site effect caused by a slow entry of the hypomodified 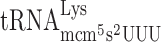 . As was suggested earlier by us (34,48) and confirmed by Rezgui et al. (51), the major function of the mcm5s2U34 nucleoside in Lys-tRNA is to improve the reading of the cognate codon. Thus, mcm5s2U34 deficiency results in slow decoding and reduced translation elongation rate but also, as shown here, induces +1 frameshifting by reducing the rate of A-site selection.
. As was suggested earlier by us (34,48) and confirmed by Rezgui et al. (51), the major function of the mcm5s2U34 nucleoside in Lys-tRNA is to improve the reading of the cognate codon. Thus, mcm5s2U34 deficiency results in slow decoding and reduced translation elongation rate but also, as shown here, induces +1 frameshifting by reducing the rate of A-site selection.
CONCLUSION
Among the tRNA isoacceptors having xm5U34 or xm5s2U34 wobble uridine nucleosides, only Lys- and Gln-tRNAs has been investigated for +1 frameshifting in both bacteria and yeast. The modified wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U34) present in yeast tRNAs specific for Gln, Lys and Glu has a chemically related form, 5-methylaminomethyl-2-thiouridine (mnm5s2U34) present in the corresponding bacterial tRNAs. In bacteria, lack of the mnm5 group in Gln-tRNA results in increased +1 frameshifting at both cognate (CAA) and near cognate (CAG) codons, whereas absence of the s2 group results in +1 frameshifting only at the cognate (CAA) codon (6). In contrast, lack of mcm5 or s2 groups in yeast Gln-tRNA does not result in increased +1 frameshifting at either CAA or CAG codons. Instead, absence of the s2 group results in reduced +1 frameshifting at the CAG codon. In bacteria, lack of mnm5 or s2 groups in Lys-tRNA cause increased +1 frameshifting at both cognate (AAA) and near cognate (AAG) codons by A- and P-site effects (6). In yeast, we also observed an increased +1 frameshifting due to lack of mcm5 or s2 groups of Lys-tRNA at AAA and AAG codons. However, we show that +1 frameshifting at the cognate (AAA) codon is induced by an A-site effect, not a P-site effect.
It has been shown that presence of modified nucleosides in tRNAs are required for tuning the decoding activity in order to maintain uniformity in translation (49). An in vitro study in yeast showed that presence of the mcm5 and s2 groups of Lys-tRNA are required for efficient A-site binding (51). Consistent with these observations, our in vivo studies show that presence of the mcm5 group of Lys-tRNA promotes its entry to ribosomal A-site and thereby avoids +1 frameshift errors. Thus, wobble uridine modifications are required to optimize the function of tRNAs and thereby promote a proper reading frame maintenance.
Supplementary Material
Acknowledgments
We acknowledge Prof. G. R. Björk, Dr. M. Johansson and Dr. S. Tuck for valuable comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Cancer Foundation [13 0301]; Swedish Research Council [621-2012-3576]; Karin and Harald Silvanders Foundation [223-2808-12 to A.S.B.]; Carl Tryggers Foundation [to G.R.B.]. Fund for open access charge: Swedish Cancer Foundation [13 0301]; Swedish Research Council [621-2012-3576]; Karin and Harald Silvanders Foundation [223-2808-12 to A.S.B.]; Carl Tryggers Foundation [to G.R.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Kurland C.G. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 2.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkins J.F., Björk G.R. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev.: MMBR. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björk G.R., Hagervall T.G. Transfer RNA Modification: Presence, Synthesis, and Function. EcoSal Plus. 2014 doi: 10.1128/ecosalplus.ESP-0007-2013. doi:10.1128/ecosalplus.ESP-0007-2013. [DOI] [PubMed] [Google Scholar]
- 5.Björk G.R., Wikström P.M., Byström A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 6.Urbonavičius J., Qian Q., Durand J.M., Hagervall T.G., Björk G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson B.A., Kwon S.Y., Chamorro M., Oroszlan S., Hatfield D.L., Lee B.J. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255:2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- 8.Carlson B.A., Mushinski J.F., Henderson D.W., Kwon S.Y., Crain P.F., Lee B.J., Hatfield D.L. 1-Methylguanosine in place of Y base at position 37 in phenylalanine tRNA is responsible for its shiftiness in retroviral ribosomal frameshifting. Virology. 2001;279:130–135. doi: 10.1006/viro.2000.0692. [DOI] [PubMed] [Google Scholar]
- 9.Waas W.F., Druzina Z., Hanan M., Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J. Biol. Chem. 2007;282:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]
- 10.Lecointe F., Namy O., Hatin I., Simos G., Rousset J.P., Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 2002;277:30445–30453. doi: 10.1074/jbc.M203456200. [DOI] [PubMed] [Google Scholar]
- 11.Lin C.A., Ellis S.R., True H.L. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 2010;30:354–363. doi: 10.1128/MCB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Yacoubi B., Hatin I., Deutsch C., Kahveci T., Rousset J.P., Iwata-Reuyl D., Murzin A.G., de Crécy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011;30:882–893. doi: 10.1038/emboj.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyauchi K., Kimura S., Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013;9:105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 14.Brierley I., Meredith M.R., Bloys A.J., Hagervall T.G. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J. Mol. Biol. 1997;270:360–373. doi: 10.1006/jmbi.1997.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Licznar P., Mejlhede N., Prère M.F., Wills N., Gesteland R.F., Atkins J.F., Fayet O. Programmed translational -1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 2003;22:4770–4778. doi: 10.1093/emboj/cdg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urbonavičius J., Stahl G., Durand J.M., Ben Salem S.N., Qian Q., Farabaugh P.J., Björk G.R. Transfer RNA modifications that alter +1 frameshifting in general fail to affect -1 frameshifting. RNA. 2003;9:760–768. doi: 10.1261/rna.5210803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maynard N.D., Macklin D.N., Kirkegaard K., Covert M.W. Competing pathways control host resistance to virus via tRNA modification and programmed ribosomal frameshifting. Mol. Syst. Biol. 2012;8:567–579. doi: 10.1038/msb.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farabaugh P.J. Programmed translational frameshifting. Annu. Revi. Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 20.Björk G.R., Durand J.M., Hagervall T.G., Leipuvienė R., Lundgren H.K., Nilsson K., Chen P., Qian Q., Urbonavičius J. Transfer RNA modification: influence on translational frameshifting and metabolism. FEBS Lett. 1999;452:47–51. doi: 10.1016/s0014-5793(99)00528-1. [DOI] [PubMed] [Google Scholar]
- 21.Farabaugh P.J., Björk G.R. How translational accuracy influences reading frame maintenance. EMBO J. 1999;18:1427–1434. doi: 10.1093/emboj/18.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallant J., Lindsley D., Masucci J. The unbearable lightness of peptidyl-tRNA. Ribosome: Struct. Func. Antibiot. Cell. Interact. 2000:385–396. [Google Scholar]
- 23.Näsvall S.J., Nilsson K., Björk G.R. The ribosomal grip of the peptidyl-tRNA is critical for reading frame maintenance. J. Mol. Biol. 2009;385:350–367. doi: 10.1016/j.jmb.2008.10.069. [DOI] [PubMed] [Google Scholar]
- 24.Jäger G., Nilsson K., Björk G.R. The phenotype of many independently isolated +1 frameshift suppressor mutants supports a pivotal role of the P-site in reading frame maintenance. PloS One. 2013;8:e60246. doi: 10.1371/journal.pone.0060246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glasser A.L., El Adlouni C., Keith G., Sochacka E., Malkiewicz A., Santos M., Tuite M.F., Desgrès J. Presence and coding properties of 2′-O-methyl-5-carbamoylmethyluridine (ncm5Um) in the wobble position of the anticodon of tRNA(Leu) (U*AA) from brewer's yeast. FEBS Lett. 1992;314:381–385. doi: 10.1016/0014-5793(92)81510-s. [DOI] [PubMed] [Google Scholar]
- 26.Johansson M.J., Esberg A., Huang B., Björk G.R., Byström A.S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keith G., Desgrès J., Pochart P., Heyman T., Kuo K.C., Gehrke C.W. Eukaryotic tRNAs(Pro): primary structure of the anticodon loop; presence of 5-carbamoylmethyluridine or inosine as the first nucleoside of the anticodon. Biochim. Biophys. Acta. 1990;1049:255–260. doi: 10.1016/0167-4781(90)90095-j. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi T., Irie T., Yoshida M., Takeishi K., Ukita T. The primary structure of yeast glutamic acid tRNA specific to the GAA codon. Biochim. Biophys. Acta. 1974;366:168–181. doi: 10.1016/0005-2787(74)90331-1. [DOI] [PubMed] [Google Scholar]
- 29.Kuntzel B., Weissenbach J., Wolff R.E., Tumaitis-Kennedy T.D., Lane B.G., Dirheimer G. Presence of the methylester of 5-carboxymethyl uridine in the wobble position of the anticodon of tRNAIII Arg from brewer's yeast. Biochimie. 1975;57:61–70. doi: 10.1016/s0300-9084(75)80110-6. [DOI] [PubMed] [Google Scholar]
- 30.Lu J., Huang B., Esberg A., Johansson M.J., Byström A.S. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith C.J., Teh H.S., Ley A.N., D'Obrenan P. The nucleotide sequences and coding properties of the major and minor lysine transfer ribonucleic acids from the haploid yeast Saccharomyces cerevisiae S288C. J. Biol. Chem. 1973;248:4475–4485. [PubMed] [Google Scholar]
- 32.Yamamoto N., Yamaizumi Z., Yokoyama S., Miyazawa T., Nishimura S. Modified nucleoside, 5-carbamoylmethyluridine, located in the first position of the anticodon of yeast valine tRNA. J. Biochem. (Tokyo) 1985;97:361–364. doi: 10.1093/oxfordjournals.jbchem.a135061. [DOI] [PubMed] [Google Scholar]
- 33.Karlsborn T., Tükenmez H., Mahmud A.K., Xu F., Xu H., Byström A.S. Elongator, a conserved complex required for wobble uridine modifications in Eukaryotes. RNA Biol. 2014;11:1519–1528. doi: 10.4161/15476286.2014.992276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esberg A., Huang B., Johansson M.J.O., Byström A.S. Elevated Levels of Two tRNA Species Bypass the Requirement for Elongator Complex in Transcription and Exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 35.Chen C., Huang B., Eliasson M., Rydén P., Byström A.S. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011;7:e1002258. doi: 10.1371/journal.pgen.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Näsvall S.J., Chen P., Björk G.R. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patil A., Chan C.T., Dyavaiah M., Rooney J.P., Dedon P.C., Begley T.J. Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol. 2012;9:990–1001. doi: 10.4161/rna.20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gietz D., St Jean A., Woods R.A., Schiestl R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burke D., Dawson D., Stearns T. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 40.Harger J.W., Dinman J.D. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belcourt M.F., Farabaugh P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajbhandary U.L., Chang S.H., Stuart A., Faulkner R.D., Hoskinson R.M., Khorana H.G. Studies on polynucleotides, lxviii the primary structure of yeast phenylalanine transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 1967;57:751–758. doi: 10.1073/pnas.57.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kisselev L.L., Buckingham R.H. Translational termination comes of age. Trends Biochem. Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 45.Huang B., Johansson M.J.O., Byström A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalhor H.R., Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazauric M.H., Dirick L., Purushothaman S.K., Björk G.R., Lapeyre B. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J. Biol. Chem. 2010;285:18505–18515. doi: 10.1074/jbc.M110.113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Björk G.R., Huang B., Persson O.P., Byström A.S. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fahlman R.P., Dale T., Uhlenbeck O.C. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 50.Percudani R., Pavesi A., Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- 51.Rezgui V.A., Tyagi K., Ranjan N., Konevega A.L., Mittelstaet J., Rodnina M.V., Peter M., Pedrioli P.G. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12289–12294. doi: 10.1073/pnas.1300781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fiorentini P., Huang K.N., Tishkoff D.X., Kolodner R.D., Symington L.S. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C., Huang B., Anderson J.T., Byström A.S. Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PloS One. 2011;6:e20783. doi: 10.1371/journal.pone.0020783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madison J.T., Kung H.K. Large oligonucleotides isolated from yeast tyrosine transfer ribonucleic acid after partial digestion with ribonuclease T1. J. Biol. Chem. 1967;242:1324–1330. [PubMed] [Google Scholar]
- 55.Nakai Y., Nakai M., Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 56.Leidel S., Pedrioli P.G., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 57.Noma A., Sakaguchi Y., Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farabaugh P., Liao X.B., Belcourt M., Zhao H., Kapakos J., Clare J. Enhancer and silencerlike sites within the transcribed portion of a Ty2 transposable element of Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:4824–4834. doi: 10.1128/mcb.9.11.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.