Abstract
Background
Attention Deficit and Hyperactivity Disorder (ADHD) is a common childhood neuropsychiatric condition. The disorder has a multifactorial background, with heritability estimates of around 76%, suggesting an important role of genetic factors. Candidate genes include those related to dopaminergic (e.g. DRD4, DRD5, SLC6A3 and DBH)and serotoninergic (e.g.HTR1B and SLC6A4) pathways.
Purpose
To explore the association of common polymorphisms in six genes (DRD4, DRD5, SLC6A3, DBH, HTR1B and SLC6A4) and the susceptibility to ADHD in a Colombian sample population.
Methods
trios and 152 healthy controls were recruited. Genotyping of the six polymorphisms was performed using described PCR-based protocols. A TDT analysis was used to test if there was preferential allelic transmission for any of the six polymorphisms. Additionally, a case-control analysis was performed to test for association of the serotoninergic (HTR1B and SLC6A4) polymorphisms with ADHD.
Results
Through the TDT analysis there was no preferential allelic transmission for any of the studied variants. Case-control analysis did not show association.
Conclusion
This is the first study in Latin America to describe six polymorphisms in a group of patients with ADHD. There was no evidence of association for any of the studied polymorphic variants in this Colombian ADHD sample. Further research, with larger sample sizes and study of endophenotypes, is needed in this population to confirm and extend the results.
Keywords: Association, Attention Deficit and Hyperactivity Disorder, Transmission Disequilibrium Test, Genes, Latin America
 Introduction
Introduction
Attention Deficit and Hyperactivity Disorder (ADHD) is a complex childhood disorder characterized by persistent symptoms of inattentiveness, hyperactivity and/or impulsivity. Its prevalence is around 5% worldwide, and varied in different population.1–3 The Diagnostic and Statistical Manual IV (DSM-IV) classify the disorder in three subtypes: hyperactive, inattentive and combined.
ADHD is a multifactorial disorder3 with heritability estimates around 76% suggesting an important role for genetic factors. Candidate variants, studied mainly in populations of European background, including genes involved in dopaminergic and serotoninergic pathways; and in neural signaling mechanisms.5
Dopaminergic genes comprise DRD4, encoding for the dopamine D4 receptor, which is highly expressed in fronto-subcortical system implicated in ADHD pathophysiology.6 A variable number tandem repeat (VNTR) located in exon 3 of DRD4 gene has been explored in ADHD,7 with the seven-repeat allele being associated with this disorder.8,9 Dopamine D5 receptor (DRD5) is also implicated in ADHD susceptibility. The dinucleotide (CA)n repeat polymorphisms; located near18.5 kb at 5’ end to the transcription start site of DRD5 has been found to be associated in several populations.10,11 The dopamine transporter gene (SLC6A3) has been implicated in ADHD pathogenesis due to the fact that some drugs used in its treatment block the dopamine transporter as their mechanism of action.12 Studies with knock-out mice have supported this hypothesis.13 The 10-repeats allele of the 148 bpVNTR located at the 3’ untranslated region (UTR) has been found to be more frequent in cases than controls.11,14,15 The dopamine beta hydroxylase gene (DBH) is responsible for the conversion of dopamine into noradrenaline. The A2 allele of the Taq1 polymorphism located in intron 5 of this gene has also been implicated in ADHD.11,16–18
Among serotoninergic genes, the serotonin receptor (HTR1B) and the serotonin transporter(SLC6A4) have been widely studied in ADHD. The associated polymorphisms include the single nucleotide polymorphism (SNP) c.861G>C (rs6296)of HTR1B.19,20 and the 44-bp insertion/deletion located in the promoter region of SLC6A4.SLC6A.21–23
Despite large evidence of these associations, controversy about results is still remaining. Studies performed in several populations with the different sample sizes methods and demonstrated contradictory results. Meta-analysis studies have also failed contributed to clarify the role of these variants.24 The study of these variants in different populations is therefore needed5 to address the genetic association with disease pathology.
The purpose of this study was to explore the possible association of six polymorphic variants located in candidate genes in a sample of Colombian ADHD patients.
Methods
Subjects
Children between 6 and 15 years old from the city of Bogotá, Colombia were included. DSM-IV criteria and Behavior Assessment System for Children (BASC) were applied to parents and teachers of patient, in order to confirm the diagnosis. The Wechsler scale of intelligence (WISC-R) was applied in order to exclude children with a score less than 70, which indicate an intellectual disability. Cases were selected among children who had at least one of the following criteria (applied to information derived from parents or teachers): 1) Six or more points, over nine, for inattention in the DSM-IV criteria and a percentile over 85 in the inattention domain of BASC; 2) Six or more points, over nine, for hyperactivity in the DSM-IV criteria and a percentile over 85 in the hyperactivity domain of BASC. The patient’s group included 86 children.
Children who did not meet ADHD criteria, did not have intellectual disability (WISC-R >70) or other neurological disorders, were included in the study as controls (n = 56). In order to have a bigger sample size, an additional 96 DNA samples were included, among children or adults who did not have a history of ADHD or any other cognitive, neurological or psychiatric disorder. Finally, the control group included 152 subjects.
Peripheral blood or buccal samples were taken from all subjects and parents of cases. Parents of cases and controls approved and signed the informed consent after a detailed explanation. The Institutional Ethical Committee approved this study (Act 117, May 29th,2008). To increase DNA samples, an amendment was presented to the ethical committee and approved by the act 165 of May 13th 2010.
DNA extraction
DNA extraction was carried out from peripheral blood or buccal samples using conventional techniques.
Genotyping
VNTRs in DRD4 and SLC6A3 as well as the insertion/deletion in the promoter region of SLC6A4 genes were genotyped by PCR followed by electrophoretic analysis as previously described.25,26 DNA amplification reactions were performed with GoTaq® Green Master Mix (Promega), using the following cycling conditions: 94°C for 10 min, followed by 30 cycles of 94°C for 45 sec, 61°C for 45 sec, 72°C for 45 sec. Amplification was completed with a final elongation step of 72°C for 10 min. Fragments were separated on 1,2% agarose gels and visualized using ethidium bromide.
Polymorphisms in DBH (Taq1 A polymorphism) and HTR1B (rs6296) genes were genotyped by PCR-RFLP. For HTR1B the PCR product was digested with HincII restriction enzyme at 37°C overnight, and the PCR product of DBH was digested with Taq1 at 61°C for 16 hours. The alleles were detected after separation on an agarose gel. Experimental approaches were used as published before.27,28
The dinucleotide (CA)n repeat polymorphism located in the DRD5 gene was genotyped using PCR with fluorescently labeled primers, followed by capillary electrophoresis in an ABI 3500 sequencer.29 Genotypes were analysed with Gene Mapper 4.1 software (Applied Biosystems).
Statistical Analysis
Statistical analysis was carried out by two different approaches. First, a transmission disequilibrium test (TDT) was done in 86 trios: corresponding to the index cases and their two biological parents. This analysis was used to test if there was preferential allelic transmission for any of the six polymorphisms. Genotype data was analyzed using PLINK.30 PLINK is a commonly used program for analysis of genetic studies, which is freely available and uses two main files as input: PED and MAP files.
A second complementary analysis was done through a case-control study where 73 cases and 152 controls were included. An analysis was performed for polymorphic variants in HTR1B and SLC6A4 genes (because these were found to be associated more frequently with the disorder24), using PLINK and SNPStat software.30,31Results for TDT and case control analysis were based in a X2 statistic and corrected for multiple testing with a Bonferroni correction.
Results
Cases were classified in subtypes according to DSM-IV criteria and BASC. Combined cases corresponded to 50.7%, 35.6% cases catergorized as inattentive and 13.7% were hyperactive.
Genotype and allele frequencies are presented in Table 1. For VNTR in DRD4, alleles of two, four, seven and eight repeats were found. For the dinucleotide of DRD5 more than ten alleles of several molecular weights were found, but a predominant allele of 148 bp was evident. Therefore alleles were grouped into the allele with a molecular weight of 148 bp and others, named “No 148 bp”. Only two alleles were found for the VNTR of SLC6A3, corresponding to alleles of nine and ten repeats. In DBH gene, the Taq1 A polymorphism gives only two alleles (A1 and A2) distinguished by the presence or not of this restriction site, respectively. The alleles of the studied SNP in HTR1B corresponded to the presence of a cytosine or a guanine in the position 861 of the gene coding region. Finally, the alleles corresponding to the 44-bp insertion/deletion of SLC6A4 are called L and S (for large and short respectively) distinguished by a difference of plus (insertion) 44-bp or minus (deletion) 44-bp respectively. We found a larger than expected deletion (approximately 100 bp) in the promoter region of SLC6A4 gene in only one patient, which we determined as an atypical shorter allele, but no further analysis was implemented in this sample.All polymorphisms studied here were in Hardy Weinberg equilibrium (p>0.05). Figure 1 shows the electrophoretic gels for the polymorphisms studied through a PCR-based technique.
Table 1: Allele and Genotype Frequencies.
| Allele frequency | Genotype Frequency | ||||
|---|---|---|---|---|---|
| Gene | Polymorphism (location) | Allele | Proportion | Genotype | Proportion |
| DRD4 | VNTR | 2 | 0,08 | 2/2 | 0,005 |
| (exon 3) | 4 | 0,69 | 2/4 | 0,112 | |
| 8 | 0,21 | 2/7 | 0,015 | ||
| 0,02 | 2/8 | 0 | |||
| 4/4 | 0,477 | ||||
| 4/7 | 0,29 | ||||
| 4/8 | 0,005 | ||||
| 7/7 | 0,091 | ||||
| 7/8 | 0,005 | ||||
| DRD5 | Dinucleotide (CA)n (18,5 kb 5’ to the start site) | 148 bp | 0,395 | 148bp/148bp | 0,140 |
| No 148 bp | 0,605 | 148bp/No 148bp | 0,368 | ||
| No 148bp/No 148bp | 0,491 | ||||
| SLC6A3 | 40 bp VNTR (3’UTR) | 9 | 0,208 | 9/9 | 0,071 |
| 10 | 0,792 | 9/10 | 0,276 | ||
| 10/10 | 0,654 | ||||
| DBH | TaqI A (intron 5) | A1 | 0,302 | A1/A1 | 0,052 |
| A2 | 0,698 | A1/A2 | 0,5 | ||
| A2/A2 | 0,448 | ||||
| HTR1B | rs6296(coding region) | C | 0,439 | C/C | 0,201 |
| G | 0,561 | C/G | 0,476 | ||
| G/G | 0,323 | ||||
| SLC6A4 | 44 bp insertion/ deletion (promoter) | L | 0,509 | S/S | 0,25 |
| S | 0,488 | S/L | 0,476 | ||
| Atypical shorter allele (ASA) | 0,003 | ASA/L | 0,006 | ||
| L/L | 0,268 | ||||
Fig. 1:
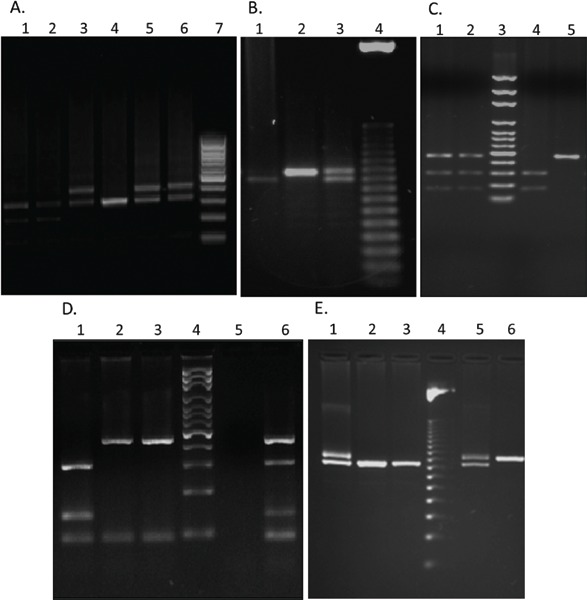
(A) Genotyping of VNTR located in exon 3 of DRD4. Lines 1 and2: Heterozygote 2/4, Lines 3, 5, 6: Heterozygote 4/7, Line 4: Homozygote 4/4, Line 7: 50 bp ladder. (B) Genotyping of VNTR located in the 3’UTR of SLC6A3. Line 1: Homozygote 9/9, Line 2: Heterozygote, Line 3: Homozygote 10/10, Line 4: 50 bp ladder. (C) Genotyping of TaqIA polymorphism in DBH. Lines 1 and 2: Heterozygote, Line 3: 50 bp ladder, Line 4: homozygote A1/A1, Line 5: homozygote A2/A2. (D) Genotyping of SNP rs6296 of HTR1B. Line 1: Homozygote C/C, Lines 2 and 3: Homozygote G/G, Line 4: 100 bp ladder, Line 5: Negative control, Line 6: Heterozygote. (E) Genotyping of the 44-bp insertion/deletion in SLC6A4. Lines 1 and 5: Heterozygote, Lines 2 and 3: Homozygote S/S, Line 6: Homozygote L/L.
As TDT study only allows two alleles to be analyzed, the two most frequent alleles were considered for polymorphisms having more than two alleles. For example, for the VNTR of DRD4, the 4 and 7-repeats allele were considered as they were the most prevalent. Similarly, for SLC6A4 where three alleles were found, only L and S alleles were included in analysis based on their higher prevalence.
Association analysis through TDT did not demonstrate a preferential allelic transmission for any of the studied polymorphisms when ADHD or its subtypes were evaluated. This is probably due to a high homozygosity percentage for some variants, leading to exclusion of several trios and therefore leads to reduced sample size in TDT analysis (Table 2).
Table 2: TDT association analysis.
| Gene | A1 | A2 | T | U | OR | CHISQ | p | Exclusion % |
|---|---|---|---|---|---|---|---|---|
| A1: Allele 1, A2: Allele 2, T: Transmitted, U: Untransmitted, OR: Odds-Ratio, CHISQ: Chi square, p: p value, Exclusion: Trio exclusion percentage. | ||||||||
| SLC6A3 | 9 | 10 | 17 | 24 | 0,708 | 1,195 | 0,2743 | 49,30% |
| DBH | A1 | A2 | 38 | 46 | 0,826 | 0,762 | 0,3827 | 30,10% |
| DRD4 | 7 | 4 | 21 | 25 | 0,832 | 0,923 | 0,4531 | 28,13% |
| DRD5 | No-148 | 148 | 32 | 33 | 0,922 | 0,015 | 0,9513 | 42,16% |
| HTR1B | C | G | 38 | 39 | 0,974 | 0,013 | 0,9093 | 16,20% |
| SLC6A4 | S | L | 33 | 43 | 0,767 | 1,316 | 0,2513 | 22,90% |
Case-control analysis for variants in HTR1B and SLC6A4 genes did not show evidence of association with ADHD susceptibility (Table 3).
Table 3: Allelic and Genotype Association using case-control approach.
| Gene | MAF | Allelic Association (p*) | Genotype Association (p*) |
|---|---|---|---|
| MAF: Minor Allele Frequency. | |||
| p: p value. | |||
| *Bonferroni correction. | |||
| HTR1B | 0,4231 | 0,2371 | 0,2919 |
| SLC6A4 | 0,4686 | 0,689 | 0,7071 |
Discussion
Although several works suggest an important influence of inherited factors in the development of ADHD, these genetic studies involving candidate polymorphisms reveal that the genetic architecture of this disorder is extremely complex.5,32
Several epidemiological and statistical approaches, in populations of different genetic background, has led to controversial results.5
Despite the fact that several candidate genes have a biological role in to ADHD pathophysiology, some studies have failed to find a significant association.17,33,34 Our TDT study performed in the population of Colombian ADHD patients also failed to find a significant relationship with several candidate genes. Since we did not have previous data for homozygosity for these variants in Colombia population, we could not determine a percentage of trio exclusion before the study, which was as high as 49.3% for some genes (SLC6A3 for example). The high percentage of excluded trios led to small sample size for several variants. A complementary case control approach also failed to find a significant association in our samples.
As ADHD is a complex disease, it is possible that each single polymorphism has a relatively small contribution to the overall risk, but all variants together could underlie a large genetic influence.35 This hypothesis presupposes large sample size in order to find the relation of a specific variant with this disorder. Therefore a large sample of well-characterized patients is a challenge for research groups in developing countries.
Additionally, it is well known that ADHD is highly heterogeneous and has a clinical overlap with other neuropsychiatric conditions,36 making it difficult to associate a single variant with a single phenotype.35 The clinical heterogeneity of ADHD and other neuropsychiatric disorders has opened the door for the study of endophenotypes as a good approach in genetic studies. Endophenotypes refer to the analysis of specific behavioral symptoms, among others, as more stable and less heterogeneousphenotypes.37 Other studies from our group, for example, have shown the contribution of a polymorphism located in SLC6A3 (DAT1) gene to the development of an ADHD endophenotype in a Colombian sample.38 We have also previously found an association between two polymorphisms of SNPA25 and ADHD.39
Conclusions
Our study has failed to demonstrate an association of six polymorphic variants located in different candidate genes and the susceptibility to ADHD in Colombian population. Our findings are in concordance with results from genetic studies other populations.24,40 Future prospects in the genetic study of ADHD might include analysis of related endophenotypes.
Authorship contributions
Dora J Fonseca: Participated in the study design, data collection, analysis and interpretation of data and drafting the manuscript, Heidi E Mateus: Participated in the study design, data collection, analysis and interpretation of data, Jubby M Gálvez: Participated in the data collection, analysis and interpretation of data, drafting the manuscript and revising the final paper, Diego A Forero: Participated in study design, statistical analysis and interpretation of data, revising the final paper, Claudia T-Gutierrez: Participated in design, study supervision and revising the final paper, Alberto V Meerbeke: Participated in study design, study supervision, statistical analysis and interpretation of data, as well as drafting the manuscript and revising the final paper
Acknowledgements
The authors thank William Donoso, Ivone Munevar and Sofia Narvaez for their contribution to the genotyping activities in this study. This work was supported by research grants from the Fundación para la Promoción de la Investigación y la Tecnología, Banco de la República and the Fondo de Investigaciones de la Universidad del Rosario (FIUR) and VCTI-UAN. Funding sources had no role in the design for their sponsorship. These institutions were not involved at any stage of the study, data collection and analysis or decision to publish.
Footnotes
This article complies with International Committee of Medical Journal editor’s uniform requirements for manuscript.
Conflict of interest: None; Funding: Fundación para la Promoción de la Investigación y la Tecnología, Banco de la República and the Fondo de Investigaciones de la Universidad del Rosario (FIUR) and VCTI-UAN.
References
- 1.Faraone SV, Sergeant J, Gillberg C et al. The worldwide prevalence of ADHD: is it an American condition? World psychiatry. 2003;2(2):104–113. [PMC free article] [PubMed] [Google Scholar]
- 2.Pineda DA, Lopera F, Palacio JD et al. Prevalence estimations of attention-deficit/hyperactivity disorder: differential diagnoses and comorbidities in a Colombian sample. Int J Neurosci. 2003;113(1):49–71. doi: 10.1080/00207450390161921. [DOI] [PubMed] [Google Scholar]
- 3.Polanczyk G, De Lima MS, Horta BL et al. The worldwide prevalence of ADHD: a systematic review and metaregression analysis. The Am J Psychiatry. 2007;164(6):942–948. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- 4.Millichap JG. Etiologic classification of attention-deficit/hyperactivity disorder. Pediatrics. 2008;121(2):e358–365. doi: 10.1542/peds.2007-1332. [DOI] [PubMed] [Google Scholar]
- 5.Faraone SV, Perlis RH, Doyle AE et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 6.Faraone SV, Biederman J. Neurobiology of attention-deficit hyperactivity disorder. Biol Psychiatry. 1998;44(10):951–958. doi: 10.1016/s0006-3223(98)00240-6. [DOI] [PubMed] [Google Scholar]
- 7.Asghari V, Sanyal S, Buchwaldt S et al. Modulation of intracellular cyclic AMP levels by different human dopamine D4 receptor variants. J Neurochem. 1995;65(3):1157–1165. doi: 10.1046/j.1471-4159.1995.65031157.x. [DOI] [PubMed] [Google Scholar]
- 8.Van Tol HH, Wu CM, Guan HC et al. Multiple dopamine D4 receptor variants in the human population. Nature. 1992;358(6382):149–152. doi: 10.1038/358149a0. [DOI] [PubMed] [Google Scholar]
- 9.Tahir E, Yazgan Y, Cirakoglu B et al. Association and linkage of DRD4 and DRD5 with attention deficit hyperactivity disorder (ADHD) in a sample of Turkish children. Mol Psychiatry. 2000;5(4):396–404. doi: 10.1038/sj.mp.4000744. [DOI] [PubMed] [Google Scholar]
- 10.Hawi Z, Lowe N, Kirley A et al. Linkage disequilibrium mapping at DAT1, DRD5 and DBH narrows the search for ADHD susceptibility alleles at these loci. Mol Psychiatry. 2003;8(3):299–308. doi: 10.1038/sj.mp.4001290. [DOI] [PubMed] [Google Scholar]
- 11.Daly G, Hawi Z, Fitzgerald M et al. Mapping susceptibility loci in attention deficit hyperactivity disorder: preferential transmission of parental alleles at DAT1, DBH and DRD5 to affected children. Mol Psychiatry. 1999;4(2):192–196. doi: 10.1038/sj.mp.4000510. [DOI] [PubMed] [Google Scholar]
- 12.Spencer T, Biederman J, Wilens T. Pharmacotherapy of attention deficit hyperactivity disorder. Child Adolesc Psychiatr Clin N Am. 2000;9(1):77–97. [PubMed] [Google Scholar]
- 13.Gainetdinov RR, Jones SR, Fumagalli F et al. Re-evaluation of the role of the dopamine transporter in dopamine system homeostasis. Brain Res. 1998;26(2–3):148–153. doi: 10.1016/s0165-0173(97)00063-5. [DOI] [PubMed] [Google Scholar]
- 14.Cook EH, Stein MA, Krasowski MD et al. Association of attention-deficit disorder and the dopamine transporter gene. Am J Hum Genet. 1995;56(4):993–998. [PMC free article] [PubMed] [Google Scholar]
- 15.Waldman ID, Rowe DC, Abramowitz A et al. Association and linkage of the dopamine transporter gene and attention-deficit hyperactivity disorder in children: heterogeneity owing to diagnostic subtype and severity. Am J Hum Genet. 1998;63(6):1767–1776. doi: 10.1086/302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comings DE, Wu S, Chiu C et al. Polygenic inheritance of Tourette syndrome, stuttering, attention deficit hyperactivity, conduct, and oppositional defiant disorder: the additive and subtractive effect of the three dopaminergic genes–DRD2, D beta H, and DAT1. Am J Med Genet B Neuropsychiatr Genet. 1996;67(3):264–288. doi: 10.1002/(SICI)1096-8628(19960531)67:3<264::AID-AJMG4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 17.Smith KM, Daly M, Fischer M et al. Association of the dopamine beta hydroxylase gene with attention deficit hyperactivity disorder: genetic analysis of the Milwaukee longitudinal study. Am J Med Genet B Neuropsychiatr Genet. 2003;119B(1):77–85. doi: 10.1002/ajmg.b.20005. [DOI] [PubMed] [Google Scholar]
- 18.Roman T, Schmitz M, Polanczyk GV et al. Further evidence for the association between attention-deficit/hyperactivity disorder and the dopamine-beta-hydroxylase gene. Am J Med Genet B Neuropsychiatr Genet. 2002;114(2):154–158. doi: 10.1002/ajmg.10194. [DOI] [PubMed] [Google Scholar]
- 19.Hawi Z, Dring M, Kirley A et al. Serotonergic system and attention deficit hyperactivity disorder (ADHD): a potential susceptibility locus at the 5-HT(1B) receptor gene in 273 nuclear families from a multi-centre sample. Mol Psychiatry. 2002;7(7):718–725. doi: 10.1038/sj.mp.4001048. [DOI] [PubMed] [Google Scholar]
- 20.Quist JF, Barr CL, Schachar R et al. The serotonin 5-HT1B receptor gene and attention deficit hyperactivity disorder. Mol Psychiatry. 2003;8(1):98–102. doi: 10.1038/sj.mp.4001244. [DOI] [PubMed] [Google Scholar]
- 21.Seeger G, Schloss P, Schmidt MH. Functional polymorphism within the promotor of the serotonin transporter gene is associated with severe hyperkinetic disorders. Mol Psychiatry. 2001;6(2):235–238. doi: 10.1038/sj.mp.4000820. [DOI] [PubMed] [Google Scholar]
- 22.Retz W, Thome J, Blocher D et al. Association of attention deficit hyperactivity disorder-related psychopathology and personality traits with the serotonin transporter promoter region polymorphism. Neurosci Lett. 2002;319(3):133–136. doi: 10.1016/s0304-3940(01)02575-7. [DOI] [PubMed] [Google Scholar]
- 23.Beitchman JH, Davidge KM, Kennedy JL et al. The serotonin transporter gene in aggressive children with and without ADHD and nonaggressive matched controls. Ann N Y Acad Sci. 2003;1008:248–251. doi: 10.1196/annals.1301.025. [DOI] [PubMed] [Google Scholar]
- 24.Forero DA, Arboleda GH, Vasquez R et al. Candidate genes involved in neural plasticity and the risk for attention-deficit hyperactivity disorder: a meta-analysis of 8 common variants. J Psychiatry Neurosci. 2009;34(5):361–366. [PMC free article] [PubMed] [Google Scholar]
- 25.Olsson C a,, Byrnes GB, Lotfi-Miri M et al. Association between 5-HTTLPR genotypes and persisting patterns of anxiety and alcohol use: results from a 10-year longitudinal study of adolescent mental health. Mol Psychiatry. 2005;10(9):868–876. doi: 10.1038/sj.mp.4001677. [DOI] [PubMed] [Google Scholar]
- 26.Vieyra G, Moraga M, Henríquez H et al. Distribución de alelos de los genes DRD4 y DAT1 del sistema dopaminérgico en la población mixta de Santiago de Chile. Rev Med Chil. 2003;131(2):135–143. [PubMed] [Google Scholar]
- 27.Bhaduri N, Sinha S, Chattopadhyay A et al. Analysis of polymorphisms in the dopamine beta hydroxylase gene: association with attention deficit hyperactivity disorder in Indian children. Indian Pediatr. 2005;42(2):123–129. [PubMed] [Google Scholar]
- 28.Corregiari F, Bernik M, Cordeiro Q et al. Endophenotypes and serotonergic polymorphisms associated with treatment response in obsessive-compulsive disorder. Clinics (Sao Paulo) 2012;67(4):335–340. doi: 10.6061/clinics/2012(04)06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Shao C, Zhang D et al. The effect of dopamine D2, D5 receptor and transporter (SLC6A3) polymorphisms on the cue-elicited heroin craving in Chinese. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(3):269–273. doi: 10.1002/ajmg.b.30264. [DOI] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Med Genet. A. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solé X, Guinó E, Valls J et al. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 32.Thapar A, O’Donovan M, Owen MJ. The genetics of attention deficit hyperactivity disorder. Hum Mol Genet. 2005;14(2):R275–282. doi: 10.1093/hmg/ddi263. [DOI] [PubMed] [Google Scholar]
- 33.Mill J, Curran S, Kent L et al. Attention deficit hyperactivity disorder (ADHD) and the dopamine D4 receptor gene: evidence of association but no linkage in a UK sample. Mol Psychiatry. 2001;6(4):440–444. doi: 10.1038/sj.mp.4000881. [DOI] [PubMed] [Google Scholar]
- 34.Todd RD, Jong YJ, Lobos EA et al. No association of the dopamine transporter gene 3’ VNTR polymorphism with ADHD subtypes in a population sample of twins. Am J Med Genet B Neuropsychiatr Genet. 2001;105(8):745–748. doi: 10.1002/ajmg.1611. [DOI] [PubMed] [Google Scholar]
- 35.Thapar A, Cooper M, Eyre O et al. What have we learnt about the causes of ADHD? J Child Psychol Psychiatry. 2013;54(1):3–16. doi: 10.1111/j.1469-7610.2012.02611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taurines R, Schwenck C, Westerwald E et al. ADHD and autism: differential diagnosis or overlapping traits? A selective review. Atten Defic Hyperact Disord. 2012;4(3):115–139. doi: 10.1007/s12402-012-0086-2. [DOI] [PubMed] [Google Scholar]
- 37.Fernández-Jaén A,, Fernández-Mayoralas DM, Calleja-Pérez B et al. [Genomic endophenotypes of attention deficit hyperactivity disorder]. Rev Neurol. 2012;54(Suppl1):S81–S87. [PubMed] [Google Scholar]
- 38.Agudelo JA, Gálvez JM, Fonseca DJ et al. Evidence of an association between 10/10 genotype of DAT1 and endophenotypes of attention deficit/hyperactivity disorder. Neurologia. 2014:1–7. [Google Scholar]
- 39.Gálvez JM, Forero DA, Fonseca DJ et al. Evidence of association between SNAP25 gene and attention deficit hyperactivity disorder in a Latin American sample. Atten Defic Hyperact Disord. 2014;6(1):19–23. doi: 10.1007/s12402-013-0123-9. [DOI] [PubMed] [Google Scholar]
- 40.Ribasés M, Ramos-Quiroga JA, Hervás A et al. Candidate system analysis in ADHD: evaluation of nine genes involved in dopaminergic neurotransmission identifies association with DRD1. World J Biol Psychiatry. 2012;13(4):281–292. doi: 10.3109/15622975.2011.584905. [DOI] [PubMed] [Google Scholar]


