Abstract
Polyreactive (natural) antibodies are primarily IgM and account for a major proportion of circulating Ig in humans. They use various V gene segments, in general, in germ line (unmutated) configuration. To analyze the VH regions of polyreactive antibodies, with particular attention at their somatically mutated status, we generated five IgG (three IgG1 and two IgG3) mAb (using B cells from a healthy subject, a patient with insulin-dependent diabetes mellitus and a patient with SLE), which bound with various efficiencies a number of different self and foreign Ag. Gene cloning experiments showed that the VH region sequences were unique to each IgG mAb. The H chain complementary determining region (CDR3) of two IgG (mAb10 and mAb426.4.2F20) displayed an identical stretch of five amino acids (RFLEW), but the other three IgG mAb CDR3 were divergent in both length and composition. The VH gene sequences of two IgG, mAb426.4.2F20 and mAb410.7.F91, were 99% identical to those of the germ line VH4.11 and VH4.21 genes, respectively. Those of the remaining three IgG mAb displayed a number of differences (93.6 to 95.9% identity) when compared with the germ line VH4.18, VH4.11, and hv1263 gene sequences. These and the VH4.21 gene have been found to encode polyreactive IgM and IgA and, in mutated configuration, monoreactive high affinity autoantibodies and antibodies induced by foreign Ag. When compared with the respective framework region, the CDR of three IgG mAb VH segment sequences displayed a significantly higher: 1) frequency of total nucleotide differences (6.1 × 10−2 vs 4.5 × 10−2 difference/base); 2) frequency of putative nucleotide changes yielding amino acid replacements (5.6 × 10−2 vs 1.4 × 10−2 replacement change/base); and 3) ratio of overall putative replacement to silent (R:S) mutations (11.0 vs 0.4). Thus, the distribution and nature of the nucleotide differences were consistent with a process of somatic mutation and Ag-dependent clonal selection. This was formally proved in IgG mAb426.12.3F1.4 and IgG mAb10 by differentially targeted polymerase chain reaction amplification and cloning and sequencing of the germ line genes that gave rise to the expressed VH segments, using DNA from polymorphonuclear cells of the same subjects whose B cells were used for the generation of these IgG mAb. Somatic mutations might have been responsible for bringing about polyreactivity in originally monoreactive antibodies or, more likely, they accumulated in originally polyreactive antibodies, which after undergoing a process of Ag selection, retained polyreactivity and may have or may have not acquired a higher affinity for the selecting Ag.
Sera of healthy humans and animals contain antibodies that react with a variety of Ag present on pathogenic microorganisms, including bacteria and viruses and with self-Ag. Because their emergence is independent of known or intentional immunization, these antibodies have been termed “natural antibodies or auto-antibodies” (1–6). Most natural mAb generated form humans and mice are polyreactive, i.e., they bind multiple Ag, dissimilar in nature, such as polysaccharides, nucleic acids, haptens, and proteins, including structural cellular and tissue components and soluble hormones (1–6). A single polyreactive mAb displays different affinities for different Ag (7–10). These are in general low, although in some polyreactive mAb, affinities of the same order of magnitude as those of specific antibodies induced by foreign Ag or those of autoantibodies found in patients with autoimmune diseases have been measured (7, 9–11).
Despite their mostly low intrinsic affinity, polyreactive antibodies display in general a high avidity for Ag due to the multivalency of their predominant Ig class, IgM (12). Because of their broad range of reactivity and high avidity, polyreactive antibodies may play a major role in primary line of defense against invading bacteria or viruses before the specific immune response is generated and in the clearance of debris, such deriving from dead cells, or, possibly some toxic substances. Analysis of the primary structure of the V regions of polyreactive primarily IgM natural antibodies has shown that these are in germ line configuration (13–17). This has led to the hypothesis that natural polyreactive antibodies do not accumulate somatic point mutations. As a consequence, their effectiveness in binding Ag would dramatically decrease, due to a decrease in overall avidity after Ig class switch and substitution of the μ, with a γH chain (18).
In these studies, we generated five polyreactive human IgG mAb with distinctive Ag binding activities. The IgG mAb VH region genes were similar to those used by polyreactive IgM and/or specific high affinity antibodies or autoantibodies. When compared with the sequences of the closest germ line genes, those of the VH genes of three IgG mAb displayed a number of differences, the distribution and nature of which suggested an Ag-dependent selection process. In two IgG mAb, such differences were formally proved to represent somatic mutations. Thus, these findings are consistent with the hypothesis that natural polyreactive IgM antibody-producing cells may class switch to IgG while undergoing an Ag-directed selection process, which is underpinned by somatic hypermutation.
Materials and Methods
Generation of the polyreactive IgG mAb-producing cell lines
PBMC obtained from a healthy subject, a patient with IDDM5, and a patient with SLE were depleted of T lymphocytes, infected with EBV, and then distributed in microcultures in presence of irradiated feeders (19, 20). After 3 wk, the microculture fluids were tested for IgG to human insulin or ssDNA or insulin, ssDNA, and tetanus toxoid (7–11). IgG mAb-producing cell lines were established from EBV-transformed lymphoblasts by sequential subculturing after fusion with the F3B6 human-mouse hybrid cells (19, 20). Concentrated mAb were prepared from culture fluids (7–11). Their Ag binding activity and Kd different Ag were measured as reported (8–11).
Cloning and sequencing of the expressed Ig VH genes
mRNA was extracted from mAb-producing cells and first strand cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (21). The degenerate sense HA1, HA3a, HA4a oligonucleotide primers, and the antisense Cγ primer were synthesized and used to amplify the VH gene cDNA. The sequence of HA1 [5′ ATGGACTGGACCTGGAGG(AG)TC(CT)TCT(GT)C 3′] was highly similar to a portion of the leader sequences of the members of VHI gene family; the sequence of HA3a [5′ ATGGAG(CT)TTGGGCTGA(CG)TTT(CT)T 3′] was highly similar to a portion of the leader sequences of members of the VHIII gene family; the sequence of HA4a [5′ ATGAA(AG)CA(TC)CTGTGGTTCTT(CT)(AC)T(CT)CT(CG)C 3′] was highly similar to a portion of the leader sequences of the VHIV family genes. The sequence of the antisense (HI-1) Cγ primer [5′ TAGTCCTTGACCAGGCAGCC 3′] was the reverse complement of a 5′-portion of the IgG constant region gene conserved in all four human IgG subclasses. PCR was performed in a 50 μl of volume using the primers described above and Taq DNA polymerase (Perkin Elmer Cetus, Norwalk, CT) under the condition previously described (21). PCR-amplified DNA fragments were separated on 2% low melting agarose gel by electrophoresis. Each amplified DNA fragment was excised, purified, and then cloned into pCR1000 plasmid vector (Invitrogen, La Jolla, CA) (21). Dideoxy sequencing was conducted using double-stranded plasmid DNA prepared from the selected bacterial clones (21). Each VH gene sequence was derived from the analysis of at least four independent clones. Nucleotide differences in sequences among different recombinant clones were observed in few cases (<0.001 difference/base). Such variants were excluded from analysis. Sequences were analyzed using the software package of the Genetics Computer Group of the University of Wisconsin, Release 6, and a Model 6000–410 VAX computer (Digital Equipment Corp., Marlboro, MA). Identity searches of the expressed VH genes were performed using the FASTA program with the GenBank database.
PCR amplification of Ig VH genes from B cell hybridoma and autologous PMN DNA
To determine whether the nucleotide differences displayed by the mAb426.12.3F1.4VH gene sequence compared with that of the closest reported germ line VH gene represented somatic point mutations, we performed PCR amplification of VH gene segments from genomic DNA of the mAb426. 12.3F1.4–producing cells and autologous PMN using combinations of the ad hoc designed 58–5, 58–3, and 12.3–3 oligonucleotide primers. The sequence of the sense 58–5 [5′ GCACTGTCTCTGGTGGCTCC 3′] primer encompassed a FR1 stretch (residues 65 to 84) shared by the mAb426.12.3F1.4 and the germ line VH4.11 genes (22). The antisense 58–3 [5′ GGCCGTGTCCGCAGCGGTCA 3′] primer was the reverse complement of a FR3 sequence (residues 253 to 273) of the germ line VH4.11 gene and differed in two bases from the expressed mAb426.12.3F1.4 VH gene. The antisense 12.3–3 [5′ TCGCGGACGTGTCCACTGACA 3′] oligonucleotide primer was the reverse complement of a part of the FR3 sequence of the expressed mAb426.12.3F1.4 VH gene (residues 206 to 226). This sequence differed from the corresponding sequence of the VH 4.11 gene in four bases. Genomic DNA (100 ng) was subjected to PCR (50 μl volume) with 100 ng of the 58–5 oligonucleotide and 100 ng of either the 12.3–3 or the 58–3 oligonucleotides. Each cycle consisted of denaturing, annealing, and extension steps of 1,2, and 2 min, respectively. Denaturing and extension temperatures were 94 and 72° C, respectively. Annealing temperatures were 61 and 57°C for the 12.3–3 and 58–3 primers, respectively. After 30 cycles, the products amplified using DNA extracted from autologous PMN or from mAb426.12.3F1.4–producing cells were fractionated on 1.7% agarose gel containing 1 μg/ml of ethidium bromide. DNA was transferred to a filter membrane and hybridized with the 32P-labeled oligonucleotide 12.3–3 at 61°C (21). Filter was washed twice with 2 × SSC/0.5% SDS at room temperature for 10 min and twice with 1 × SSC/0.5 × SDS at 61°C for 20 min before exposure to Kodak XAR film (Eastman Kodak, Rochester, NY). The PCR product amplified from autologous PMN DNA using the 58–5 and 58–3 primer pair and that amplified from the mAb426.12.3F1.4–producing cell DNA using the 58–5 and 12.3–3 primer pair were also fractionated on 2% low melting agarose gel. The DNA fragments were isolated, cloned, and sequenced.
To determine whether the nucleotide differences displayed by the mAb10 VH gene sequence compared with that of the closest reported germ line VH gene represented somatic point mutations, we performed PCR amplification of VH gene segments from genomic DNA of the mAb10–producing B cell hybridoma and autologous PMN using combinations of the ad hoc designed VH4.18/V2–1FR1-CDR1 and mAb10VHCDR1 oligonucleotide primers, and the HBL-3 FR3 primer. The sequence of the sense VH4. 18/V2–1 FR1-CDR1 [5′ GCTCCATCAGCAGTAGTAGT 3′] primer encompassed a FR1-CDR1 stretch (residues 80 to 99) of the germ line VH4.18 gene (22), which differed by four nucleotides from that of the corresponding area of the mAb10 VH gene. The sequence of the sense mAb10VHCDR1 oligonucleotide primer [5′ GCTCCATCAGTACCAGTACT 3′] encompassed a FR1-CDR1 stretch (residues 80 to 99) of the mAb10 VH gene, which was different by four nucleotides from that of the corresponding area (same as that of VH4.18/V2–1 FR1-CDR1) of the germ line VH4.18 gene (Fig. 2A). The antisense HBL-3 oligonucleotide belonged to our collection of primers and was the reverse complement [5′ TCGCGGACGTGTCCACTGACA 3′] of a FR3 sequence (residues 256 to 275) identical in the germ line VH4.18 gene and the mAb10 VH gene except for a G instead of an A at position 270 (Fig. 2A). Genomic DNA (100 ng) was subjected to PCR (50 μl vol) with 100 ng of the HBL-3 FR3 and 100 ng of either the VH4.18/V2–1 FR1-CDR1 or the mAb10VH CDR1 oligonucleotide primer. Each cycle consisted of denaturing, annealing, and extension steps of 1, 2, and 2 min, respectively. Denaturing and extension temperatures were 94° and 72°C, respectively. Annealing temperatures were 60°C for both the VH4.18/V2–1 FR1-CDR1 and the mAb10VH CDR1 oligonucleotide primers. After 30 cycles, the products amplified using DNA extracted from autologous PMN or from mAb10–producing B cell hybridoma were fractionated on 1.7% agarose gel containing 1 μg/ml of ethidium bromide or on 2% low melting agarose gel for cloning and sequencing.
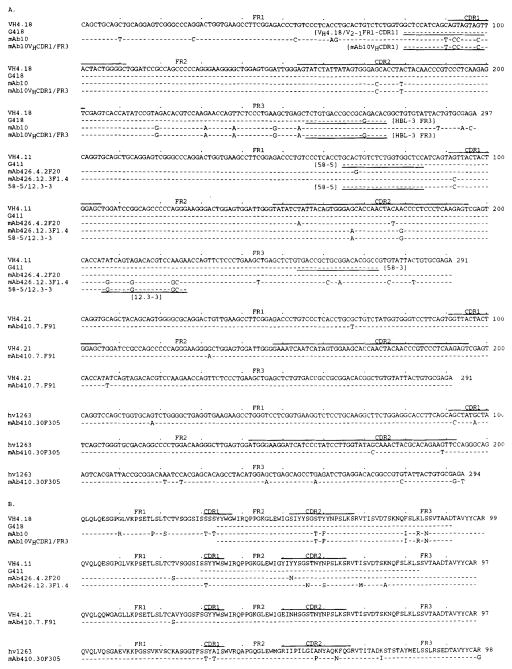
FIGURE 2.
Nucleotide (A) and deduced amino acid (B) sequences of the VH genes used by the polyreactive IgG mAb. In each cluster, the top sequence is given for comparison and represents the closest published germ line VH gene. Dashes indicate identities. Solid lines above each cluster depict CDR. G418 and G411 are the germ line sequences derived from healthy subject A and IDDM patient B, respectively (see text). The sequences or complementary sequences of the primers used for PCR amplification of genomic DNA are underlined. The present sequences are available from the EMBL/GenBank under accession numbers L23515, L23516, L23517, L23518, and L23519.
Results
Establishment of the IgG mAb-producing cell lines and mAb Ag binding features
EBV-transformed B cells from a healthy subject and two autoimmune patients (one with IDDM and one with SLE) were selected for production of IgG to human recombinant insulin (mAb10) (23) or ssDNA (mAb410.7.F91 and mAb410.30F305) or ssDNA, insulin, and tetanus toxoid (mAb426.4.2F20 and mAb426.12.3F1.4) and used to generate mAb-producing cell lines. All IgG mAb were poly-reactive. Their IgG subclasses, L chain types, and Kd values for insulin, ssDNA, and tetanus toxoid are listed in Table I. Their dose-saturable bindings to a panel of self and foreign Ag are depicted in Figure 1.
Table I.
Ag binding features and VH, D, and JH genes of human polyreactive IgG mAb
| Clone | Source | Chains
|
Kd (M)
|
VH Segment
|
Nucleotide Differences
|
R:S Mutation Ratio
|
D Geneb | JH Genec | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Closest Gene
|
%
|
|||||||||||||
| HL | Insulin | ssDNA | Tetanus Toxoid | Family | Membera | Identity | CDR | FR | CDR | FR | ||||
| mAb10 | Healthy subject A | γ3, λ | 1.0 × 10−5 | 2.0 × 10−7 | 1.0 × 10−5 | VHIV | VH4.18 | 93.6 | 5 | 14 | 4:1 | 6:8 | DKP4 | JH4 |
| mAb426.4.2F20 | IDDM patient B | γ1, κ | 2.0 × 10−5 | 6.8 × 10−7 | 3.9 × 10−7 | VHIV | VH4.11 | 99.0 | 2 | 1 | 1:1 | 1:0 | DKP4 | JH6 |
| mAb426.12.3F1.4 | IDDM patient B | γ1, κ | 4.0 × 10−5 | 1.7 × 10−8 | 1.0 × 10−6 | VHIV | VH4.11 | 95.9 | 3 | 9 | 3:0 | 2:7 | DK4 | JH3 |
| mAb410.7.F91 | SLE patient C | γ1, κ | 1.9 × 10−6 | 1.0 × 10−8 | 1.0 × 10−6 | VHIV | VH4.21 | 99.0 | 0 | 3 | 0:0 | 1:2 | DXP′1 | JH4 |
| mAb410.30F305 | SLE patient C | γ3, κ | 2.5 × 10−4 | 8.3 × 10−8 | 1.7 × 10−6 | VHI | hv1263 | 95.9 | 4 | 8 | 4:0 | 2:6 | DK4 | JH4 |
FIGURE 1.
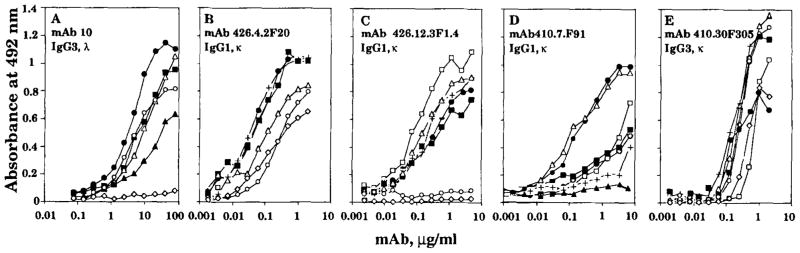
Dose-saturable binding of the polyreactive IgG mAb. The Ag binding activity of each mAb is expressed as optical absorbance at 492 nm. The following Ag were used: human recombinant insulin (●), tetanus toxoid (■), ssDNA (△), phosphorylcholine (+), human recombinant TNF-α (○), thyroglobulin (▲), and BSA (◇).
Sequences of the IgG mAb VH segments
The nucleotide and deduced amino acid sequences of the five IgG mAb VH genes are depicted in Figure 2A and B, respectively. Their comparisons with the sequences of the closest germ line VH genes are summarized in Table I. The mAb10 VH gene sequence displayed the highest degree of identity with that of the germ line VH4.18 gene (22). Nucleotide differences were five in the CDR and 14 in the FR, yielding four and six amino acid differences, respectively. The mAb426.4.2F20 and mAb426.12.3F1.4 VH gene sequences displayed the highest degree of identity with that of the germ line VH4.11 gene (22). The mAb426.4.2F20 VH gene contained three nucleotide differences, two in the CDR2 and 1 in the FR1, resulting in one and one amino acid differences, respectively. The mAb426.12.3F1.4 VH gene displayed 12 nucleotide differences, of which three were in the CDR and nine in the FR. All three differences in the CDR resulted in amino acid differences, whereas only three of the nine in the FR did. The mAb410.7.F91 VH gene sequence differed from that of the germ line VH4.21 gene in three nucleotides. These yielded a single amino acid difference. The mAb410.30F305 VH gene sequence displayed the highest similarity to that of the germ line (VHI) hv1263 gene (24). The four differences in the CDR and the eight in the FR resulted in three and two amino acid differences, respectively.
Thus, although the mAb426.4.2F20 and mAb410.7.F91 VH gene sequences were virtually identical with those of the respective germ line genes, the VH genes of the IgG mAb10, mAb426.12.3F1.4, and mAb410.30F305 displayed a high number of differences when compared with the respective germ line gene sequences. Table II shows the total number of putative changes and R:S mutation ratios in the VH genes of the five polyreactive IgG mAb, in those of the highly substituted three IgG, mAb10, mAb426.12.3F1.4, and mAb410.30F305, and in those of the virtually unmutated two IgG, mAb426.4.2F20 and mAb410.7.F91. In the three highly substituted IgG VH segments, the overall putative nucleotide changes were 12 in the CDR and 31 in the FR, yielding a frequency of 6.1 × 10−2 and 4.5 × 10−2 nucleotide change/base, respectively. These putative changes entailed a significantly higher number of replacement mutations in the CDR than the FR, 5.6 × 10−2 vs 1.4 × 10−2 R mutation/base, respectively (p < 0.01, χ2 test), and a significantly higher overall R:S mutation ratio (11.0 in CDR vs 0.4 in FR) (p < 0.01, χ2 test).
Table II.
Mutations in human polyreactive IgG mAb VH segments
| CDR | FR | |
|---|---|---|
| 5 (all) IgG mAb | ||
| Total (R and S) mutationsa | 14 | 35 |
| Total mutations (mutation/base) | 4.3 × 10−2 | 3.0 × 10−2 |
| R mutations (mutation/base) | 3.7 × 10−2 | 1.0 × 10−2 |
| R:S mutation ratio | 6.0 | 0.5 |
| 3 IgG mAb (mAb10.mAb426.12.3F1.4, and mAb410.30F305) | ||
| Total (R and S) mutationsa | 12 | 31 |
| Total mutations (mutation/base) | 6.1 × 10−2 | 4.5 × 10−2 |
| R mutations (mutation/base) | 5.6 × 10−2 | 1.4 × 10−2 |
| R:S mutation ratio | 11.0 | 0.4 |
| 2 IgG mAb (mAb426.4.2F20 and mAb410.7.F91) | ||
| Total (R and S) mutationsa | 2 | 4 |
| Total mutations (mutation/base) | 1.6 × 10−2 | 0.7 × 10−2 |
| R mutations (mutation/base) | 0.8 × 10−2 | 0.4 × 10−2 |
| R:S mutation ratio | 1.0 | 1.0 |
In mAb10, mAb426.12.3F1.4, and mAb426.4.2F20 VH segments mutations were formally verified.
D and JH gene sequences and CDR3 configuration
The nucleotide and deduced amino acid sequences of the D and JH segments of the polyreactive IgG mAb are depicted in Figure 3A and B, respectively. The mAb410.7.F91 D segment contained a stretch of nucleotides highly similar to most of the DXP′1 gene sequence (25); mAb10 and mAb426.4.2F20 D segments contained nucleotide sequence stretches identical to portions of the germ line DXP′4 gene (25); mAb426.12.3F1.4 and mAb410.30F305 D segments contained short nucleotide sequences identical with areas of the DK4 gene (25). The numbers of putative N segment additions at the 5′- and 3′-flanks of the D genes were highly variable. The overall lengths of the D segments ranged from 17 to 58 nucleotides. The nucleotide sequence of mAb426.12.3F1.4 JH segment was identical, except for one base to that of JH3 (26); those of the mAb10, mAb410.7.F91, and mAb410.30F305 JH segments consisted of truncated forms of the putatively predominant haplotype of the JH4 germ line gene, JH4b (27), with two replacement mutations in the mAb10 JH sequence. The mAb426.4.2F20 JH segment consisted of a truncated form of the putatively major haplotype of the germ line JH6 gene (26), JH6c (27).
FIGURE 3.
Nucleotide (A and B) and deduced amino acid (C) sequences of the D and JH segments of the polyreactive IgG mAb. The top sequence in each cluster is given for comparison. Dashes indicate identities. Solid lines above the deduced amino acid sequences depicts the CDR3. The present sequences are available from the EMBL/GenBank under accession numbers L23515, L23516, L23517, L23518, and L23519.
The deduced amino acid sequences of the expressed D-JH segments were segregated into CDR3 and FR4 according to Kabat et al. (28) and are depicted in Figure 3C. The IgG mAb CDR3 were divergent in length (8 to 24 amino acids; mean value, 16.4 ± 6.5 SD) and structure, with the exception of those of the two IgG mAb using the DXP′4 gene. In these IgG (mAb10 and mAb426.4.2F20), the DXP′4 gene was used in the same reading frame, yielding an identical H chain CDR3 stretch of five amino acids (RFLEW). The FR4 structure of the five polyreactive IgG mAb was conserved.
Somatic point mutations in the polyreactive IgG mAb426.12.3F1.4 VH segment
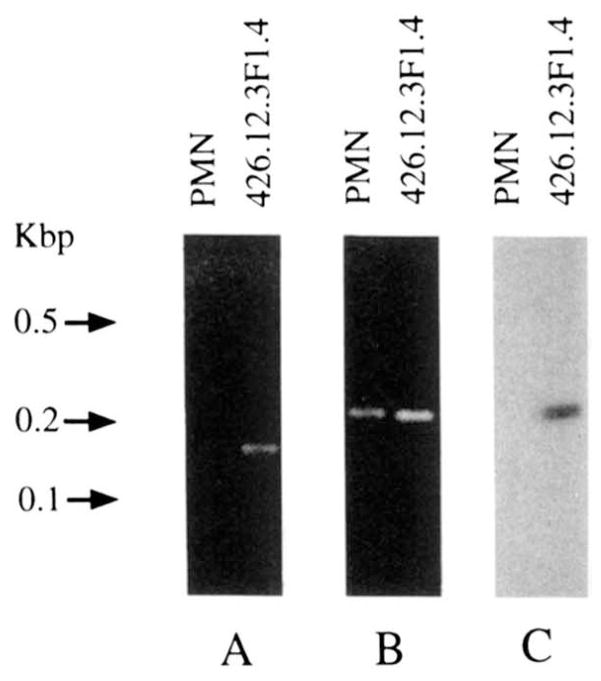
The polyreactive mAb426.12.3F1.4 VH gene sequence displayed 12 differences when compared with that of the germ line VH4.11 gene. To assess whether these nucleotide differences resulted from somatic mutation or were due to the use of a germ line VH gene related to but not identical with VH4.11, we performed the following experiments. We synthesized the sense 58-5 oligonucleotide primer encompassing a FR1 sequence shared by the VH4.11 and mAb426.12.3F1.4 VH genes (Fig. 2A). We also synthesized the antisense 12.3-3 primer, the sequence of which was identical with the reverse complement of a 5prime;-portion of the FR3 region of mAb 426.12-3F1.4 VH sequence, but differed in four nucleotides from the reverse complement of the corresponding VH4.11 sequence. We chose this FR3 area because it was the only one throughout the mAb426.12.3F1.4 VH gene that contained at least four nucleotide differences when compared with the VH4.11 gene sequence. The theoretically preferable VH CDR2 area contained only two nucleotide differences (Fig. 2A). Somatic mutations have been found to commonly occur in the FR3 5′-area concomitant with mutations in the CDR2, as suggested by the analysis of mAb generated through the response against various Ag in mice (29–34) and humans (21, 35–38). In addition, a relative lack of conservation in this FR3 sequence among VHIV gene family members has been shown (22, 39–41), suggesting that the use of the 12.3-3 primer would have preferentially targeted those VHIV genes different in this area from VH4.11, thus maximizing the discrimination power of our analysis. The PCR involving the 58-5 and 12.3-3 primers resulted in amplification of DNA from the mAb426.12.3F1.4–producing B cell hybridoma DNA, but not from autologous PMN DNA. The molecular size of the amplified product (~160 bp) was consistent with that of the sequence spanning residues 65 to 226 (161 bp) of the expressed VH gene (Fig. 4A). The sequence of this DNA fragment was identical with the corresponding sequencing of the mAb426.12.3F1.4 VH gene (Fig. 2A, 58-5/12.3-3 sequence). This suggested that the expressed VH gene was somatically mutated. This was confirmed by the results of the subsequent experiments. We synthesized the antisense 58-3 oligonucleotide primer, the sequence of which was identical with the reverse complement of a VH4.11 FR3 sequence and differed in two bases from the reverse complement of the corresponding mAb426.12.3F1.4 VH gene sequence. PCR involving the 58-5 and 58-3 primer pair yielded products of the expected and identical size (~200 bp) when applied to the amplification of not only B cell hybridoma, but also autologous PMN DNA (Fig. 4B). This demonstrated that the failure to amplify any DNA from autologous PMN DNA using the 12.3-3 primer was not due to flaws inherent to the DNA preparation and provided the material necessary for differential hybridization with the 12.3-3 oligonucleotide as a probe. The 12.3-3 nucleotide hybridized with DNA amplified from B cell hybridoma but not with that amplified from PMN (Fig. 4C). The DNA fragment PCR amplified from the PMN DNA was used to identify, by cloning and sequencing, the putative germ line gene that gave rise to the expressed mAb426.12.3F1.4 VH gene. Five independent clones were analyzed and found to contain an identical VH gene sequence. This (G411) was identical with the sequence of the VH4.11 gene throughout the overlapping area (169 bp). These experiments proved that the expressed mAb 426.12.3F1.4 VH gene was somatically mutated and strongly suggested that VH4.11 was the germ line gene that gave rise to it.
FIGURE 4.
PCR and Southern blot analysis of somatic mutations in the mAb 426.12.3F1.4 VH gene. A, ethidium bromide staining of DNA PCR amplified using the 58-5 and 12.3-3 primers and genomic DNA extracted from autologous PMN (left lane) or the mAb426.12.3F1.4–producing B cell hybridoma (right lane). B, ethidium bromide staining of the DNA PCR amplified using the 58-5 and 58-3 primers and genomic DNA extracted from autologous PMN (left lane) or the mAb426.12.3F1.4–producing B cell hybridoma (right lane). C, Southern blot hybridization of the PCR-amplified products shown in B with the 32P-labeled 12.3-3 oligonucleotide probe.
Somatic point mutations in the polyreactive IgG mAb10 VH segment
The polyreactive mAb10 VH gene sequence displayed 19 differences when compared with that of the germ line VH4.18 gene. To assess whether these nucleotide differences resulted from somatic mutation or were due to the use of a germ line VH gene related to but not identical with VH4.18, we performed the following experiments. We synthesized the sense VH4.18/V2-1 FR1-CDR1 oligonucleotide primer encompassing a FR1-CDR1 sequence of the germ line VH4.18 gene, which differed by four residues from that of the corresponding area of the mAb10 VH gene (Fig. 2A). We used this primer in conjunction with the antisense HBL-3 FR3 primer, the sequence of which was the reverse complement of a FR3 sequence shared by the germ line VH4.18 gene and the mAb10 VH gene, except for a G instead of an A at position 270. The PCR involving this pair of primers yielded a DNA amplification product when using DNA from autologous PMN but not when using DNA from the mAb10–producing B cell hybridoma. The molecular size of the amplified DNA (~ 195 bp) was consistent with that of the sequence spanning residues 80 to 275 of the VH4.18 gene (Fig. 5A). In addition, the sequence (from five independent clones) of the amplified DNA fragment was identical with that of residues 80 through 275 of the germ line VH4.18 gene, except for the G of the HBL-3 primer (Fig. 2A, G418 sequence). The results of the above experiments were consistent with the hypothesis that the mAb10 VH segment was somatically mutated. To demonstrate that the failure to amplify DNA from the mAb10-producing B cell hybridoma DNA when using the VH4.18/V2-1 FR1 primer was not due to flaws inherent to the genomic DNA preparation, and to provide a “positive” finding in favor of mutations in the mAb10 VH segment, we synthesized the sense mAb10VHCDR1 oligonucleotide primer encompassing a FR1-CDR1 sequence of the mAb10 VH segment that differed from that of the VH4.18/V2-1 FR1-CDR1 oligonucleotide by four bases (Fig. 2A). The PCR involving the mAb10VHCDR1 primer in conjunction with the antisense HBL-3 FR3 primer yielded an amplification DNA product using DNA from the mAb10-producing B cell hybridoma but not DNA from autologous PMN. The molecular size of the amplified fragment (~195 bp) was consistent with that of the sequence spanning residues 80 to 275 of the mAb10 VH gene (Fig. 5B). In addition, the sequence (from five independent clones) of the amplified fragment was identical with that of residues 80 through 275 of the mAb10 VH gene, except for the G of the HBL-3 primer (Fig. 2A, mAb10VHCDR1/FR3 sequence). These experiments showed that the expressed mAb10 VH gene was somatically mutated and strongly suggested that VH4.18 was the germ line gene that gave rise to it. The somatically mutated status of the mAb10 VH segment was further emphasized by the two mutations, a T replacing an I and a T replacing a S at the seventh and fourth but last residues, respectively, in the mAb10 JH4b FR4 (Fig. 3).
FIGURE 5.
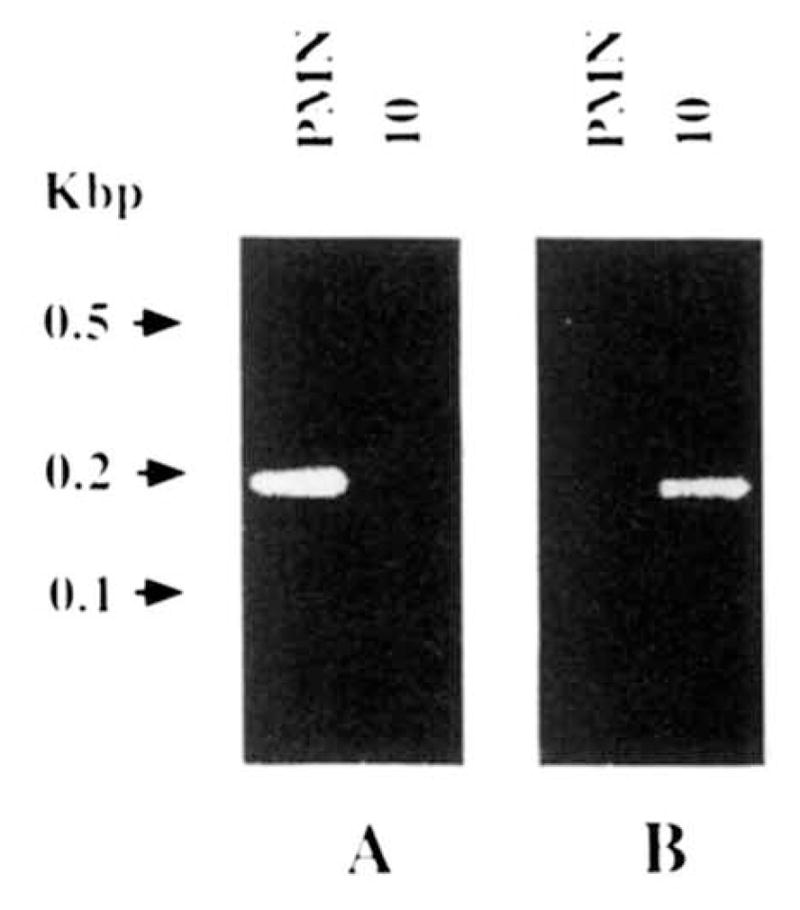
PCR analyses of somatic mutations in the mAb10 VH gene. A, ethidium bromide staining of DNA PCR amplified using the VH4.18/V2–1 FR1-CDR1 and HBL-3 FR3 primers and genomic DNA extracted from autologous PMN (left lane) or the mAb10-producing B cell hybridoma (right lane). B, ethidium bromide staining of the DNA PCR amplified using the mAb10VH CDR1 and HBL-3 primers and genomic DNA extracted from autologous PMN (left lane) or the mAb10-producing B cell hybridoma (right lane).
Discussion
Polyreactive natural antibodies are primarily IgM and are thought to rely on the use of V regions in germ line configuration for the recognition of multiple Ag. To test the hypothesis that polyreactivity may be also a function of somatically mutated antibodies, we generated five human polyreactive IgG mAb and sequenced the genes encoding their VH segments. The choice of the IgG class was dictated by the hypothesis that if polyreactive somatically mutated antibodies existed, these would be more numerous among IgG than IgM. Thus, selection for IgG would increase the probability of generating polyreactive mAb with mutated V regions. The results of our experiments showed that: 1) the VH genes used by the five polyreactive IgG mAb were identical with or closely related to those used by natural polyreactive IgM antibodies, as well as high affinity specific autoantibodies and antibodies induced by foreign Ag; 2) the Ag binding features and the VH-D-JH sequences were distinctive in each polyreactive IgG mAb; and 3) the VH segments of three IgG mAb contained somatic mutations, which displayed a distribution and a nature similar to those found in Ig V regions that underwent a process of Ag-dependent selection.
Findings in mice and humans have suggested that the Ag binding properties of individual polyreactive IgM mAb are distinctive (4–12, 14). Like polyreactive IgM, the present polyreactive IgG mAb display discrete Ag binding activities for different self and foreign Ag. The functional “uniqueness” of each polyreactive IgG probably reflects the structural heterogeneity of the Ag binding sites of these mAb. Previous observations have shown that while polyreactive, mainly IgM, human antibodies can use various arrays of different VH and VL segments, as well as different D, JH, or JL segments (5, 13, 15–17, 21, 37, 38, 42–44), they may display some preference for the use of the VHIV family genes (15, 17, 44a). Accordingly, four of the five polyreactive IgG mAb used three different VHIV genes, the sequences of which are similar to those of the germ line VH4. 11, VH4.18, and VH4.21 genes. These VH genes, however, along with the hv1263 (VHI) gene used by the fifth polyreactive IgG mAb are used also by monoreactive high affinity mAb to encode reactivities for different self and foreign Ag (5, 17, 21, 37, 38, 43, 44). For example, germ line and variously mutated forms of the VH4.21 gene are used by various polyreactive and monoreactive antibodies and autoantibodies (44); variously mutated forms of the VH4.18 gene are used by the monoreactive RF IgM mAb 61 (17) and the H3 IgG1 mAb, which neutralizes both HSV-1 and HSV-2 (45). Variously mutated forms of the hv1263 gene are used by monoreactive high affinity IgG RF mAb (46), the IgG1 mAb 57 (21), which neutralizes rabies virus in vivo and in vitro (9), and by the high affinity IgG1 mAb 49 to insulin generated from an IDDM patient (23) (H. Ikematsu et al., manuscript in preparation). Thus, the pool of VH genes used by the present polyreactive IgG autoantibodies overlaps, at least partially, with that of VH genes recruited in high affinity immune responses to foreign Ag in healthy subjects or to self Ag in autoimmune patients (4, 5, 43).
Some unmutated VH segments are inherently capable of binding certain foreign Ag, as exemplified by the frequent selection of the VHOx1 segment in the BALB/c mouse response to 2-phenyl-5-oxazolone (29, 47) and that of the S107 VH segment in the response of the same mouse strain to phosphorylcholine (32) or self Ag, as exemplified by the VH11 segment use of the autoantibodies to DNA in BALB/c mice (48) and the 3H9 VH segment use of DNA-selected autoantibodies in SLE-prone MRL/lpr mice (49). The somatically generated H chain CDR3, however, appears to be the major contributor to the overall function of the Ag binding site, at least in the case of relatively large proteinic Ag (50, 51). The H chain CDR3 structures of two IgG, mAb10 and mAb426.4.2F20, which were generated from different subjects, shared an identical five amino acid stretch, RFLEW, but those of the other three IgG mAb were highly divergent in composition and length and did not allow for the identification of any obvious common motif. Thus, if consistent with what has been suggested by the in vitro expression of a human IgM RF H and L chain genes (52) and by a census of 84 natural polyreactive and Ag-induced monoreactive murine mAb (53), the H chain CDR3 provides the correlate for polyreactivity in our five different IgG mAb, it does so by virtue, in most cases, of discrete primary structures. A thorough evaluation, however, of the contribution of the different H chain regions to the overall structure and activity of a polyreactive IgG Ag binding site must take into account also the configuration of the VL segment.
This study is one of the first evidences that the VH regions of polyreactive antibodies can be somatically mutated. The relatively high degree of allelic polymorphism of members of certain human VH multigene families and the presence of different but highly homologous members in a single haplotype can make it difficult to assess the somatically mutated status of expressed VH genes in the outbred human population by mere inspection of the already reported closest germ line genes. Some individual Ig VH genes, however, including VH4.18 and VH4.21 (VHIV family) and hv1263 (VHI family) are highly conserved among humans (22, 24, 40, 43, 54). The data obtained from differential PCR amplification, Southern hybridization, and genomic DNA sequencing confirmed that the mAb426.12.3F1.4 and mAb10 VH segments contained more than eight and seven somatic point mutations, respectively. The patient B (Table I) source of the germ line G411 gene also provided the B cells used for the generation of the IgG mAb426.4.2F20, which probably used the same VH4.11 or VH4.11-like gene used by the mAb426.12.3F1.4. Thus, at least two of the three nucleotide differences (from the germ line G411 gene) found in the polyreactive IgG mAb426.4.2F20 VH gene also represented somatic point mutations (Fig. 2A and B). Finally, the somatically mutated status of the IgG mAb410.30F305 VH segment was suggested not only by the high degree of conservation of the hv1263 gene (24, 43), but also by the fact that none of the six amino acid differences displayed by the mAb410. 30F305 VH gene deduced amino acid sequence (compared with that of the germ line hv1263 gene) are shared by the 51P1 sequence, possibly the closest gene to hv1263 and by the fact that only one (residue 33) of these six amino acid differences is shared by any of the reported expressed hv1263 segment sequences, including that of the mAb57 (21).
Thus, the present experiments suggested that although some polyreactive IgG antibodies can be virtually unmutated, possibly reflecting the germ line configuration of the IgM-producing clones from which they arose, other polyreactive IgG antibodies bear the imprints characteristic of a selective antigenic pressure. They, however, could provide little clue to the natural history of the polyreactive somatically mutated IgG-producing cell clones. High R:S mutation ratios in the Ig V segment CDR and low R:S ratios in the FR are features characteristic of the clonotypes that have been recruited and positively selected during the maturation of an antibody response to a foreign Ag or to a self Ag in the course of a chronic autoimmune disease (37, 38, 46, 47, 55–60). At different stages of such antibody and autoantibody responses, similar features, however, are also displayed by relatively large numbers of clones with a reduced affinity for the selecting Ag (47, 61, 62). These clones are possibly negatively selected (by Ag), do not contribute to affinity maturation, and are part of the tremendous (clonotypic) “wastage” accompanying any Ag-directed affinity maturation process (47, 61, 62). Other clones with reduced affinity may emerge as a result of a positive selection by stimuli, other than those provided by the Ag driving the overall response, such as an anti-idiotypic interaction. Thus, the predominant distribution of the mutations in the three polyreactive IgG mAb410.30F305, mAb10, and mAb426.12.3F1.4 VH segment CDR and high R:S ratio (11.0 in CDR vs 0.8 in FR) suggest that these antibodies underwent a process of Ag-directed selection. They, however, do not allow for any inferences as to whether the clones producing these IgG participated in vivo to an affinity maturation to any of the Ag tested or to any other undetermined Ag. In this respect, the relatively high affinity for ssDNA displayed by the three “highly substituted” IgG mAb possibly reflects the strong ssDNA binding activity in general displayed by natural IgM, IgA, and IgG polyreactive autoantibodies (5–9, 63), including the remaining two “minimally” substituted IgG mAb (mAb426.4.2F20 and mAb410.7.F91), rather than suggesting that DNA was the in vivo selecting Ag.
Somatic mutations could accumulate in the V segments of an originally polyreactive antibody without significantly affecting its ability to bind multiple Ag. An alternative but not mutually exclusive possibility, is that somatic mutations accumulated in a inherently monoreactive “specific” antibody and conferred it the ability to bind multiple Ag. Although we cannot formally rule out this possibility, we believe the first to be far more likely. To the best of our knowledge, accumulation of somatic mutations has never been reported to result in acquisition of polyreactivity by antibodies undergoing an Ag-directed selection in the course of various murine responses to foreign or self Ag (47, 55–60). Rather, polyreactivity has been shown to be a function, in general, of unmutated IgM VH segments. It has been speculated that application of antigenic pressure to polyreactive natural antibodies in unmutated configuration can result in the accumulation of somatic point mutations, somatic selection, loss of polyreactivity, and, possibly, class switch (52). In humans, a strong evidence for somatic hypermutation of clonally related VH-D-JH sequences expressed with IgM, IgG, and IgA in the spleen has been recently provided (64). The present findings are consistent with the hypothesis that application of antigenic pressure to germ line natural IgM autoantibodies can lead to the accumulation of somatic point mutations and class switch to IgG without significantly affecting the antibody ability to bind multiple Ag. A similar mechanism has been suggested to underlie the generation of some IgG clones emerging, after the second injection with Ag, during the affinity maturation of the response to p-azophenyl arsonate (Ars) from (originally germ line) IdCR+ (also called CRI-A) polyreactive natural IgM autoantibody-producing cells, as shown by Naparstek et al. (65; see clones hVH65-219 and 31–62 in group III of Table I). It also has been called into question to account for the polyreactivity of the human IgM and IgG mAb specific for blood group Ag, emerging in vivo after alloimmunization (66).
These and our present findings would be consistent with our preliminary data suggesting that in a polyreactive antibody different, although partially overlapping, V segment structures can mediate the binding to different Ag (Ichiyoshi et al., manuscript in preparation).
Acknowledgments
We are grateful to Dr. John Hill for help with and continuous improvement of the N.Y.U. School of Medicine central VAX service and nucleic acid sequence data bank. We thank the Lilly Research Laboratories (Indianapolis, IN) for providing us with recombinant human insulin. We also thank Drs. Minoru Nakamura and Megumi Hiida and Thomas E. Steger for their collaboration in some of these experiments.
Footnotes
This work was supported by U. S. Public Health Service Grant AR-40908. This is publication 19 from The Jeanette Greenspan Laboratory for Cancer Research.
Abbreviations used in this paper: IDDM, insulin-dependent diabetes mellitus; CDR, complementary determining region; FR, framework region; Kd, dissociation constant; PMN, polymorphonuclear cells; PCR, polymerase chain reaction; R, replacement (mutation); RF, rheumatoid factor; S, silent (mutation).
References
- 1.Boyden S. Natural antibodies and the immune response. Adv Immunol. 1965;5:1. doi: 10.1016/s0065-2776(08)60271-0. [DOI] [PubMed] [Google Scholar]
- 2.Michael JG. Natural antibodies. Curr Top Microbiol Immunol. 1969;48:43. doi: 10.1007/978-3-642-46163-7_3. [DOI] [PubMed] [Google Scholar]
- 3.Dighiero G, Lymberi P, Guilbert B, Ternynch T, Avrameas S. Natural autoantibodies constitute a substantial part of normal circulating immunoglobulins. Ann NY Acad Sci. 1986;475:135. doi: 10.1111/j.1749-6632.1986.tb20863.x. [DOI] [PubMed] [Google Scholar]
- 4.Avrameas S. Natural autoantibodies: from “horror autotoxicus” to “gnothi seuton”. Immunol Today. 1991;12:154. doi: 10.1016/S0167-5699(05)80045-3. [DOI] [PubMed] [Google Scholar]
- 5.Riboldi P, Kasaian MT, Mantovani L, Ikematsu H, Casali P. Natural antibodies. In: Bona CA, Siminovitch K, Zanetti M, Theofilopoulos AN, editors. The Molecular Pathology of Autoimmune Diseases. Gordon & Breach Science Publishers; Philadelphia: 1993. pp. 45–65. [Google Scholar]
- 6.Casali P, Kasaian MT, Haughton G. B-1 (CD5 B) cells. In: Coutinho A, Kazatchkine MD, editors. Autoimmunity. John Wiley & Sons, Inc; New York: 1993. In press. [Google Scholar]
- 7.Nakamura M, Burastero SE, Notkins AL, Casali P. Human monoclonal rheumatoid factor-like antibodies from CD5 (Leu-1)+ B cells are polyreactive. J Immunol. 1988;140:4180. [PubMed] [Google Scholar]
- 8.Nakamura M, Burastero SE, Ueki Y, Larrick JW, Notkins AL, Casali P. Probing the normal and autoimmune B cell repertoire with Epstein-Barr Virus: frequency of B cells producing monoreactive high affinity autoantibodies in patients with Hashimoto’s disease and systemic lupus erythematosus. J Immunol. 1988;141:4165. [PubMed] [Google Scholar]
- 9.Ueki Y, I, Goldfarb S, Harindranath N, Gore M, Koprowski H, Notkins AL, Casali P. Clonal analysis of a human antibody response: quantitation of precursors of antibody-producing cells and generation and characterization of monoclonal IgM, IgG, and IgA to rabies virus. J Exp Med. 1990;171:19. doi: 10.1084/jem.171.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasaian MT, Ikematsu H, Casali P. Identification and analysis of a novel human CD5− B lymphocyte subset producing natural antibodies. J Immunol. 1992;148:2690. [PMC free article] [PubMed] [Google Scholar]
- 11.Casali P, Burastero SE, Balow JE, Notkins AL. High affinity autoantibodies to ssDNA are produced by CD5− B cells in Systemic Lupus Erythematosus patients. J Immunol. 1989;143:3476. [PubMed] [Google Scholar]
- 12.Casali P. Immunoglobulin M. In: Roitt IM, Delves PJ, editors. Encyclopaedia of Immunology. II. Academic Press Ltd; London: 1992. pp. 743–747. [Google Scholar]
- 13.Chen PP, Liu MF, Sinha S, Carson DA. A 16/6 idiotype-positive anti-DNA antibody is encoded by a conserved VH gene with no somatic mutation. Arthritis Rheum. 1988;31:1429. doi: 10.1002/art.1780311113. [DOI] [PubMed] [Google Scholar]
- 14.Baccala R, Quang TV, Gilbert M, Ternynck T, Avrameas S. Two murine natural polyreactive autoantibodies are encoded by nonmutated germ-line genes. Proc Natl Acad Sci USA. 1989;86:4624. doi: 10.1073/pnas.86.12.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanz I, Casali P, Thomas JW, Notkins AL, Capra JD. Nucleotide sequences of eight human natural autoantibodies VH region reveal apparent restricted use of VH families. J Immunol. 1989;142:4054. [PubMed] [Google Scholar]
- 16.Siminovitch KA, Misener V, Kwong PC, Song Q-L, Chen PP. A natural autoantibody is encoded by germline heavy and lambda light chain variable region genes without somatic mutation. J Clin Invest. 1989;84:1675. doi: 10.1172/JCI114347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harindranath N, I, Goldfarb S, Ikematsu H, Burastero SE, Wilder RL, Notkins AL, Casali P. Complete sequence of the genes encoding the VH and VL regions of low-and high-affinity monoclonal IgM and IgA1 rheumatoid factors produced by CD5+ B cells from a rheumatoid arthritis patient. Int Immunol. 1991;3:865. doi: 10.1093/intimm/3.9.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McHeyzer-Williams MG, Nossal GJV. Clonal analysis of autoantibody-producing cell precursors in the pre-immune B cell repertoire. J Immunol. 1988;141:4118. [PubMed] [Google Scholar]
- 19.Larrick JW, Chiang YL, Sheng-Dong R, Senyk G, Casali P. Generation of specific monoclonal antibodies by in vitro expansion of human B cells. A novel recombinant DNA approach. In: Borrebaek CAK, editor. In Vitro Immunization in Hybridoma Technology. Elsevier Science Publishers B. V; Amsterdam: 1988. pp. 231–246. [Google Scholar]
- 20.Ikematsu H, I, Goldfarb S, Harindranath N, Kassaian MT, Casali P. Generation of human monoclonal antibody-producing cell lines by Epstein-Barr virus (EBV)-transformation and somatic cell hybridization techniques. J Tissue Cult Methods. 1992;14:9. [Google Scholar]
- 21.Ikematsu H, Harindranath N, Ueki Y, Notkins AL, Casali P. Clonal analysis of a human antibody response. II. Sequences of the VH genes of human IgM, IgG, and IgA to rabies virus reveal preferential utilization of VHIII segments and somatic hypermutation. J Immunol. 1993;150:1325. [PMC free article] [PubMed] [Google Scholar]
- 22.Sanz I, Kelly P, Williams C, Scholl S, Tucker PW, Capra JD. The smaller human VH gene families display remarkably little polymorphism. EMBO J. 1989;8:3741. doi: 10.1002/j.1460-2075.1989.tb08550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casali P, Nakamura M, Ginsberg-Fellner F, Notkins AL. Frequency of B cells committed to the production of antibodies to insulin in newly diagnosed patients with insulin-dependent diabetes mellitus and generation of high affinity monoclonal IgG to insulin. J Immunol. 1990;144:3741. [PubMed] [Google Scholar]
- 24.Chen PP, Liu MF, Glass CA, Sinha S, Kipps TJ, Carson DA. Characterization of two immunoglobulin VH genes that are homologous to human rheumatoid factors. Arthritis Rheum. 1989;32:72. doi: 10.1002/anr.1780320112. [DOI] [PubMed] [Google Scholar]
- 25.Ichihara Y, Matsuoka H, Kurosawa Y. Organization of human immunoglobulin heavy chain diversity gene loci. EMBO J. 1988;7:4141. doi: 10.1002/j.1460-2075.1988.tb03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravetch JV, Siebenlist U, Korsmeyer S, Waldmann T, Leder P. Structure of the human immunoglobulin m locus: characterization of embryonic and rearranged J and D genes. Cell. 1981;27:583. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- 27.Yamada M, Wasserman R, Richard BA, Shane S, Caton AJ, Rovera G. Preferential utilization of specific immunoglobulin heavy chain diversity and joining segments in adult human peripheral blood B lymphocytes. J Exp Med. 1991;173:395. doi: 10.1084/jem.173.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequence of Proteins of Immunological Interest. 5. U. S. Department of Health and Human Services; Washington D.C: 1991. [Google Scholar]
- 29.Griffiths GM, Berek C, Kaartinen M, Milstein C. Somatic mutation and the maturation of immune response to 2-phenyl oxazolone. Nature. 1984;312:271. doi: 10.1038/312271a0. [DOI] [PubMed] [Google Scholar]
- 30.McKean D, Huppi K, Bell M, Staudt L, Gerhard W, Weigert MG. Generation of antibody diversity in the immune response of Balb/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci USA. 1984;81:3180. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke SH, Huppi K, Ruezinsky D, Staudt L, Gerhard W, Weigert M. Intra- and intraclonal diversity in the antibody response to influenza hemagglutinin. J Exp Med. 1985;161:687. doi: 10.1084/jem.161.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy NS, Malipiero UV, Lebecque SG, Gearhart PJ. Early onset of somatic mutation in immunoglobulin VH genes during the primary immune response. J Exp Med. 1989;169:2007. doi: 10.1084/jem.169.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manser T. Evolution of antibody structure during the immune response. J Exp Med. 1989;170:1211. doi: 10.1084/jem.170.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss U, Rajewsky K. The repertoire of somatic mutants accumulating in the memory compartment after primary immunization is restricted through affinity maturation and mirrors that expressed in the secondary response. J Exp Med. 1990;172:1681. doi: 10.1084/jem.172.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adderson EE, Shackelford PG, Quinn A, Caroll WL. Restricted Ig H chain V gene usage in the human antibody response to Haemophilus influenzae type b capsular polysaccharide. J Immunol. 1991;147:1667. [PubMed] [Google Scholar]
- 36.Andris JS, Johnson S, Zolla-Paztner S, Capra JD. Molecular characterization of five human anti-human immunodeficiency virus type 1 antibody heavy chains reveals extensive somatic mutation typical of an antigen-driven immune response. Proc Natl Acad Sci USA. 1991;88:7783. doi: 10.1073/pnas.88.17.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olee T, Liu EW, Huang DF, Soto-Gil RW, Deftos M, Kozin F, Carson DA, Chen PP. Genetic analysis of self-associating immunoglobulin G rheumatoid factors from two rheumatoid synovia implicates an antigen driven response. J Exp Med. 1992;172:831. doi: 10.1084/jem.175.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani L, Wilder RL, Casali P. Human rheumatoid B-1a (CD5+ B) cells make somatically hyper-mutated high affinity IgM rheumatoid factors. J Immunol. 1993:151. In press. [PMC free article] [PubMed] [Google Scholar]
- 39.Lee K, Matsuda F, Kinashi T, Kodaira M, Honjo T. A novel family of variable region genes of the human immunoglobulin heavy chain. J Mol Biol. 1987;195:761. doi: 10.1016/0022-2836(87)90482-7. [DOI] [PubMed] [Google Scholar]
- 40.van Es JH, Heutink M, Aanstoot H, Logtenberg T. Sequence analysis of members of the human Ig VH4 gene family derived from a single VH locus. J Immunol. 1992;149:492. [PubMed] [Google Scholar]
- 41.Weng N, Snyder JG, Lee LY-, Marcus DM. Polymorphism of human immunoglobulin VH4 germ-line genes. Eur J Immunol. 1992;22:1075. doi: 10.1002/eji.1830220430. [DOI] [PubMed] [Google Scholar]
- 42.Hoch S, Schwaber J. Identification and sequence of the VH gene elements encoding a human anti-DNA antibody. J Immunol. 1987;139:1689. [PubMed] [Google Scholar]
- 43.Pascual V, Capra JD. Human immunoglobulin heavy-chain variable region genes: organization, polymorphism, and expression. Adv Immunol. 1991;49:1. doi: 10.1016/s0065-2776(08)60774-9. [DOI] [PubMed] [Google Scholar]
- 44.Pascual V, Capra JD. VH4.21, a human VH gene segment overpresented in the autoimmune repertoire. Arthritis Rheum. 1992;35:11. doi: 10.1002/art.1780350103. [DOI] [PubMed] [Google Scholar]
- 44.Chai S, Kasaian MT, Ikematsu H, Casali P. Sequences of the VH-D-JH genes expressed by human B-1a, B-1b and B-2 cells: lack of N segment additions in the VH genes of natural antibodies produced by B-1b cells. J Immunol. 1993;150:253A. [Google Scholar]
- 45.Huang DF, Olee T, Masuhiko Y, Matsumoto Y, Carson DA, Chen PP. Sequence analyses of three immunoglobulin G anti-virus antibodies reveal their utilization of autoantibody-related immunoglobulin VH genes, but not Vλ genes. J Clin Invest. 1992;90:2197. doi: 10.1172/JCI116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pascual V, Randed I, Thompson K, Sioud M, Forre O, Natvig J, Capra JD. The complete nucleotide sequence of the heavy chain variable regions of six monospecific rheumatoid factors derived from Epstein-Barr virus-transformed B cells isolated from synovial tissue of patients with rheumatoid arthritis. J Clin Invest. 1990;86:1320. doi: 10.1172/JCI114841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berek C, Milstein C. Mutation drift and repertoire shift in the maturation of the immune response. Immunol Rev. 1987;96:23. doi: 10.1111/j.1600-065x.1987.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 48.Shefner R, Kleiner G, Turken A, Papazian L, Diamond B. A novel class of anti-DNA antibodies identified in Balb/c mice. J Exp Med. 1991;173:287. doi: 10.1084/jem.173.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radic MZ, Mascelli MA, Erikson J, Shan H, Weigert MG. Ig H and L chain contribution to autoimmune specificities. J Immunol. 1991;146:176. [PubMed] [Google Scholar]
- 50.Amit AG, Mariuzza RA, Phillips SE, Poliak RJ. Three-dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science. 1986;233:747. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- 51.Stanfield RL, Fieser TM, Lerner RA, Wilson IA. Crystal structure of an antibody to a peptide and its complex with peptide antigen at 2.8 A. Science. 1990;248:712. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- 52.Martin T, Duffy SF, Carson DA, Kipps TJ. Evidence for somatic selection of natural antibodies. J Exp Med. 1992;175:983. doi: 10.1084/jem.175.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C, Stenzel-Poore MP, Rittenberg MB. Natural and polyreactive antibodies differing from antigen-induced antibodies in the H chain CDR3. J Immunol. 1991;147:2359. [PubMed] [Google Scholar]
- 54.van der Maarel S, van Dijk KW, Alexander CM, Sasso EH, Bull A, Milner EC. Chromosomal organization of the human VH4 gene family. J Immunol. 1993;150:2858. [PubMed] [Google Scholar]
- 55.Weigert GM. The influence of somatic mutation on the immune response. In: Cinader B, Miller RG, editors. Progress in Immunology VI. Academic Press, Inc; New York: 1986. pp. 138–144. [Google Scholar]
- 56.Kocks C, Rajewsky K. Stable expression and somatic hypermutation of antibody V regions in B-cell developmental pathways. Ann Rev Immunol. 1989;7:537. doi: 10.1146/annurev.iy.07.040189.002541. [DOI] [PubMed] [Google Scholar]
- 57.French DL, Laskov R, Scharff MD. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989;244:1152. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- 58.Shlomchik MJ, Marshak-Rothstein A, Wolfowicz CB, Rothstein TL, Weigert MG. The role of clonal selection and somatic mutation in autoimmunity. Nature. 1987;328:805. doi: 10.1038/328805a0. [DOI] [PubMed] [Google Scholar]
- 59.Shlomchik M, Mascelli M, Shan H, Radic MZ, Pisetsky D, Marshak-Rothstein A, Weigert MG. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171:265. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Es JH, Aanstoot H, Gmelig-Meyling FHJ, Derksen RHWM, Logtenberg T. A human systemic lupus erythematosus-related anti-cardiolipin/single-strand DNA autoantibody is encoded by somatically mutated variant of the developmentally restricted 51P1 VH gene. J Immunol. 1992;149:2234. [PubMed] [Google Scholar]
- 61.Manser T. Evolution of antibody structure during the immune response: the differentiative potential of a single B lymphocyte. J Exp Med. 1989;170:1211–1230. doi: 10.1084/jem.170.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C, V, Robert A, Rittenberg MB. Generation and analysis of random point mutations in an antibody CDR2 sequence: many mutated antibodies lose their ability to bind antigen. J Exp Med. 1992;176:855. doi: 10.1084/jem.176.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zouali M, Stollar BD, Schwartz RS. Origin and diversification of anti-DNA antibodies. Immunol Rev. 1988;105:137. doi: 10.1111/j.1600-065x.1988.tb00770.x. [DOI] [PubMed] [Google Scholar]
- 64.Varade WS, Insel RA. Isolation of germinal centerlike events form human spleen RNA. Somatic hypermutation of a clonally related VH6DJH rearrangement expressed with IgM, IgG, and IgA. J Clin Invest. 1993;91:1838. doi: 10.1172/JCI116397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naparstek Y, Andre-Schwartz J, Manser T, Wysocki LJ, Breitman L, Stollar D, Gefter M, Schwartz RS. A single germline VH gene segment of normal A/J mice encodes autoantibodies characteristic of systemic lupus erythematosus. J Exp Med. 1986;164:614. doi: 10.1084/jem.164.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson KM, Sutherland J, Barden G, Melamed MD, Wright MG, Bailey S, Thorpe SJ. Human monoclonal antibodies specific for blood group antigens demonstrate multispecific properties characteristic of natural antibodies. Immunology. 1992;76:146–157. [PMC free article] [PubMed] [Google Scholar]