SUMMARY
Hedgehog (Hh) signaling plays essential roles in animal development and tissue homeostasis, and its misregulation causes congenital diseases and cancers. Regulation of the ubiquitin/proteasome-mediated proteolysis of Ci/Gli transcription factors is central to Hh signaling, but whether deubiquitinase is involved in this process remains unknown. Here, we show that Hh stimulates the binding of an ubiquitin-specific protease Usp7 to Ci, which positively regulates Hh signaling activity through inhibiting Ci ubiquitination and degradation mediated by both Slimb-Cul1 and Hib-Cul3 E3 ligases. Furthermore, we find that Usp7 forms a complex with GMP-synthetase (GMPS) to promote Hh pathway activity. Finally, we show that the mammalian counterpart of Usp7, HAUSP, positively regulates Hh signaling by modulating Gli ubiquitination and stability. Our findings reveal a conserved mechanism by which Ci/Gli is stabilized by a deubiquitination enzyme and identify Usp7/HUASP as a critical regulator of Hh signaling and potential therapeutic target for Hh-related cancers.
INTRODUCTION
The Hh pathway plays key roles in controlling embryonic development and adult tissue homeostasis (Briscoe and Therond, 2013; Ingham and McMahon, 2001; Jiang and Hui, 2008). Deregulation of Hh pathway activity has been implicated in numerous human diseases including birth defects and cancers (Jiang and Hui, 2008; Pasca di Magliano and Hebrok, 2003). The core components and regulatory systems of the Hh pathway are conserved from invertebrate to human with few exceptions (Jiang and Hui, 2008; Wilson and Chuang, 2010). In Drosophila wing discs, Hh protein acts as a morphogen, which is produced by posterior (P) compartment cells and move into anterior (A) compartment to form a concentration gradient. In A compartment cells near the A/P boundary, secreted Hh protein binds the 12-span transmembrane receptor Patched (Ptc) to relieve an inhibitory effect of Ptc on the GPCR family protein Smoothened (Smo), leading to the activation of the transcription factor Cubitus interruptus (Ci) and thereby the expression of Hh target genes, such as decapentaplegic (dpp), ptc, knot (kn) and engrailed (en) (Jiang and Hui, 2008).
Many important insights into the regulatory mechanisms of the Hh pathway come from a wealth of studies in Drosophila. For instance, the key transcription effector Ci is regulated by multi-faceted mechanisms, one of which is the ubiquitin-mediated proteolysis. In the absence of Hh, full-length Ci (CiFL) is sequentially phosphorylated by PKA, GSK3 and CK1, which targets Ci ubiquitination mediated by Slimb-Cul1 E3 ligase (Jia et al., 2002; Jia et al., 2005; Jiang and Struhl, 1998; Ou et al., 2002; Smelkinson and Kalderon, 2006). Slimb-Cul1-mediated ubiquitination causes partial degradation of Ci depending on a Ter94 ATPase complex to generate a truncated repressor form of Ci (CiR) that actively inhibits a subset of Hh target genes including dpp and hh itself (Aza-Blanc et al., 1997; Methot and Basler, 1999; Zhang et al., 2013b). In the presence of Hh, Slimb-Cul1-mediated Ci processing is inhibited, at least in part, due to Hh-induced dissociation of Ci-Cos2-kinase complexes and thereby inhibition of Ci phosphorylation (Zhang et al., 2005). In response to high levels of Hh, CiFL is converted into an active but labile form of Ci (CiA) that turns on the expression other Hh target genes, including ptc, kn, en and hib (Kent et al., 2006; Methot and Basler, 1999; Ou et al., 2007; Zhang et al., 2006). As a negative feedback control of the pathway, Hib together with Cul3 forms a Hib-Cul3 E3 ligase complex to ubiquitinate Ci, leading to complete degradation of Ci and termination of Hh pathway activity (Kent et al., 2006; Ou et al., 2007; Zhang et al., 2009; Zhang et al., 2006). Although it is clear that Ci is degraded by dual ubiquitin pathways (Jiang, 2006), the mechanism by which Ci proteolysis is opposed by Hh signal to achieve appropriate pathway activity remains poorly understood.
Ubiquitination is an enzymatic process by which proteins are modified with ubiquitin chains (Hochstrasser, 1995). A major function of ubiquitination is to target proteins for degradation by the proteasome. However, the process of ubiquitination is reversible modification due to the action of deubiquitinases, which remove ubiquitin chains from target proteins (Wilkinson, 2000). The deubiquitinases comprise two major groups: the ubiquitin C-terminal hydrolase (Uch) family and the ubiquitin-specific protease (Usp) family. Usp7 is an evolutionarily conserved protease initially isolated as a partner of the herpesvirus protein (Everett et al., 1997). Several substrates of Usp7 have been identified, including P53 (Li et al., 2002), FOXO4 (van der Horst et al., 2006), PTEN (Song et al., 2008) and H2B (van der Knaap et al., 2005), indicating that Usp7 plays roles in multiple cellular processes. Usp7 often forms a stable heteromeric complex with guanosine 5’-mono-phosphate synthetase (GMPS) (van der Knaap et al., 2005), which strongly stimulates Usp7 deubiquitinating activity (Faesen et al., 2011).
To determine whether deubiquitinase is involved in the regulation of Hh signaling, we systematically screened the Drosophila deubiquitinases by transgenic RNAi and identified Usp7 as a positive regulator of the Hh pathway. We provided evidence that Usp7 binds Ci and recruits GMPS to form a trimetric complex that decreases Ci ubiquitination and increases Ci level. Usp7-mediated deubiquitination can counteract Ci proteolysis conducted by both Slimb-Cul1 and Hib-Cul3 E3 ligases. Furthermore, the mammalian homologue, HAUSP, has a similar role in the regulation of Gli stability and Hh pathway activity.
RESULTS
Loss of usp7 Compromises Hh Signaling in Drosophila
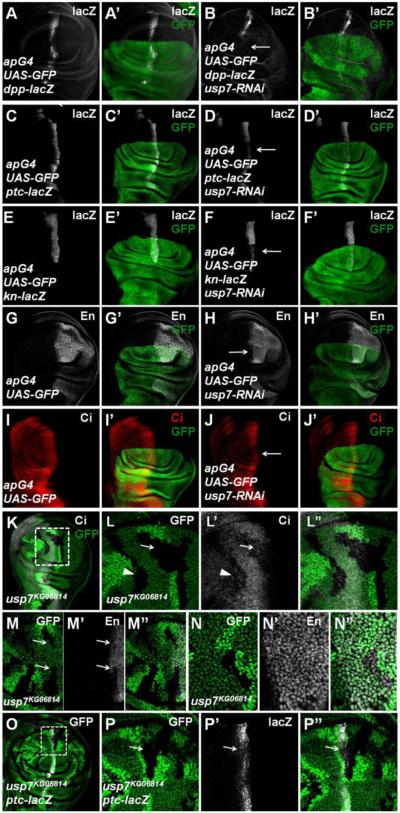
To explore a possible role of deubiquitinase in the regulation of Ci, we performed a RNAi-based genetic screen, in which individual transgenic RNAi lines targeting potential deubiquitinases were expressed in wing discs using the MS1096 gal4 driver, and RNAi wing discs were examined for Hh target gene expression by immunostaining (Table S1; see Method for detail). From this screen, we identified Usp7 as a positive regulator of Hh pathway. As shown in Figures S1A-B, the usp7 transcription, which was visualized by in situ hybridization, was ubiquitous throughout the wing disc. Compared with control discs (Figures 1A-A’, 1C-C’, 1E-E’ and 1G-G’), knockdown of usp7 by expressing usp7-RNAi (v18231) with a dorsal compartment specific gal4 driver ap-Gal4 (apG4>usp7-RNAi) down-regulated the expression of Hh target genes including dpp-lacZ (Figures 1B-B’), ptc-lacZ (Figures 1D-D’), kn-lacZ (Figures 1F-F’) and En (Figures 1H-H’), suggesting that Usp7 is required for the optimal expression of both low and high threshold Hh responsive genes.
Figure 1. Loss of usp7 Represses Hh Signaling and Decreases Ci Protein Level.
All wing imaginal discs shown in this study were oriented with anterior on the left and ventral on the top.
(A-A’) UAS-GFP (green) marks the apG4-mediated gene expression pattern. apG4 drives UAS transgenes to be specifically expressed in the dorsal region of wing discs.
(B-B’) Knockdown of usp7 by apG4 attenuated the expression of dpp-lacZ (arrow).
(C-J’) Knockdown of usp7 with apG4 attenuated the expression of ptc-lacZ (compared D-D’ with C-C’), kn-lacZ (compared F-F’ with E-E’), En (compared H-H’ with G-G’) and Ci (compared J-J’ with I-I’). Arrows indicate the decrease of ptc-lacZ, kn-lacZ, En and Ci.
(K-L”) Low (K) and high (L-L”) magnifications of wing disc carrying usp7KG06814 clones was immunostained to show the expression of GFP (green) and Ci (red). usp7KG06814 clones are recognized by the lack of GFP.
(L-L”) are enlarged views of the region marked by dashed lines in (K).
(M-N”) Wing discs carrying usp7KG06814 clones were immunostained to show the expression of GFP (green) and En (white). The clones in A compartment near A/P boundary showed decrease of En (M-M”, arrows), while the clones in P compartment did not show En decrease (N-N”).
(O-P”) usp7KG06814 clones showed a decrease of ptc-lacZ expression (arrow). (P-P”) are enlarged views of the region marked by dashed lines in (O).
See also Figure S1.
To determine whether Usp7 regulates Hh signaling through modulating Ci levels, we examined CiFL protein distribution using a rat anti-Ci antibody in apG4>usp7-RNAi wing discs. Compared with control discs (Figures 1I-I’), knockdown of usp7 downregulated CiFL levels in A-compartment cells near the A/P boundary (Figures 1J-J’), but did not affect ci transcription as shown by the expression of ci-lacZ, an enhancer trap that mimics ci transcription (Figures S1E-F’), indicating that Usp7 may affect Ci through regulating Ci protein stability. This notion was confirmed by Cycloheximide (CHX) treatment experiments (Figures S1I-K’).
To confirm the results obtained by RNAi, we generated mutant clones for a strong allele of usp7, usp7KG06814 (van der Knaap et al., 2010), using FLP/FRT-mediated mitotic recombination and examined Ci levels and ptc and en expression. usp7KG06814 clones, which were marked by the lack of GFP expression, showed decreased CiFL levels, especially in A-compartment cells near the A/P boundary of the wing discs (Figures 1K-L” and Figures S1L-M”). usp7KG06814 mutant cells near A/P boundary showed a decrease in the level of en expression (indicated by arrows in Figures 1M-M’), which is normally induced by high levels of Hh (Chen et al., 2010), while usp7KG06814 mutation did not affect Hh-independent en expression in the P compartment cells (Figures 1N-N”). Furthermore, the expression of ptc-lacZ, which is induced by intermediate levels of Hh signaling activity, was also attenuated in usp7 mutant cells (Figures 1O-P”).
To determine the specificity of Usp7, we examined whether Usp7 regulates other signaling pathways involved in wing development, including Hippo (Hpo) and Wingless (Wg) pathways. Compared with control disc (Figures S1N-N”), knockdown of usp7 did not affect the expression of Hpo pathway target gene ex-lacZ (Figures S1O-O”). Furthermore, the Wg pathway transcription effector Arm (Figures S1P-P’) and target gene Vg (Figures S1Q-Q”) were not affected in usp7 mutant clones. Collectively, these findings suggest that Usp7 specifically regulates Hh signaling likely by stabilizing Ci.
Usp7 Interacts with Ci
Since Usp7 is a deubiquitinase, it may promote Ci protein stabilization through binding and deubiquitinating Ci. To test this possibility, we co-expressed Myc-tagged CiFL (Myc-Ci) with Flag (Fg)-tagged Usp7 (Fg-Usp7) in S2 cells and carried out co-immunoprecipitation (Co-IP) experiments. The results revealed that Fg-Usp7 could bind Myc-Ci (Figure 2C). In addition, the endogenous Ci could pull down exogenously expressed Fg-Usp7 transfected into Clone8 (Cl8) cells (Figure 2D). By contrast, Fg-Usp7 did not pull down Myc-Arm when coexpressed in S2 cells (Figure S1R), suggesting that Usp7 specifically interacts with Ci.
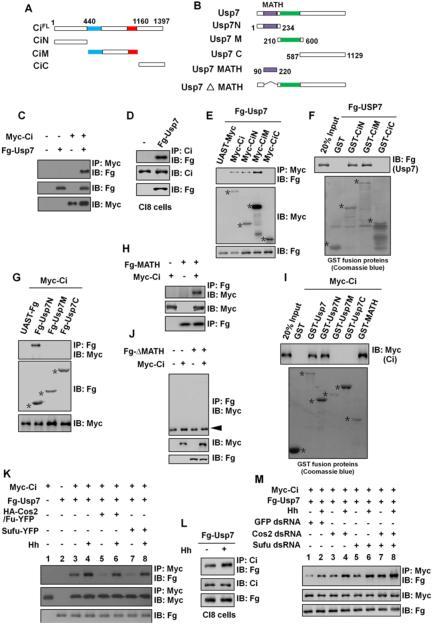
Figure 2. Usp7 binds Ci through Its MATH Domain.
(A and B) Schematic drawings show the domains or motifs in Ci and Usp7 and their truncated fragments used in subsequent Co-IP and pull-down assays. Blue and red bars denote the Zn finger DNA binding and dCBP binding domains of Ci. Purple and green bars represent MATH domain and core catalytic domain of Usp7.
(C) Fg-Usp7 interacted with Myc-Ci in S2 cells.
(D) Fg-Usp7 interacted with endogenous Ci in Clone8 cells.
(E) Fg-Usp7 associated with Myc-tagged N- and M-fragments of Ci. Asterisks indicate expressed Ci deletion mutants.
(F) Extracts from S2 cells expressing Fg-Usp7 were incubated with GST or indicated GST fusion proteins. The bound proteins were analyzed by western blot. Asterisks mark GST fusion proteins.
(G) Usp7 interacted with Ci through its N-region in S2 cells. Asterisks mark expressed Usp7 truncated fragments.
(H) Fg-Usp7-MATH associated with Myc-Ci in S2 cells.
(I) GST pull-down between Myc-tagged Ci and GST or GST-tagged Usp7 fragments. Asterisks indicate expressed GST fusion proteins.
(J) MATH domain-deleted form of Usp7 did not bind Ci in S2 cells. The arrowhead indicates IgG.
(K) Cos2-Fu and Sufu competed Usp7 to bind Ci with or without Hh-conditioned medium treatment. S2 cells were treated with MG132 for 4hrs prior to harvesting the cells.
(L) Hh-conditioned medium treatment promoted endogenous Ci binding to Fg-Usp7 in Clone8 cells.
(M) Knockdown of cos2 or/and sufu promoted Ci binding to Usp7 in absence or in presence of Hh-conditioned medium treatment. S2 cells were treated with MG132 for 4hrs prior to harvesting the cells.
See also Figure S2 and S3.
To map the Usp7-interacting domain(s) in Ci, various Myc-Ci truncated mutants (Figure 2A) were transfected into S2 cells with Fg-Usp7, followed by Co-IP assays. As shown in Figure 2E, both N- and M- region of Ci (Myc-CiN and Myc-CiM) pulled down Fg-Usp7, whereas the C-terminal Ci fragment (Myc-CiC) did not. We confirmed these results through GST pull-down assay (Figure 2F).
Previous studies revealed that Usp7 binds the regions on P53 and MDM2 containing P/AxxS motifs (Hu et al., 2006; Saridakis et al., 2005). Ci protein harbors eighteen potential Usp7-binding motifs, which we named S1 to S18 (Table S2); therefore, we went on to determine whether these sites contribute to Ci interaction with Usp7. S17 and S18 were not tested since they are located in the C-terminal region of Ci (CiC) that does not bind Usp7. Through Co-IP assay, we found that Ci-1-225, Ci-441-900 and Ci-901-1106 could bind strongly to Usp7 (Figures S2A-A’) whereas Ci-226-440 showed weak interaction with Usp7 (Figure S2A lane4). By contrast, Ci-1-177 and Ci-441-550, which do not harbor any P/AxxS sequence, did not show any detectable interaction with Usp7 (Figure S2A lane2 and lane5). Further Co-IP experiments showed that S3, S4, S8, S12 and S16 may play a role in Ci-Usp7 interaction (Figures S2B-B’). Consistently, a mutant CiFL, Ci−5S, which has these five sites mutated (S212A, S219A, S555A, S888A, S1113A), interacted with Usp7 less effectively compared with wild type CiFL (Figure S2C). These data suggest that multiple P/AxxS motifs in Ci may contribute to Ci-Usp7 interaction.
To determine which region in Usp7 was responsible for binding to Ci, we generated truncated forms of Usp7 (Figure 2B). Through Co-IP and GST pull-down experiments, we found that the N-terminally located MATH was both necessary and sufficient to mediated Ci binding (Figures 2G-J). Ci-Usp7 interaction did not show apparent changes by coexpression of Hib (Figure S3A), an E3 ligase that also binds Ci via its MATH domain (Zhang et al., 2006). This is consistent with our previous finding that Hib binds Ci through multiple S/T-rich motifs (Zhang et al., 2009), which are distinct from the Usp7 binding motifs.
To examine whether ubiquitin modification of Ci affects its association with Usp7, we employed Ub-K0, in which all the Lysines were replaced by Arginines to block the formation of polyubiquitin chains. Expression of Ub-K0 blocked Slimb-Cul1 and Hib-Cul3-mediated polyubiquitination of Ci (Figures S3B-C) but did not influence the binding affinity between Ci and Usp7 (Figure S3D).
Hh promotes Ci interaction with Usp7
Usp7 could bind Ci constitutively to counteract Ci degradation regardless whether Hh is present or not. Alternatively, Ci-Usp7 association could be regulated by Hh signaling. To distinguish these two possibilities, we carried out Co-IP experiments in S2 cells treated with a Hh-conditioned medium or control medium. Our result showed that Hh treatment increased the binding affinity between Ci and Usp7 (Figure 2K, lane3-4). In Cl8 cells, endogenous Ci pulled down more Fg-Usp7 protein when treated with the Hh-conditioned medium (Figure 2L). These results suggest that Ci-Usp7 association is promoted by Hh signaling.
In the absence of Hh, Ci forms multiple complexes with Costal2 (Cos2)-Fused (Fu) and Suppressor of fused (Sufu), and these complexes dissociate in response to Hh (Lum et al., 2003; Methot and Basler, 2000; Robbins et al., 1997; Ruel et al., 2007; Shi et al., 2011; Wang et al., 2000). Therefore, it is possible that Cos2-Fu and Sufu may compete with Usp7 to bind Ci. Indeed, we found that coexpression of Cos2-Fu or Sufu could suppress the basal association between Usp7 and Ci (Figure 2K, compare lanes 5 and 7 with lane3), and this suppression could be overcome by Hh treatment (Figure 2K, compare lanes 6 and 8 with lanes 5 and 7). On the other hand, knockdown of cos2 or/and sufu promoted Ci-Usp7 association both in the absence or presence of Hh stimulation (Figure 2M). Furthermore, Usp7 coexpression attenuated Ci-Cos2 and Ci-Sufu interactions in a dose-dependent manner (Figures S3E-F). These results suggest that Usp7 may compete with Cos2 and Sufu for binding Ci.
The observation that Hh still promoted Ci binding to Usp7 when both cos2 and sufu were knocked down implied that additional mechanism(s) might regulate Ci-Usp7 association. Indeed, when transfected S2 cells were treated with okadaic acid (OA, a phosphatase inhibitor) before cell harvesting, Ci exhibited an increased binding to Usp7 (Figures S3G-H), implying that Ci-Usp7 association is regulated by phosphorylation. Upon OA treatment, Usp7 exhibited little if any mobility shift, which is indicative of phosphorylation, either on SDS PAGE or on phosphor-tag gel even in the presence of Hh stimulation (Figure S3I). By contrast, Ci exhibited an extensive mobility shift, which was further enhanced by Hh treatment (Figure S3J). Hh-induced Ci phosphorylation did not occur on previously indentified PKA/CK1 phosphorylation clusters (Wang et al., 1999) because mutating the PKA sites (S838A, S856A and S892A) in these clusters (Ci−PKA) did not affect Hh-induced mobility shift of Ci−PKA (compared Figure S3K with Figure S3J). Furthermore, Ci−PKA still formed a complex with Usp7 in a manner regulated by OA treatment (Figure S3L) and Hh stimulation (Figure S3M). Taken together, these results suggest that Hh may promote Ci-Usp7 interaction in part through stimulating Ci phosphorylation.
Usp7 Antagonizes Ci Degradation Caused by Both Slimb-Cul1 and Hib-Cul3 E3 Ligases
Because Usp7 interacts with Ci and inactivation of Usp7 destabilizes Ci, we next examined whether gain-of-function of Usp7 could stabilize Ci. Compared with control disc (Figure 3A), overexpression of usp7 increased Ci protein levels but not ci transcription in A-compartment cells (Figures 3B-B’, 3C-C” and S1G-H’). Given that Ci is subjected to dual degradation in the cytoplasm by Slimb-Cul1 and in the nucleus by Hib-Cul3 (Jiang, 2006), we went on to determine the subcellular localization of a Fg-Usp7 in S2 cells, salivary gland cells (which have large nuclei), and wing disc cells. We found that Fg-Usp7 protein was mainly localized in the nucleus with a small proportion localized in the cytoplasm (Figures 3D-F’ and Figures S4A-B”). The subcellular localization of Usp7 indicates that it may influence both cytoplasmic and nuclear Ci.
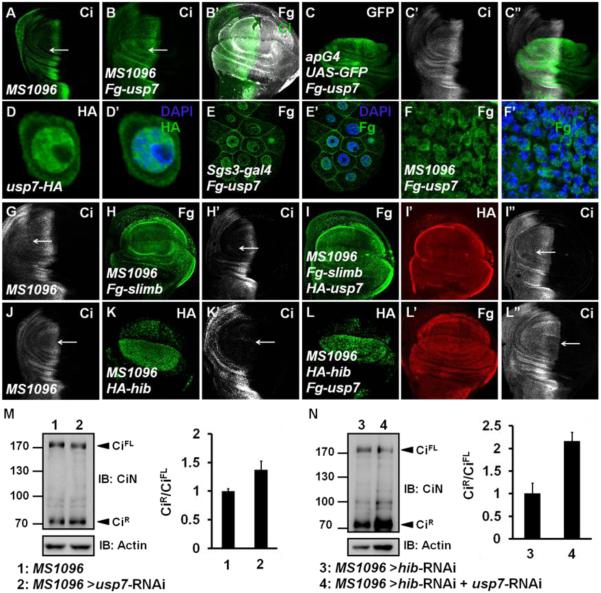
Figure 3. Usp7 Counteracts Ci Degradation Mediated by Both Slimb-Cul1 and Hib-Cul3 E3 ligases.
UAS transgene expression with MS1096 gal4 is usually stronger in the dorsal region than in the ventral of wing pouch.
(A) A wing disc of MS1096 line was stained to show full-length Ci (green).
(B-B’) A wing discs expressing Fg-usp7 with MS1096 was immunostained with Fg (white) and Ci (green) antibodies. Ci was accumulated when usp7 was overexpressed (arrow).
(C-C”) A wing disc expressing Fg-usp7 with apG4 was immunostained to show the expression of GFP (green) and Ci (white).
(D-D’) Expressed Usp7-HA (green) in S2 cells mainly localized in nuclei.
(E-E’) Fg-Usp7 protein (green) expressed by sgs3-gal4 in salivary glands mainly localized in nuclei.
(F-F’) Fg-Usp7 protein (green) expressed by MS1096 in wing discs mainly localized in nuclei. From D to F’, the nuclei were marked by DAPI (blue)
(G-I”) A MS1096 wing disc (G) and the discs expressing Fg-slimb alone (H-H’) or Fg-slimb/HA-usp7 together (I-I”) by MS1096 were immunostained to show Fg tag (green), Ci (white), and HA tag (red). Usp7 could counteract Ci degradation by Slimb-Cul1 E3 ligase (arrows).
(J-L”) A MS1096 wing disc (J) and the discs expressing HA-hib alone (K-K’) or HA-hib/Fg-usp7 (L-L”) together by MS1096 were immunostained with HA tag (green), Ci (white), and Fg tag (red) antibodies. Usp7 could attenuate Ci degradation by Hib-Cul3 E3 ligase (arrows).
(M-N) Western blot analysis of lysates from control wing discs or wing discs expressing indicated RNAi using MS1096 driver. Approximately 40 discs were dissected, lysed and blotted with rabbit anti-CiN antibody, respectively. Intensity ratio of CiR to CiFL of each lane was shown on the right. The ratio result was presented as means±SD of values from three independent experiments.
See also Figure S4.
To examine whether Usp7 can block both Slimb-Cul1 and Hib-Cul3-mediated Ci degradation, we coexpressed usp7 with slimb or hib and examine CiFL levels by immunostaining. Compared with the control disc in Figure 3G, overexpression of slimb alone down regulated Ci level in A-compartment cells away from the A/P boundary (Figures 3H-H’) while simultaneous expression of slimb and usp7 led to an accumulation of Ci (Figures 3I-I”). Similarly, overexpression of hib strong decreased Ci in A-compartment cells both distant from and closed to the A/P boundary (Figures 3J-K’) but usp7 coexpression could impede Hib-induced Ci degradation (Figures 3L-L”).
We further verified these results in S2 cells treated with CHX to prevent protein synthesis. The results showed that Usp7 significantly prevented Slimb-Cul1 and Hib-Cul3-mediated Ci degradation (Figures S4C-D). In addition, Usp7 could attenuate Hib-Cul3-mediated destabilization of a processing-resistant form of Ci, Ci−PKA (Figure S4E). Consistent with the notion that Usp7 may antagonize both Slimb-Cul1 and Hib-Cul3 mediated Ci degradation, Ci levels decreased in usp7 mutant clones located both away from (Figures 1L-L”, arrowhead) and near the A/P boundary of wing discs (Figures 1L-L”, arrow).
Slimb-Cul1-mediated Ci ubiquitination results in partial degradation to generate a truncated repressor form of Ci (CiR). To examine whether Usp7 regulates CiR formation, we employed a rabbit anti-CiN antibody (Zhang et al., 2013b), which could recognizes both CiFL and CiR. The relative ratio of CiR to CiFL was mildly increased (from 1.00 to 1.36) upon knockdown of usp7 in wing discs (Figure 3M). To remove the effect of Hib-Cul3, we simultaneously knocked down hib using hib-RNAi. The relative ratio of CiR to CiFL was dramatically increased (from 1.00 to 2.15) in usp7 and hib double knockdown in wing discs compared to hib single knockdown in wing discs (Figure 3N), indicating that Usp7 inhibits Slimb-Cul1-mediated Ci processing.
Usp7 Deubiquitinase Activity is Essential for Ci Stabilization
Usp7 belongs to the ubiquitin-specific processing protease family of deubiquitination enzymes and contains the characteristic Cys motif at the core enzymatic domain (Li et al., 2002). Mutation of the catalytic Cys often leads to an inactive form of the enzyme. To examine whether Usp7 stabilizes Ci through its deubiquitinase activity, we generated a deubiquitination-defective substitution mutant of Usp7 with Cys250 changed to Ala (Fg-Usp7CA). Although this point mutation did not affect Usp7 binding to Ci (Figure 4A), it does affect Usp7 activity as revealed by the rescue experiments shown in Figure 4. The characteristic adult wing phenotypes caused by Hh pathway attenuation due to expression of a dominant negative form of Smo (Smo−PKA) is a reduction of the intervein space between vein 3 and vein 4 (Figure 4B) (Chen et al., 2010; Jia et al., 2010). Knockdown of usp7 further narrowed the width between vein 3 and vein 4 in wing expressing Smo−PKA (Figures 4B-C), and this enhancement was suppressed by overexpression of wild type usp7 (Figure 4D). By contrast, overexpression of usp7CA failed to rescue the phenotype caused by usp7 knockdown (Figure 4E), but instead, deteriorated the wing defect (compare Figures 4E with 4C). Consistently, overexpression of usp7CA did not upregulate Ci levels in wing discs (Figures 4F-F”), but instead, resulted in decreased of dpp-lacZ expression (Figures 4G-G”). These observations suggest that Usp7CA acts as a dominant negative mutant and that the deubiquitinase activity of Usp7 is essential for Ci stabilization.
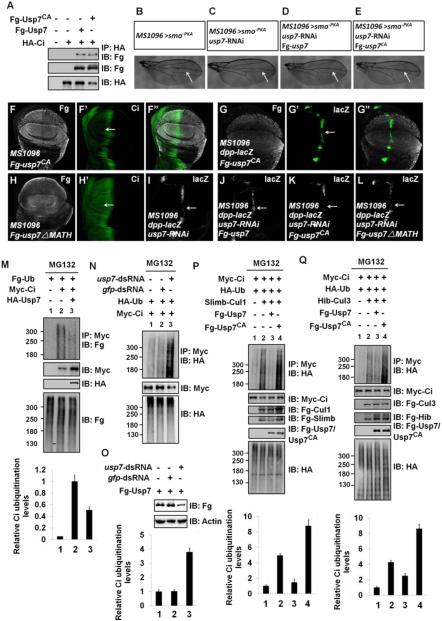
Figure 4. Usp7 Attenuates Slimb-Cul1- and Hib-Cul3-mediated Ci Ubiquitination through Its Deubiquitinating Activity.
(A) Both Fg-Usp7 and Fg-Usp7CA could bind Myc-Ci in S2 cells.
(B-E) Comparison of adult wing phenotypes from control flies (B), usp7 knockdown (C), Fg-usp7 expression in usp7 knockdown background (D), Fg-usp7CA expression in usp7 knockdown background (E). Arrows marked the space between vein3 and vein4.
(F-F”) A wing disc expressing usp7CA with MS1096 was immunostained to show Fg tag (white) and Ci (green). Ci level was not upregulated by usp7CA expression (arrow).
(G-G”) A wing disc expressing usp7CA with MS1096 was stained for Fg tag (white) and dpp-lacZ (green). usp7CA overexpression decreased dpp-lacZ expression (arrow).
(H-H’) A wing disc expressing usp7-ΔMATH with MS1096 was stained with Fg tag (white) and Ci (green) antibodies. Ci level was not altered by usp7-ΔMATH expression (arrow).
(I-L) Wing discs expressing usp7 RNAi (I), Fg-usp7/usp7 RNAi (J), Fg-usp7CA/usp7 RNAi (K) or Fg-usp7-ΔMATH/usp7 RNAi (L) were stained for dpp-lacZ expression (white). The attenuation of dpp-lacZ caused by usp7 knockdown (I, arrow) could be restored by the expression of Fg-usp7 (J, arrow), but not Fg-usp7CA (K, arrow) and Fg-usp7-ΔMATH (L, arrow).
(M) Western blots of immunoprecipitates (top) or lysates (bottom three panels) from S2 cells expressing indicated proteins and treated with MG132.
(N) Knockdown of usp7 using usp7-dsRNA promoted Ci ubiquitination in S2 cells.
(O) usp7-dsRNA could effectively knock down Fg-usp7 expression in S2 cells. Actin acts as a loading control.
(P-Q) Transfected S2 cells were treated with MG132 for 4hrs before cell harvesting. Fg-Usp7 decreased, but Fg-Usp7CA promoted Ci ubiquitination mediated by Slimb-Cul1 (P) and Hib-Cul3 (Q). From M-Q, quantification analyses of the ubiquitination levels of Ci were shown below each autoradiogram. Data are means±SD from three independent experiments.
Overexpression of usp7-ΔMATH, which fails to interact with Ci, did not elevate Ci levels (Figures 4H-H’), suggesting that Usp7 binding Ci is essential for Ci stabilization. While usp7 RNAi reduced dpp-lacZ expression (Figure 4I), simultaneous expression of usp7 and usp7-RNAi could effectively restore dpp-lacZ level (Figure 4J). However, neither usp7CA nor usp7-ΔMATH rescued the decrease of dpp-lacZ caused by usp7 knockdown (Figures 4K-L). In sum, Usp7 modulates Ci and Hh signaling through its interaction with Ci and its deubiquitinase activity.
Usp7 Counteracts Both Slimb-Cul1 and Hib-Cul3-dependent Ubiquitination of Ci
We next determined whether Usp7 stabilizes Ci through deubiquitinating Ci using a cell-based ubiquitination assay (Zhang et al., 2006). We found that coexpression with Usp7 decreased whereas knockdown of usp7 increased Ci ubiquitination (Figures 4M-N). Knockdown of usp7 was confirmed by co-transfection of Fg-Usp7 with usp7-dsRNA (Figure 4O).
Because Usp7 could suppress both Slimb-Cul1 and Hib-Cul3-mediated Ci degradation, we then asked whether Usp7 could attenuate Ci ubiquitination mediated by both Slimb-Cul1 and Hib-Cul3 E3 ligases. Overexpression of Slimb-Cul1 E3 ligase promoted Ci ubiquitination (Figure 4P lane2 compared with lane1), which was suppressed by co-expression of Usp7 (Figure 4P lane3 compared with lane2). By contrast, co-expression Usp7CA enhanced the Slimb-Cul1-mediated ubiquitination of Ci (Figure 4P lane4 compared with lane2). Similarly, Usp7 prevented Hib-Cul3-mediated Ci ubiquitination, whereas Usp7CA promoted Hib-Cul3-mediated Ci ubiquitination (Figure 4Q). Taken together, these findings suggest that Usp7 can oppose both Slimb-Cul1 and Hib-Cul3-mediated ubiquitination of Ci.
Usp7 Regulates Ci through Usp7-GMPS Complex
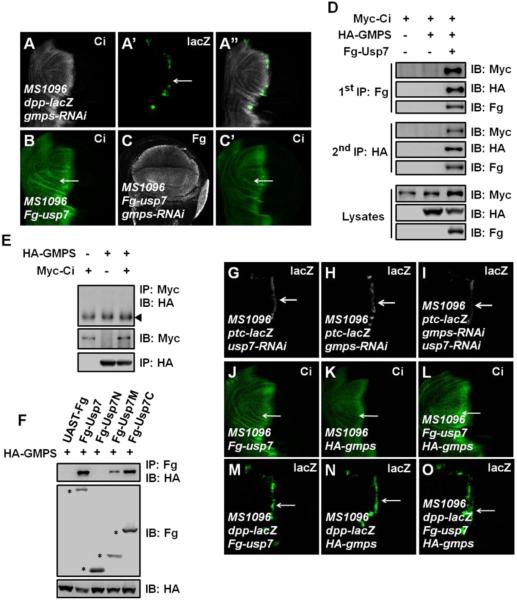
In both Drosophila and mammal, GMPS associates with Usp7 to enhance its deubiquitination activity (Faesen et al., 2011; van der Knaap et al., 2010). Given that the deubiquitination activity of Usp7 is essential for Ci regulation, we next determined whether GMPS cooperates with Usp7 to modulate Ci levels. Similar to usp7, gmps was also ubiquitously expressed throughout the wing disc as revealed by in situ hybridization (Figures S1C-D). Knockdown of gmps using transgenic RNAi (v24152 or 9242R-1) attenuated dpp-lacZ expression (Figures 5A-A”), and prevented Ci upregulation induced by overexpression of usp7 (Figures 5C-C’ compared with Figure 5B), suggesting that GMPS is required for Usp7-mediated Ci stabilization.
Figure 5. Usp7 Regulates Hh Signaling through Usp7-GMPS Complex.
(A-A”) A wing disc expressing gmps RNAi by MS1096 was immunostained to show the expression of Ci (white) and dpp-lacZ (green). gmps knockdown resulted in a decrease of dpp-lacZ (arrow).
(B-C’) Wing discs overexpressing usp7 alone (B) or usp7 plus gmps RNAi (C-C’) were immunostained by Fg tag (white) and Ci (green) antibodies. gmps knockdown suppressed the elevation of Ci by Usp7 expression (arrows).
(D) S2 cells expressing the indicated proteins were harvested for the two-step immunoprecipitation and analyzed by western blot.
(E) GMPS did not bind Ci. The indicated proteins were expressed in S2 cells. The arrowhead indicates IgG.
(F) Usp7 interacted with GMPS through its M- and C- regions in S2 cells. Asterisks mark positions of each protein.
(G-I) Wing discs expressing usp7 RNAi alone (G), gmps RNAi alone (H) or usp7 RNAi plus gmps RNAi (I) were immunostained to show ptc-lacZ expression (white). ptc-lacZ expression decreased more apparently when both usp7 and gmps were knocked down (arrows).
(J-L) Wing discs overexpressing usp7 alone (J), gmps alone (K) or usp7 plus gmps (L) were immunostained for Ci expression (green). Overexpression of usp7 plus gmps resulted in a more dramatic increase of Ci (arrows).
(M-O) Wing discs overexpressing usp7 alone (M), gmps alone (N) or usp7 plus gmps (O) were immunostained to show dpp-lacZ expression (green). Overexpression of usp7 plus gmps resulted in a more dramatic increase of dpp-lacZ (arrows).
We performed a two-step immunoprecipitation experiment and found that Myc-Ci, Fg-Usp7 and HA-GMPS could form a trimeric complex when co-expressed in S2 cells (Figure 5D). Furthermore, HA-GMPS did not pull down Myc-Ci in the absence of Fg-Usp7 coexpression (Figure 5E), suggesting that Usp7 most likely bridges Ci and GMPS. Given that Usp7 binds Ci through its N-terminal MATH domain, we envisioned that Usp7 binds GMPS via other regions. Indeed, both the M- and C-regions (Usp7M and Usp7C) but not the N-region (Usp7N) of Usp7 interacted with GMPS in the Co-IP assay (Figure 5F).
We next examined whether Usp7 synergizes with GMPS to positively regulate Ci and Hh signaling, Double knockdown of usp7 and gmps caused a more dramatic decrease of ptc-lacZ expression than usp7 or gmps single knockdown (Figures 5G-I). On the other hand, simultaneous overexpression of usp7 and gmps caused a more dramatic stabilization of Ci than overexpression of usp7 or gmps alone (Figures 5J-L). Furthermore, co-expression of usp7 and gmps resulted in an anterior expansion of the dpp-lacZ expression domain (Figure 5O), which was not observed when usp7 or gmps was overexpressed alone (Figures 5M-N). Given that both usp7 and gmps are ubiquitously expressed in wing discs, these results suggest that Usp7 most likely form a complex with GMPS to modulate Hh pathway in vivo.
HAUSP Deubiquitinates Gli and Promotes Hh Signaling Activity in Mammalian Systems
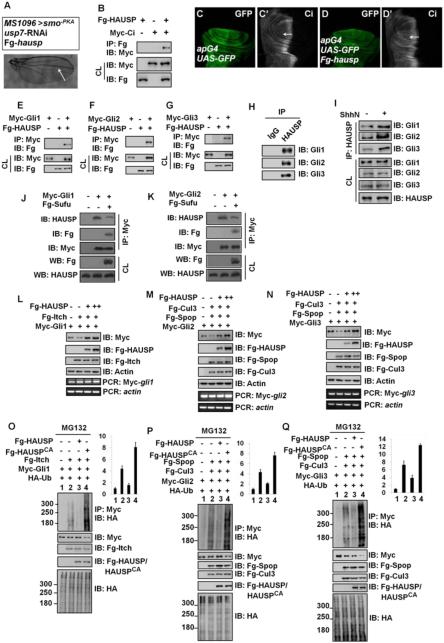
The mammal homologs of Ci, Gli1, Gli2 and Gli3 also undergo ubiquitination-mediated degradation (Huntzicker et al., 2006; Pan et al., 2006; Wang and Li, 2006), however whether Gli proteins are regulated by deubiquitinases has remained unknown. We found that overexpression of the mammalian homolog of usp7, hausp, restored the wing defect caused by usp7 knockdown (Figure 6A compared with Figures 4B-C). Furthermore, HAUSP could bind Ci (Figure 6B), and misexpression of hausp by apG4 resulted in an increase of Ci levels (Figures 6D-D’ compared with 6C-C’), implying that HAUSP could functionally replace Usp7 in the regulation of Ci.
Figure 6. HAUSP Binds and Deubiquitinates Gli Proteins.
(A) Adult wing that expressing usp7 RNAi and Fg-tagged hausp. Arrow marked the space between vein3 and vein4.
(B) Fg-HAUSP could bind Myc-Ci in S2 cells.
(C-C’) A wing disc of apG4 was stained to show GFP (green) and Ci (white).
(D-D’) Overexpression of hausp by apG4 resulted in an increase of Ci (white, arrow) in wing discs.
(E-G) Fg-HAUSP could bind Myc-Gli1, Myc-Gli2 and Myc-Gli3 in 293T cells, respectively. The expression of corresponding proteins from cell lysis (CL) was shown below.
(H) Endogenous HAUSP pulled down endogenous Gli1, Gli2 and Gli3 in 293T cells. The cells from one 10cm plate were lysed and the lysis was equivalently divided two parts for IP with control IgG or HAUSP antibody.
(I) ShhN treatment increased the interaction between endogenous HAUSP and endogenous Gli proteins in 293T cells.
(J-K) Sufu expression inhibited the interaction between HAUSP and Myc-tagged Gli1 (J) and Gli2 (K) in Sufu−/− MEF cells. The transfected cells were treated with MG132 for 4hrs prior to cell harvesting.
(L-N) HAUSP blocked Gli protein degradation mediated by corresponding E3 ligases. 293T cells were transfected with indicated plasmids. Actin is shown as a loading control. RT-PCR analysis was performed to test the levels of Myc-gli mRNA. The forward primer was on Myc tag, whereas the reverse primer was on gli. actin acts as a control. (O-Q) HAUSP inhibited, but HAUSPCA promoted the ubiquitination of Gli proteins mediated by according E3 ligases. 293T were transfected with indicated plasmids and treated with MG132 for 4hrs before cell harvesting. Quantification analyses of the ubiquitination levels of Gli proteins were shown on right. The results were presented as means±SD of values from three independent experiments.
See also Figure S5.
We next determined whether HAUSP is involved in the regulation of Gli stability in mammalian cells. First, we determined whether HAUSP interacted with different Gli proteins by expressing Myc-tagged Gli constructs and Fg-tagged HAUSP in 293T cells and carried out Co-IP experiments. The results revealed that HAUSP could bind Gli1 (Figure 6E), Gli2 (Figure 6F) and Gli3 (Figure 6G). In addition, endogenous HAUSP could pull down endogenous Gli proteins in 293T cells (Figure 6H). Treatment with Shh-conditioned medium promoted the interaction between HAUSP and Gli proteins (Figure 6I and Figures S5A-C). In addition, mSufu overexpression decreased Gli-HAUSP interactions in Sufu−/− MEFs (Figures 6J-K). These results suggest that Gli-HAUSP interactions are stimulated by Shh but inhibited by Sufu.
Previous studies have demonstrated that Gli1 is subject to ubiquitination by Itch E3 ligase (Di Marcotullio et al., 2006; Di Marcotullio et al., 2011), whereas Gli2 and Gli3 are ubiquitinated by Spop-Cul3 E3 ligase (Zhang et al., 2009; Zhang et al., 2006). We found that HAUSP blocked Itch-mediated degradation of Gli1 and Spop-Cul3-mediated degradation of Gli2/Gli3 (Figures 6L-N, top panels) without affecting Gli mRNA levels (Figures 6L-N, bottom panels). Knockdown of hausp also promoted Gli3 processing in 293T cells (Figure S5D). Furthermore, we found that HAUSP dramatically inhibited whereas HAUSPCA (a dominant negative with Cys223 changed to Ala), promoted Itch-mediated Gli1 ubiquitination (Figure 6O). Similarly, HAUSP also counteracted whereas HAUSPCA promoted Spop-Cul3-mediated ubiquitination of Gli2 and Gli3 (Figures 6P-Q). Taken together, these results suggest that HAUSP binds all the three Gli proteins to modulate their levels by counteracting the ubiquitination mediated by their cognate E3 ligases.
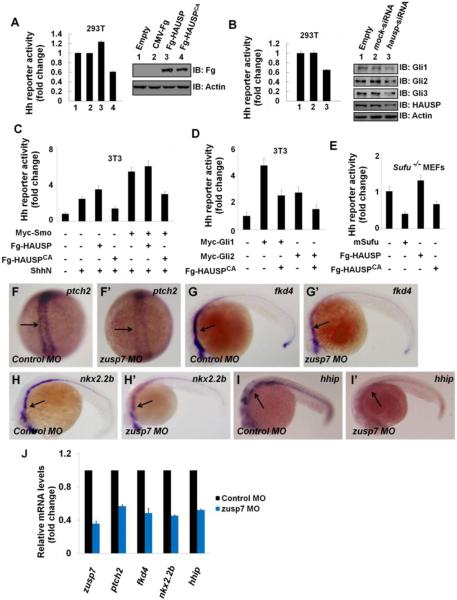
The ability of HAUSP to suppress Gli proteins ubiquitination implies that HAUSP may affect Shh pathway activity through modulating Gli protein levels. To test that, we performed Gli-luciferase reporter assay in 293T cells, which express Shh pathway components and target genes (Figures S5E-F). HAUSP is also expresses in 293T cells (Figure S5E) and the endogenous HAUSP protein mainly localizes in the nucleus (Figures S5G-H’”). Overexpression of HAUSP promoted whereas HAUSPCA decreased Shh pathway activity (Figure 7A). Knockdown of endogenous hausp suppressed the expression of Shh reporter gene and attenuated endogenous Gli levels (Figure 7B). The Gli-luciferase reporter assays in NIH3T3 cells and Sufu−/− MEF cells indicated that HAUSP positively regulated Shh pathway downstream of Sufu and at levels of Gli proteins (Figures 7C-E). Taken together, these results suggest that HAUSP plays a conserved role in Shh pathway.
Figure 7. The Regulation of Usp7 on Hh Signaling Is Evolutionally Conserved in Mammalian Cells and Zebrafish.
(A) Gli-luciferase (Gli-luc) reporter assay in 293T cells transfected with indicated constructs. Gli luciferase activities were normalized to Renilla Luciferase activities. The expression of indicated constructs was shown.
(B) Gli-luc reporter assay in 293T cells transfected with indicated siRNAs. Knockdown of hausp decreased Hh pathway activity. The protein levels of HAUSP and Gli were tested through western blotting analysis. Actin acts as a loading control.
(C-D) Gli-luc reporter assay in 3T3 transfected with indicated constructs.
(E) Gli-luc reporter assay in Sufu−/− MEFs transfected with indicated constructs. Of note, HAUSP regulated Hh pathway downstream of Sufu.
(F-I’) Expression of Hh target genes in zebrafish embryos that were injected with the indicated MOs at 10hpf (F-F’) or 24hpf (G-I’). Changes of the in situ staining are marked by arrows.
(J) Relative mRNA levels of zusp7, ptch2, fkd4, nkx2.2b and hhip from zebrafish embryos indicated in F-I’ were revealed by real-time PCR.
See also Figure S6.
zUsp7 Regulates Hh Signaling in Zebrafish
We then turn to zebrafish to determine whether Usp7 plays a conserved role in Shh signaling in vivo. Through Co-IP experiments, we confirmed that zebrafish Usp7 (zUsp7) could bind zebrafish Gli proteins (zGli1, zGli2 and zGli3) (Figures S6A-C). Next, we examined the expression levels of several Hh-responsive genes, including patched2 (ptch2), forkhead4 (fkd4), NK2 homeobox 2b (nkx2.2b), and hedgehog interacting protein (hhip), in control or zusp7 morpholino treated zebrafish embryos 24h postfertilization (hpf) by in situ hybridization and real-time PCR (Zhang et al., 2013a). Compared with control morphants (Figures 7F, G, H, I), Hh target gene expression was notably down-regulated in zusp7 morphants (Figures 7F’, G’, H’, I’). The real-time PCR assays further confirmed the results (Figure 7J). The zusp7 MO could effectively knock down the expression of endogenous zusp7 (Figure 7J, lane1). Collectively, these data indicate that zUsp7 plays an evolutionarily conserved role as a positive regulator of Hh pathway in zebrafish.
DISCUSSION
In this study, we identified the deubiquitinase Usp7/HUASP as a positive regulator of the Hh pathway. We provided both genetic and biochemical evidence that Usp7/HUASP interacts with Ci/Gli and removes the ubiquitin moiety from Ci/Gli to promote their stabilization. Our findings thus unveil a conserved mechanism by which a deubiquitinating enzyme antagonizes multiple E3 ligases to achieve optimal Hh pathway activity.
Previous studies have demonstrated that Ci is controlled by dual ubiquitination systems that employ Slimb-Cul1 and Hib-Cul3 to regulate CiR and CiA, respectively (Jiang, 2006). In the absence of Hh, Ci forms a complex with Cos2-Fu that recruits multiple kinases to phosphorylate Ci, targeting Ci for Slimb-mediated proteolytic processing to generate CiR that inhibits dpp expression. Hh stimulates dissociation of the Ci-Cos2-Fu-kinase complexes (Zhang et al., 2005) as a consequence, Ci phosphorylation and Slimb recruitment are compromised, leading to blockage of Ci processing. Here, we show that Hh-stimulated Ci-Cos2 dissociation may also facilitate the binding of Usp7 to Ci, leading to deubiquitination of Ci. Indeed, inactivation of Usp7 increased the ratio of CiR/CiFL and caused reduction of dpp expression. Hence, by promoting the binding of Usp7 in addition to inhibiting Slimb recruitment, Hh signaling can effectively block Ci ubiquitination and processing.
It has been shown that high levels of Hh convert CiFL into a labile but active form (CiA) by dissociating Ci from its inhibitor Sufu (Ohlmeyer and Kalderon, 1998; Shi et al., 2011). Dissociation of Sufu from Ci facilitates binding of Hib to Ci (Zhang et al., 2006), which explains why CiA is unstable. Hib expression is induced by Hh signaling in both embryos and imaginal discs, thus Hib-mediated degradation of Ci serves as a negative feedback to fine tune or terminate Hh pathway activity depending on the context (Kent et al., 2006; Ou et al., 2007; Zhang et al., 2006). However, excessive degradation of CiA by Hib may cause premature loss of Hh signaling activity, leading to developmental defects. Therefore, it is likely that Hib-mediated Ci degradation is subjected to tight control to ensure appropriate pathway activity. One such mechanism is the attenuation of Hib binding by CK1-mediated phosphorylation of Ci at multiple Hib degrons (Shi et al., 2014). Here, we show that Hh-stimulated Usp7 binding to Ci may provide another mechanism to attenuate Hib-mediated degradation of CiA. Our results also suggest that Ci-Usp7 interaction is stimulated by Hh-induced Ci phosphorylation at sites distinct from previously identified PKA/CK1/GSK3 phosphorylation clusters (Figure S4), some of which may occur at the Hib degrons (Shi et al., 2014). Interestingly, a recent study also revealed that Hh inhibited phosphorylation at PKA sites while induced phosphorylation at other sites to control graded Gli activities (Niewiadomski et al., 2014). We propose that Hh-induced phosphorylation may positively regulate Ci/Gli activity, at least in part, by promoting their association with Usp7/HAUSP. Hh-induced phosphorylation of Ci/Gli proteins could cause their dissociation from Cos2/Kif7 and/or Sufu, thus facilitating their association with Usp7/HAUSP, as Usp7 appears to compete with Cos2 and Sufu to bind Ci. Alternatively but not mutual exclusively, Hh-induced phosphorylation of Ci/Gli proteins may play a more active role in recruiting Usp7/HAUSP. Further study is needed to determine the underlying mechanism.
Interestingly, other key components of Hh signaling pathway, including Smo and Ptc, are also subjected to the regulation by ubiquitination (Fan et al., 2013; Huang et al., 2013; Li et al., 2012; Xia et al., 2012; Yue et al., 2009). In addition, it has been reported that the deubiquitinase Usp8 could deubiquitinate Smo to influence Hh signaling activity (Li et al., 2012; Xia et al., 2012). Based on these and our observation, we predict that other deubiquitinases, for example, deubiquitinase for Ptc, might be involved in Hh pathway regulation.
The deubiquitinating function of HAUSP on Gli proteins we uncovered here suggests that HAUSP may play a role in regulating Hh pathway in mammalian systems. hausp knockout mice die during early embryonic development between embryonic days E6.5 and E7.5 (Kon et al., 2010). Therefore, it would be interesting to determine whether hausp mutant embryos exhibit any defects in Hh signaling. HAUSP also plays an important role in a number of pathologies, in particular an oncogenic role in neoplastic diseases. For example, hausp overexpression has been tightly linked to human prostate cancer (Song et al., 2008), in which the Hh/Gli signaling has also been implicated (Chen et al., 2011). It should be interesting to determine whether Gli activity contributes to hausp overexpression-induced tumors. Several HAUSP inhibitors, such as P045204 and P5091, have been developed for preclinical study (Chauhan et al., 2012; Nicholson and Suresh Kumar, 2011). Our finding that HAUSP regulates Hh pathway through Gli proteins raises a possibility of using HAUSP inhibitors to treat Hh-related cancers.
EXPERIMENTAL PROCEDURES
Immunostaining and in situ hybridization of wing discs, GST-fusion protein pull-down, cell culture, transfection, immunoprecipitation, western blot, and real-time PCR were performed according to standard protocols. For detailed procedures, fly strains and constructs used in this study, see Supplemental Experimental Procedures.
Generating usp7 Mutation Clones
Clones of mutant cells were generated by FLP/FRT-mediated mitotic recombination as described (Theodosiou and Xu, 1998). Genotype for generating usp7 clones is as follows: usp7KG06814 FRT19A/hs-flp ubi-GFP FRT19A.
Luciferase Reporter Assay
In mammal cells, Hh reporter assay was performed as previously described (Di Marcotullio et al., 2006). All luciferase activity data are presented as means±SD of values from at least three experiments, each performed in triplicate.
RNA Interference
To knock down genes in S2 cells, the double-stranded RNA (dsRNA) was generated by MEGAscript High Yield Transcription Kit (Ambion) according to the manufacturer’s instructions. DNA templates targeting usp7 (aa34-267 and aa981-1097), cos2 (aa1-234 and aa870-1102) and sufu (aa1-233) were generated by PCR and used for generating dsRNA. dsRNA targeting the GFP full-length coding sequence was used as a control. The dsRNA-mediated gene knockdown was performed as previous described (Li et al., 2012; Liu et al., 2014). To silence hausp expression in 293T cells, a mixture of two different siRNAs were used. The siRNA sequences were as follows: hausp-siRNA-1 (5’-ACCCUUGGACAAUAUUCCUdTdT), hausp-siRNA-2 (5’-AGUCGUUCAGUCGUCGUAUdTdT), mock-siRNA (5’-UUCUCCGAACGUGUCACGUdTdT). All siRNA duplexes were transfected at a final concentration of 100nM using lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Ci Protein Stability Assay and Ubiquitination Assay
S2 cells were plated in 10-cm dishes and transfected with indicated plasmids after18-24hrs. After another 24hrs, the cells were transferred into 6-well cell culture plates at equivalent densities. Cells were treated with 20μg/ml CHX (Calbiochem) for the indicated times before harvesting. S2 or 293T cells were transiently transfected with the indicated combinations of expression vectors. 4hrs before cells harvesting, MG132 (Calbiochem) was added to the media at a final concentration of 10μM. The ubiquitination assays were then carried out based on the previously described protocol (Zhang et al., 2006). Briefly, cells were lysed with denaturing buffer (1% SDS, 50mM Tris-base, pH 7.5, 0.5mM EDTA, and 1mM DTT) and incubated at 100 ºC for 5 min. The lysates were then diluted 10-fold with regular lysis buffer and subject to immunoprecipitation and western blot analysis. After western blots, the band intensity was measured by Image J. For quantitation, data are presented as means±SD of values from at least three experiments.
MO Knockdown
Antisense MOs (Gene Tools) were microinjected into one to four cell stage embryos according to the standard protocols. A 4-nl volume of MOs was injected at the concentration of 0.15 mM. MO sequences used were usp7-MO (5’-TAATGGCAGTAACTGCTTACGTGGA-3’) and standard control-MO (Gene Tools).
In Situ Hybridization of Zebrafish Embryos
The in situ hybridization of zebrafish embryos were carried out based on the previously described protocol (Thisse and Thisse, 2008; Zhang et al., 2013a). In brief, zebrafish embryos were obtained by natural spawning of adult AB strain zebrafish. Embryos were maintained at 28.5 ºC on a 14-h light/10-h dark cycle. To avoid variation of in situ hybridization, we injected the MOs, collected the embryos, performed the in situ, and took pictures at the same time under the same conditions. The RNA probe used was labeled with digoxigenin. The hhip probe encompassed the 1193-2112bp regions (start from ATG) of mRNA. Other probes used were fkd4 (Tay et al., 2005), nkx2.2b and ptch2 (Concordet et al., 1996).
Supplementary Material
ACKNOWLEDGMENTS
We thank Fly Stocks of National Institute of Genetics of Japan (NIG-Fly), Vienna Drosophila RNAi Center (VDRC), the Bloomington Stock Center and Developmental Studies Hybridoma Bank at the University of Iowa for providing fly stocks and reagents. This work was supported by grants from the National Key Scientific Program of China (2011CB943902, 2010CB945102), the “Strategic Priority Research Program” of the Chinese Academy of Sciences (XDA01010405), and also from the National Natural Science Foundation of China (30971679, 31071264, 31271531 and 31371492). Jin Jiang is supported by grants from NIH (GM061269, GM067045, and GM106188), NSFC (31328017), and Welch foundation (I-1603).
REFERENCES
- Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews Molecular cell biology. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov MP, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Carkner R, Buttyan R. The hedgehog/Gli signaling paradigm in prostate cancer. Expert review of endocrinology & metabolism. 2011;6:453–467. doi: 10.1586/EEM.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li S, Tong C, Zhao Y, Wang B, Liu Y, Jia J, Jiang J. G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes & development. 2010;24:2054–2067. doi: 10.1101/gad.1948710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, Lewis KE, Moore JW, Goodrich LV, Johnson RL, Scott MP, Ingham PW. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development. 1996;122:2835–2846. doi: 10.1242/dev.122.9.2835. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nature cell biology. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Greco A, Mazza D, Canettieri G, Pietrosanti L, Infante P, Coni S, Moretti M, De Smaele E, Ferretti E, et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene. 2011;30:65–76. doi: 10.1038/onc.2010.394. [DOI] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. The EMBO journal. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Molecular cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Fan J, Jiang K, Liu Y, Jia J. Hrs promotes ubiquitination and mediates endosomal trafficking of smoothened in Drosophila hedgehog signaling. PloS one. 2013;8:e79021. doi: 10.1371/journal.pone.0079021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Current opinion in cell biology. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS biology. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhang Z, Zhang C, Lv X, Zheng X, Chen Z, Sun L, Wang H, Zhu Y, Zhang J, et al. Activation of Smurf E3 ligase promoted by smoothened regulates hedgehog signaling through targeting patched turnover. PLoS biology. 2013;11:e1001721. doi: 10.1371/journal.pbio.1001721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzicker EG, Estay IS, Zhen H, Lokteva LA, Jackson PK, Oro AE. Dual degradation signals control Gli protein stability and tumor formation. Genes & development. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & development. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, Jia J. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. The Journal of biological chemistry. 2010;285:37218–37226. doi: 10.1074/jbc.M110.174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416:548–552. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, Amanai K, Jiang J. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Developmental cell. 2005;9:819–830. doi: 10.1016/j.devcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Jiang J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell cycle. 2006;5:2457–2463. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Developmental cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–2010. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Li S, Chen Y, Shi Q, Yue T, Wang B, Jiang J. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS biology. 2012;10:e1001239. doi: 10.1371/journal.pbio.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhou Z, Yao X, Chen P, Sun M, Su M, Chang C, Yan J, Jiang J, Zhang Q. Hedgehog signaling downregulates suppressor of fused through the HIB/SPOP-Crn axis in Drosophila. Cell research. 2014;24:595–609. doi: 10.1038/cr.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Molecular cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell biochemistry and biophysics. 2011;60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, Rohatgi R. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell reports. 2014;6:168–181. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes & development. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CY, Wang CH, Jiang J, Chien CT. Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Developmental biology. 2007;308:106–119. doi: 10.1016/j.ydbio.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Molecular and cellular biology. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nature reviews Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Ruel L, Gallet A, Raisin S, Truchi A, Staccini-Lavenant L, Cervantes A, Therond PP. Phosphorylation of the atypical kinesin Costal2 by the kinase Fused induces the partial disassembly of the Smoothened-Fused-Costal2-Cubitus interruptus complex in Hedgehog signalling. Development. 2007;134:3677–3689. doi: 10.1242/dev.011577. [DOI] [PubMed] [Google Scholar]
- Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, Lee W, Edwards AM, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Molecular cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Shi Q, Li S, Jia J, Jiang J. The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development. 2011;138:4219–4231. doi: 10.1242/dev.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Li S, Li S, Jiang A, Chen Y, Jiang J. Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E5651–5660. doi: 10.1073/pnas.1416652111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smelkinson MG, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Current biology : CB. 2006;16:110–116. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay SY, Ingham PW, Roy S. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 2005;132:625–634. doi: 10.1242/dev.01606. [DOI] [PubMed] [Google Scholar]
- Theodosiou NA, Xu T. Use of FLP/FRT system to study Drosophila development. Methods. 1998;14:355–365. doi: 10.1006/meth.1998.0591. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature cell biology. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kozhevnikova E, Langenberg K, Moshkin YM, Verrijzer CP. Biosynthetic enzyme GMP synthetase cooperates with ubiquitin-specific protease 7 in transcriptional regulation of ecdysteroid target genes. Molecular and cellular biology. 2010;30:736–744. doi: 10.1128/MCB.01121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Molecular cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Wang B, Li Y. Evidence for the direct involvement of {beta}TrCP in Gli3 protein processing. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes & development. 2000;14:2893–2905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes & development. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Seminars in cell & developmental biology. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- Wilson CW, Chuang PT. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development. 2010;137:2079–2094. doi: 10.1242/dev.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Jia H, Fan J, Liu Y, Jia J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS biology. 2012;10:e1001238. doi: 10.1371/journal.pbio.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S, Chen Y, Cheng SY. Hedgehog signaling promotes the degradation of tumor suppressor Sufu through the ubiquitin-proteasome pathway. Oncogene. 2009;28:492–499. doi: 10.1038/onc.2008.403. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Shi Q, Chen Y, Yue T, Li S, Wang B, Jiang J. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21191–21196. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Developmental cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Developmental cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Feng J, Pan C, Lv X, Wu W, Zhou Z, Liu F, Zhang L, Zhao Y. Atrophin-Rpd3 complex represses Hedgehog signaling by acting as a corepressor of CiR. The Journal of cell biology. 2013a;203:575–583. doi: 10.1083/jcb.201306012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lv X, Yin WC, Zhang X, Feng J, Wu W, Hui CC, Zhang L, Zhao Y. Ter94 ATPase complex targets k11-linked ubiquitinated ci to proteasomes for partial degradation. Developmental cell. 2013b;25:636–644. doi: 10.1016/j.devcel.2013.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.