Cellular signaling by endogenously generated nitric oxide (NO) was initially characterized as dependent upon the ability of NO to bind heme, and in particular the heme of guanylate cyclase, thereby enhancing production of cGMP1. However, it is now recognized that protein S-nitrosylation, the oxidative modification by NO of the thiol side chain of cysteine (Cys) to generate an S-nitrosothiol (SNO) (first described in 1992)2 conveys the large part of the cellular influence of NO. The critical distinction between earlier and contemporary models of NO biology is the novel and unanticipated redox chemistry of NO that underlies a ubiquitous redox-based mechanism for both allosteric and active-site regulation of protein function (note that NO binding to heme in guanylate cyclase does not entail redox chemistry). S-nitrosylation operates ubiquitously across phylogeny from plants to microbes to man, and in all cell types, to regulate the function of proteins in most or all classes, and provides by far the most sophisticated and broadly operative system for redox regulation and signaling. Further, dysregulated S-nitrosylation has been implicated in an increasingly broad spectrum of human pathophysiologies, including prominently cardiovascular disorders. Notably, SNOs in proteins are represented by several different forms, with both biological and methodological implications.
Computational algorithms predict that ~70% of proteins undergo S-nitrosylation3, but only ~3000 have been identified experimentally. Indeed, elucidation of the targets and regulation of S-nitrosylation has posed substantial methodological hurdles. These reflect, as in the case of phosphorylation, the sometimes low population stoichiometry of modification4 and the dynamic and ongoing reversibility of modification in the cellular milieu by specific denitrosylating mechanisms5. However, in addition, S-nitrosylation is intrinsically relatively labile, so that, for example, mass spectrometric analysis in most cases requires conversion of SNO to a more stable form.
A major advance was provided by the development of the “biotin-switch” method in 20016. In this approach, free thiols are blocked by a disulfide-forming reagent, the NO group is cleaved from S-nitrosylated Cys by treatment with ascorbate, and the newly available Cys thiols are coupled by a disulfide to biotin for subsequent analysis. Variants of this method have proliferated. Alternative blockers of free thiols including alkylating agents, alternative reducers of the S-NO bond7, and multiple alternative reagents for labeling nascent thiols following SNO-specific reduction have been described8–10. In addition, phenylmercury has been employed to label SNOs without prior reduction11, and phosphine-based approaches show great promise12. Recent advances in redox proteomics and the use of isobaric reagents in conjunction with mass spectrometry have led to not only the identification of an increasing number of SNO-proteins but has also permitted relative quantification across various samples, allowing for greater insight into the regulation of signaling by protein S-nitrosylation.
Unexpectedly, proteomic analyses including the new study of Chung et al.13 that have employed alternative labels for nascent thiols have generated large disparities between identified sets of SNO-proteins. Rigorous controls,11, 14 combined with accumulating evidence that alternative redox-based modifications target non-overlapping sets of thiols15, 16, indicate that “specificity” (or lack thereof) is not the cause of disparity. Rather, as described below, these disparities likely highlight the fact that all biological SNOs are not alike, and in particular that the SNO linkage can vary substantially in chemical character. Additionally, the free thiols generated from the reduction of SNOs are also not identical in terms of reactivity.
Chung et al.13 performed a modified biotin-switch on mice hearts and S-nitrosoglutathione (GSNO) treated human embryonic kidney (HEK) cells using two different thiol-reactive reagents to label thiols generated by ascorbate. Although the isobaric reagents used, cysteine-reactive tandem mass tag (cysTMT) and iodoacetyl tandem mass tag (iodoTMT), differed only in their cysteine-reactive groups, and the number of SNO-proteins identified by the two methods was similar, the SNO-proteomes identified overlapped by only ~30%. In combination, Chung et al.13 identified 1008 SNO-sites (371 sites by both reagents) in GSNO-treated HEK 293 cell lysates, as well as 315 endogenous SNO-sites (98 sites by both reagents) in untreated samples from mouse hearts. A number of previously undescribed targets of S-nitrosylation were found in cardiac samples including SERCA2A, alpha-actinin-2 and isoform 3 of LIM domain-binding protein 3. Their analysis suggests that studies identifying SNO-proteomes using only a single label will detect incomplete sets of SNO-proteins.
The discrepancy in the populations of SNO-proteins identified by different methods can be ascribed to two aspects of SNOs: first, all SNOs are not identical because they vary in their structure (chemical nature), and second, the reactivity and local environment of all S-nitrosylated cysteines residues are not identical. With regard to structural differences between SNOs, the chemical structure of the SNO group is determined by a number of factors, which reflect its reactivity17. The R-SNO bond dihedral angle may be present in cis- or trans-configuration. Further, “SNOs” can have varying degrees of double bond character (R-S-N=O to R-S+=N-O−) or might carry an unpaired electron (RSNO·− or RSNO·H)17. SNOs can also interact with another thiol to form a nitroxyl disulfide [(RS)2NO−]18. Moieties in the near vicinity of SNOs including the aromatic amino acid tyrosine can mediate π-π-interactions, and hemes can also influence the character of the SNO (i.e., heme/thiol/NO redox coupling). SNOs of various forms cannot be differentiated from each other with current detection methods. Indeed, as far as is known, the sensitivity and specificity of current methods has only been determined for the simple cis-dihedral R-S-N=O. It should be noted that Chung et al.13 employed somewhat dissimilar conditions to reduce SNOs prior to labeling: 1 mM ascorbate/1 mM copper for cysTMT and 5 mM ascorbate for iodoTMT. Because inclusion of copper changes the chemical mechanism by which NO is liberated from S-NO19, it seems likely that the reactivities and chemistries of varied SNO-forms would differ under these disparate conditions. Moreover, the thermodynamic grounds for specificity of the biotin-switch for SNO vs. alternative oxidative modifications of thiols (sulfenic acid and mixed disulfides)14 does not hold in the presence of copper and so caution is warranted.
Another recent study used two different methods to enrich for isoproterenol-stimulated S-nitrosylation of calcium handling proteins in the heart20. Although the phenylmercury and biotin-switch methods identified similar numbers of SNO-proteins under resting conditions, the proportion of those that showed increases following isoproterenol was greater with phenylmercury. Again, the disparity in targets identified by the two methods can most likely be attributed to the existence of multiple SNO-populations.
Although not completely understood, the factors that determine the target specificity of S-nitrosylation include the local environment of the cysteine residue and the nature of the NO donor/source21, 22. Transnitrosylation from a SNO-protein or from low-molecular-weight SNOs including GSNO or S-nitroso-coenzyme A (SNO-CoA) is an important mode of S-nitrosylation. Distinct sets of proteins are S-nitrosylated by GSNO or SNO-CoA21, 23 demonstrating differences in accessibility/reactivity of cysteines. By the same rationale, it seems likely that labeling and subsequent identification of SNO-sites reflect the nature of the thiol generated by ascorbate-dependent reduction, even when proteins are denatured (see below). In this regard, it is of interest that, compared to iodoTMT labeled cysteines, cysTMT labeled cysteines tended to be surrounded by more positively charged residues and cysTMT labeled SNO-proteins also had a higher aliphatic index13. In addition, enhanced labeling of samples from GSNO reductase knockout (GSNOR−/−) mice, which would preferentially promote SNO-sites that are regulated by GSNO levels, was preferentially observed with cysTMT.
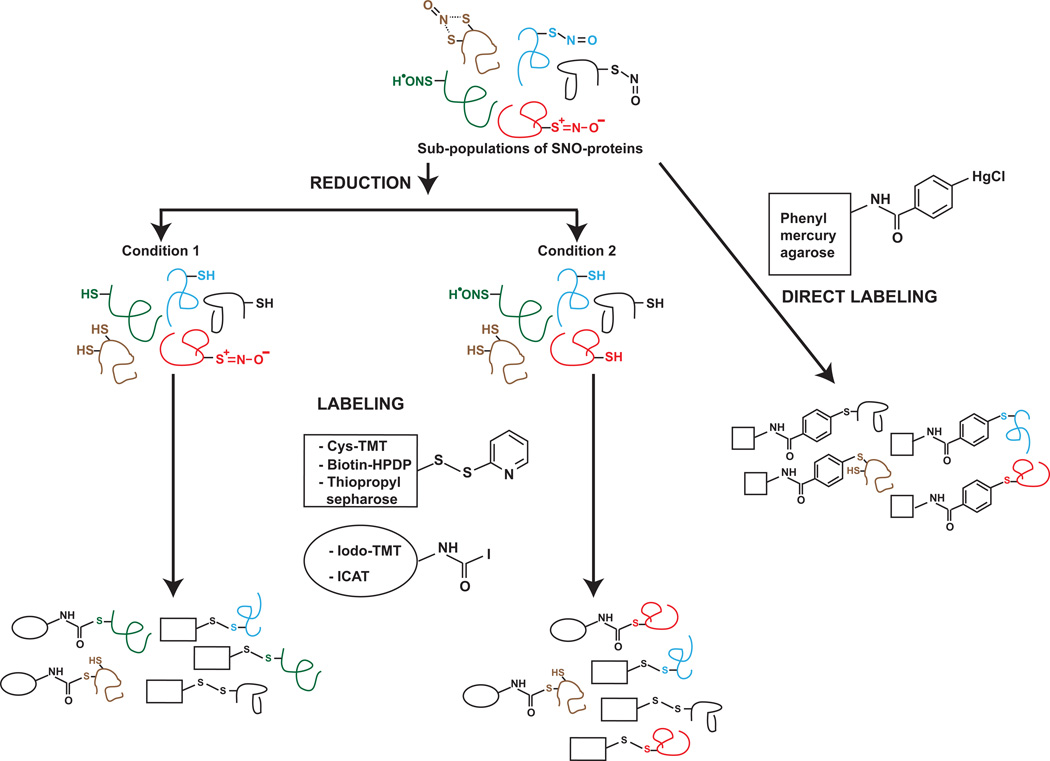
Cysteine labeling reagents used for proteomics fall mainly into two classes: pyridyldithiol-based reagents that reversibly label thiols via a disulfide bond (including cysTMT, biotin-HPDP and thiopropyl sepharose) and iodoacetyl-based reagents that irreversibly alkylate cysteines by nucleophilic substitution of iodine with the sulfur atom (including iodoTMT and isotope-coded affinity tag (ICAT)). Other methods circumvent the reduction step and use reagents that directly react with SNO-cysteine. One such approach employs the reaction of phenylmercury compounds with SNO-cysteines (following blocking of free thiols) to form a tight bond with mercury to capture SNO-proteins15. Direct reaction of SNOs with triarylphosphine has also been used to quantify GSNO in cells12 and early reports from the Tannenbaum group indicate that this reagent may provide unique molecular signatures for SNO-proteins as well. Considering the diverse chemistries involved in the labeling of cysteines using different reagents, it is easy to envision that the same level of sensitivity of detection will not be achieved with all reagents across all subpopulations of SNO-proteins. Thus, the use of more than one label, preferably involving different chemistries, would optimize capture of the SNO-proteome, a large proportion of which may otherwise be missed (Figure 1).
Figure 1.
Hypothetical situations leading to differential identification of sub-populations of SNO by different methods.
Some of the differences among SNO-proteins that might be expected to influence detectability will be negated by the fact that the detection assays are carried out under denaturing conditions (and certain SNO populations that are stabilized by secondary, tertiary or quaternary structure would be lost). On the other hand, commonly employed denaturing detergents (SDS) are highly charged and might themselves influence the efficacy of reducing and labeling reactions. Also, it is possible that denaturation of proteins might be incomplete. Several examples of SDS-resistant protein complexes are known, which are dissociated only at high temperatures. Again, current methods to detect and identify SNO-proteins are based on the chemistry of a simple S-nitrosothiol and are aimed at differentiating between SNOs and other cysteine redox modifications, in particular sulfenics and disulfides. In light of these new studies13, 20, SNO-detection protocols should be modified to optimize identification of multiple SNO-populations that exist in biological samples.
Cellular S-nitrosylation levels, and hence the SNO-proteome, are regulated in substantial part by denitrosylases. GSNO reductase (GSNOR) shifts the equilibrium towards protein denitrosylation by reducing GSNO and thereby regulates cardiovascular function and cardioprotection24, 25. A GSNOR−/− mouse has inherently higher levels of endogenous protein S-nitrosylation and consequently can be used in proteomic studies to facilitate the identification of SNO-sites. Chung et al.13 identified 493 SNO-sites in GSNOR−/− hearts, and many of these were hypernitrosylated compared to WT hearts. Irie et al.20 established the specificity of a subset of SNO-proteins identified in their study by comparing SNO-levels in WT with SNO-levels in transgenic mice in which GSNOR was overexpressed. As expected, S-nitrosylation of target proteins was diminished in the transgenic mouse samples. That levels of SNO-proteins are increased upon knock out of GSNOR and decreased by overexpression of GSNOR as assessed by both biotin-switch and phenylmercury methods represents a powerful genetic demonstration of the specificity of the current methodologies toward SNOs.
S-nitrosylation in the cardiac system is precisely regulated, but the factors influencing the cardiac SNO-proteome are not fully understood. Chung et al.13 found not only hypernitrosylated SNO-sites in GSNOR-knockout mouse hearts, but also a significant number of hyponitrosylated SNO-sites. S-nitrosylation is known to regulate transcription and to modulate stability of proteins. There are numerous examples of S-nitrosylation facilitating degradation of proteins including the phosphatase PTEN26, iron-regulatory protein-227 and Pias328. On the other hand, S-nitrosylation also increases the stability of many proteins including membrane repair protein TRIM724, HIF-1α29 and Bcl230. Because S-nitrosylation can alter protein production and stability and thereby protein abundance, it may be informative, although often overlooked, to evaluate SNO-protein levels relative to protein abundance under conditions of altered S-nitrosylation.
The issues faced in the identification of the SNO-proteome are largely reminiscent of the challenges faced in identification of the phosphoproteome. Different phospho-sites are identified preferentially by different methods31 due to multiple factors (e.g. differential enrichment of diverse subsets of phosphoproteins and interference with proteolytic digestion by phosphorylation itself). The use of multiple proteases and enrichment methods has been proposed to facilitate identification of the phosphoproteome. The methodologies used to identify the SNO-proteome are rapidly evolving and have led to the identification of thousands of SNO-proteins in various tissues and conditions. The results of Chung et al.13 emphasize the wisdom of employing more than one strategy to assure adequate coverage of the SNO-proteome. More generally, however, inasmuch as SNOs differing with respect to structure and source of NO group can be predicted to have different effects on protein function32, 33, more specific methods that can distinguish between different SNO forms will be needed for understanding the broader repertoire of NO-based protein modification and the molecular code through which alternative redox-based modifications elicit different effects. Careful validation of current methods with respect to different SNO subtypes and new approaches to detect specific SNOs will potentially reveal the full extent of regulation of cell signaling by S-nitrosylation and thus elucidate its essential role in normal physiology and disease causation.
Acknowledgments
Sources of funding
This work was supported by National Institutes of Health grants HL075443 and GM099921.
Footnotes
Disclosures
None
References
- 1.Murad F. Cyclic guanosine monophosphate as a mediator of vasodilation. J Clin Invest. 1986;78:1–5. doi: 10.1172/JCI112536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: Synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abunimer A, Smith K, Wu TJ, Lam P, Simonyan V, Mazumder R. Single-nucleotide variations in cardiac arrhythmias: Prospects for genomics and proteomics based biomarker discovery and diagnostics. Genes (Basel) 2014;5:254–269. doi: 10.3390/genes5020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy E, Kohr M, Menazza S, Nguyen T, Evangelista A, Sun J, Steenbergen C. Signaling by S-nitrosylation in the heart. J Mol Cell Cardiol. 2014;73:18–25. doi: 10.1016/j.yjmcc.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: Enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 6.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 7.Kallakunta VM, Staruch A, Mutus B. Sinapinic acid can replace ascorbate in the biotin switch assay. Biochim Biophys Acta. 2010;1800:23–30. doi: 10.1016/j.bbagen.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Murray CI, Uhrigshardt H, O'Meally RN, Cole RN, Van Eyk JE. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.013441. M111 013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan KT, Chen YY, Pu TH, Chao YS, Yang CY, Bomgarden RD, Rogers JC, Meng TC, Khoo KH. Mass spectrometry-based quantitative proteomics for dissecting multiplexed redox cysteine modifications in nitric oxide-protected cardiomyocyte under hypoxia. Antioxid Redox Signal. 2014;20:1365–1381. doi: 10.1089/ars.2013.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smolka MB, Zhou H, Purkayastha S, Aebersold R. Optimization of the isotope-coded affinity tag-labeling procedure for quantitative proteome analysis. Anal Biochem. 2001;297:25–31. doi: 10.1006/abio.2001.5318. [DOI] [PubMed] [Google Scholar]
- 11.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seneviratne U, Godoy LC, Wishnok JS, Wogan GN, Tannenbaum SR. Mechanism-based triarylphosphine-ester probes for capture of endogenous rsnos. J Am Chem Soc. 2013;135:7693–7704. doi: 10.1021/ja401565w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung HS, Murray CI, Venkatraman V, Crowgey EL, Rainer PP, Cole RN, Bomgarden RD, Rogers JC, Balkan W, Hare JM, Kass DA, Van Eyk JE. Dual labeling biotin switch assay to reduce bias derived from different cysteine subpopulations: A method to maximize S-nitrosylation detection. Circ Res. 2015 doi: 10.1161/CIRCRESAHA.115.307336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 15.Doulias PT, Tenopoulou M, Raju K, Spruce LA, Seeholzer SH, Ischiropoulos H. Site specific identification of endogenous S-nitrosocysteine proteomes. J Proteomics. 2013;92:195–203. doi: 10.1016/j.jprot.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gould NS, Evans P, Martinez-Acedo P, Marino SM, Gladyshev VN, Carroll KS, Ischiropoulos H. Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem Biol. 2015;22:965–975. doi: 10.1016/j.chembiol.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: Purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 18.Houk KN, Hietbrink BN, Bartberger MD, McCarren PR, Choi BY, Voyksner RD, Stamler JS, Toone EJ. Nitroxyl disulfides, novel intermediates in transnitrosation reactions. J Am Chem Soc. 2003;125:6972–6976. doi: 10.1021/ja029655l. [DOI] [PubMed] [Google Scholar]
- 19.Holmes AJ, Williams DLH. Reaction of ascorbic acid with S-nitrosothiols: Clear evidence for two distinct reaction pathways. Journal of the Chemical Society-Perkin Transactions 2. 2000:1639–1644. [Google Scholar]
- 20.Irie T, Sips PY, Kai S, Kida K, Ikeda K, Hirai S, Moazzami K, Jiramongkolchai P, Bloch DB, Doulias PT, Armoundas AA, Kaneki M, Ischiropoulos H, Kranias E, Bloch KD, Stamler J, Ichinose F. S-nitrosylation of calcium-handling proteins in cardiac adrenergic signaling and hypertrophy. Circ Res. 2015;117 doi: 10.1161/CIRCRESAHA.115.307157. xxx-xxx [in this issue] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster MW, Forrester MT, Stamler JS. A protein microarray-based analysis of S-nitrosylation. Proc Natl Acad Sci U S A. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seth D, Stamler JS. The sno-proteome: Causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand P, Hausladen A, Wang YJ, Zhang GF, Stomberski C, Brunengraber H, Hess DT, Stamler JS. Identification of S-nitroso-coa reductases that regulate protein S-nitrosylation. Proc Natl Acad Sci U S A. 2014;111:18572–18577. doi: 10.1073/pnas.1417816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beigi F, Gonzalez DR, Minhas KM, Sun QA, Foster MW, Khan SA, Treuer AV, Dulce RA, Harrison RW, Saraiva RM, Premer C, Schulman IH, Stamler JS, Hare JM. Dynamic denitrosylation via S-nitrosoglutathione reductase regulates cardiovascular function. Proc Natl Acad Sci U S A. 2012;109:4314–4319. doi: 10.1073/pnas.1113319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lima B, Lam GK, Xie L, Diesen DL, Villamizar N, Nienaber J, Messina E, Bowles D, Kontos CD, Hare JM, Stamler JS, Rockman HA. Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci U S A. 2009;106:6297–6302. doi: 10.1073/pnas.0901043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwak YD, Ma T, Diao S, Zhang X, Chen Y, Hsu J, Lipton SA, Masliah E, Xu H, Liao FF. NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol Neurodegener. 2010;5:49. doi: 10.1186/1750-1326-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Wing SS, Ponka P. S-nitrosylation of irp2 regulates its stability via the ubiquitin-proteasome pathway. Mol Cell Biol. 2004;24:330–337. doi: 10.1128/MCB.24.1.330-337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu J, Liu GH, Wu K, Han P, Wang P, Li J, Zhang X, Chen C. Nitric oxide destabilizes pias3 and regulates sumoylation. PLoS One. 2007;2:e1085. doi: 10.1371/journal.pone.0001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li CY. Regulation of hif-1alpha stability through S-nitrosylation. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azad N, Vallyathan V, Wang L, Tantishaiyakul V, Stehlik C, Leonard SS, Rojanasakul Y. S-nitrosylation of bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J Biol Chem. 2006;281:34124–34134. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- 31.Solari FA, Dell'Aica M, Sickmann A, Zahedi RP. Why phosphoproteomics is still a challenge. Mol Biosyst. 2015;11:1487–1493. doi: 10.1039/c5mb00024f. [DOI] [PubMed] [Google Scholar]
- 32.Kim SO, Merchant K, Nudelman R, Beyer WF, Jr, Keng T, DeAngelo J, et al. OxyR: A molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 33.Seth D, Hausladen A, Wang YJ, Stamler JS. Endogenous protein S-nitrosylation in E. coli: Regulation by OxyR. Science. 2012;336:470–473. doi: 10.1126/science.1215643. [PubMed: 22539721] [DOI] [PMC free article] [PubMed] [Google Scholar]