Abstract
In this review we present recent developments in the analysis of Plasmodium vivax clinical trials and ex vivo drug-susceptibility assays, as well approaches currently being used to identify molecular markers of drug resistance. Clinical trials incorporating the measurement of in vivo drug concentrations and parasite clearance times are needed to detect early signs of resistance. Analysis of P. vivax growth dynamics ex vivo have defined the criteria for acceptable assay thresholds for drug susceptibility testing, and their subsequent interpretation. Genotyping and next-generation sequencing studies in P. vivax field isolates are set to transform our understanding of the molecular mechanisms of drug resistance.
Keywords: Plasmodium vivax, drug resistance, malaria, clinical trials, chloroquine, in vitro drug susceptibility test, genotyping, molecular markers
The emerging threat of drug resistant Plasmodium vivax
Over the past decade, substantial investments in malaria research and control activities have had a significant impact in reducing the burden of falciparum malaria. The successful control and ultimate elimination of P. vivax is more challenging, a consequence of its transmission dynamics and ability to form dormant liver stages (hypnozoites), which can result in recurrent blood-stage infections (relapses) weeks to months after initial infection. These characteristics help the parasite to survive in environments that are hostile to the mosquito vector for much of the year.
The first-line treatment of vivax malaria has changed little over the past 60 years. In the 1930s a drug discovery program resulted in the identification and development of several potent antimalarial drugs, including chloroquine (CQ), the antifolates, and primaquine [1]. Intensive research over the next two decades demonstrated that CQ was highly effective and safe, and by the 1950s it became the first-line treatment for P. vivax infection, coadministered with primaquine to prevent relapses from dormant liver-stages of the parasite. However, this combination is increasingly threatened by the emergence and spread of drug-resistant strains of P. vivax.
The first reports of CQ resistant (CQR) P. vivax began to emerge in 1989 [2,3], 30 years after the documentation of CQR Plasmodium falciparum. Intrinsic biological differences between these two species may account for much of this discrepancy, particularly the early development of gametocytes and the transmission potential of relapsing infections [4]. Two non-biological factors are also likely to play an important role: complacency and a lack of robust diagnostic tools. The former reflects the perception of vivax malaria as a benign infection of low public health relevance. The latter reflects the inherent complexity of distinguishing resistant from relapsing infections.
P. vivax infection is not a benign disease; on the contrary, it has a major impact on the health of vulnerable populations in poorly resourced communities, particularly pregnant women and young children [5–8]. It is also associated with severe and fatal disease and substantial morbidity [9–11]. On the island of New Guinea, clinical trials have demonstrated unequivocal evidence of high-grade resistance to CQ, with early clinical deterioration requiring hospitalization, delayed parasite clearance, and early recurrent parasitaemia [12–14]. Partially effective drug treatment has been proposed as an important contributing factor to associated reports of severe vivax malaria [15]. Evidence for declining CQ efficacy against P. vivax, albeit less pronounced than that observed in New Guinea, has now been reported from across the vivax endemic world, threatening the public health of a much larger population (Figure 1) [15,16]. The effective detection of drug-resistant P. vivax parasites may help to combat this threat if timely changes in treatment policy can be implemented. In this article we review the difficulties in characterising drug-resistant P. vivax and current strategies being applied to overcome these.
Figure 1.
Reports of CQR Plasmodium vivax in 2009. Red stars highlight areas of confirmed resistance as defined by greater than 10% recurrence (and greater than five clinical treatment failures) by day 28 with or without measurement of chloroquine drug concentration; orange diamonds locate area suggestive of resistance as defined by less than 10% recurrence (or less than five clinical treatment failures) by day 28, with chloroquine drug concentrations; yellow circles locate sites of possible resistance as defined by less than 10% recurrence (or less than five clinical treatment failures) by day 28, without drug concentrations. Adapted from [16].
The clinical response
World Health Organization (WHO) protocols for the evaluation of antimalarial efficacy focus primarily on the treatment of P. falciparum. These guidelines have undergone extensive revision over the past 20 years, the more recent versions extending their scope to investigate therapeutic efficacy against P. vivax infections. Declining antimalarial efficacy is manifested by a reduction in the speed of initial parasite clearance (early treatment failure, ETF) and by the inability of what should be a curative treatment regimen to eliminate all blood-stage parasites from the body (late treatment failure, LTF, or late parasitological failure, LPF). ETF is usually a marker of advanced clinical drug resistance, whereas an earlier indicator of declining drug efficacy is the reappearance of recrudescent infections many days after the initial treatment. The timing of the recurrence is dependent upon the pharmacology of the initial treatment regimen, the degree of drug resistance, and the level of host immunity. In P. vivax infections the interpretation of the LPF outcome is confounded not only by reinfection (in patients remaining within an area of ongoing transmission), but also the occurrence of relapses, arising from activation of the dormant liver-stage parasites.
The use of genotyping to distinguish the origin of recurrent infections
The development of molecular genotyping in the 1990s provided a useful tool for discriminating true recrudescence from reinfection, and was a crucial factor in refining our ability to characterise drug-resistant P. falciparum. Although a standardised genotyping protocol was published by the WHO in 2007, no recommendations were made for its application to P. vivax [17]. A variety of methodologies have been developed for P. vivax with as few as three polymorphic markers proving to be sufficient to discriminate homologous from heterologous infections [18–21]. Although this raises the potential for a standardised approach for PCR-adjustment in P. vivax clinical trials, the application of these molecular approaches and their interpretation are less straightforward. Recurrence of P. vivax genetically identical to the pretreatment isolate can occur from either a true recrudescence of the initial infection or from a relapse from hypnozoites generated by the prior blood-stage infection [19,22]; current molecular methods are unable to distinguish between these alternatives. A high prevalence of multiple clones is often observed with P. vivax infections, even in areas of low transmission [23], undermining further the comparison of paired isolates. Finally, parasitaemia in P. vivax infections can be an order of magnitude lower than those with P. falciparum, reducing the sensitivity to detect minority clones and increasing the likelihood of indeterminate genotyping results. The confounding effect of relapsing infections varies considerably between geographical locations, both for the absolute risk of relapse and the timing at which these occur. In equatorial regions, 50–80% of patients can have a relapse starting within 3 weeks of the initial infection, whereas in patients infected by temperate strains, the risk of relapse may fall to 5–20%, recurrences occurring months after the initial infection [24].
Complex interpretation of post-treatment prophylaxis
The timing of recurrence is also determined by the treatment administered. Long-acting antimalarial drugs, such as CQ, mefloquine, and piperaquine, delay the time of the first observed recurrence. Expansion of P. vivax parasite biomass will occur only once drug concentrations in the blood have fallen below the minimum inhibitory concentration (MIC) of the infecting parasite. Clinical studies of CQ-sensitive parasites have demonstrated that if a total dose of 25 mg/kg is well absorbed, then the first relapse should not be detectable for approximately 35 days following treatment, by which time the mean whole-blood concentrations of CQ will have fallen to below 100 ng/ml [25]. Therefore, any P. vivax recurrence within 28 days raises suspicion of a CQR infection, although pharmacokinetic analysis is required to confirm adequate drug absorption and the ability of the parasite to grow in the presence of blood concentrations known to exceed the MIC of a sensitive parasite [25].
Although the current 28-day therapeutic efficacy study for CQ efficacy in P. vivax has proven useful in identifying populations of emerging drug resistance, it has notable shortcomings. The test is insensitive in detecting parasites at an early stage of resistance because it only detects recurrent parasites capable of recurring in the presence of relatively high blood concentrations of CQ, whereas early stages of clinical resistance to long-acting antimalarial drugs may not manifest until 63 days or more after treatment.
Relation of biomarkers to clinical outcomes
Caution is needed when attempting to extrapolate the biomarkers of resistance to the clinical response. In the current study protocol, treatment failures predominantly represent the ability of the recurrent parasite to grow in blood concentrations above in vivo MIC of a sensitive parasite, and not necessarily the therapeutic response of the initial parasite biomass. If the recurrent parasite infection originated from relapse, reinfection, or from outgrowth of an originally minor population, then no correlation between a putative molecular marker of resistance and the pretreatment parasite isolate would be expected. Hence, additional analysis should be placed on correlates of the initial speed of parasite clearance after treatment. Emerging drug resistance in P. falciparum is manifest by a delay in parasite clearance as parasite MIC begins to rise and the parasite reduction slows [26]. This situation is currently the focus of surveillance for artemisinin resistance, but has been shown also to be a manifestation of CQR P. vivax [13]. For example, in a recent clinical study conducted on the Thai Myanmar border, patients still parasitaemic 48 h after their first dose of CQ were at a fourfold greater risk of recurrence at day 28 compared to those with faster initial parasite clearance [27]. It should be noted that primaquine has both blood and tissue schizontocidal activity, and hence its early coadministration can augment initial asexual parasite clearance, thus masking early indications of declining CQ efficacy. Where possible, clinical efficacy trials should delay the use of primaquine for radical cure until after the end of the follow-up period.
Current pragmatic approach
In view of the difficulties in confirming true recrudescence, many clinical efficacy studies of P. vivax have adopted a pragmatic approach, reporting and comparing the overall risk of recurrent P. vivax infection within a defined period of time. These assessments reflect both blood-stage schizontocidal and post-treatment prophylaxis. In equatorial regions the risk of recurrence within 28 days of treatment with artemether–lumefantrine may exceed 20% [28], but this reflects high blood schizontocidal efficacy combined with a short period of post-treatment prophylaxis afforded by lumefantrine (~16 days); the recurrent infections are likely to be due almost entirely to relapsing infections rather than to true recrudescence. Paradoxically, a much lower recurrence rate within 28 days (e.g., 10%) following CQ should raise greater alarm for emerging drug resistance because neither relapse, reinfection, nor recrudescent sensitive parasites would be expected to recur in the presence of suppressive CQ concentrations. Hence, the unadjusted risk of recurrence needs to be interpreted in light of the half-life of the treatment regimen, concomitant use of primaquine, the timing of recurrence, available molecular data, schizontocidal drug concentrations, entomological inoculation rates, and the epidemiology of relapse in the study area.
The ex vivo response
For P. falciparum, the in vitro assessment of parasite drug susceptibility, involving short-term culture of parasites in the presence of serial drug concentrations, has proved to be extremely useful in assessing intrinsic susceptibility to antimalarial drugs. The development of a continuous in vitro culture system has allowed analysis of laboratory-adapted isolates that can be genetically manipulated and assessed repeatedly over time to elucidate mechanisms of drug resistance. The in vitro test has been applied to monitoring P. falciparum drug resistance in field isolates, although heterogeneity of methodologies results in significant difficulties in comparing results between different laboratories and across time [29].
The analysis of P. vivax drug susceptibility in vitro is more challenging. Unlike P. falciparum, the parasite preferentially invades young red blood cells, limiting its reproductive capacity and ability to adapt in continuous in vitro culture [30,31]. There are relatively few parasite strains available for laboratory investigations; these have been obtained by adapting and propagating clinical isolates in non-human primate hosts such as Aotus and Saimiri monkeys and chimpanzees. Without culture adaptation the ex vivo assessment of drug susceptibility in P. vivax field isolates has been limited to clinical samples derived directly from the human host and subjected to short-term culture and drug exposure, as exemplified by the schizont maturation test [32–35]. However, such an approach is vulnerable to confounding factors of the clinical scenario, such as time from venipuncture to processing, a priori administration of antimalarial medication, and synchronicity of infection [36]. Furthermore, the inability to sustain in vitro growth restricts analysis of field isolates to a single time-point, making assessment of reproducibility difficult.
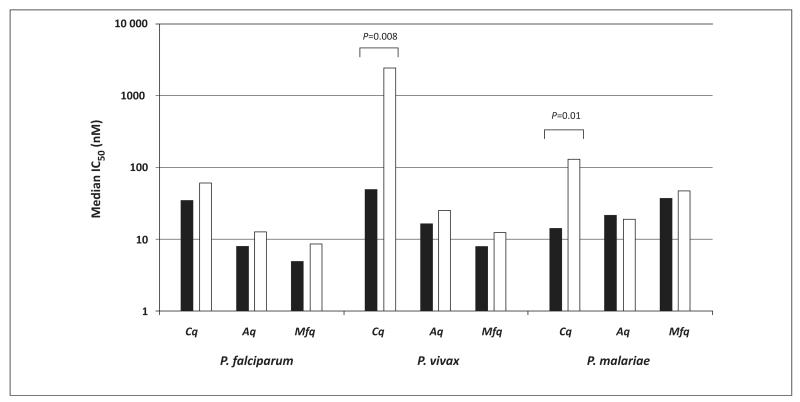
A variety of methods have been used to quantify parasite growth in the schizont maturation test, including microscopy [36], enzymatic assays [35], and nucleic acid staining followed by fluorometry or cytometry [37,38]. These different approaches are likely to explain some of the discrepancies between results from different laboratories [35,39,40]. The evaluation of the ex vivo drug response in P. vivax is further confounded by a marked stage-specificity of drug activity. Stage specificity is particularly apparent for CQ activity in P. vivax, the trophozoite stages being almost completely resistant to CQ [36]. These observations are potentially confounded by a difference in the duration of drug exposure, which is inherently shorter for trophozoite stages than for earlier ring stages. However, the stage specificity remains even when antimalarial drug exposure is equalised by shortening the exposure time for ring-stage parasites [41]. Irrespective of the duration of exposure, stage-specific activity is markedly different between classes of drugs and different Plasmodium species (Figure 2).
Figure 2.
Ex vivo drug susceptibility for isolates initially at majority ring stage (black bars) and majority trophozoite stage (white bars). Marked reduced susceptibility of P. vivax and P. malariae trophozoites to chloroquine (CQ) compared to ring stages. This difference is not apparent in P. falciparum or following exposure to amodiaquine (AQ) or mefloquine (MFQ). Data derived from [36,49,90].
Whereas clinical isolates of P. falciparum almost always present as synchronous infections (i.e., all peripheral parasites at early ring stage), P. vivax parasitaemia includes most stages of development. The degree of synchronicity correlates with host factors, such as the age of the patient, geographical location, and transmission intensity. The schizont maturation test is generally terminated once a particular proportion of asexual parasites have become schizonts (usually about 40%), thus variation in the initial composition of parasite developmental stages determines the assay duration. Shorter-duration assays are likely to contain predominantly late trophozoite or schizont stages, and they will thus result in unrealistically high IC50 values, indicative of drug exposure after the window of susceptibility [36]. The ex vivo drug response, therefore, needs to be interpreted according to the initial stage of the parasite and the duration of the assay. Recent threshold analysis suggests that parasites need to be exposed to the drug for at least 30 h before they mature to schizonts to ensure derivation of robust IC50 values [42].
Recent funding opportunities have stimulated renewed efforts aimed at generating laboratory methods capable of sustaining P. vivax in continuous in vitro culture. If successful, such systems will transform the current ex vivo assay, accommodating cryopreservation of field isolates; thereby reducing reliance on the analysis of fresh isolates and enabling repeated in vitro testing of the same parasite isolate. In the meantime, despite its limitations, the current schizont maturation assay has demonstrated utility in discriminating between parasite populations with different degrees of CQ resistance. There was a fivefold difference in IC50 values observed between Papua Indonesia, where clinical CQ resistance is high, compared to Thailand, where CQ retains reasonable efficacy [34,40,43]. The current assay has also been able to characterise drug susceptibility profiles of P. vivax to commonly used antimalarial drugs [44,45], as well as screening of P. vivax susceptibility to novel therapeutic agents [46–50].
Genetic basis of P. vivax drug resistance
The molecular mechanism of P. vivax CQ resistance remains elusive. Earlier studies focusing on genes known to be key determinants of CQ resistance in P. falciparum have failed to demonstrate a strong correlation between pvcrt-o and pvmdr1 genotypes and the CQR phenotype. Nomura et al. compared pfcrt homologues from several Plasmodium species, and although synteny was observed between P. falciparum and P. vivax, they found no relationship between mutations in pvcrt-o and clinical response to CQ [51]. Sa et al. transfected pvcrt-o into two heterologous systems: in P. falciparum, pvcrt-o expression resulted in a 2.2-fold increase in CQ susceptibility which could be reversed by verapamil. They also showed that expression of pvcrt-o engineered to have either a lysine or a threonine at position 76 in a Dictyostelium discoideum model was associated with impaired accumulation of CQ. These results suggested a potential role for pvcrt-o in CQ transport and accumulation in P. vivax, independent of the analogous lysine to threonine mutation at position 76 in pfcrt [52]. Field studies of more than 330 isolates from Thailand, Indonesia, and Madagascar, found no relationship between any pvcrt-o sequence polymorphisms and either clinical or in vitro susceptibility phenotype [40,53]. Transcription levels of pvcrt-o have only been assessed in a study of a returning traveller with severe vivax malaria contracted from the Brazilian Amazonia [54]. This study demonstrated 21-fold and twofold increases in transcript levels of pvcrt-o and pvmdr1, respectively, compared to a P. vivax isolate from a patient with non-severe disease. Further studies are required to determine the role of overexpression of pvcrt-o in CQR P. vivax.
A stronger correlation has been demonstrated between ex vivo CQ resistance and sequence polymorphisms in pvmdr1. A sequence polymorphism in pvmdr1 conferring Y976F was present in all patients presenting to a clinic in Papua Indonesia, where CQR P. vivax is both at high prevalence (~65%) and high level [40]. By contrast, the same polymorphism was identified in only 25% of Thai isolates from an area where CQ retains efficacy. The ubiquitous presence of Y976F in Papua precluded correlation with ex vivo drug susceptibility to CQ, but in the Thai isolates, the Y976F substitution was associated with a 1.7-fold higher IC50 to CQ. In a complementary study from Papua New Guinea, the pvmdr1 Y976F substitution together with quadruple mutant pvdhfr has been shown to correlate with treatment failure following amodiaquine plus sulphadoxine–pyrimethamine (SP) combination therapy [55]. However, because CQ resistance can occur in isolates with wild type pvmdr1 [40,56,57], pvmdr1 mutations are likely to be, at best, minor determinants of CQ susceptibility.
In contrast to CQ resistance, mechanisms of P. vivax resistance to mefloquine and SP are similar to those observed in P. falciparum. Pvmdr1 amplification was reported in 21% of Thai isolates (maximum three copies), with increased copy-number being associated with a twofold rise in mefloquine IC50 values [44]. Other studies have confirmed amplification of pvmdr1 in P. vivax isolates from Thailand and Laos, although this was not apparent in Indonesian and Myanmar isolates [44,58]. These findings suggest that pvmdr1 amplification may play a role in P. vivax susceptibility to mefloquine, with selection for isolates with increased copy-number occurring in areas with high mefloquine pressure. Candidate molecular markers for monitoring drug resistance in P. vivax are listed in Table 1.
Table 1.
Candidate molecular markers for monitoring drug resistance in P. vivaxa
| Molecular markers | CQ | Mefloquine | Antifolate | Amodiaquine + SP |
|---|---|---|---|---|
| Pvcrt-o | Overexpression [52] |
– | – | – |
| Pvmdr1 | Y976F, F1076L [40,88] |
Gene amplification [44,58] |
– | Y976F [55] |
| Pvdhfr | – | – | F57L, S58R, T61M, S117T, S117N [55,59–63] |
F57L, S58R, T61M, S117T [55] |
| Pvdhps | – | – | A383G, A553G [67,89] |
– |
Each marker is listed as: the amino acid position with the amino acid of the wild type allele (left) and the amino acid substitution caused by the mutation (right).
Abbreviations: CQ, chloroquine; SP, sulphadoxine–pyrimethamine.
The roles of pfdhfr and pfdhps have been well characterized in P. falciparum and the relationship with clinical and in vitro drug susceptibility has been established. P. vivax isolates exposed to intense antifolate pressure revealed sequential acquisition of mutations in pvdhfr and pvdhps [14,55,59–63]. Episomal or stable P. falciparum transfectants expressing mutant pvdhfr have demonstrated a clear role of pvdhfr mutations in conferring in vitro resistance to conventional antifolate drugs [64–66]. Several mutations have also been identified in pvdhps where a similar effect has been predicted; however, data on the phenotypic correlation with these genotypes are limited [67].
Strategies for identifying new resistance loci
Analysis of orthologous genes highlights significant differences in the molecular basis of drug resistance between Plasmodium species. Therefore, the detection of novel drug-resistance loci in P. vivax will require a less constrained approach. The classical genetic cross of CQ sensitive and CQ resistant isolates, followed by linkage analysis, was pivotal in identifying the pfcrt locus in P. falciparum; however, a similar approach in P. vivax will have to overcome the considerably greater difficulties generated by the inability to culture continuously ex vivo. The need to clone and cultivate the progeny in classical linkage studies renders this approach dependent upon propagation in non-human primate hosts. Alternatively, linkage group selection, which has been applied to uncloned progeny in murine models, may offer potential for mapping resistance in P. vivax [68,69].
With ongoing technological advances and declining costs, whole genome sequencing (WGS) is now feasible for hundreds of Plasmodium isolates, permitting population-genetic approaches for detecting candidates in ‘natural’ isolates sourced directly from patients [70]. Proof-of-principle for the genome-wide association study (GWAS) approach has been demonstrated in several P. falciparum studies, with the detection of validated, as well as novel determinants of resistance [71–73]. Genome-wide scans offer potential to identify resistance loci by detecting regions under recent positive selection. When an advantageous (i.e., drug resistant) mutation spreads rapidly through a population, flanking neutral polymorphisms are carried along with it. This ‘hitchhiking’ effect creates ‘signatures of selection’ that include reduced genetic variation and increased linkage disequilibrium at flanking loci [71,72,74,75]. Methods to detect these signatures include the long-range haplotype (LRH) test, and the cross-population extended haplotype homozygosity (XP-EHH) test, which identifies contrasting patterns between populations under differing drug pressure [74,76].
WGS platforms require moderately high DNA quantities with limited human DNA ‘contamination’. These prerequisites are particularly challenging in P. vivax infections, which typically exhibit low peripheral parasitaemias and are difficult to propagate in culture. Recent studies have led to the development of a method to facilitate effective P. vivax sample preparation using a two-step process of cellulose (CF11) filtration [77,78] followed by short-term (42–48 h) ex vivo culture [36]. This approach can generate high P. vivax DNA yields (median 3.1 ng μl−1 packed red blood cells) with low human DNA contamination (median 4.2%) (Auburn et al., unpublished), but requires access to a fresh venous blood sample from a patient with modest peripheral parasitaemia (at least 1000 μl−1 in 1 ml whole blood). Whole-genome amplification methods, such as multiple-displacement amplification, may enable samples with low yield to be salvaged, but at the risk of introducing errors and coverage bias during the amplification. A recently described whole-genome capture method enables WGS analysis of archival P. vivax samples which were not subject to leukocyte filtration or short-term culture [79].
Even when quality parasite DNA is available, the high rates of polyclonality in P. vivax field isolates confounds accurate mapping of short-read sequence data, identification of authentic polymorphisms, and effective genotype calling. In P. falciparum, the extreme A+T bias particularly in non-coding genomic regions (~90%) renders these regions beyond accurate assessment using current WGS methods [80]. However, in the less A+T-biased (~58%) P. vivax genome [81,82], highly uniform coverage can be observed across both coding and non-coding regions, improving the ability to detect single-nucleotide polymorphism (SNPs) and copy-number variants. Insights from large-scale WGS efforts in P. falciparum patient isolates should greatly facilitate effective variant detection and genotyping methods for P. vivax [80].
Population substructure remains a challenge for GWASs, potentially leading to false-positive associations. Microsatellite-based studies have demonstrated a range of structures in P. vivax-endemic populations, with complex patterns presumably reflecting relapse dynamics [20,22,83,84]. WGS will enable detection of more subtle patterns that are not revealed by microsatellite analysis. Using capillary-based sequencing of a 100 Kb region on chromosome 4, Orjuela-Sanchez and colleagues demonstrated high SNP diversity in Cambodia, Vietnam, Sri Lanka, and Brazil, with evidence of substructure in the latter [85]. However, parasite population structure has yet to be defined from areas highly prevalent for drug-resistant P. vivax.
Unconstrained approaches are likely to generate a number of candidate loci. Several criteria may be used to prioritise these candidates for downstream validation, including biological plausibility, high-frequency derived alleles, hits with GWAS, and signatures of selection. Ultimately, validation of putative molecular markers and their clinical utility will require demonstration that the genotype called at these loci effectively predicts the phenotypic response to treatment as defined by appropriate in vivo and in vitro characterisation.
Concluding remarks
The last decade has witnessed a major reappraisal on the public health importance of P. vivax, and this is beginning to focus greater attention and resources to this parasite [5,86,87]. Clinical and epidemiological studies have highlighted that delays in diagnosis, the application of partially effective treatment regimens, and our inability to reliably cure the dormant stages of the parasite, all contribute to P. vivax infections – causing significant morbidity and a huge socio-economic burden [15]. High-grade drug-resistant P. vivax is a major determinant of partially effective treatment, and as such poses a significant threat to endemic countries; this threat must be quantified and contained if there is to be any realistic prospect of the global elimination of malaria.
In the absence of evidence to the contrary, there is a tendency to assume that current antimalarial treatment regimens continue to retain efficacy long after declining antimalarial activity has begun to emerge. Inadequate surveillance tools compounded by complacency delayed the detection and containment of chloroquine-resistant P. falciparum, with devastating public health consequences. If a repetition of history is to be avoided, then the threat of CQ-resistant P. vivax needs to be acknowledged and greater resources applied for developing standardised, validated, and reproducible tools for the in vivo, in vitro, and molecular characterisation of drug-resistant P. vivax.
Acknowledgements
We thank Nick White, Francois Nosten, Kevin Baird, Nick Anstey, Bruce Russell, and Lorenz von Seidlein for informative discussions that have shaped this Review. R.N.P. is a Wellcome Trust Senior Research Fellow in Clinical Science (091625) and J.M. is supported by a Career Development Fellowship from the Swiss National Science Foundation.
References
- 1.Berliner RW, et al. In: A Survey of Antimalarial Drugs, 1941–1945. Wiseogle FY, editor. Edwards; 1946. [Google Scholar]
- 2.Rieckmann KH, et al. Plasmodium vivax resistance to chloroquine? Lancet. 1989;2:1183–1184. doi: 10.1016/s0140-6736(89)91792-3. [DOI] [PubMed] [Google Scholar]
- 3.Baird JK, et al. Resistance to chloroquine by Plasmodium vivax in Irian Jaya, Indonesia. Am. J. Trop. Med. Hyg. 1991;44:547–552. doi: 10.4269/ajtmh.1991.44.547. [DOI] [PubMed] [Google Scholar]
- 4.Baird JK, et al. Diagnosis and treatment of Plasmodium vivax malaria. Adv. Parasitol. doi: 10.1016/B978-0-12-397900-1.00004-9. (in press) [DOI] [PubMed] [Google Scholar]
- 5.Price RN, et al. Vivax malaria: neglected and not benign. Am. J. Trop. Med. Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 6.Poespoprodjo JR, et al. Vivax malaria: a major cause of morbidity in early infancy. Clin. Infect. Dis. 2009;48:1704–1712. doi: 10.1086/599041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poespoprodjo JR, et al. Adverse pregnancy outcomes in an area where multidrug-resistant Plasmodium vivax and Plasmodium falciparum infections are endemic. Clin. Infect. Dis. 2008;46:1374–1381. doi: 10.1086/586743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poespoprodjo JR, et al. Severe congenital malaria acquired in utero. Am. J. Trop. Med. Hyg. 2010;82:563–565. doi: 10.4269/ajtmh.2010.09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anstey NM, et al. The pathophysiology of vivax malaria. Trends Parasitol. 2009;25:220–227. doi: 10.1016/j.pt.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Tjitra E, et al. Multidrug-resistant Plasmodium vivax Associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med. 2008;5:e128. doi: 10.1371/journal.pmed.0050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genton B, et al. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med. 2008;5:e127. doi: 10.1371/journal.pmed.0050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sumawinata IW, et al. Very high risk of therapeutic failure with chloroquine for uncomplicated Plasmodium falciparum and Plasmodium falciparum malaria in Indonesian Papua. Am. J. Trop. Med. Hyg. 2003;68:416–420. [PubMed] [Google Scholar]
- 13.Ratcliff A, et al. Therapeutic response of multidrug-resistant Plasmodium falciparum and Plasmodium falciparum to chloroquine and sulfadoxine–pyrimethamine in southern Papua, Indonesia. Trans. R. Soc. Trop. Med. Hyg. 2007;101:351–359. doi: 10.1016/j.trstmh.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karunajeewa HA, et al. A trial of combination antimalarial therapies in children from Papua New Guinea. N. Engl. J. Med. 2008;359:2545–2557. doi: 10.1056/NEJMoa0804915. [DOI] [PubMed] [Google Scholar]
- 15.Price RN, et al. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr. Opin. Infect. Dis. 2009;22:430–435. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 16.Douglas NM, et al. Artemisinin combination therapy for vivax malaria. Lancet Infect. Dis. 2010;10:405–416. doi: 10.1016/S1473-3099(10)70079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization and Medicines for Malaria Venture . Methods and Techniques for Clinical Trials on Antimalarial Drug Efficacy. World Health Organization; 2008. Genotyping to identify parasite populations. ( http://www.who.int/malaria/publications/atoz/9789241596305/en/) [Google Scholar]
- 18.Imwong M, et al. Practical PCR genotyping protocols for Plasmodium vivax using Pvcs and Pvmsp1. Malar. J. 2005;4:20. doi: 10.1186/1475-2875-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen N, et al. Relapses of Plasmodium vivax infection result from clonal hypnozoites activated at predetermined intervals. J. Infect. Dis. 2007;195:934–941. doi: 10.1086/512242. [DOI] [PubMed] [Google Scholar]
- 20.Koepfli C, et al. Evaluation of Plasmodium vivax genotyping markers for molecular monitoring in clinical trials. J. Infect. Dis. 2009;199:1074–1080. doi: 10.1086/597303. [DOI] [PubMed] [Google Scholar]
- 21.Barnadas C, et al. Characterization of treatment failure in efficacy trials of drugs against Plasmodium vivax by genotyping neutral and drug resistance-associated markers. Antimicrob. Agents Chemother. 2011;55:4479–4481. doi: 10.1128/AAC.01552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imwong M, et al. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J. Infect. Dis. 2007;195:927–933. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- 23.Imwong M, et al. Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int. J. Parasitol. 2007;37:1013–1022. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 24.White NJ. Determinants of relapse periodicity in Plasmodium vivax malaria. Malar. J. 2011;10:297. doi: 10.1186/1475-2875-10-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird JK, et al. Diagnosis of resistance to chloroquine by Plasmodium vivax: timing of recurrence and whole blood chloroquine levels. Am. J. Trop. Med. Hyg. 1997;56:621–626. doi: 10.4269/ajtmh.1997.56.621. [DOI] [PubMed] [Google Scholar]
- 26.White NJ. The assessment of antimalarial drug efficacy. Trends Parasitol. 2002;18:458–464. doi: 10.1016/s1471-4922(02)02373-5. [DOI] [PubMed] [Google Scholar]
- 27.Phyo AP, et al. Dihydroartemisinin–piperaquine versus chloroquine in the treatment of Plasmodium vivax malaria in Thailand: a randomized controlled trial. Clin. Infect. Dis. 2011;53:977–984. doi: 10.1093/cid/cir631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratcliff A, et al. Two fixed-dose artemisinin combinations for drug-resistant falciparum and vivax malaria in Papua, Indonesia: an open-label randomised comparison. Lancet. 2007;369:757–765. doi: 10.1016/S0140-6736(07)60160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bacon DJ, et al. World Antimalarial Resistance Network (WARN) II: in vitro antimalarial drug susceptibility. Malar. J. 2007;6:120. doi: 10.1186/1475-2875-6-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udomsangpetch R, et al. Short-term in vitro culture of field isolates of Plasmodium vivax using umbilical cord blood. Parasitol. Int. 2007;56:65–69. doi: 10.1016/j.parint.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Golenda CF, et al. Continuous in vitro propagation of the malaria parasite Plasmodium vivax. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6786–6791. doi: 10.1073/pnas.94.13.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tasanor O, et al. Clinical-parasitological response and in-vitro sensitivity of Plasmodium vivax to chloroquine and quinine on the western border of Thailand. Trans. R. Soc. Trop. Med. Hyg. 2006;100:410–418. doi: 10.1016/j.trstmh.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Russell BM, et al. Simple in vitro assay for determining the sensitivity of Plasmodium vivax isolates from fresh human blood to antimalarials in areas where Plasmodium falciparum is endemic. Antimicrob. Agents Chemother. 2003;47:170–173. doi: 10.1128/AAC.47.1.170-173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chotivanich K, et al. In vitro efficacy of antimalarial drugs against Plasmodium vivax on the western border of Thailand. Am. J. Trop. Med. Hyg. 2004;70:395–397. [PubMed] [Google Scholar]
- 35.Druilhe P, et al. Improved assessment of Plasmodium vivax response to antimalarial drugs by a colorimetric double-site Plasmodium lactate dehydrogenase antigen capture enzyme-linked immunosorbent assay. Antimicrob. Agents Chemother. 2007;51:2112–2116. doi: 10.1128/AAC.01385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell B, et al. Determinants of in vitro drug susceptibility testing of Plasmodium vivax. Antimicrob. Agents Chemother. 2008;52:1040–1045. doi: 10.1128/AAC.01334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosaisavee V, et al. Plasmodium vivax: isotopic, PicoGreen, and microscopic assays for measuring chloroquine sensitivity in fresh and cryopreserved isolates. Exp. Parasitol. 2006;114:34–39. doi: 10.1016/j.exppara.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Malleret B, et al. A rapid and robust tri-color flow cytometry assay for monitoring malaria parasite development. Sci. Rep. 2011;1:118. doi: 10.1038/srep00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Congpuong K, et al. Sensitivity of Plasmodium vivax to chloroquine in Sa Kaeo Province, Thailand. Acta Trop. 2002;83:117–121. doi: 10.1016/s0001-706x(02)00090-6. [DOI] [PubMed] [Google Scholar]
- 40.Suwanarusk R, et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS ONE. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharrock WW, et al. Plasmodium vivax trophozoites insensitive to chloroquine. Malar. J. 2008;7:94. doi: 10.1186/1475-2875-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerlin DH, et al. An analytical method for assessing stage-specific drug activity in Plasmodium vivax malaria: implications for ex vivo drug susceptibility testing. PLoS Negl. Trop. Dis. 2012;6:e1772. doi: 10.1371/journal.pntd.0001772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasugian AR, et al. In vivo and in vitro efficacy of amodiaquine monotherapy for treatment of infection by chloroquine-resistant Plasmodium vivax. Antimicrob. Agents Chemother. 2009;53:1094–1099. doi: 10.1128/AAC.01511-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suwanarusk R, et al. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J. Infect. Dis. 2008;198:1558–1564. doi: 10.1086/592451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chotivanich K, et al. Antimalarial drug susceptibility of Plasmodium vivax in the Republic of Korea. Am. J. Trop. Med. Hyg. 2009;80:902–904. [PMC free article] [PubMed] [Google Scholar]
- 46.Lek-Uthai U, et al. Stronger activity of human immunodeficiency virus type 1 protease inhibitors against clinical isolates of Plasmodium vivax than against those of P. falciparum. Antimicrob. Agents Chemother. 2008;52:2435–2441. doi: 10.1128/AAC.00169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marfurt J, et al. Ex vivo activity of histone deacetylase inhibitors against multidrug-resistant clinical isolates of Plasmodium falciparum and Plasmodium falciparum. Antimicrob. Agents Chemother. 2011;55:961–966. doi: 10.1128/AAC.01220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marfurt J, et al. Ex vivo drug susceptibility of ferroquine against chloroquine-resistant isolates of Plasmodium falciparum and Plasmodium falciparum. Antimicrob. Agents Chemother. 2011;55:4461–4464. doi: 10.1128/AAC.01375-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price RN, et al. In vitro activity of pyronaridine against multidrug resistant Plasmodium falciparum and Plasmodium falciparum. Antimicrob. Agents Chemother. 2010;51:5146–5150. doi: 10.1128/AAC.00801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marfurt J, et al. Artemisinin-based combination therapy against Plasmodium falciparum: Comparative ex vivo activity of next-generation endoperoxides in multidrug resistant field isolates. Antimicrob. Agents Chemother. 2012 doi: 10.1128/AAC.00283-12. http://dx.doi.org/10.1128/AAC.00283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nomura T, et al. Evidence for different mechanisms of chloroquine resistance in 2 Plasmodium species that cause human malaria. J. Infect. Dis. 2001;183:1653–1661. doi: 10.1086/320707. [DOI] [PubMed] [Google Scholar]
- 52.Sa JM, et al. Expression and function of pvcrt-o, a Plasmodium vivax ortholog of pfcrt, in Plasmodium falciparum and Dictyostelium discoideum. Mol. Biochem. Parasitol. 2006;150:219–228. doi: 10.1016/j.molbiopara.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Barnadas C, et al. Plasmodium vivax resistance to chloroquine in Madagascar: clinical efficacy and polymorphisms in pvmdr1 and pvcrt-o genes. Antimicrob. Agents Chemother. 2008;52:4233–4240. doi: 10.1128/AAC.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandez-Becerra C, et al. Increased expression levels of the pvcrt-o and pvmdr1 genes in a patient with severe Plasmodium vivax malaria. Malar. J. 2009;8:55. doi: 10.1186/1475-2875-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marfurt J, et al. Molecular markers of in vivo Plasmodium vivax resistance to amodiaquine plus sulfadoxine-pyrimethamine: mutations in pvdhfr and pvmdr1. J. Infect. Dis. 2008;198:409–417. doi: 10.1086/589882. [DOI] [PubMed] [Google Scholar]
- 56.Sa JM, et al. Plasmodium vivax: allele variants of the mdr1 gene do not associate with chloroquine resistance among isolates from Brazil, Papua, and monkey-adapted strains. Exp. Parasitol. 2005;109:256–259. doi: 10.1016/j.exppara.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 57.Kim YK, et al. Therapeutic efficacy of chloroquine in Plasmodium vivax and the pvmdr1 polymorphisms in the Republic of Korea under mass chemoprophylaxis. Am. J. Trop. Med. Hyg. 2011;84:532–534. doi: 10.4269/ajtmh.2011.10-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imwong M, et al. Gene amplification of Plasmodium vivax multidrug resistance 1 gene in Thailand, Laos, and Myanmar. Antimicrob. Agents Chemother. 2008;52:2657–2659. doi: 10.1128/AAC.01459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Auliff A, et al. Amino acid mutations in Plasmodium vivax DHFR and DHPS from several geographical regions and susceptibility to antifolate drugs. Am. J. Trop. Med. Hyg. 2006;75:617–621. [PubMed] [Google Scholar]
- 60.Hastings MD, et al. Novel Plasmodium vivax dhfr alleles from the Indonesian Archipelago and Papua New Guinea: association with pyrimethamine resistance determined by a Saccharomyces cerevisiae expression system. Antimicrob. Agents Chemother. 2005;49:733–740. doi: 10.1128/AAC.49.2.733-740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rungsihirunrat K, et al. Geographical distribution of amino acid mutations in Plasmodium vivax DHFR and DHPS from malaria endemic areas of Thailand. Am. J. Trop. Med. Hyg. 2008;78:462–467. [PubMed] [Google Scholar]
- 62.Zakeri S, et al. Molecular characterization of antifolates resistance-associated genes, (dhfr and dhps) in Plasmodium vivax isolates from the Middle East. Malar. J. 2009;8:20. doi: 10.1186/1475-2875-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tjitra E, et al. Therapeutic efficacies of artesunate–sulfadoxine–pyrimethamine and chloroquine–sulfadoxine–pyrimethamine in vivax malaria pilot studies: relationship to Plasmodium vivax dhfr mutations. Antimicrob. Agents Chemother. 2002;46:3947–3953. doi: 10.1128/AAC.46.12.3947-3953.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Auliff AM, et al. Defining the role of mutations in Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene using an episomal Plasmodium falciparum transfection system. Antimicrob. Agents Chemother. 2010;54:3927–3932. doi: 10.1128/AAC.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Neil MT, et al. A novel Plasmodium falciparum expression system for assessing antifolate resistance caused by mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase. J. Infect. Dis. 2007;196:467–474. doi: 10.1086/519286. [DOI] [PubMed] [Google Scholar]
- 66.Auliff AM, et al. Functional analysis of Plasmodium vivax dihydrofolate reductase-thymidylate synthase genes through stable transformation of Plasmodium falciparum. PLoS ONE. 2012;7:e40416. doi: 10.1371/journal.pone.0040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imwong M, et al. Limited polymorphism in the dihydropteroate synthetase gene (dhps) of Plasmodium vivax isolates from Thailand. Antimicrob. Agents Chemother. 2005;49:4393–4395. doi: 10.1128/AAC.49.10.4393-4395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carter R, et al. Linkage Group Selection – a fast approach to the genetic analysis of malaria parasites. Int. J. Parasitol. 2007;37:285–293. doi: 10.1016/j.ijpara.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 69.Hunt P, et al. Experimental evolution, genetic analysis and genome re-sequencing reveal the mutation conferring artemisinin resistance in an isogenic lineage of malaria parasites. BMC Genomics. 2010;11:499. doi: 10.1186/1471-2164-11-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Auburn S, et al. An effective method to purify Plasmodium falciparum DNA directly from clinical blood samples for whole genome high-throughput sequencing. PLoS ONE. 2011;6:e22213. doi: 10.1371/journal.pone.0022213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mu J, et al. Recent progress in functional genomic research in Plasmodium falciparum. Curr. Genomics. 2010;11:279–286. doi: 10.2174/138920210791233081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park DJ, et al. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13052–13057. doi: 10.1073/pnas.1210585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Tyne D, et al. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum. PLoS Genet. 2011;7:e1001383. doi: 10.1371/journal.pgen.1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 75.Amambua-Ngwa A, et al. SNP genotyping identifies new signatures of selection in a deep sample of West African Plasmodium falciparum Malaria Parasites. Mol. Biol. Evol. 2012 doi: 10.1093/molbev/mss151. http://dx.doi.org/10.1093/molbev/mss151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dharia NV, et al. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20045–20050. doi: 10.1073/pnas.1003776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sriprawat K, et al. Effective and cheap removal of leukocytes and platelets from Plasmodium vivax infected blood. Malar. J. 2009;8:115. doi: 10.1186/1475-2875-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bright AT, et al. Whole genome sequencing analysis of Plasmodium vivax using whole genome capture. BMC Genomics. 2012;13:262. doi: 10.1186/1471-2164-13-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012 doi: 10.1038/nature11174. http://dx.doi.org/10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carlton JM, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neafsey DE, et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat. Genet. 2012 doi: 10.1038/ng.2373. http://dx.doi.org/10.1038/ng.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ferreira MU, et al. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J. Infect. Dis. 2007;195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- 84.Karunaweera ND, et al. Extensive microsatellite diversity in the human malaria parasite Plasmodium vivax. Gene. 2008;410:105–112. doi: 10.1016/j.gene.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 85.Orjuela-Sanchez P, et al. Single-nucleotide polymorphism, linkage disequilibrium and geographic structure in the malaria parasite Plasmodium vivax: prospects for genome-wide association studies. BMC Genet. 2010;11:65. doi: 10.1186/1471-2156-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baird JK. Neglect of Plasmodium vivax malaria. Trends Parasitol. 2007;23:533–539. doi: 10.1016/j.pt.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 87.Mueller I, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect. Dis. 2009;9:555–566. doi: 10.1016/S1473-3099(09)70177-X. [DOI] [PubMed] [Google Scholar]
- 88.Brega S, et al. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J. Infect. Dis. 2005;191:272–277. doi: 10.1086/426830. [DOI] [PubMed] [Google Scholar]
- 89.Korsinczky M, et al. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob. Agents Chemother. 2004;48:2214–2222. doi: 10.1128/AAC.48.6.2214-2222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siswantoro H, et al. In vivo and in vitro efficacy of chloroquine against Plasmodium malariae and P. ovale in Papua, Indonesia. Antimicrob. Agents Chemother. 2011;55:197–202. doi: 10.1128/AAC.01122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]