Abstract
The Encyclopedia of DNA Elements (ENCODE) Project aims to identify all functional sequence elements in the human genome sequence by use of high-throughput DNA/cDNA sequencing approaches. To aid the standardization, comparison, and integration of data sets produced from different technologies and platforms, the ENCODE Consortium selected several standard human cell lines to be used by the ENCODE Projects. The Tier 1 ENCODE cell lines include GM12878, K562, and H1 human embryonic stem cell lines. GM12878 is a lymphoblastoid cell line, transformed with the Epstein-Barr virus, that was selected by the International HapMap Project for whole genome and transcriptome sequencing by use of the Illumina platform. K562 is an immortalized myelogenous leukemia cell line. The GM12878 cell line is attractive for the ENCODE Projects, as it offers potential synergy with the International HapMap Project. Despite the vast amount of sequencing data available on the GM12878 cell line through the ENCODE Project, including transcriptome, chromatin immunoprecipitation-sequencing for histone marks, and transcription factors, no small interfering siRNA-mediated knockdown studies have been performed in the GM12878 cell line, as cationic lipid-mediated transfection methods are inefficient for lymphoid cell lines. Here, we present an efficient and reproducible method for transfection of a variety of siRNAs into the GM12878 and K562 cell lines, which subsequently results in targeted protein depletion.
Keywords: RNAi studies, transfection, siRNA, protein knockdown
INTRODUCTION
Rational and wide-spread use of RNA interference (RNAi)-mediated knockdown of gene transcripts and encoded proteins has enabled facile studies of gene function and has led to the characterization of many new genes in their respective native host. Developed in 1998, siRNAs continue to be an easy and effective method for dramatically reducing the mRNA and protein products from a gene of interest within a chosen cell line.1, 2 Although a powerful technique, mRNA and protein knockdown efficiency is limited by the effective concentration of siRNA that can be introduced into the cytoplasm of the cell. For gene knockdown studies, one widely used strategy for RNAi knockdown studies makes use of a precursor miRNA (pre-miRNA) mimic encoded by a short hairpin (sh)RNA.3–6 This method uses a plasmid expression vector with an RNA polymerase III U6 small nuclear RNA promoter to drive expression of the shRNA, which is then processed in the cytoplasm to generate siRNAs that target their specific gene transcripts.7–10 Here, the siRNA exists in the cell, as long as the plasmid vector continues to be expressed, generally by selection of drug-resistant cell lines expressing the U6-shRNA-encoding gene. This strategy results in a steady and relatively long-lasting supply of siRNA but comes with some key disadvantages. The expressed shRNA must compete against endogenous microRNA processing pathways in the cell, thereby limiting the concentration of shRNA produced and the effective knockdown efficiency. This can make shRNA-mediated RNAi knockdown an ineffective technique for highly expressed transcripts. Additionally, gene characterization via transgenic shRNA knockdown becomes difficult when one is targeting essential proteins, as this procedure may select for stably transformed cell lines that yield low mRNA and protein knockdown within the isolated cell clones.
Transient electroporation of chemically synthesized, preprocessed, short siRNA duplexes directly into the cell lines of interest may be faster and lead to more efficient mRNA and protein knockdown, especially for hard-to-transfect cell lines. This method has advantages over shRNA-mediated knockdown, as it does not require generation of stable cell lines and bypasses use of endogenous microRNA processing pathways, while providing higher effective concentrations of siRNA delivered to the cell. Electroporation also exhibits a more immediate knockdown effect, as it does not rely on cellular transcriptional machinery for expression. Furthermore, the use of fluorescently labeled siRNA duplexes can allow an estimate of the efficiency with which cells have taken up the siRNAs. For these reasons, an electroporation-based approach is well suited for knockdown characterization of essential and highly expressed proteins in the cell, particularly with lymphoid cell lines that are resistant to standard, cationic, lipid-mediated transfection protocols.
In contrast to commonly used human cell lines, such as human embryonic kidney (HEK)293 and HeLa, the GM12878 and K562 cell lines have more stable chromosome karyotypes. HEK293 and HeLa have undergone extensive chromosome aneuploidy events and are, respectively, hypotriploidy and hypertriploidy. The relatively normal karyotype of the GM12878 and K562 was 1 feature that led to their use in research-intensive projects, such as ENCODE.11, 12 Whereas GM12878 and K562 have favorable karyotypic properties, experimental work has shown a significant barrier to standard, lipid-based transfection methods. The optimization of transfection of siRNAs into the ENCODE cell lines GM12878 and K562 is important for downstream functional characterization and hypothesis testing with a broad subset of genes, including those involved in RNA splicing, such as the RNA-binding proteins heterogeneous nuclear (hn)RNP A1 and Asp-Glu-Ala-Asp box protein 5 (DDX5). Expression data suggest that hnRNP A1 and DDX5 genes are highly expressed in human cells and are essential for proper RNA splicing and cell survival, making both hnRNP A1 and DDX5 potentially problematic candidates for RNAi-mediated mRNA and protein knockdown studies. The improvement of the efficiency of siRNA transfection in these ENCODE cell lines with normal karyotypes will allow more rigorous functional characterization of highly expressed and/or essential transcripts, thus paving the way for future studies with relevance to medicine, as well as basic science. Here, we present an efficient transfection method that can be used for mRNA and protein knockdown studies in ENCODE Tier 1 cell lines GM12878 and K562 for gene expression, mRNA splicing isoform pattern, and functional studies.
MATERIALS AND METHODS
Cells
Human GM12878 (Coriell Institute, Camden, NJ, USA) cells were maintained in RPMI media, supplemented with 15% FBS and 1 mM sodium pyruvate (Gibco, Life Technologies, Grand Island, NY, USA), at 3.5 × 105 cells/ml cell density. Human K562 cells (American Type Culture Collection, Manassas, VA, USA) were maintained in RPMI media, supplemented with 15% FBS, 1 mM sodium pyruvate (Gibco, Life Technologies), and 1× nonessential amino acids (Gibco, Life Technologies), at 1 × 105 cells/ml cell density.
GFP Fluorescence Microscopy Imaging Assays
Cells were electroporated by use of the Amaxa Nucleofector II instrument with 0.8 or 2 μg TurboGFP plasmid (Lonza, Basel, Switzerland)/sample by use of the indicated Nucleofector kits, according to the manufacturer’s instructions. Different Nucleofector programs for electroporation were tested for GM12878 cells. Twenty-four hours after transfection, cells were treated with 1 μg/ml Hoechst nuclear DNA stain (33342; Cell Signaling Technology, Danvers, MA, USA) to visualize nuclear DNA for 1 h before imaging on a fluorescence microscope. Transfection efficiency was calculated by counting the fraction of green fluorescent protein (GFP)-expressing cells by use of the ImageJ software program (NIH, Bethesda, MD, USA).
siRNA Transfection and Protein Knockdown
siRNA duplexes (Ambion, Life Technologies) were generated against the following target sequences or commercially purchased: hnRNP A1 siRNA 1: CAGCTGAGGAAGCTCTTCA (sequence provided by Dr. James Manley); hnRNP A1 siRNA 2 (s6710; Ambion, Life Technologies); DDX5 siRNA 1 (s4009; Ambion, Life Technologies); and DDX5 siRNA 2 (s4008; Ambion, Life Technologies). Nonspecific control siRNA duplexes 1 [scrambled siRNA (scr si)] were purchased from Life Technologies (4390843). For all knockdown experiments, combinations of hnRNP A1 siRNA 1 and 2 (hnRNP A1 combo siRNA) or DDX5 siRNA 1 and siRNA 2 (DDX5 combo siRNA) were used.
GM12878 cells were maintained at a cell density of 3.5 × 105 cells/ml before transfection. GM12878 cells were subcultured 48 and 24 h before transfection. Culture medium (1.5 ml) was divided in aliquots into each well in a 6-well plate and equilibrated to 37°C in the incubator. For transfection of GM12878 cells, 4 × 106 cells/reaction (DDX5 knockdown) or 6 × 106 cells/reaction (knockdown of highly abundant proteins, e.g., hnRNP A1) were used. The cells were washed twice with 1× PBS. For each electroporation reaction, 100 μl Nucleofector V-Kit and 10 μl of 50 μM hnRNP A1 combo siRNA, DDX5 combo siRNA, or scr si were prepared. The cell pellets were resuspended with the siRNA duplex suspension; then, cells/siRNA duplex oligo suspensions were transferred into cuvettes and electroporated. Immediately after electroporation, 400 μl of the pre-equilibrated culture medium to the cuvette was added and transferred to a 6-well plate. Twenty-four hours posttransfection, the medium was changed with fresh medium; 48 h post-transfection, cells were subjected with a second round of siRNA transfection; and 24 h post-second siRNA transfection, the media were changed again. The cells were harvested for protein immunoblot analysis or RNA isolation, 72 h postsecond siRNA transfection.
For K562 cells, the cells were maintained at a density of 1 × 105 cell/ml and subcultured 48 and 24 h before transfection. For the electroporation-based protocol, 4.5 × 105 cells were used per reaction in a 6-well plate. The same siRNA transfection protocol was used as in GM12878 cell electroporation-mediated transfection, with the exception of use of a Nucleofector program (T-016) for K562 cell transfection. For the cationic, liposome-based transfection protocol, 5 × 105 cells were plated/reaction. The transfection mixture was prepared containing 150 μl Opti-MEM media (Gibco, Life Technologies)/10 μl Lipofectamine RNAiMAX (Invitrogen, Life Technologies) in a 1.5 ml centrifuge tube and 150 μl Opti-MEM (Gibco, Life Technologies)/5 μl of 50 μM siRNA/reaction in a separate 1.5 ml centrifuge tube. The transfection mix was incubated at room temperature for 5 min. Diluted siRNA duplexes were added to diluted Lipofectamine RNAiMAX reagent and incubated at room temperature for 15 min. The 300 μl siRNA/lipid complexes were added to freshly plated 5 × 105 cells in a 6-well plate. Twenty-four hours post-transfection, the medium was changed with fresh media. Samples were harvested for protein lysates and immunoblotting or for RNA isolation, 72 h post-transfection.
HEK293 cells were subcultured 24 h before transfection. Cells (1 × 106) were used/transfection on a 6-well plate with 250 nM siRNA duplex. The same siRNA transfection protocol was used as in GM12878 cell electroporation-mediated transfection, with the exception of the use of a Nucleofector program (A-023) for transfection in HEK293 cells. For the cationic liposome-based transfection protocol, 5 × 105 cells were plated/reaction. The same cationic liposome-based transfection protocol was used in HEK293 cells, as described for K562 cells. The transfection mixture was prepared containing 150 μl Opti-MEM media (Gibco, Life Technologies)/10 μl Lipofectamine RNAiMAX (Invitrogen, Life Technologies) in a 1.5 ml centrifuge tube and 150 μl Opti-MEM (Gibco, Life Technologies)/5 μl of 50 μM siRNA/reaction in a separate 1.5 ml centrifuge tube. The transfection mix was incubated at room temperature for 5 min. Diluted siRNA duplexes were added to diluted Lipofectamine RNAiMAX reagent and incubated at room temperature for 15 min. The 300 μl siRNA/lipid complexes were added to freshly plated 5 × 105 cells in a 6-well plate. Twenty-four hours post-transfection, the media were changed with fresh media. Samples were harvested for protein lysates and immunoblotting or for RNA isolation, 72 h post-transfection.
Cell Extracts and Immunoblots
Cellular protein lysates were prepared in lysis buffer [50 mM Tris-Cl, pH 8.0, 100 mM NaCl, 1% (v/v) Nonidet P-40, 0.1% (w/v) SDS] containing protease inhibitors and quantified by use of the Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA). Equivalent protein amounts of each sample were subjected to SDS-polyacrylamide and immunoblotting with the following antibodies: TurboGFP (PIPA522688; 1:1000 dilution; Thermo Fisher Scientific, Waltham, MA, USA), hnRNP A1 (4B10; 1:1000 dilution; Sigma, St. Louis, MO, USA), DDX5 (NB200-351; 1:1000 dilution; Novus Biologicals, Littleton, CO, USA), and β-actin (AC-74; 1:3000 dilution; Sigma).
RNA Isolation, RT, and PCR
Total cellular RNA was isolated for RT-PCR by use of TRIzol (Life Technologies). Total RNA (1 μg) was reverse transcribed (170-8891; Bio-Rad Laboratories), according to the manufacturer’s instructions, and subjected to RT-PCR by use of the following temperature cycles: 98°C for 30 s (1 cycle); 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s (35 cycles); and 72°C for 5 min (1 cycle). The following primers sets were used for RT-PCR validation: synaptotagmin-binding, cytoplasmic RNA-interacting protein (SYNCRIP) cDNA was amplified by use of forward: 5′-CGATACCATCGGACAGGATT-3′ and reverse: 5′-TTGAAGAACTGCCAATGCAC-3′ primers; zinc finger, CCHC domain-containing 17 (ZCCHC17) cDNA was amplified by use of forward: 5′-AAGGCCTGAGACCATGGAA-3′ and reverse: 5′-GCCCCATAGTCTGTCACCAT-3′ primers; neurofibromin 1 (NF1) cDNA was amplified by use of forward: 5′-CGAAGTGTGTGCCACTGTTT-3′ and reverse: 5′-GGTGAGACAATGGCAGGATT-3′ primers; ubiquitin‐specific protease‐like 1 (USPL1) cDNA was amplified by use of forward: 5′-ACGTTGCAACTAGGGTGGAG-3′ and reverse: 5′-CAAGCAGGGCAATACTCATCT-3′ primers; eukaryotic translation initiation factor 4E-binding protein 3 (eIF4EBP3) cDNA was amplified by use of forward: 5′-GGAGAACTCTCCTGCCCTTT-3′ and reverse: 5′-AGGATACACAGGAGGCATCG-3′ primers; Las 1-like protein (Las1L) cDNA was amplified by use of forward: 5′-TTGAACAGTTGGCAGCTTTG-3′ and reverse: 5′-AACTGAGAGAACGGCTTTGG-3′ primers; and aspartyl/asparaginyl β-hydroxylase (ASPH) cDNA was amplified by use of forward: 5′-TCGAAGATGAAGCAAAAGAACA-3′ and reverse: 5′-CTTCCACGTGGTAACTATGCTC-3′ primers. PCR products were resolved, visualized, and quantitated by use of an Agilent Technologies (Santa Clara, CA, USA) Bioanalyzer.
RESULTS
Efficient and reproducible delivery of gene or shRNA/siRNA into the cell is essential for gene expression, gene function, and differential mRNA splicing expression studies. For instance, RNA-sequencing (seq) experiments often use RNAi technologies to knock down mRNA expression and protein levels of the gene of interest to study gene regulation and mRNA isoform expression. However, the experimental conditions of RNAi-knockdown experiments are dependent on cell type, and gene expression levels need to be optimized correspondingly.
The human GM12878 lymphoblastoid cell line is used extensively by the ENCODE Project and is particularly attractive because of the vast amount of sequencing data, including multiple RNA immunoprecipitation (RIP)-chromatin immunoprecipitation (ChIP), RIP-seq, and ChIP-seq experiments, readily available.12–20 Yet, RNAi studies have not been routinely performed in this line as a result of technical difficulties with nucleic acid delivery by cationic lipid-mediated transfection. In this study, efforts were made to optimize the transfection conditions in GM12878 cells by use of electroporation for RNAi studies. Electric-field strength and pulse duration are crucial parameters for maximum transfection efficiency and highest cell viability. Here, we report a transfection protocol by use of electroporation of siRNAs that results in efficient and reproducible RNAi knockdown of genes of interest in human GM12878 cells.
K562 cells, a myelogenous leukemia cell line that also belongs to Tier 1 of the ENCODE cell line, are readily transfectable via cationic, lipid-based transfection reagents,21, 22 such as Lipofectamine 2000 or RNAiMax (Invitrogen, Life Technologies), as well as electroporation.23 In contrast, numerous lipid-based transfection reagents and conditions tested in GM12878 cells were not successful at different cell densities (5 × 105, 1 × 106–4 × 106 GM12878 cells) by use of 150 pmol DDX5 or hnRNP A1 siRNAs (data not shown). Often, electroporation is used for transfection for cell lines, particularly lymphoid cells, which are difficult to transfect by use of lipid-based methods. The two electrical parameters that impact the success of electroporation are electric-field strength and pulse duration during transfection. These parameters need to be modulated carefully to maximize the transfection efficiency while maintaining cell viability. Here, we report a transfection protocol that uses electroporation of siRNAs, resulting in efficient and reproducible RNAi knockdown of genes of interest in human GM12878 cells.
Optimization of GM12878 Cell Transfection Efficiency by Electroporation
Cell density is crucial when working with human lymphoblastoid cell lines, such as GM12878. It is critical to maintain the cell density between 2 × 105 and 1 × 106 cells/ml for GM12878 cell viability. In these experiments, the GM12878 cells were maintained at 3.5 × 105 cells/ml before transfection. Cells (2 × 106) were transfected with 2 μg GFP-expressing plasmid in a 6-well plate. The Cell Line Optimization Nucleofector Kit, which includes Lonza Kit L and Kit V, was first tested in GM12878 cells by use of 7 programs (A-020, T-020, T-030, X-001, X-005, L-029, and D-023), spanning different electrical parameters for electric-field strength and pulse duration, according to the manufacturer’s instructions. Twenty-four hours post-transfection, the cells were stained with 1 μg/ml Hoechst fluorescent DNA stain for 1 h at 37°C. GFP-expressing cells were observed and quantitated by use of an inverted fluorescence microscope (University of California Berkeley Biologic Imaging Facility) and normalized to the total number of cells by use of Hoechst-stained cells to calculate the percentage of GFP-expressing cells by use of ImageJ software (NIH). Based on these initial tests, it was determined that the X-001 program resulted in the highest transfection efficiency among the programs tested.
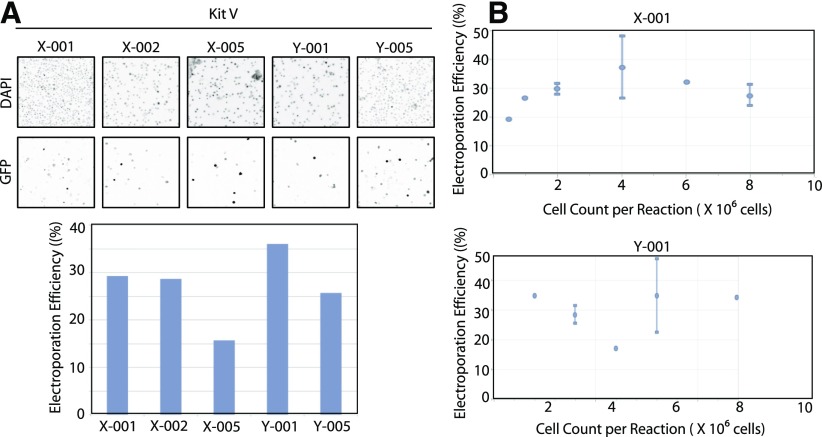
To further optimize the transfection in GM12878 cells, different programs, including X-002, X-005, Y-001, and Y-005, with similar electric-field strengths and pulse durations to that of the X-001 programs were then tested (Fig. 1A). Again, 2 μg GFP-expressing plasmid was transfected into 2 × 106 cells and harvested 24 h post-transfection. Cells were then stained with Hoechst nuclear DNA stain, and the transfection efficiency was determined. Among the programs tested, the highest transfection efficiency was observed with programs X-001 and Y-001. The transfection efficiency was still extremely low (<40%) and not sufficient to achieve an efficient delivery of nucleic acids into the GM12878 cell line. Cell density during electroporation was noted as a crucial parameter; hence, different numbers of cells or cell densities were subjected to electroporation, varying from 5.5 × 105 to 8 × 106 cells for both the X-001 and Y-001 programs (Fig. 1B). GM12878 cells (4 × 106), transfected by use of the X-001 program, resulted in 37.2% GFP-expressing cells, whereas 8 × 106 GM12878 cells transfected with the Y-001 program resulted in 42.1% GFP-expressing cells.
Figure 1.
Optimization of electroporation of GM12878 cell transfection efficiency by use of GFP-expressing plasmids. A) GM12878 cells (2 × 106) were transfected with GFP-expressing plasmid by use of Lonza Kit V with the indicated channels. Twenty-four hours post-transfection, the cells were stained with Hoechst nuclear DNA stain (1 μg/ml) for 1 h. All cells were imaged with an inverted fluorescence microscope (upper), and the percentage of GFP-expressing cells was quantitated (lower). Images shown in A have been inverted for easier viewing. B) GM12878 cells at varying cell densities were transfected with 2 μg GFP-expressing plasmid by use of Lonza Kit V with the indicated channels. Twenty-four hours post-transfection, the cells were stained with Hoechst nuclear DNA stain (1 μg/ml) for 1 h. The percentage of GFP-expressing cells was quantified by use of the ImageJ software program (NIH).
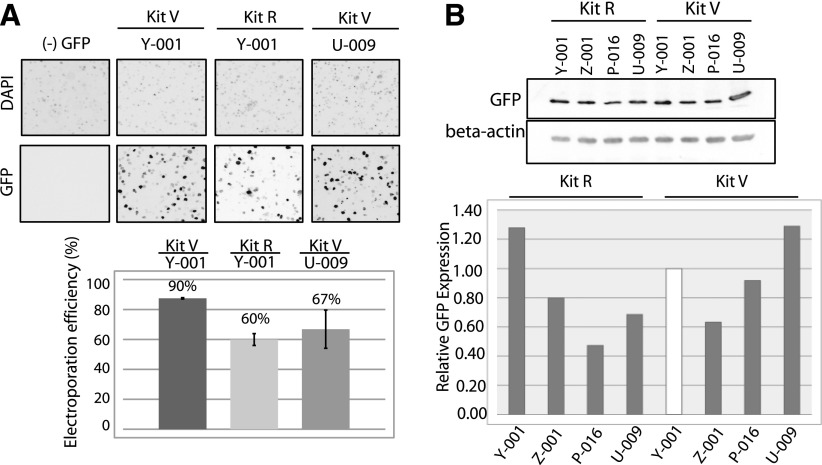
After searching for other Lonza programs used for other human B cell lines, we also tested programs Z-001, P-016, and U-009, in addition to Y-001, which gave the highest GFP transfection efficiency in the initial testing. GM12878 cells (4 × 106) were transfected with 0.8 μg GFP-expressing plasmid by use of Lonza Kit R or Kit V. Twenty-four hours post-transfection, the cells were harvested for fluorescence imaging (Fig. 2A) or protein immunoblot analysis (Fig. 2B). Cell lysates (15 μg) were loaded for each sample, resolved in 10% SDS-polyacrylamide gel, and immunoblotted by use of anti-GFP or anti-β-actin antibodies. GFP protein expression levels were quantified from the immunoblots by use of Image Lab software (Bio-Rad Laboratories) and normalized against β-actin, used as a loading control. The relative GFP expression levels were calculated by comparing the GFP/actin expression level from each sample with the GFP/actin expression level observed from the sample electroporated by use of the Y-001 program. These data indicated that the highest level of GFP expression in GM12878 cells was achieved by transfecting 4 × 106 cells by use of the U-009 program (Fig. 2B) and the buffers in Lonza Kit V. Very comparable results were obtained by transfecting 4 × 106 GM12878 cells by use of Lonza Kit R in the Y-001 program. Once the transfection conditions were optimized in GM12878 cells by use of the GFP-expressing plasmid, we proceeded to attempt siRNA-mediated mRNA and protein knockdown of two genes of interest: hnRNP A1 and DDX5.
Figure 2.
Further optimization of electroporation of GM12878 cell transfection efficiency by use of GFP-expressing plasmids and immunoblotting. A) GM12878 cells (4 × 106) were transfected with 0.8 μg GFP-expressing plasmid by use of Lonza Kit V and Kit R with the indicated channels. The cells were stained 24 h after electroporation with Hoechst nuclear stain (1 μg/ml) for 1 h. All cells were imaged with an inverted fluorescence microscope, and the percentage of GFP-expressing cells was quantified (lower). B) Transfection efficiency was measured by protein immunoblot analysis by use of anti-GFP antibodies. The GFP expression levels were normalized by comparison of β-actin protein levels as a loading control.
siRNA-Mediated Protein Knockdown of hnRNP A1 and DDX5 in GM12878 and K562 Cells
Alternative pre-messenger RNA (pre-mRNA) splicing is one of the major mechanisms used by metazoans to regulate gene expression and to increase the functional diversity of the eukaryotic proteomes. In humans, >95% of multiexon genes are alternatively spliced, and these alternative RNA processing events have implications for health and disease, as disease gene mutations that affect the splicing process result in human genetic disorders.
RNA chaperone proteins alter RNA secondary structure in vitro through RNA–RNA annealing or unwinding and by RNP remodeling. These proteins have also been shown to be involved in pre-mRNA splicing and transcription in vivo. hnRNP A1 belongs to the A/B subfamily of ubiquitously expressed hnRNPs, which are RNA-binding proteins that form complexes with hnRNA, shown to play a role in splicing. DDX5 belongs to DEX(D/H) box-containing RNA helicases that restructure RNAs or RNPs in an ATP-dependent manner and has been shown to be involved in alternative pre-mRNA splicing. The roles and targets transcripts of hnRNP A1 and the p68/DDX5 RNA helicase in pre-mRNA splicing can be studied by comparing alternative splicing patterns by RNA-seq after RNAi knockdown of these factors.
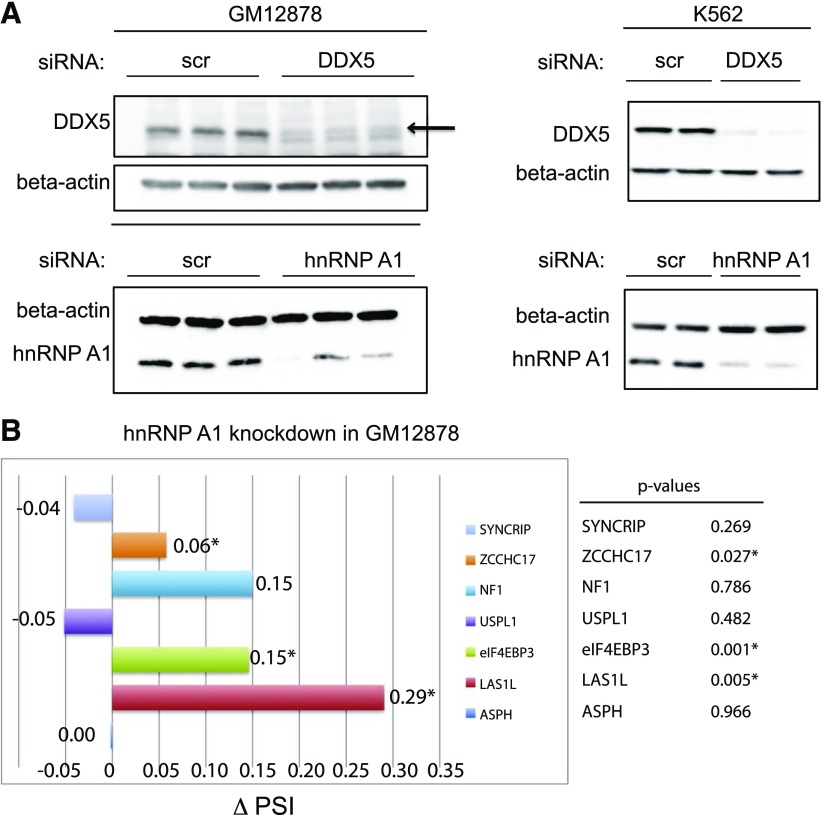
For knockdown experiments, 4 × 106 cells (DDX5 knockdown) or 6 × 106 cells (highly abundant proteins, e.g., hnRNP A1 knockdown) were subjected to electroporation of siRNAs. hnRNP A1 and DDX5 siRNAs were electroporated into GM12878 cells in parallel with scr si as controls. Forty-eight hours post-transfection, the cells were electroporated again with a second round of siRNAs. The cells were harvested 72 h after the final round of siRNA transfection for immunoblot analysis and RT-PCR analysis to gauge knockdown efficiencies. This transfection methodology results in 83% DDX5 knockdown and 75% hnRNPA1 knockdown efficiencies at the protein level, as measured by immunoblot (Fig. 3A). In data not shown here, we also attempted to electroporate 4 × 106 GM12878 cells in the presence of 100 pmol siRNA by use of the Gene Pulser (220 V, 975 μF; Bio-Rad Laboratories) for the knockdown studies. This specific voltage and capacitance were selected after testing different electroporation parameters to achieve ∼50% cell survival 24 h post-electroporation. Yet, we did not observe an efficient knockdown of hnRNP A1 or DDX5 in GM12878 cells under this condition.
Figure 3.
siRNA-mediated knockdown of hnRNP A1 and DDX5 proteins in GM12878 and K562 cells. A) GM12878 (left) and K562 cells (right) were transfected with nonspecific control siRNA oligos (scr si) or hnRNP A1 or DDX5 siRNA duplexes. After a second round of siRNA transfection, the cells were harvested for preparation of protein lysates, which were subjected to immunoblotting with anti-hnRNP A1 or anti-DDX5 antibodies to assess the efficiency of siRNA-mediated protein knockdown. Immunoblot analysis of the β-actin protein serves as a loading control. B) RT-PCR analysis of several pre-mRNA splicing target mRNAs reveals changes in hnRNP A1-dependent, alternative splicing events, recapitulated by RNAi in GM12878 cells. GM12878 cells were electroporated with nonspecific control siRNAs (scr si) or hnRNP A1 duplex siRNA duplexes. After a 2nd round of siRNA transfection, the cells were harvested for total RNA isolation and subjected to RT-PCR analysis. *P (P < 0.05), calculated by use of a 2-tailed Student's t test, statistically significant. PSI, Percent spliced in.
hnRNP A1 belongs to the A/B subfamily of ubiquitously expressed hnRNPs and has been shown to play a role in splicing/regulation of alternative splicing. There are five basic modes of alternative splicing that include the following: alternative 5′ splice site (alternative donor site), alternative 3′ splice site (alternative acceptor site), mutually exclusive exons, exon skipping (cassette exon), and intron retention. In mammalian, exon skipping is the most common mode of alternative splicing.24 To verify the effect of depletion of hnRNP A1 on pre-mRNA splicing patterns, total RNA from the knockdown experiments were reverse transcribed and subjected to RT-PCR with previously published sets of primers,25 designed to detect specifically alternative cassette exon-splicing events. RT-PCR products were analyzed by use of a Bioanalyzer (Agilent Technologies), and the exon-skipped and -included PCR products were quantified to calculate Percentage Spliced In (PSI) (or Ψ) values, where Ψ is between [0,1]. ∆Ψ, or change in PSI values upon hnRNP A1 knockdown samples, is shown in Fig. 3B, where the scr si-transfected samples served as the control. Experimental results demonstrate that Las1L (∆Ψ = 0.27; P = 0.005) and eIF4EBP3 (∆Ψ = −0.15; P = 0.001) are alternatively spliced as a result of 75% hnRNP A1 knockdown in GM12878 cells. Additional mRNA transcripts, including eIF4EBP3, ASPH, USPL1, and SYNCRIP, showed only very slightly spliced isoform changes.
To validate siRNA electroporation protocol, DDX5 and hnRNP A1 knockdown experiments were performed in the K562 (Fig. 3A, right) and HEK293 cell lines (data not shown). These cell lines are easily electroporated, and we were able to achieve an efficient knockdown of hnRNP A1 and DDX5 proteins (>85%, as determined by immunoblot analysis). Splicing analysis that uses RT-PCR reactions was also performed from RNA isolated from the hnRNP A1 siRNA knockdown samples, and compared with the control samples from scr si-transfected samples. Although some differences in values for the isoforms tested for each of the specific target genes were observed, the effects of hnRNP A1 knockdown on different targets were generally conserved across different cell lines tested (data not shown). These results demonstrate that our siRNA electroporation protocol works well in the ENCODE K562 and GM12878 cell lines, resulting in an efficient knockdown of the target proteins, and that this method can be used to study gene expression or pre-mRNA splicing changes.
DISCUSSION
With the ENCODE project aiming to identify all functional elements encoded in the human genome, the project has generated and made publicly available vast amounts of high-throughput sequencing data for transcription profiles, chromatin marks, and transcription factors in ChIP-seq.12–20 Both GM12878 and K562 cells possess a relatively normal karyotype, unlike HEK293 and HeLa cells that have undergone extensive chromosomal aneuploidy. For this reason the GM12878 and K562 cell lines were chosen for use in the Tier 1 ENCODE Project, along with H1 human embryonic stem cells. However, as a result of limitations in transfection efficiency, no RNAi-mediated mRNA and protein knockdown studies have been reported by use of the GM12878 cell line.
siRNA- and shRNA-mediated transcript and protein knockdown experiments are used routinely to study gene expression and changes in alternative pre-mRNA isoforms. For these experiments, it is imperative to optimize transfection conditions to deliver efficiently nucleic acid molecules of interest, while maintaining cell viability. Electroporation conditions for different human cell lines need to be determined empirically. Once RNAi-mediated knockdown conditions are optimized, the downstream analysis can then be performed to study gene expression and key interactions of splicing factors with their target pre-mRNAs. There are multiple methods to deliver nucleic acid materials into a desired human cell line, from chemical transfection (e.g., cationic liposome-mediated transfection or calcium phosphate) and nonchemical transfection (e.g., electroporation). RNAi can be achieved without much difficulty for easily transfectable cells lines, such as HEK293 and HeLa, by use of liposome-mediated transfection methods to deliver siRNAs to host cells. However, for hard-to-transfect human cell lines, such as embryonic stem cells, lymphoid, or other lines, electroporation-mediated transfection yields higher transfection efficiencies. Electroporation generally involves permeabilizing cellular membranes with high electric voltage, allowing nucleic acids to enter the cells.
In this report, we present an optimized set of parameters for efficient electroporation of GM12878 and K562 cells that result in an efficient RNAi-mediated knockdown of two different RNA-binding proteins of interest. For the purposes of this study, electroporation channels and cell densities were optimized with the highest electroporation efficiencies by use of Lonza Program U-009 with 4 × 106 cells (Kit V) and Program Y-001 (Kit R). Fluorescence microscopy and successive protein immunoblot analysis demonstrated a relatively efficient knockdown, leading to a 75% reduction of the hnRNP A1 protein and an 83% reduction of the DDX5 protein, an efficiency previously undocumented for the ENCODE Project GM12878 cell line. In addition, efficient knockdowns of the hnRNP A1 and DDX5 proteins (>90%) were achieved in K562 cells by use of electroporation. In data not shown here, we also performed siRNA-mediated knockdown of DDX5 protein by use of a cationic liposome-mediated transfection protocol and achieved a >90% knockdown efficiency in K562 cells. The same protocol with varying numbers of cells was tested for hnRNP A1 protein knockdown in K562 cells, but this led to only a 50% protein knockdown, probably as a result of a high expression level of hnRNP A1 protein in the cells. Again, this demonstrates that electroporation is a more efficient way of delivering nucleic acids into hard to transfect cells or to deplete the activity of very highly expressed genes.
Experimental results demonstrate that the Las1L (∆Ψ = 0.27; P = 0.005) and eIF4EBP3 (∆Ψ = −0.15; P = 0.001) pre-mRNAs are alternatively spliced as a result of 75% hnRNP A1 protein knockdown in GM12878 cells.Additional, pre-mRNA transcripts, including eIF4EBP3, ASPH, USPL1, and SYNCRIP, showed very slightly spliced mRNA isoform changes. Similar magnitudes of changes in these splicing events were observed in HEK293 and K562 cells after siRNA treatment (>90% protein knockdown efficiency). These data demonstrate that the transfection methods we developed result in an efficient knockdown of several genes of interest and resulted in changes in the pattern of several known hnRNP A1 splicing target pre-mRNAs to be detected by RT-PCR. The same protocol reported here can be used to examine the effects of RNAi depletion on cells after knockdowns of any other gene of interest. This method will pave the way for the study of hypotheses generated regarding the function of genes and proteins that result from the examination of the vast amount of ENCODE Project data readily available and that will be generated in the future with the use of GM12878 and K562 cells.
Acknowledgments
The authors thank Dr. James Manley (Columbia University) for providing the hnRNP A1 siRNA target sequence. The authors also thank George Ghanim and Malik Francis for critical review of the manuscript. The authors thank University of California Berkeley Biological Imaging Facility for the use of the fluorescence microscope and the facility's assistance. This work was supported by the U.S. National Institutes of Health Grants P50 GM102706 (to the Center of RNA Systems Biology, J. Cate, PI) and GM097352 (to D.C.R.).
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806–811. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999;286:950–952. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494–498. [DOI] [PubMed] [Google Scholar]
- 4.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA 2001;98:9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zamore PD. RNA interference: listening to the sound of silence. Nat Struct Biol 2001;8:746–750. [DOI] [PubMed] [Google Scholar]
- 6.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 2002;16:948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannon GJ. RNA interference. Nature 2002;418:244–251. [DOI] [PubMed] [Google Scholar]
- 8.Devroe E, Silver PA. Retrovirus-delivered siRNA. BMC Biotechnol 2002;2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubinson DA, Dillon CP, Kwiatkowski AV, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 2003;33:401–406. [DOI] [PubMed] [Google Scholar]
- 10.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol 2002;20:1006–1010. [DOI] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 2011;9:e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baroni TE, Chittur SV, George AD, Tenenbaum SA. Advances in RIP-chip analysis : RNA-binding protein immunoprecipitation-microarray profiling. Methods Mol Biol 2008;419:93–108. [DOI] [PubMed] [Google Scholar]
- 14.George AD, Tenenbaum SA. MicroRNA modulation of RNA-binding protein regulatory elements. RNA Biol 2006;3:57–59. [DOI] [PubMed] [Google Scholar]
- 15.Jain R, Devine T, George AD, et al. RIP-ChIP analysis: RNA-binding protein immunoprecipitation-microarray (ChIP) profiling. Methods Mol Biol 2011;703:247–263. [DOI] [PubMed] [Google Scholar]
- 16.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 2002;26:191–198. [DOI] [PubMed] [Google Scholar]
- 17.Arvey A, Agius P, Noble WS, Leslie C. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res 2012;22:1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundaje A, Kyriazopoulou-Panagiotopoulou S, Libbrecht M, et al. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res 2012;22:1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landt SG, Marinov GK, Kundaje A, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012;22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho JW, Jung YL, Liu T, et al. Comparative analysis of metazoan chromatin organization. Nature 2014;512:449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Song Y, Ren J, Qu X. Knocking-down cyclin A(2) by siRNA suppresses apoptosis and switches differentiation pathways in K562 cells upon administration with doxorubicin. PLoS One 2009;4:e6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su WX, Chen YH, Zeng W, Liu WL, Sun HY. [Effect of Gli1 gene silencing on proliferation of K562 cells and its mechanisms]. Zhonghua Xue Ye Xue Za Zhi 2012;33:570–573. [PubMed] [Google Scholar]
- 23.Delgado-Cañedo A, Santos DG, Chies JA, Kvitko K, Nardi NB. Optimization of an electroporation protocol using the K562 cell line as a model: role of cell cycle phase and cytoplasmic DNAses. Cytotechnology 2006;51:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sammeth M, Foissac S, Guigó R. A general definition and nomenclature for alternative splicing events. PLOS Comput Biol 2008;4:e1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huelga SC, Vu AQ, Arnold JD, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Reports 2012;1:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]