Abstract
We report GWAS results for the levels of A1, A2 and fetal hemoglobins, analyzed for the first time concurrently. Integrating high-density array genotyping and whole-genome sequencing in a large general population cohort from Sardinia, we detected 23 associations at 10 loci. Five are due to variants at previously undetected loci: MPHOSPH9, PLTP-PCIF1, FOG1, NFIX, and CCND3. Among those at known loci, 10 are new lead variants and 4 are novel independent signals. Half of all variants also showed pleiotropic associations with different hemoglobins, which further corroborated some of the detected associations and revealed features of coordinated hemoglobin species production.
INTRODUCTION
The provision of oxygen to tissues depends on hemoglobin, requiring the coordinated expression of several globin chains that form functional tetramers. An index of the importance of hemoglobin function is the evolutionary duplication and divergence of regulation of globin gene copies to adapt to stages of development and buffer the effects of mutational loss. In particular, at birth, a switch occurs from fetal hemoglobin (HbF) toward hemoglobin A2 (HbA2) and hemoglobin A1 (HbA1), so that during adult life the hemoglobin forms comprise ~1 % HbF, ~3 % HbA2 and ~96 % HbA1. The different hemoglobins all contain α-globin chains, encoded by two eponymous genes on chromosome 16. Those aggregate with non-α-globin chains encoded, respectively, by the γ (for HbF), δ (for HbA2) and β-globin (for HbA1) genes in the “β-globin gene cluster” on chromosome 11 (Figure 1). The molecular switch between fetal and adult hemoglobin occurs via the binding of transcription factors to regulatory DNA sequences controlling the expression of globin genes. In particular, the various genes in the β-globin cluster are sequentially activated during ontogeny, so that time-specific expression patterns follow their genomic order1.
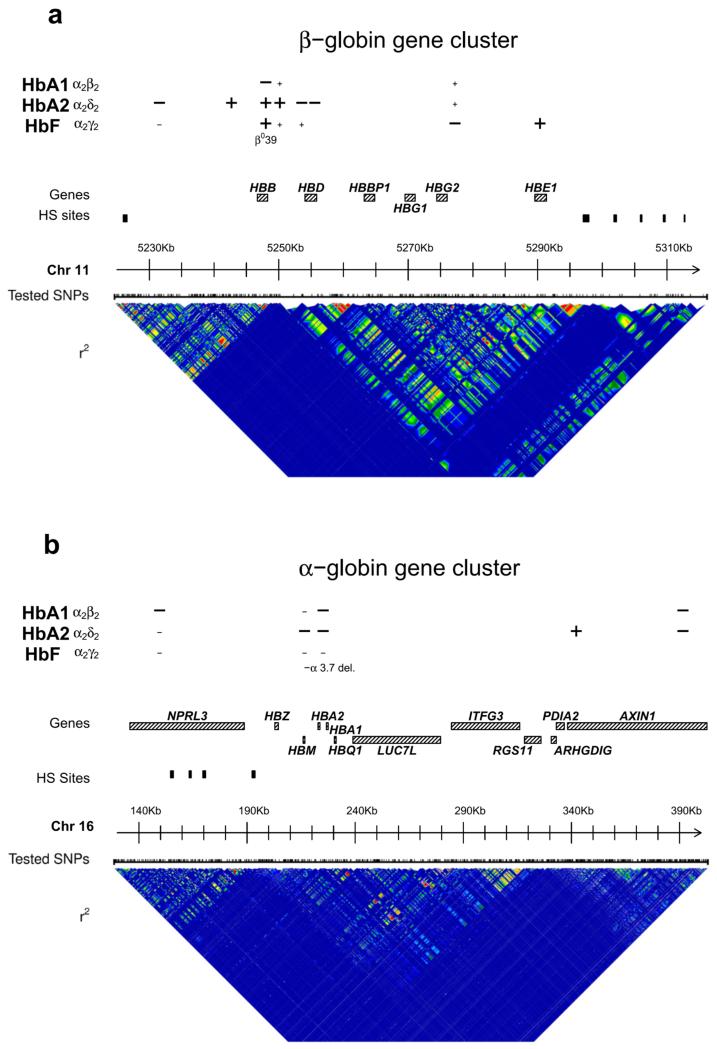
Figure 1. Association at the globin clusters.
Schematic representation of association results in the genomic context of the β-globin (panel a) and α-globin (panel b) gene clusters. For each hemoglobin, the markers associated are positioned with + or – corresponding to an increase or decrease in the corresponding trait by the effective allele (as in Table 1). Symbol is larger if the marker is associated at genome-wide level or smaller if it results from the analysis of pleiotropic effects. The β039 mutation and –α 3.7 type I deletion as well as relevant genes and the locus control region hypersensitivity sites (HS) are indicated. Finally, at the bottom of each panel is represented the linkage disequilibrium (r2) profile for the region in Sardinia, with colors ranging from high (red), to intermediate (green), and low (blue).
Inherited disorders of hemoglobin, such as β-thalassemia caused by mutations at the hemoglobin β (HBB) locus, represent the most common monogenic disorders worldwide2. Prevalence is highest in areas where malaria was or remains endemic3. The severity of inherited hemoglobin disorders is also variable, from severe life-long transfusion-dependent anemia to mild anemia that does not require transfusion, depending on the molecular defect and genotype status as well as ameliorating variants in modifier genes. Therefore, studying the genetic regulation of hemoglobin levels might reveal new factors and mechanisms to optimize strategies for the therapy of the disorders.
The large heritable contribution to phenotypic variance of HbA2 and HbF in the general population (0.728 and 0.633 respectively; see Online Methods and previous report4) indicates that genetic analyses could lead to new insights. In genome-wide association studies (GWAS), two genomic regions, the β-globin gene cluster locus and the HBS1L-MYB locus, have been associated at a genome-wide significant level with variations in the amount of HbA25, and only those loci and BCL11A have been associated with HbF levels6,7. Variants at all four loci are powerful modifiers of the severity of β-thalassemia and sickle-cell disease7–10. Notably, none of the variants associated with HbA2 or HbF have been found associated with total hemoglobin, even in the largest meta-analysis of over 135,000 individuals11. This indicates that in analyses of total hemoglobin levels, association signals for subtypes are diluted and possibly obscured by opposite directions of effects. Currently, most of the HbF and HbA2 heritability also remains to be explained, and HbA1 variation has never been specifically assessed by GWAS at all.
A promising source to extend analyses is the founder Sardinian population, in which previous associations have been detected in a large cohort through the analysis of genotyping arrays bearing common/ubiquitous variants7. Here, we extend these analyses to rarer and Sardinian-specific variants inferred from whole-genome population sequencing in the same cohort (see Supplementary Note and Supplementary Figure 1). Furthermore, analyzing variants modulating HbA1, HbA2 and HbF levels concurrently in a single cohort provides a route to assess associations that overlap for different hemoglobin forms without the need to account for differences in study size, ethnic background or measurements.
RESULTS
To test for genetic associations with the levels of HbA1, HbA2 and HbF, we interrogated ~10.9 million single nucleotide polymorphisms (SNPs), genotyped or imputed in 6,602 general population volunteers of the SardiNIA longitudinal study4 (see Online Methods and Supplementary Table 1).
Initial analyses showed a predominant role for the HBB:c.118C>T stop-codon mutation -- Q40X, better known as β039 mutation -- a variant common in Sardinia (rs11549407, allele frequency 4.8 %). It results in complete absence of β-globin chain synthesis (β0) and consequent β-thalassemia in homozygous individuals, and in a decrease of HbA1 and increase of HbA2 and HbF in heterozygous individuals (with p-values < 1.0×10−200). Because its effect has been established previously7,12, we considered this mutation and other rarer β0-thalassemia mutations known in Sardinia as covariates (see Online Methods and Supplementary Table 2). The assessed individuals in the cohort include 664 healthy heterozygous carriers but no β0-thalassemia patients.
The genome-wide scan revealed 23 unique variants at 10 loci at the classical 5×10−08 threshold. Of note, 21 are significant even considering a more stringent threshold of p = 1.4×10−8, calculated based on an empirical estimate of the number of independent tests in the Sardinian genome (see companion paper13).
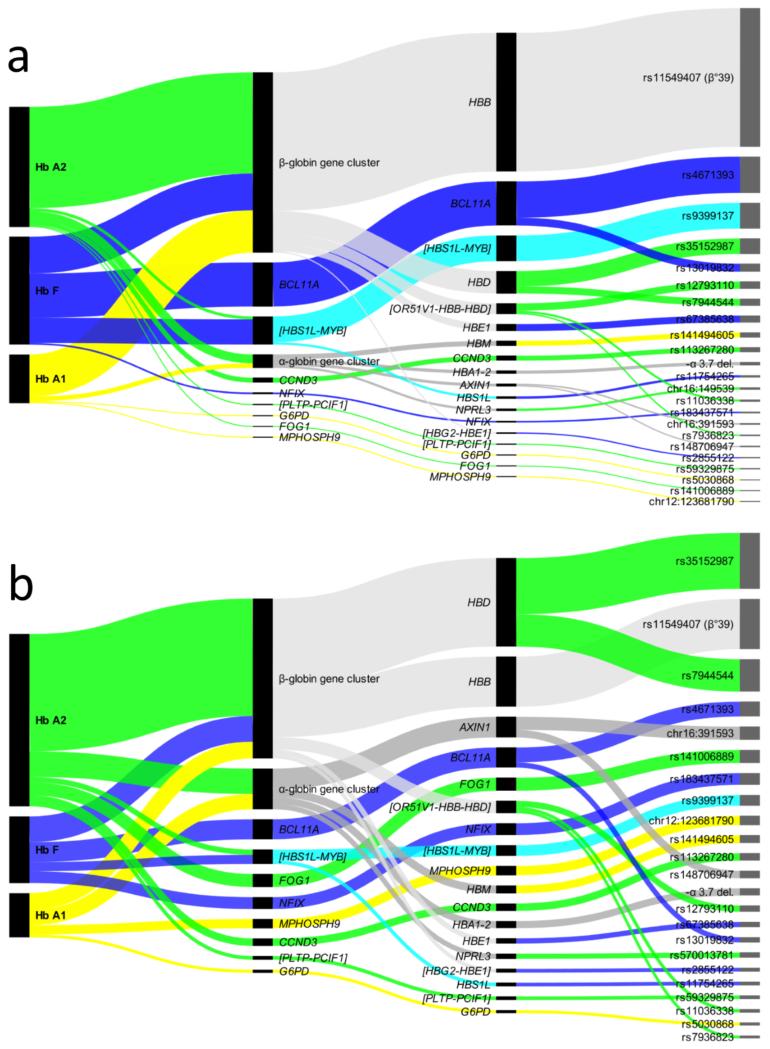
Five variants are at previously undetected loci, 4 are new independent signals at known loci, and 10 refine previously described associations to new lead polymorphisms that may have functional effects (Table 1). Six, 14 and 8 independent genome-wide significant signals were seen for HbA1, HbA2 and HbF respectively (Supplementary Figure 2). Hence, some of the associated variants significantly affected more than one hemoglobin, resulting in 28 variant-trait associations (see Table 1, Figure 2 and Supplementary Table 3). Variants resulting from imputation and not supported by linked genotyped markers were experimentally validated (Supplementary Table 4)
Table 1. Most significant independent association results from single variant tests for hemoglobin A1, A2 and fetal.
The table shows the most significant association results (all results are corrected for β0 mutations observed in the HBB gene, and results on the α-globin gene cluster are adjusted for the −α 3.7 deletion type I, see Online Methods). Novel signals are shown in bold while variants refining previously reported signals are in italic. At each locus, we indicated the chromosome and genomic position (hg19 build), the rs ID when available, the effect allele tested for association (EA) and the other allele at the SNP (OA), the imputation accuracy (RSQR), the SNP effect allele frequency (EAF) and the regression coefficients. We then indicated whether the SNP is also linked the other hemoglobin forms (p < 0.01), and indicated the direction of the effect allele (+ for increasing the levels of Hb, - for decreasing). The candidate genes likely to be modulated by the lead SNP are also reported along with their inclusion criteria, as described in Online Methods (p = position, c = coding, e = eQTL, o = OMIM, b = biological). Where “α-globin gene cluster” is mentioned we refer to NPRL3, HBZ, HBQ1, HBA1, HBA2 and HBM genes; while for “β-globin gene cluster” we refer to HBB, HBD, HBBP1, HBG1, HBG2 and HBE1 genes. Association coefficients for males and females are reported in Supplementary Table 11.
| Shared effects |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Traits (units) and loci # | Candidate genes | chr:position | rsID from dbsnp142 | Alleles (EA/OA) | RSQR | EAF | Effect (StdErr) | p-value | HbA1 | HbA2 | HbF |
| HbA1 (g/dl) | |||||||||||
| locus1 1 | α-globin gene cluster(p,o,b); MPG(p) | 16:149539 1,4 | rs570013781 | A/G | 0.98 | 0.136 | −0.1995 (0.023) | 5.86×10−18 | − | − | − |
| α-globin gene cluster (p,o,b); AXIN1(p) | 16:391593 1,3,5 (cond.) | − | T/C | 0.94 | 0.012 | −0.4028 (0.058) | 3.28×10−12 | − | − | ||
|
|
|||||||||||
| locus2 | FAM3A(p); G6PD(p,c,o,b); IKBKG(p) | X:153762634 4 | rs5030868 | A/G | Genotyped | 0.085 | −0.1256 (0.019) | 2.78×10−11 | − | ||
|
|
|||||||||||
| locus3 2 | MPHOSPH9(p) | 12:123681790 2 | − | A/C | 0.96 | 0.010 | −0.3606 (0.064) | 1.68×10−08 | − | − | |
|
| |||||||||||
| HbA2 | |||||||||||
|
|
|||||||||||
| locus1 4 (%) | β-globin gene cluster(p,o,b); HBD(c) | 11:5255582 4 | rs35152987 | A/C | Genotyped | 0.004 | −2.182 (0.109) | 4.35×10−86 | − | ||
| β-globin gene cluster (p,o,b); HBD(c) | 11:5251849 4 (cond.) | rs7944544 | T/G | 0.98 | 0.005 | −1.26 (0.097) | 3.90×10−38 | − | + | ||
| β-globin gene cluster (p,o,b); HBB(c); HBG1/HBG2(e); OR51V1(p) | 11:5231565 4 (cond.) | rs12793110 | T/C | 1.00 | 0.181 | −0.2408 (0.019) | 5.75×10−36 | − | − | ||
| β-globin gene cluster (p,o,b); OR51V1(p) | 11:5242698 4 (cond.) | rs11036338 | C/G | 0.99 | 0.381 | 0.1282 (0.017) | 2.03×10−14 | + | |||
| β-globin gene cluster (p,o,b); HBG1/HBG2(e) | 11:5250168 4 (cond.) | rs7936823 | G/A | 0.96 | 0.466 | 0.1117 (0.015) | 5.00×10−13 | + | + | + | |
|
|
|||||||||||
| locus2 1,3,5 (g/dl) | α-globin gene cluster (p,o,b); HBM (c); LUC7L(p) | 16:216593 1,3 | rs141494605 | C/T | 0.97 | 0.149 | −0.3080 (0.025) | 3.94×10−35 | − | − | − |
| α-globin gene cluster (p,o,b); AXIN1(p) | 16:391593 1,3,5 (cond.) | − | T/C | 0.94 | 0.012 | −0.5112 (0.063) | 6.48×10−16 | − | − | ||
| α-globin gene cluster (p,o,b); ARHGDIG(p); AXIN1(p); ITFG3(p); PDIA2(p); RGS11(p) | 16:342218 1,3,5 (cond.) | rs148706947 | T/C | 0.93 | 0.021 | 0.2892 (0.051) | 1.04×10−08 | + | |||
|
|
|||||||||||
| locus3 2 (%) | CCND3(p,b) | 6:41952511 2 | rs113267280 | G/T | 0.99 | 0.101 | 0.2923 (0.026) | 1.11×10−29 | + | + | |
|
|
|||||||||||
| locus4 (%) | MYB(b) | 6:135418916 | rs7776054 | G/A | Genotyped | 0.210 | 0.1762 (0.020) | 3.71×10−19 | + | + | |
|
|
|||||||||||
| locus5 2 (%) | CTSA(p); PCIF1(p,c); PLTP(p,e); MMP9(e); TNNC2(e) | 20:44547672 2 | rs59329875 | C/T | 1.00 | 0.134 | −0.1399 (0.024) | 3.64×10−09 | − | ||
|
|
|||||||||||
| locus6 2 (%) | FOG1(p,b,c); C16orf85(p) | 16:88601281 2 | rs141006889 | G/A | Genotyped | 0.007 | −0.5074 (0.087) | 5.33×10−09 | − | ||
|
| |||||||||||
| HbF (g/dl) | |||||||||||
| locus1 | BCL11A(p,o,b) | 2:60720951 | rs4671393 | A/G | 1.00 | 0.136 | 0.578 (0.023) | 2.60×10−130 | + | ||
| BCL11A(p,o,b) | 2:60710571 4 (cond.) | rs13019832 | A/G | 1.00 | 0.484 | −0.2024 (0.017) | 9.12×10−33 | − | |||
|
|
|||||||||||
| locus2 | MYB(b) | 6:135419018 | rs9399137 | C/T | Genotyped | 0.205 | 0.4202 (0.020) | 1.09×10−93 | + | + | |
| HBS1L(p,c,e); ALDH8A1(e) | 6:135356216 3 (cond.) | rs11754265 | C/G | 1.00 | 0.367 | −0.1421 (0.021) | 5.04×10−12 | − | |||
|
|
|||||||||||
| locus3 4 | β-globin gene cluster (p,o,b); HBG1/HBG2(e) | 11:5290370 4 | rs67385638 | G/C | 1.00 | 0.236 | 0.2038 (0.019) | 1.09×10−25 | + | ||
| β-globin gene cluster (p,o,b); HBG1/HBG2(e) | 11:5277236 4 (cond.) | rs2855122 | C/T | 1.00 | 0.395 | −0.1458 (0.022) | 2.57×10−11 | + | + | − | |
|
|
|||||||||||
| locus4 2,5 | NFIX(p) | 19:13121899 2,5 | rs183437571 | T/C | 0.97 | 0.010 | 0.4607 (0.081) | 1.61×10−08 | + | ||
= association results locally corrected for the −α 3.7 deletion type I (NG_000006.1:g.34164_37967del3804) (see Supplementary Note).
= first time associated to the trait and in a novel locus.
= first time associated to the trait in a previously reported locus.
= signal refining a previously reported signal.
= result not found using the 1000 Genomes reference panel.
cond. = obtained by conditional analysis on variants reported on the upper rows for the considered locus.
Figure 2. Diagram of genome-wide associated loci.
Representation of genome-wide significant findings on hemoglobin levels in relation to their contribution to the phenotypic variation (variance explained, panel a) or to their individual impact (effect size, panel b). At each step, the length of the black bar represents the magnitude of variance explained (panel a) or effect size (panel b) for each trait, locus, gene and variant. The bars are connected by colored bands to their sub-components (loci for each trait, genes for each locus, variants for each gene). Three colors (yellow, green and blue) represent the 3 hemoglobin forms (HbA1, HbA2 and HbF respectively), and for loci or genes affecting more than one hemoglobin: gray combines HbA1 and HbA2, cyan combines HbA2 and HbF, and light gray represents effects common to all 3 hemoglobin forms. Each panel is drawn to show loci in order of their importance, i.e. from the largest to smallest amount of explained phenotypic variance (panel a) or effect size (panel b). The variance explained by each locus was calculated fitting a regression model including all variants at that locus, while the effect size for a locus is the sum of effect sizes of all variants in that locus (Supplementary Table 3 reports effect sizes for such joint models). For variants associated with more than one trait the maximum value is used. Markers are reported as chromosome:position when an rs ID was not available; and when an intergenic region is involved instead of a single gene, we show nearby genes within brackets.
Novel associations at new loci
Novel associations were detected for all 3 hemoglobin forms. For HbA1, we observed a signal led by chr12:123681790 (in an intron of MPHOSPH9), encompassing several SNPs in complete linkage disequilibrium (LD) in a region encoding several genes (see Supplementary Figure 3). Which gene is truly associated, and how it affects hemoglobin production, remains unclear, although among the top associated SNPs, a variant in an intron of ARL6IP4 (chr12:123465483) falls in a highly conserved region rich in putative transcription factor binding sites and has the highest score for insilico prediction of deleterious impact on function (CADD score)14 as detailed in Supplementary Table 2. Although this association is just below the more stringent empirical threshold of significance, it is further strengthened by independent association with another hemoglobin form (HbA2, p = 5.9×10−5), as detailed in Table 1.
For HbA2, we identified 3 novel signals. One, rs141006889, is a missense variant located in ZFPM1, a gene also known as FOG1 that encodes a cofactor of the hematopoietic transcription factors GATA1 and GATA215 (Supplementary Figure 4). The complexes formed by FOG1 and GATA proteins are essential for normal erythroid differentiation15, as demonstrated by pathogenetic mutations that abrogate the FOG-GATA interaction to cause familial dyserythropoietic anemia and thrombocytopenia16. Another signal is defined by a pair of statistically indistinguishable variants, rs113267280 and rs112233623 (p-values: 1.11×10−29 and 1.29×10−29), located in CCND3 gene, whose product, cyclin D3, is thought to be critical for erythropoiesis17. Knockdown of cyclin D3 correlates with reduction in the number of cell divisions during terminal erythropoiesis, thereby producing fewer and larger red blood cells18. These variants are also in partial LD with rs9349205 (r2 = 0.40), a SNP previously associated with mean red blood cell volume and number (see Supplementary Table 6), which falls 160bp away from rs112233623 in the same erythroid specific enhancer functionally associated with CCND318–20. The latter is also the associated variant with highest CADD score (see Supplementary Table 5).
An additional variant related to HbA2, rs59329875, was observed for the first time in this study. It is situated between PLTP, which has been associated with several plasma lipoprotein and triglyceride levels21–24, and PCIF1, which is thought to negatively regulate gene expression by RNA polymerase II25.
As for HbF, we identified one new variant associated with its level: rs183437571, located on chromosome 19 in an intron of NFIX, which encodes a CCAAT-binding transcription factor. This variant is just below the empirical significance threshold of p = 1.4×10−8 but is supported by considerable biological evidence implicating the gene and the surrounding region in hemoglobin regulation. Specifically, rs183437571 falls in a CpG region that is differentially methylated in fetal and adult red blood cell progenitors26. In mice, Nfix was recently identified as one of the regulatory factors with relatively restricted expression in hematopoietic stem cells,27 and required for the survival of hematopoietic stem and progenitor cells during stress hematopoiesis28. Intriguingly, NFIX is situated in a region of ~300 Kb that encompasses a number of genes involved in erythropoiesis (DNASE2 and KLF1)29–33 or otherwise associated with red blood cell traits, including mean corpuscular hemoglobin (SYCE2, FARSA and CALR)11 (Supplementary Figure 5 and Supplementary Table 6). KLF1 is a particularly interesting candidate gene33,34, but mutations observed in previous studies35 were not found and the gene itself is situated in an LD block distinct from our association signal. However, long distance regulatory interactions remain a possibility.
Of the 5 novel signals, the discovery of chr12:123681790 for HbA1, rs141006889 for HbA2, and rs183437571 for HbF were strongly influenced by the assessment of variants from Sardinian whole-genome sequencing. Specifically, chr12:123681790 was missing in 1000 Genomes phase III36, and using this public reference panel the signal was misplaced to another variant ~1Mb away; rs141006889 was included in the design of one genotyping array (ExomeChip) after it was identified through our sequencing effort, but is currently not detected in sequenced 1000 Genomes samples; and rs183437571 was poorly imputed with 1000 Genomes phase III, with a resulting signal that was not genome-wide significant (see Table 1 and Supplementary Table 7).
Overall, the amount of variance explained by markers associated at the genome-wide level (Table 1) account for a fraction of the estimated genetic component of each trait (from 46 % for HbA1 to 68 % for HbA2, see Online Methods), supporting inheritance models that include small effect size and/or rare variants. For instance, 21 additional genes with suggestive significance signals (p<1.×10−04, minor allele frequency [MAF] > 0.5 %) were related to genome-wide significant loci listed here, either in the scientific literature (Pubmed before 2006) or by expression levels (Human Expression Atlas37) or Gene Ontology38 categories, using GRAIL software39 (see Supplementary Note and Supplementary Table 8). Four of the suggestive signals most strongly linked to genome-wide association findings were located in NFE2, which encodes Erythroid Nuclear Factor 240; ADGB, which encodes a recently discovered globin of unknown physiological function41; and SPTB and ANK1, both of which encode proteins affecting the stability of erythrocyte membranes42.
To test for replication of the associations at new loci detected in Sardinia, we used the largest independent sample reported to date, which measured HbA2 and HbF as well as F-cells (see Online Methods) in 4,131 individuals from the TwinsUK cohort enrolled from the United Kingdom (UK) general population43. For two loci, both associated with HbA2, we successfully replicated the association seen in Sardinia. In particular, we observed a p-value of 6.98×10−06 for rs59329875 in the PLTP-PCIF1 intergenic region (MAF of 0.18) and a p-value of 1.73×10−04 for rs113267280 in CCND3 (MAF of 0.01). The rarity of other variants precluded replication. The MPHOSPH9 and FOG1 variants associated with HbA1 and HbA2, respectively, are missing in publicly available imputation panels (as detailed above), and rs183437571 in NFIX associated with HbF was imputed as monomorphic in the TwinsUK cohort (see Table 2 and Online Methods).
Table 2. Replication of novel loci.
The table describes association in the TwinsUK cohort (N = 4,131 individuals). For each SNP, we indicated the associated hemoglobin tested, the number of samples analysed, the imputation accuracy according to the IMPUTE-INFO metric, the effect allele tested for association (EA) and the other allele at the SNP (OA), the SNP effect allele frequency (EAF) and the regression coefficients. The last column explains the reason for the SNPs not being tested.
| Traits (units) and loci # from Table 1 | SNP | Candidate genes | INFO score | Alleles (EA/OA) | EAF | Effect (StdErr) | p-value | Notes |
|---|---|---|---|---|---|---|---|---|
| HbA1 (g/dl) | ||||||||
| locus3 | chr12:123681790 | MPHOSP9 | - | - | - | - | - | Not imputable because absent in 1000 Genomes; at the moment, Sardinian specific. |
|
| ||||||||
| HbA2 (%) | ||||||||
| locus3 | rs113267280 | CCND3 | 0.843 | G/T | 0.011 | 0.442 (0.118) | 1.73×10−04 | . |
| locus5 | rs59329875 | PLPT-PCIF1 | 0.994 | C/T | 0.185 | 0.132 (0.029) | 6.98×10−06 | |
| locus6 | rs141006889 | FOG1 | - | - | - | - | - | Not imputable because absent in 1000 Genomes; detected in the NHLBI GO Exome Sequencing Project (ESP). |
|
| ||||||||
| HbF (%) | ||||||||
| locus4 | rs183437571 | NFIX | 0.294 | T/C | 0.000 | - | - | Imputed as monomorphic in TwinsUK cohort. |
Fine mapping at known loci
The integration of whole-genome sequence variants in the scan was also instrumental to refine signals at previously known loci, either identifying a better lead variant or indicating novel independent signals. Specifically, as detailed below, we refined the association within the α and β-globin gene clusters with all 3 hemoglobins; the association of the HBS1L-MYB intergenic region with HbA2 and HbF; and the association of the BCL11A gene with HbF.
Associations within the β-globin gene cluster were intricate. As reported above, the strongest modifier in this region is the HBB β039 variant, acting on all 3 hemoglobin types (see Figure 1, Online Methods and Supplementary Table 2). Multiple additional independent signals were observed in conditional analyses for HbA2 and HbF, but they were distinct for each hemoglobin type, highlighting different regulatory patterns within the β-globin gene cluster. Specifically, for HbA2, we confirmed 2 known independent associations at missense mutations in the HBD gene (rs35152987 and rs35406175, the latter perfectly tagged by our lead signal, see Supplementary Table 2). In addition, we identified 3 novel independent signals (rs12793110, rs11036338 and rs7936823) within a block of LD around the HBB gene, confirming a controlling role of this region in HbA2 production5 (see Figure 1 and Supplementary Figure 4). For HbF levels, 2 new independent signals were detected in a separate LD-block of the β-globin gene cluster (see Figure 1 and Supplementary Figure 5). The first, situated in an intron of the HBE1 gene (rs67385638), remained associated even when taking into account 43 other variants in the β-globin gene cluster associated with hemoglobin variation (see Supplementary Note). The second was located in a cyclic AMP response element upstream from HBG2 (rs2855122) already implicated in drug-mediated HbF induction by butyrate44 : different features of this marker make it a strong candidate for fetal to adult hemoglobin switching modulation (see Supplementary Note).
At the α-globin gene cluster, 2 variants were associated with HbA1 and 3 with HbA2, of which one affected both traits (Table 1 and Figure 1). All results at this locus were corrected for any effect of the most frequent α-globin gene deletion present in Sardinia (NG_000006.1:g.34164_37967del3804, known as –α 3.7 deletion type I), directly genotyped in a subset of the volunteers and imputed for the rest of the cohort (see Online Methods). This deletion was associated at the genome-wide level with both HbA1 and HbA2 and only nominally with HbF (see Table 1 and Supplementary Table 2). The most strongly associated signals (rs570013781 and rs141494605) were situated within the NPRL3 and HBM genes, affecting HbA1 and HbA2 respectively. NPRL3 contains several hypersensitive sites involved in the regulation of α-globin gene. HBM encodes a globin member of the avian α-D family45 and its expression is highly regulated in human erythroid cells, although the protein has not been detected in human erythroid tissues. These observations suggest a possible regulatory function for which high-level protein expression is not required45. An independent variant associated with HbA1 and HbA2 (chr16:391593) was observed within the AXIN1 gene, in which a further independent SNP (rs148706947) was found associated with HbA2 alone (Supplementary Figure 3 and Supplementary Figure 4).
We also examined variants in the HBS1L-MYB intergenic region known to be associated with HbF and HbA2 levels5. We confirmed the role of the known variant (rs66650371, a TAC deletion) on the expression of both forms of hemoglobin46,47 (see Supplementary Note). A further novel independent signal for HbF was found at rs11754265 in an intron of HBS1L, which has been shown to be a much stronger eQTL than rs66650371 for HBS1L and the neighboring ALDH8A1 in monocytes48.
In line with previous studies6–8,49,50 the second intron of BCL11A gave multiple signals associated with HbF levels. They are explicable by the joint action of variants in each of two independent groups of statistically indistinguishable SNPs: one group formed by rs4671393, rs766432 and rs1427407, with p-values between 2.6×10−130 and 5.6×10−129, and the other by rs13019832 and rs7606173, with p-values of 6.1×10−33 and 9.1×10−33 in our cohort. The most likely causal candidate in the first group is rs1427407, a variant already associated with HbF in other population cohorts and functionally associated with BCL11A regulation51. In the second group we can instead point to rs13019832, which shows the highest functional CADD score (Supplementary Table 5). This variant has also been correlated, in adipose tissue, with the methylation of a CpG site (cg23678058) in a region that is functionally associated with BCL11A expression52 and shows evidence of an effect on GATA-1 binding in peripheral blood-derived erythroblasts53,54.
Pleiotropic effects
Among our 23 lead variants, 6 were associated (at least with p<0.01) with a second hemoglobin type, and another 6 were associated with all 3 (including β039 and –α 3.7 deletion type I) (Figure 1 and Table 1). Overall, all but 3 pleiotropic variants modulate different hemoglobins in the same manner, i.e., with the same allele increasing the levels of all associated hemoglobins. The 3 exceptions include the β039 variant, which decreases HbA1 while increasing HbA2 and HbF, and 2 SNPs mapping in the β-globin gene cluster, both affecting HbA2 and HbF but in opposite directions (Figure 1 and Table 1). In addition, many of the additional suggestive signals are associated with more than one hemoglobin type, increasing the likelihood that they are true signals (see Online Methods). In fact, 14 of these variants – all sharing effects on HbA1 and HbA2, but none with HbF – showed between-trait combined p-values that were genome-wide significant (Supplementary Table 9) and hint at additional pathways of potential interest in hemoglobin dynamics.
In general, the extended number of genetic variants showing joint association with HbA1 and HbA2 rather than HbF is consistent with high correlations of levels of adult hemoglobins HbA1 and HbA2 but only partial correlations of these hemoglobin forms with levels of HbF (see Online Methods).
Given the central role of hemoglobin in providing oxygen to the body tissues and the substantial fraction of total body cells accounted for by circulating red cells, factors impacting hemoglobin production and red cell count unsurprisingly have pleiotropic effects on other non-hematological traits. This is exemplified by the strong impact of the major β039 mutation on cholesterol and LDL-cholesterol (see companion paper13). Here we extended the analysis for this mutation to 69 non-hematological quantitative traits selected from among those assessed in the SardiNIA cohort4 (see Supplementary Note). We found the variant also significantly associated with increased total white blood cell counts (p = 3×10−7) -- with the major contribution coming from neutrophil counts (p = 1×10−6) -- and platelet counts (p = 9×10−5) (see Supplementary Table 10)
DISCUSSION
We provide evidence for 23 associated variants at 10 loci influencing the levels of one or more of the 3 hemoglobin species measurable in post-natal life. Our results are based on a cohort from the Sardinian founder population that is much larger than previously described GWAS for HbF and HbA2 and interrogates a high resolution genetic map, based on population sequencing that expands the assessed spectrum of allelic variants 10-fold compared to previous studies. The finding that 2 of the 5 newly reported loci were not detectable without using the SardiNIA reference panel, and the others were misplaced (Table 1 and Supplementary Table 7), further highlights how large-scale sequencing efforts in this founder population can reveal functionally relevant variants that may be very rare and hence missed in other populations.
For the same reasons, however, replication of results for such variants or translation of findings directly to other populations is difficult. For example, the other currently reported sample of comparable size, from the United Kingdom, could provide replication only for the two variants present there. Similar limitations will likely be found in other GWAS designed to detect effects of rare and founder variants. However, additional corroboration of our findings for such variants comes from their independent associations with other hemoglobin species and hematological traits in Sardinians, and also from the biological function of the genes involved. For instance, variant chr12:123681790 within MPHOSP9, associated with HbA1, also shows suggestive evidence of association with HbA2. The variant in FOG1, very rare in Europeans (MAF 0.4 %), is a missense variant in a gene implicated in erythropoiesis; and the variant in NFIX, absent in other European populations, falls within a cluster of genes involved in erythropoiesis and in a CpG region differentially methylated in fetal and adult red blood cell progenitors26.
By carrying out GWAS for HbA1, HbA2 and HbF assessed for the first time in the same individuals, we see a wide range of pleiotropic effects of variants across the 3 hemoglobin types (Table 1). Strikingly, HbA2 harbors more than half of the loci discovered here (see Figure 2), with many pleiotropic effects on HbA1 and some on HbF. Thus, although it has a minor role in the transport of oxygen to tissues55, variations in HbA2 participate in pathways that regulate the levels of the other hemoglobins active in postnatal life.
The direction of pleiotropic effects among the different hemoglobin types provides some additional clues to mechanism. Within the α-globin gene cluster, in agreement with the presence of α-globin chains in HbA1, HbA2 and HbF, all variants affecting more than one hemoglobin showed the same direction of effect for all. The regulation of globin chains from the β-globin gene cluster, however, is more complicated. It involves variants with the same direction of effect for all hemoglobins (rs7936823) and other variants most likely involved in switching mechanisms that affect fetal and adult hemoglobins in opposite directions (rs2855122). Still other variants change the kinetics of competition among non-α globin chains; for example, the β039 mutation decreases β-globin levels and thereby increases the availability of α-globin chains to combine with δ and γ-globins, leading to higher levels of HbA2 and HbF.
Variants influencing only 2 forms of hemoglobin acted mainly in the same direction and never jointly affected HbA1 and HbF. As for variants shared only between HbA2 and HbF, they can be attributed to specific cis-regulatory mechanisms in the β-globin gene cluster (rs12793110 and rs7944544) or to loci with a role in erythroid differentiation (CCND3 and MYB). By contrast, variants shared between HbA2 and HbA1 were either trans-acting (in MPHOSPH9) or localized in the α-globin gene cluster but with effect sizes probably too small to impact HbF production. Consistent with the latter possibility, the –α 3.7 deletion type I, which has strong genome-wide significant effects on HbA1 and HbA2, had much smaller, only suggestive, effects on HbF (see Supplementary Table 2).
Our analyses also detected broader pleiotropic impacts, most strikingly for the β039 variant. In addition to effects on LDL-c described in the companion paper13, we report for the first time that β039 is also significantly associated with increased total counts of white blood cells (and some subsets) as well as platelet counts. This suggests that in heterozygous carriers this variant drives a broader increase in bone marrow-derived blood cells. Speculatively, some of these, such as augmented leukocyte and neutrophil counts, may have provided protection against pathogens other than malaria, thus increasing selection for the balanced polymorphism.
The detected variants provide candidate modifiers influencing the clinical status of patients with monogenic hemoglobin disorders. For example, we carried out a preliminary analysis of a small sample of 306 β-thalassemia patients homozygous for the β039 stop codon mutation but showing very great heterogeneity in disease presentation and course. In addition to those described previously7–10, some variants detected in this study showed possible effects as modifiers of disease severity (see Supplementary Note). However, the potential of these variants to help predict disease severity remains tentative without studies of larger sample sets. Nevertheless, the variants already add to the candidate targets for therapeutic intervention in the widely prevalent inherited β-thalassemia and other hemoglobinopathies2.
ONLINE METHODS
Sample description
The population studied here includes 6,921 individuals, representing > 60 % of the adult population of 4 villages in the Lanusei Valley in Sardinia, Italy. They are part of the SardiNIA project, a longitudinal study including genetic and phenotypic data of 1,257 multigenerational families with more than 37,000 relative pairs. Details of phenotype assessments for these samples have been published previously4. All participants gave informed consent to study protocols, which were approved by the institutional review board of the University of Cagliari, the National Institute on Aging, and the University of Michigan.
For whole-genome sequencing, we selected 1,122 individuals from the SardiNIA study and 998 individuals enrolled in case–control studies of Multiple sclerosis and Type I Diabetes in Sardinia. Genomes were sequenced to an average coverage of 4.16-fold. Details on sequencing protocol, data process and variant calling can be found elsewhere56 and in the companion paper13. The 2,120 sequenced samples consist of 695 complete and incomplete trios; to avoid over-representation of rare haplotypes during imputation process we considered only parents for each trio – totaling 1,488 samples – to build our reference panel56 (see companion paper13 for details). Part of the sequencing data used in this study are available through dbGap, under “SardiNIA Medical Sequencing Discovery Project”, Study Accession: phs000313.v3.p2.
Genotyping and Imputation
The 4 micro-arrays used for genotyping the entire SardiNIA cohort were the Illumina® Infinium HumanExome BeadChip, ImmunoChip, Cardio-MetaboChip and HumanOmniExpress BeadChip. Genotyping was carried out according to manufacturer protocols at the SardiNIA Project Laboratory (Lanusei, Italy), at the Technological Center - Porto Conte Ricerche (Alghero, Italy) and at the National Institute on Aging Intramural Research Program Laboratory of Genetics (Baltimore, MD). Genotypes were called using GenomeStudio (version 1.9.4) and refined using Zcall (version 3)57. We applied standard per sample quality control filters to remove samples with low call rates or for which reported relationships and/or gender disagreed with genetic data. Details on quality controls were described elsewhere56. Altogether, 890,542 autosomal markers and 16,325 X-linked markers were genotyped across SardiNIA study samples. We selected for phasing and imputation only the 6,602 samples for which all 4 arrays were successfully genotyped.
Genotypes were phased using MACH software58, using 30 iterations of the haplotyping Markov chain and 400 states per iteration. We performed imputation using Minimac software59 and a reference panel including haplotypes of 1,488 Sardinian whole-genomes56 (see companion paper13). Variants with estimated imputation quality (RSQR) <= 0.3 or <0.8 were discarded if the estimated MAF was >= 1 % or between 0.5 % and 1 % respectively; variants with MAF < 0.5 % were kept only if genotyped. RSQR thresholds for rare and low frequency variants were more stringent than those proposed for other traits56 as they led to better genomic control parameters (1.001, 0.993 and 0.985 for HbA1, A2 and fetal, respectively). We also performed imputation using the 1000 Genomes Project Phase III (version 5)60 haplotype set, and used the same thresholds to discard variants. Genomic control parameters for 1000 Genomes imputation were 1.050, 0.997 and 0.984 for HbA1, A2 and fetal, respectively.
Association analysis
We performed association analyses of all 3 hemoglobins in grams per deciliter (g/dl) as well as percentage (%) for HbA2 and HbF. HbA2 (%) and HbF (%) were directly measured from high-performance liquid chromatography, while HbA1 (g/dl), HbA2 (g/dl) and HbF (g/dl) were derived from total hemoglobin measured by Coulter counter. As expected, measurements in % and g/dl were highly correlated for HbF (Spearman’s Rho = 0.99) and for HbA2 (Rho = 0.85). HbA1 (%) was not considered for genetic association because it was too highly correlated with both HbA2 (%) and HbF (%) as a consequence of their derivation formula (Rho = −0.803 and −0.757, respectively, p < 1×10−20). Considering only non-carriers of β0-mutations, HbA1 (g/dl) was highly correlated with HbA2 (g/dl) (Rho = 0.662, p < 1×10−20) and poorly with HbF (g/dl) (Rho = −0.055, p = 3.44×10−5). Likewise, HbA2 and HbF were weakly positively correlated as percentage measures (Rho = 0.108, p = 4.08×10−16) and even less as g/dl (Rho = 0.066, p = 5.81×10−5), consistent with previous findings5. Measurements were available for a subset of 6,305 individuals; descriptive statistics are reported in Supplementary Table 1. Association results were considered genome-wide significant when p-value was less than 5×10−08, however we also noted in the text variants that would not meet a threshold of 1.4×10−8 we introduce for sequencing based GWAS carried out in Sardinians for variants with MAF > 0.5 % (see companion paper13).
Before association analyses, traits were normalized using inverse normal transformation; for HbF we also removed outliers with values above 5 %. Analyses were adjusted for age, age2, and gender as well as for the presence of at least one of the 3 β0 mutations (β039 (rs11549407), HBB:c.20delA (rs63749819) and HBB:c.315+1G>A (rs33945777)), all directly genotyped or sequenced (see Characterization of β0 mutations paragraph). Regression coefficients for β039 – the most common in Sardinia with 10.3 % of carriers – are reported in the Supplementary Table 2.
Association was performed using the q.emmax test in EPACTS61, which implements a linear mixed model procedure to correct for cryptic relatedness and population stratification by incorporating a genomic-based kinship matrix. Associations reported in the table refer to the best p-value obtained with either percentage or original units for HbA2 and HbF. Notably, HbF signals always resulted in lower p-values considering g/dl, whereas for HbA2 analysis, this was only the case for rs141494605. All loci passed the genome-wide significance threshold of p<5×10−08 for both % and g/dl except for rs59329875, which was genome-wide significant only for the HbA2 measure reported in Table 1.
To identify independent signals we performed regional conditional analysis, using forward selection procedure adding, at each step, the most associated variant as covariate in the model. In this sequential analysis, we tested only SNPs lying in a region of 2Mb centered on the lead variant. The same genome-wide significance threshold used for primary signals was also considered for independent signals. For loci where different independent signals were found, we also report model parameters of jointly associated variants in Supplementary Table 3. Finally, the lead variants and their surrogates (r2 > 0.90) were annotated using Combined Annotation Dependent Depletion (CADD) score14 and reported in Supplementary Table 5.
Heritability and variance explained
We estimated heritability for the 3 hemoglobins using Merlin-regress62 on the same sample used for the GWAS study. Estimates for normalized levels of hemoglobins were respectively 0.520 for HbA1 (g/dl), 0.728 for HbA2 (%) (0.700 for g/dl) and 0.633 for HbF (%) (0.624 for g/dl). We then calculated for each hemoglobin form the proportion of phenotypic variance explained by the associated lead variants. We measured that as the difference of R2-adjusted observed between the full and the basic model, where the basic model includes only phenotypic covariates (age, age2 and gender) and the full model also includes all the independent SNPs associated with the specific trait. R2-adjusted values were calculated using a linear mixed model procedure from lmekin() function in the “Kinship” R package63. Estimates were 0.240 for HbA1 (g/dl), 0.492 for HbA2 (%) and 0.383 for HbF (%).
Characterization of β0 mutations
For the present study we designed a Taqman custom assay for the HBB:c.118C>T nonsense mutation (rs11549407, also known as β039), and genotyped 6,602 samples. Comparison of Taqman genotypes and imputation results (rs11549407, RSQR = 0.92) produced an overall concordance of 98.8 %. Also, we further sequenced all samples discordant between red blood cell index-based diagnosis (using MCV, MCH, HbF % and HbA2 %) and Taqman genotypes, using Sanger sequencing to determine any additional β-globin mutations different from β039, thus identifying 3 carriers for the HBB:c.20delA (rs63749819) and one for the HBB:c.315+1G>A (rs33945777) mutations.
Characterization of the deletion at the α-globin gene cluster
In Sardinia 3 variants are known to be mainly responsible for α-thalassemia: SNPs rs111033603 and rs41474145, and the deletion NG_000006.1:g.34164_37967del3804; the latter, known as the –α 3.7 deletion type I, is by far the most common64. We did not observe the rarer rs111033603 or rs41474145 in our sequencing effort. To establish genotypes at the deletion site in the full cohort, we used an inference strategy combined with experimental data. Specifically, we first characterized the structural variant by PCR in 260 unrelated sequenced individuals randomly selected in the SardiNIA cohort. We calculated the relative coverage of the deleted region in the whole-genome sequenced samples by considering the ratio of read count in the potentially deleted region (223,450 to 226,953 bp – excluding 150 bp boundaries) with read count in the nearby region not subject to deletion (227,254 to 230,757 bp). We then identified coverage ratio thresholds that best predicted PCR genotypes at the deletion and used these thresholds to infer genotypes for the 2,120 sequenced individuals. We then inserted genotypes in the Sardinian reference panel and imputed the deletion on the total SardiNIA cohort. To assess accuracy of imputation we considered the best guess genotypes and searched for Mendelian errors in families. The observed rate was 0.58 % over 1,193 parent-offspring pairs, consistent with high imputation precision. Association results reported in the manuscript at this locus are corrected for the inferred –α 3.7 deletion type I dosages.
Variants validation
We validated all variants that showed genome-wide significant p-values in the primary or conditional analysis that were not directly genotyped or had no surrogates (r2 > 0.90) that were directly genotyped. We did not validate variant rs13019832 at BCL11A for HbF, which was highly linked with findings of previous reports (rs7606173)49,51. Validation was performed using Sanger sequencing or Taqman, depending on variant frequency, for 5 variants. We selected for each variant all individuals carrying the minor allele (heterozygous and homozygous) plus a random subset of subjects homozygous for the other allele (in all, 3,084 subjects were genotyped), except for rs141494605 and chr16:391593, for which we specifically selected worse imputation dosages (borderline RSQR). In addition, for rs17525396, we used independent genotypes available for a subset of the cohort65, derived from Affymetrix 6.0 (see Supplementary Table 4).
Replication of variant effects
Replication was performed in the TwinsUK cohort43. Genotyping was performed using a combination of Illumina arrays (HumanHap300, HumanHap610Q, 1M-Duo and 1.2MDuo 1M), and imputation performed using the IMPUTE software package (v2) and 1,000 Genomes haplotypes released on 16 Jun 2014-- Phase I integrated variant set release36,66. Details on quality controls are provided as Supplementary Note. HbA2 levels and HbF percentage were obtained by HPLC, and F-cells were enumerated after intracellular HbF staining and subsequent flow cytometry67. Measurements were available in 4,131 samples. Association analyses were performed with merlinoffline package in Merlin, to account for relatedness62. To be consistent with analyses performed in the SardiNIA study, age, age squared and gender were used as covariates and the traits transformed using quantile normalization.
Selection of candidate genes
At each locus, we defined a list of genes to be considered as plausible candidates if they satisfied one of the following: 1) genes that were +/− 25Kb of the lead SNP, indicated (p) in Table 1; 2) genes with exonic variants (frame-shift, stop-codon, non-synonymous and synonymous) along with splice-site and 5′/3′ UTR variants in LD (r2≥0.8) with the lead SNP (c); 3) genes whose expression was modulated by the SNP itself or by an eQTL in LD (r2≥0.8) with the top SNP (e); 4) genes with clear biological function connected to the traits (b); or 5) genes harboring variants responsible for which Mendelian diseases, as reported in OMIM (o). Candidate genes from eQTL data were searched using an automatized pipeline querying 16 eQTL public repositories48,68–82, including the Pritchard eQTL browser; only top SNP eQTLs or any SNP with FDR < 0.05 were considered.
Pleiotropy and gene connections analysis
To characterize genome-wide significant results and to identify suggestively significant ones, we searched for effects shared between the different hemoglobin forms as well as evidence of connections between both. Specifically, for genome-wide significant markers, we simply reported the effect direction for all traits with p < 0.01 when a marker is associated at genome-wide level for one trait (see Table 1). To identify candidates with suggestive p-values between 1.00×10−04 and 5.00×10−08, we selected among these:
- markers with MAF > 0.5 % and showing 2-trait combined p-values < 5×10−08; p-values were combined using inverse variance weighted meta-analysis, as implemented in Metal software83;
- markers falling in or nearby genes that demonstrated evidence of connections with genome-wide significant loci, either in Pubmed (using the 2006 data set to avoid confounding by subsequent GWAS discoveries), or in Human Expression Atlas37 and Gene Ontology38 databases using GRAIL39 and considering genes reported with multiple hypothesis corrected p-values < 0.05.
Using these criteria, we identified 21 further genes with biological connections to genome-wide significant loci reported in Supplementary Table 8 and 14 variants with combined p-values between 2.08×10−08 and 1.18×10−11, reported in Supplementary Table 9.
Supplementary Material
ACKNOWLEDGEMENTS
This work is dedicated to Prof. Antonio Cao, Prof. Renzo Galanello and Prof. Maurizio Longinotti, who devoted their scientific lives to understanding, preventing and treating hematological diseases in Sardinia. We are also grateful to Maria Serafina Ristaldi and Maria Giuseppina Marini for knowledge and insight that they freely shared with us. Finally, we thank all the volunteers who generously participated in this study and made this research possible. The SardiNIA study was funded in part by the National Institutes of Health (National Institute on Aging, National Heart Lung and Blood Institute, and National Human Genome Research Institute). This research was supported by National Human Genome Research Institute grants HG005581, HG005552, HG006513, and HG007022; by National Heart Lung and Blood Institute grant HL117626; by the Intramural Research Program of the NIH, National Institute on Aging, with contracts N01-AG-1-2109 and HHSN271201100005C; by Sardinian Autonomous Region (L.R. no. 7/2009) grant cRP3-154; by grant FaReBio2011 “Farmaci e Reti Biotecnologiche di Qualità”; and by PB05 InterOmics MIUR Flagship Project. TwinsUK study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013); National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. SNP Genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR. S.L.T was supported by the Medical Research Council, UK (Grant G0000111, ID51640) and S.Men. received funding from The British Society for Heamatology (start-up grant).
URLS
SardiNIA project: https://sardinia.irp.nia.nih.gov
1000 Genomes project: http://www.1000genomes.org
HumanExome BeadChip design: http://genome.sph.umich.edu/wiki/Exome_Chip_Design
ImmunoChip, Cardio-MetaboChip and HumanOmniExpress BeadChip: http://www.illumina.com
GenomeStudio software: http://www.illumina.com/applications/microarrays/microarray-software/genomestudio.html
MACH software: http://csg.sph.umich.edu/abecasis/MACH
Minimac software: http://genome.sph.umich.edu/wiki/Minimac
Zcall software: https://github.com/jigold/zCall
IMPUTE v2 software: http://mathgen.stats.ox.ac.uk/impute/impute_v2.1.0.html
Merlin (including Merlin-regress and Merlin-offline): http://csg.sph.umich.edu/abecasis/merlin
Epacts software: http://genome.sph.umich.edu/wiki/EPACTS
Metal software: http://csg.sph.umich.edu/abecasis/metal
GWAS Catalog: http://www.genome.gov/gwastudies
Grail software: https://www.broadinstitute.org/mpg/grail
Gene Ontology: http://geneontology.org
Human Expression Atlas: http://symatlas.gnf.org
Pritchard eQTL browser: http://eqtl.uchicago.edu
R project: http://www.R-project.org
AUTHOR CONTRIBUTIONS
G.A, D.S. and F.C. conceived the study.
F.D., D.S., S.S. and F.C. drafted the manuscript.
F.D., M.Z., M.U., P.M., S.L.T., G.A., D.S., S.S. and F.C. revised the manuscript.
F.B., A.M. and A.A. performed sequencing experiments.
M.Pi., G.A. and S.S. selected samples for sequencing.
F.D., C.S., M.S., E.P., G.P. and S.S. carried out genetic association analyses in the SardiNIA cohort.
C.S. analyzed DNA sequence data.
M.Z., F.B. and A.Mu. carried out SNP array genotyping.
M.Z. designed the validation strategy and M.Z., F.B. and A.Mu. verified genotypes by Sanger sequencing and Taqman genotyping.
L.P. performed genotyping of the –α 3.7 deletion type I.
M.Pa. created an automatized pipeline to query the eQTLs public repositories.
P.M. and R.G. provided thalassemia patients genotypes and phenotypic data.
S.B. and R.G. supervised hemoglobins’ characterisation in the Sardinia cohort.
F.D. analyzed the thalassemia patients cohort.
S.Men., T.D.S. and S.L.T., provided replication samples.
S.Met. analyzed replication samples.
L.L. provided IT support for sequencing and genotype data processing and analyses.
D.S. and F.C. supervised the study.
All authors reviewed and approved the final manuscript.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Sankaran VG, Xu J, Orkin SH. Advances in the understanding of haemoglobin switching. Br. J. Haematol. 2010;149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Modell B, Darlison M. Global epidemiology of haemoglobin disorders and derived service indicators. Bull. World Health Organ. 2008;86:480–487. doi: 10.2471/BLT.06.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malaria Genomic Epidemiology Network & Malaria Genomic Epidemiology Network Reappraisal of known malaria resistance loci in a large multicenter study. Nat. Genet. 2014;46:1197–1204. doi: 10.1038/ng.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilia G, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menzel S, Garner C, Rooks H, Spector TD, Thein SL. HbA2 levels in normal adults are influenced by two distinct genetic mechanisms. Br. J. Haematol. 2013;160:101–105. doi: 10.1111/bjh.12084. [DOI] [PubMed] [Google Scholar]
- 6.Bae HT, et al. Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood. 2012;120:1961–1962. doi: 10.1182/blood-2012-06-432849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uda M, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lettre G, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danjou F, et al. Genetic modifiers of β-thalassemia and clinical severity as assessed by age at first transfusion. Haematologica. 2012;97:989–993. doi: 10.3324/haematol.2011.053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danjou F, et al. A genetic score for the prediction of beta-thalassemia severity. Haematologica. 2014 doi: 10.3324/haematol.2014.113886. haematol.2014.113886. doi:10.3324/haematol.2014.113886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trecartin RF, et al. beta zero thalassemia in Sardinia is caused by a nonsense mutation. J. Clin. Invest. 1981;68:1012–1017. doi: 10.1172/JCI110323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidore C, et al. Genome sequencing elucidates Sardinian genetic architecture and augments GWAS findings: the examples of lipids and blood inflammatory markers. Nat. Genet. 2015 doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freson K, et al. Molecular cloning and characterization of the GATA1 cofactor human FOG1 and assessment of its binding to GATA1 proteins carrying D218 substitutions. Hum. Genet. 2003;112:42–49. doi: 10.1007/s00439-002-0832-1. [DOI] [PubMed] [Google Scholar]
- 16.Nichols KE, et al. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat. Genet. 2000;24:266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozar K, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 18.Sankaran VG, et al. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev. 2012;26:2075–2087. doi: 10.1101/gad.197020.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soranzo N, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamatani Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 21.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvik GP, et al. Genetic and nongenetic sources of variation in phospholipid transfer protein activity. J. Lipid Res. 2010;51:983–990. doi: 10.1194/jlr.M000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lettre G, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kettunen J, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirose Y, et al. Human phosphorylated CTD-interacting protein, PCIF1, negatively modulates gene expression by RNA polymerase II. Biochem. Biophys. Res. Commun. 2008;369:449–455. doi: 10.1016/j.bbrc.2008.02.042. [DOI] [PubMed] [Google Scholar]
- 26.Lessard S, Beaudoin M, Benkirane K, Lettre G. Comparison of DNA methylation profiles in human fetal and adult red blood cell progenitors. Genome Med. 2015;7:1. doi: 10.1186/s13073-014-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riddell J, et al. Reprogramming Committed Murine Blood Cells to Induced Hematopoietic Stem Cells with Defined Factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmfeldt P, et al. Nfix is a novel regulator of murine hematopoietic stem and progenitor cell survival. Blood. 2013;122:2987–2996. doi: 10.1182/blood-2013-04-493973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawane K, et al. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science. 2001;292:1546–1549. doi: 10.1126/science.292.5521.1546. [DOI] [PubMed] [Google Scholar]
- 30.Porcu S, et al. Klf1 affects DNase II-alpha expression in the central macrophage of a fetal liver erythroblastic island: a non-cell-autonomous role in definitive erythropoiesis. Mol. Cell. Biol. 2011;31:4144–4154. doi: 10.1128/MCB.05532-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou D, Liu K, Sun C-W, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and [gamma]- to [beta]-globin gene switching. Nat Genet. 2010;42:742–744. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]
- 32.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118:2044–2054. doi: 10.1182/blood-2011-03-331371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satta S, et al. Compound heterozygosity for KLF1 mutations associated with remarkable increase of fetal hemoglobin and red cell protoporphyrin. Haematologica. 2011;96:767–770. doi: 10.3324/haematol.2010.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borg J, et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet. 2010;42:801–805. doi: 10.1038/ng.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perseu L, et al. KLF1 gene mutations cause borderline HbA2. Blood. 2011;118:4454–4458. doi: 10.1182/blood-2011-04-345736. [DOI] [PubMed] [Google Scholar]
- 36.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raychaudhuri S, et al. Identifying Relationships among Genomic Disease Regions: Predicting Genes at Pathogenic SNP Associations and Rare Deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews NC. The NF-E2 transcription factor. Int. J. Biochem. Cell Biol. 1998;30:429–432. doi: 10.1016/s1357-2725(97)00135-0. [DOI] [PubMed] [Google Scholar]
- 41.Hoogewijs D, et al. Androglobin: a chimeric globin in metazoans that is preferentially expressed in Mammalian testes. Mol. Biol. Evol. 2012;29:1105–1114. doi: 10.1093/molbev/msr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iolascon A, Perrotta S, Stewart GW. Red blood cell membrane defects. Rev. Clin. Exp. Hematol. 2003;7:22–56. [PubMed] [Google Scholar]
- 43.Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: TwinsUK and Healthy Ageing Twin Study. Int. J. Epidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sangerman J, et al. Mechanism for fetal hemoglobin induction by histone deacetylase inhibitors involves gamma-globin activation by CREB1 and ATF-2. Blood. 2006;108:3590–3599. doi: 10.1182/blood-2006-01-023713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goh S-H, et al. A newly discovered human alpha-globin gene. Blood. 2005;106:1466–1472. doi: 10.1182/blood-2005-03-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farrell JJ, et al. A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood. 2011;117:4935–4945. doi: 10.1182/blood-2010-11-317081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadhouders R, et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest. 2014;124:1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeller T, et al. Genetics and Beyond – The Transcriptome of Human Monocytes and Disease Susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhatnagar P, et al. Genome-wide association study identifies genetic variants influencing F-cell levels in sickle-cell patients. J. Hum. Genet. 2011;56:316–323. doi: 10.1038/jhg.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bauer DE, Orkin SH. Update on fetal hemoglobin gene regulation in hemoglobinopathies. Curr. Opin. Pediatr. 2011;23:1–8. doi: 10.1097/MOP.0b013e3283420fd0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grundberg E, et al. Global Analysis of DNA Methylation Variation in Adipose Tissue from Twins Reveals Links to Disease-Associated Variants in Distal Regulatory Elements. Am. J. Hum. Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenbloom KR, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinberg MH, Adams JG. Hemoglobin A2: origin, evolution, and aftermath. Blood. 1991;78:2165–2177. [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 56.Pistis G, et al. Rare variant genotype imputation with thousands of study-specific whole-genome sequences: implications for cost-effective study designs. Eur. J. Hum. Genet. EJHG. 2014 doi: 10.1038/ejhg.2014.216. doi:10.1038/ejhg.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldstein JI, et al. zCall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinforma. Oxf. Engl. 2012;28:2543–2545. doi: 10.1093/bioinformatics/bts479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Consortium T. 1000 G. P. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 63.R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 64.Origa R, et al. Complexity of the alpha-globin genotypes identified with thalassemia screening in Sardinia. Blood Cells. Mol. Dis. 2014;52:46–49. doi: 10.1016/j.bcmd.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 65.Naitza S, et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8:e1002480. doi: 10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Menzel S, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat. Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 68.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 69.Stranger BE, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veyrieras J-B, et al. High-Resolution Mapping of Expression-QTLs Yields Insight into Human Gene Regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimas AS, et al. Common Regulatory Variation Impacts Gene Expression in a Cell Type-Dependent Manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fehrmann RSN. Trans-eQTLs Reveal That Independent Genetic Variants Associated with a Complex Phenotype Converge on Intermediate Genes, with a Major Role for the HLA. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Innocenti F, et al. Identification, Replication, and Functional Fine-Mapping of Expression Quantitative Trait Loci in Primary Human Liver Tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montgomery SB, Lappalainen T, Gutierrez-Arcelus M, Dermitzakis ET. Rare and Common Regulatory Variation in Population-Scale Sequenced Human Genomes. PLoS Genet. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaffney DJ, et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright FA, Shabalin AA, Rusyn I. Computational tools for discovery and interpretation of expression quantitative trait loci. Pharmacogenomics. 2012;13:343–352. doi: 10.2217/pgs.11.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Westra H-J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma. Oxf. Engl. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.