Abstract
Protein kinases are key players in a large number of cellular signaling pathways. Dysregulated kinase activity has been implicated in a number of diseases, and members of this enzyme family are of therapeutic interest. However, due to the fact that most inhibitors interact with the highly conserved ATP-binding sites of kinases, it is a significant challenge to develop pharmacological agents that target only one of the greater than 500 kinases present in humans. A potential solution to this problem is the development of bisubstrate and bivalent kinase inhibitors, in which an active site-directed moiety is tethered to another ligand that targets a location outside of the ATP-binding cleft. Because kinase signaling specificity is modulated by regions outside of the ATP-binding site, strategies that exploit these interactions have the potential to provide reagents with high target selectivity. This review highlights examples of kinase interaction sites that can potentially be exploited by bisubstrate and bivalent inhibitors. Furthermore, an overview of efforts to target these interactions with bisubstrate and bivalent inhibitors is provided. Finally, several examples of the successful application of these reagents in a cellular setting are described.
Keywords: bivalent, bisubstrate, protein kinase, inhibitor, SNAP-tag
Introduction
Intra-cellular signaling pathways are utilized by eukaryotic organisms in order to regulate cellular behavior. Extra-cellular signals are received and propagated through tightly regulated intra-cellular signaling cascades that are mediated by spatially and temporally controlled intra-cellular interactions. A number of these interactions are regulated by protein phosphorylation, which is catalyzed by protein kinases. Protein kinases are a large subset of the human proteome, consisting of over 500 members (Manning et al 2002). These enzymes catalyze the transfer of the γ-phosphate of adenosine triphosphate (ATP) to a protein substrate at a serine, threonine, or tyrosine residue. These phosphorylation events mediate important cellular processes such as development, differentiation, and transformation. Aberrant protein kinase activity has been implicated in a variety of diseases including diabetes, cancer, and chronic inflammation, making members of this enzyme family attractive drug targets (Blume-Jensen and Hunter 2001, Cohen 2002, Hunter 2000). Despite widespread interest in this enzyme family, the cellular function of many protein kinases is not well understood. Chemical genetic tools can be used to better elucidate the function of protein kinases. In order to provide clear insight into kinase function, these tools must be selective for a particular kinase of interest, which is still a major challenge in kinase reagent development.
Current chemical methods for studying the function of protein kinases involve the use of ATP-competitive small molecule inhibitors (Knight and Shokat 2005). These reagents allow for high temporal control over kinase function in cells. There are, however, several disadvantages to the use of ATP-competitive molecules as functional probes of protein kinases. The active sites of protein kinases are highly conserved, which makes achieving specificity for a single kinase target difficult. Additionally, as these molecules can diffuse freely throughout the cell, spatial control cannot be readily achieved. Techniques such as RNAi and genetic knockouts can also provide the ability to study the cellular function of kinases. In contrast to small molecule inhibition of protein kinases, genetic methods remove the entire kinase of interest from the cell, and thus, the catalytic function of a kinase cannot be differentiated from its non-catalytic functions (Knight and Shokat 2007). Furthermore, these methods lack spatial and temporal control.
A pharmacological method that allows the rapid identification of potent and selective kinase inhibitors has the potential to overcome many of these problems. Bisubstrate and bivalent inhibitors are attractive in this regard because they can exploit the same interactions that kinases use for obtaining specificity in signaling (Figure 1). Bisubstrate and bivalent reagents are generated by linking an ATP-competitive small molecule to a ligand that targets a region outside of the ATP-binding cleft of a kinase of interest. By targeting two distinct binding sites on a particular kinase, the potency and selectivity of inhibition can be increased. Numerous reports of bisubstrate and bivalent inhibitors of protein kinases have emerged in the literature over the last two decades, providing insight into their design and functional properties. In this review, we highlight a number of signaling interactions that are used by kinases in signaling events and that can be targeted by bisubstrate and bivalent inhibitors. Furthermore, this review provides an overview of efforts to develop and apply bisubstrate and bivalent inhibitors of protein kinases, highlighting the application of these reagents in a cellular setting.
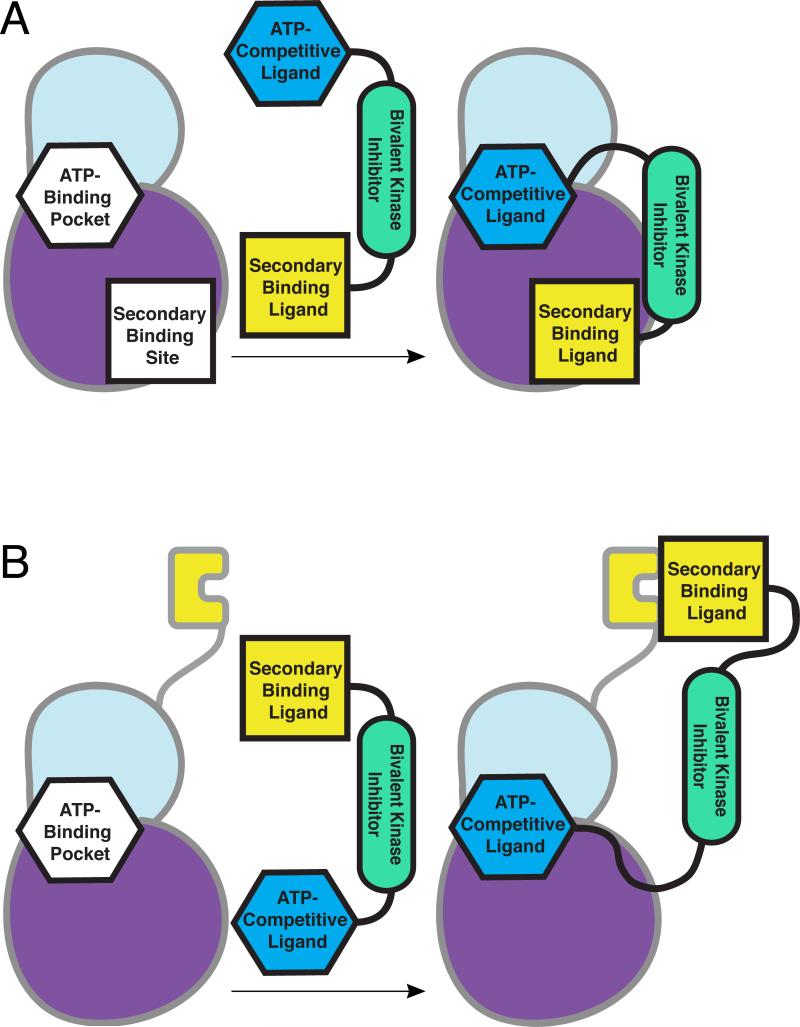
Figure 1.
Bivalent inhibition of a protein kinase. (a) A single inhibitor is generated by fusing an ATP-competitive ligand to a secondary binding ligand. The ATP-competitive ligand interacts with the kinase active site, directing the bivalent inhibitor to the kinase. The secondary binding ligand introduces selectivity for a particular kinase. In the case of bisubstrate inhibitors, this ligand targets the protein substrate site of the protein kinase. (b) Regulatory domains of protein kinases present an avenue for bivalent inhibition where secondary-binding ligands can target domains distal to the kinase domain.
Protein kinases and substrate specificity
The conserved protein kinase fold consists of an N-terminal lobe composed mainly of β-strands and an α-helical C-terminal lobe (Hanks and Hunter 1995, Manning et al 2002, Taylor et al 1988, Turk 2008, Ubersax and Ferrell 2007). At the junction of these two lobes lies the ATP-binding site. All protein kinases are bisubstrate enzymes that recognize both the cofactor ATP and specific protein substrates. Amidst the complex environment of the cell, kinases select specific substrates for phosphorylation, which is necessary for the proper alignment of signaling cascades. Various mechanisms play a role in the ability of a protein kinase to properly identify a specific substrate within the cell. One of the major mechanisms is the interaction of the kinase with amino acid residues immediately surrounding the modified serine, threonine, or tyrosine (Bhattacharyya et al 2006, Blume-Jensen and Hunter 2001, Cohen 2002, Hunter 2000, Turk 2008, Ubersax and Ferrell 2007). The epitope displayed by the consensus phosphorylation sequence is complemented by the protein kinase active site. This recognition event conveys selectivity at the primary sequence level. Beyond the consensus sequence, protein kinases make interactions with the substrate that are distal to the site of phosphorylation (Bhattacharyya et al 2006, Chen and Turk 2010, Hutti et al 2004, Knight and Shokat 2005, Mok et al 2010). These interactions can be mediated by distant binding sites on the catalytic domain or by modular binding domains.
Specificity determinants within the kinase domain
For most protein substrates, the four amino acids N-terminal and C-terminal to the phosphorylation site interact extensively with the kinase catalytic domain (Ubersax and Ferrell 2007). The substrate preferences of kinases for residues surrounding the phosphorylation site have been mapped with a number of profiling techniques. One such method, described by Turk and coworkers, involves the radiometric analysis of biotinylated peptide substrates that have been treated with a purified kinase of interest (Chen and Turk 2010, Hutti et al 2004, Knight and Shokat 2007, Mok et al 2010). With this method, libraries of peptides are analyzed where amino acids surrounding the phosphorylation site are systematically varied. From studies of this type, general trends regarding the preferences of protein kinases for substrates have been observed. While many kinases can generally be classified as acidophilic, basophilic, or proline-directed with respect to their phosphorylation sequence preference, the presence of particular residues at fixed positions plays a role in sequence selectivity as well (Mok et al 2010). For some, determinants of this nature are one of the major mechanisms that are exploited to select substrates in the cell. However, for many protein kinases, protein substrate interactions are low-affinity, which facilitates substrate turnover. Therefore, in most cases, the phosphorylation site consensus motif is not the main driver of substrate selection in the cell.
Distinct from the active site, protein kinases interact with substrates through other binding sites and modular domains. Within the kinase domain, certain serine/threonine kinases possess docking sites that are located outside of the active site (Biondi and Nebreda 2003). Docking sites interact with protein substrates through a binding motif that is distinct from the phosphoacceptor site, introducing an additional selectivity determinant for substrate interactions. In some cases, this site exhibits allosteric control over kinase function, either positively or negatively regulating kinase activity (Bhattacharyya et al 2006, Sheridan et al 2008, Ubersax and Ferrell 2007). The mitogen-activated protein kinases (MAPKs) are one example of a kinase family that utilizes a docking site to enhance substrate specificity. Interestingly, the three subfamilies of MAPKs, extracellular signal-regulated kinases (ERK), p38 MAPKs (p38) and c-Jun N-terminal kinases (JNK), display a striking lack of selectivity regarding their phosphorylation site consensus motif. Peptide library screening has shown that a necessary proline residue following the phosphoacceptor serine or threonine is the main factor dictating the phosphorylation consensus sequence for ERK2, p38α, p38δ, and JNK2 (Sheridan et al 2008). This suggests that MAPKs likely use docking interactions to achieve proper phosphorylation of substrates within the cell. For example, a docking site for substrates that contain a DEF (docking site for ERK, FXF) motif, which recognizes an FX(F/Y)P sequence located approximately 6-20 amino acids C-terminal to the phosphoacceptor site, is a major determinant of ERK substrate selectivity (Mok et al 2010, Sheridan et al 2008). While both ERK2 and p38α show a similar and strong preference for aromatic residues at positions one and three of this motif, p38δ displays a striking preference for aliphatic residues at the first position. Binding determinants of this nature play a crucial role in the ability of MAPKs to successfully recognize and phosphorylate their proper targets in cells.
Obtaining specificity through modular domains
Through the interaction of protein substrates with recognition sites within the kinase domain, the efficiency and stringency of substrate phosphorylation is enhanced. This can occur through an increase in local substrate concentration or proper alignment of residues for phosphorylation (Barr et al 2004, Kothe et al 2007, Lowery et al 2005, Ubersax and Ferrell 2007). Beyond the catalytic kinase domain, a number of kinases use separate modular binding domains to obtain substrate specificity. Modular binding domains have been shown to interact with unaltered substrate sequence motifs, as well as motifs that have been post-translationally modified (Bhattacharyya et al 2006). Modular binding domains not only deliver kinase catalytic domains to substrates in specific locations, but also serve a regulatory role. For example, the Src homology 3 (SH3) domains of Src-family kinases (SFKs) play a major role in the localization and regulation of the catalytic domains of these kinases. This domain regulates kinase function through both inter- and intra-molecular protein-protein interactions. SFK SH3 domains interact with substrate polyproline motifs inter-molecularly, facilitating substrate recognition, and intra-molecularly with sub-optimal linker sequences, to regulate kinase catalytic activity (Qiu and Miller 2003). SH3 domains are used by a number of additional non-receptor tyrosine kinases in a similar manner (Yadav and Miller 2008, Zarrinpar et al 2003).
A variety of modular domains are utilized by protein kinases to recognize post-translationally modified, mainly phosphorylated, recognition motifs within the cell. These binding events can serve both as substrate specificity determinants and regulatory interactions. For example, the Src homology 2 (SH2) domains of SFKs recognize a phospho-tyrosine residue in their own C-terminal tail, which results in a closed, auto-inhibited kinase with low catalytic activity (Schlessinger and Lemmon 2003, Yadav and Miller 2008). The same SFK SH2 domains also recognize substrates that contain phosphorylated tyrosine residues. Furthermore, the SH2 domains of other tyrosine kinases have been shown to recognize phospho-tyrosine residues in their substrates. This primed interaction facilitates multi-site phosphorylation of substrates, indicating further regulatory roles of the SH2 domain (Mayer et al 1995). Similarly, the polo-box domain of the mitotic serine/threonine polo-like kinase (PLK) family is a phospho-recognition domain that acts to both properly localize PLK during cell division and modulate PLK's kinase activity (Barr et al 2004, Kothe et al 2007, Lowery et al 2005). These interactions not only provide greater phosphorylation fidelity, but also facilitate temporal control over signaling events.
Additional phospho-recognition domains include forkhead-associated (FHA) and conserved domain 2 (C2) domains. FHA domains are generally selective for phosphothreonine-containing motifs, and proteins containing these domains have been implicated in many cellular processes, such as DNA repair, signaling, transcription, and protein transport and degradation (Durocher and Jackson 2002). The kinase CHK2 is involved in the DNA damage checkpoint response, and two tumor-associated mutations of this kinase have been mapped to the FHA domain, demonstrating its importance for proper cellular function of the kinase. The C2 domain of the kinase PKC was originally discovered as a modulator of catalytic activity through binding phospholipids and is found in a variety of human protein kinases (Bhattacharyya et al 2006). This domain has also been shown in the specific case of PKC, to bind a phosphotyrosine residue of the protein CDCP1, which has been primed by Src (Benes et al 2005). Crystallographic data indicate that the sequence specificity of the C2 domain for a substrate peptide is determined by residues located both upstream and downstream of the phosphotyrosine residue, which may point toward a highly selective interaction (Benes et al 2005).
Pleckstrin homology (PH) domains, like C2 domains, recognize phospholipids, which are often the products of agonist-stimulated phosphoinositide 3-kinases (PI3-kinases) (Lemmon and Ferguson 2000). Additionally, a pathway has been described where the non-receptor tyrosine kinase BTK is recruited to PI3-kinase products in the plasma membrane through a PH domain. This allows BTK to act on substrates in a location-specific manner (Lemmon and Ferguson 2000, Schlessinger and Lemmon 2003). While BTK's PH domain does not interact with protein binding partners, it facilitates proper substrate selection by restricting the number of potential substrates available to this kinase. Therefore, this domain, like the protein-protein interaction motifs and modular domains described above, is an attractive target for the development of new and potentially selective inhibitors. While the affinities of the domains described above for their respective ligands are often weak, these interactions can, in many cases, be very selective. The exploitation of known substrate selection mechanisms of kinases represents an interesting avenue for the development of inhibitors with very high target specificity.
Overview of protein kinase inhibitors
Currently, most potent kinase inhibitors target the highly conserved ATP-binding site, which makes the generation of mono-selective pharmacological agents extremely challenging (Fedorov et al 2007, Krishnamurty and Maly 2007). While it is possible to obtain ATP-competitive inhibitors with exquisite selectivity, this process requires iterative rounds of synthesis and testing and is quite laborious (Knight and Shokat 2005). As a result, there has been a high level of interest in the development of inhibitors that disrupt the non-conserved binding interactions described above. Presumably, pharmacological tools that target these sites will have the same level of selectivity that kinases display in signaling cascades. However, most pharmacological agents that target sites outside of the ATP-binding pocket are of low affinity, with a majority of these ligands possessing dissociation constants in the micromolar to millimolar range. For this reason, bivalent ligands that target two distinct surfaces of a protein kinase have been pursued by a number of research groups. Many bivalent ligands are able to obtain selectivity by interacting with the low affinity, heterogeneous binding interfaces of protein kinases, while maintaining potency by engaging kinase active sites. If proper ligands and linkers are selected, bivalent reagents can possess incredibly high potency and selectivity. Bivalent inhibitors of protein kinases can be separated into two groups depending on the interactions that they make with their kinase target. Bisubstrate kinase inhibitors are composed of an ATP-competitive ligand covalently tethered through a linker to a pseudosubstrate peptide of the protein kinase of interest. On the other hand, bivalent kinase inhibitors display an active site-directed ligand tethered to a second ligand that targets a region outside of the active site. Both classes of inhibitors are described in this review.
Bisubstrate inhibitors of serine/threonine protein kinases
An early report of a bisubstrate protein kinase inhibitor was described in 1991, by Ricouart and coworkers (Table 1) (Ricouart et al 1991). In this study, the serine/threonine protein kinase C (PKC) was targeted. In mammals, the PKC family is made up of over ten isozymes shown to be involved in numerous cellular processes (Rosse et al 2010). Elucidating the specific function of each PKC isoform has proven difficult due to high sequence homology within the kinase domain and their largely overlapping substrate specificities. Probes that are able to differentiate between PKC family members would be highly valuable for achieving this goal. To this end, Ricouart and co-workers developed a PKC-directed bisubstrate inhibitor consisting of a pseudosubstrate peptide containing a single serine followed by six arginine residues, which targets the protein substrate site of PKC, linked to an ATP-competitive 5-isoquinolinylsulfonyl inhibitor, which targets the ATP-binding site. The 5-isoquinolinylsulfonyl inhibitor selected is a derivative of the known PKC inhibitor H7. The peptide and ATP-competitive ligands were covalently tethered through a 16 Å linker, a distance predicted to be optimal based on a crystal structure of phosphoglycerate kinase, which was the most similar kinase structure available at that time. To test the potency of this inhibitor, a PKC activity assay using histone as an in vitro substrate was developed. With this assay, it was observed that the bisubstrate inhibitor (IC50 = 0.3 μM) was 67-fold more potent against PKC activity than the 5-isoquinolinylsulfonyl ATP-competitive inhibitor alone (IC50 = 20 μM). Importantly, a bivalent effect was verified by testing kinase inhibition in the presence of a mixture of both unlinked monovalent ligands. Enhancement of potency was only observed when the two components of the bisubstrate inhibitor were covalently tethered together. Unfortunately, the potency of the bisubstrate inhibitor was 100-fold greater for closely-related PKA than for PKC, despite the pseudosubstrate peptide component of the bisubstrate inhibitor being selected for PKC specificity. Despite the lack of desired selectivity for PKC over PKA, this early example demonstrated that two distinct kinase ligands can be tethered together into a single bisubstrate molecule with enhanced potency.
Table 1.
A summary of the bisubstrate and bivalent inhibitors of protein kinases described in this review.
| Kinase | Specificity | Ligand A (active-site directed) | Fold Inc Over A | Ligand B | Fold Inc Over B | Linker | Reference |
|---|---|---|---|---|---|---|---|
| PKA/PKC | Ser/Thr | isoquinolinylsulfonyl | 670/67 | Ser-Arg6 (substrate site) | N/A | -NH[CH2]2NH[CH2]2CO- | Ricouart et al. (1991) |
| PKCS | Ser/Thr | bisindolyl maleimide | >43 | CREB peptide (substrate site) | >43 | alkylated Arg | van Ameijde et al. (2010) |
| PKC0 | Ser/Thr | bisindolyl maleimide | 17 | CREB peptide (substrate site) | >58 | alkylated Arg | van Ameijde et al. (2010) |
| PKA | Ser/Thr | ATPγS | N/R | Kemptide (substrate site) | N/R | acetyl | Hines and Cole (2004) |
| PKA | Ser/Thr | indolocarbazole | 0.038 | PKA specificity epitope of PKI (substrate site) | 32 | -NH[CH2]6NHCOCH2- | Schneider et al. (2005) |
| PKA | Ser/Thr | AdoC | 1200 | Arg6 (substrate site) | >830 | -NH(CH2)5C(O)- | Loog et al. (1999) |
| cIRK | Tyr | ATPγS | 310 | IRS727 peptide (substrate site) | N/R | acetyl | Parang et al. (2001) |
| CSK | Tyr | ATPγS | N/R | SRC (aa83-524) (substrate site) | > 17 | -CONH-CQ-Aph-NHCOCH2S- | Shen and Cole (2003) |
| JNK1 | Ser/Thr | indazole | 20000 | JIP1-derived peptide (docking site) | > 70000 | -NH[CH2]3CONH-GlyGly- | Stebbins et al. (2011) |
| JNK1 | Ser/Thr | indazole | 780 | JIP1 TAT peptide (docking site) | > 2700 | -NH[CH2]3CONH-GlyGly- | Stebbins et al. (2011) |
| PKA | Ser/Thr | staurosporine | 93 | cyclo(CTFRVFGC)G | 22000 | PEG3 | Meyer et al. (2007) |
| SRC | Tyr | EELL-(F5)Phe-amide | 230 | coumarin-pYEEIE (SH2 domain) | N/R | Abu8 | Profit et al. (1999) |
| SRC | Tyr | (Ba)EEEIFGEFDap(Hna) | 48 | pYEEIE (SH2 domain) | 50 | βAla3 | Hah et al. (2006) |
The upper panel shows bisubstrate inhibitors (categorized by kinase specificity), while the bottom panel shows bivalent inhibitors. Fold increase refers to the potency of kinase inhibition. (N/A = not applicable; N/R = not reported)
More recent efforts have been described to develop isozyme-selective PKC inhibitors. A PKC pseudosubstrate (EILSRRPAYRKIL), derived from the cAMP-responsive element binding protein (CREB), was tethered to a staurosporine mimetic to generate an isoform-selective PKC bisubstrate inhibitor (van Ameijde et al 2010). Tethering was achieved by modifying an arginine residue to present an azide moiety that was subsequently derivatized with an alkyne-displaying ATP-competitive small molecule using click chemistry. The resulting bisubstrate inhibitor was analyzed for its ability to prevent phosphorylation of substrates CREB1, KPCB, and MARCS by three clinically relevant and highly homologous PKC isoforms, PKCα, PKCζ, and PKCθ. The bisubstrate inhibitor exhibited modest potency against PKCζ (IC50 = 0.23-0.59 μM) and PKCθ (IC50 = 0.17-0.81 μM) but no inhibition of PKCα, while the staurosporine mimic only inhibited PKCα (IC50 = 1.8-2.9 μM) and PKCθ (IC50 = 2.3-2.9 μM). The pseudosubstrate alone was unable to inhibit any of the PKC isoforms, but microarray analysis of the parent peptide substrate showed that it is most strongly phosphorylated by PKCζ, followed by PKCθ and PKCα. Thus, the selectivity profile of the bisubstrate inhibitor tracks with that of the original substrate, suggesting that the secondary ligand acts to steer the affinity of this construct. Interestingly, a bisubstrate inhibitor previously studied by the same group which contained a neutral amino acid residue in place of the positively charged modified arginine, did not show inhibitory activity against PKCζ, but was selective for PKCθ (Poot et al 2009). This could point to the importance of maintaining contacts made by the original substrate when generating a pseudosubstrate, as well as proper linkage of the ATP-competitive small molecule.
In addition to the unintended inhibition of PKA described above, bisubstrate inhibitors have been designed to specifically target this important serine/threonine protein kinase. The cAMP-dependent protein kinase A (PKA) is activated by a number of signaling pathways within the cell (Carnegie et al 2009). Activated PKA then acts in a variety of distinct pathways, the differentiation of which is likely mediated through scaffolding complexes. Selective inhibition of PKA has the potential to illuminate the complex signaling networks that this kinase is involved in. One bisubstrate inhibitor of PKA that has been described used a pseudosubstrate peptide analog of the well-defined substrate kemptide fused through an acetyl linker to a non-hydrolyzable ATP derivative, ATPγS (Hines and Cole 2004). By using kemptide as a selectivity determinant, this bisubstrate inhibitor showed a greater than 20-fold preference for PKA over closely-related PKC. Alternatively, Schneider and coworkers developed a bisubstrate inhibitor consisting of a PKA specificity epitope from the heat-stable Protein Kinase Inhibitor protein (PKI) linked to the high-affinity ATP-competitive inhibitor K252a (Figure 2 a) (Schneider et al 2005). Rather than using a linear peptide to display the PKI epitope, a designed miniature protein with the stable fold from avian pancreatic polypeptide (aPP) was employed as a scaffold. Miniature proteins based on this scaffold have been designed to present recognition epitopes from the α-helix of aPP and can often lead to potent binders of target proteins. Interestingly, the bisubstrate inhibitor displayed from the miniature protein was 60-fold more potent (IC50 = 3.7 nM) than a bisubstrate inhibitor consisting of K252a directly conjugated to PKI (IC50 = 220 nM). This may be due to differences in the binding modes between the two inhibitors or suboptimal linkage between K252a and PKI in the absence of the miniature protein. While the K252a-PKI (miniprotein) bisubstrate inhibitor was 26-fold less potent than K252a (IC50 = 0.14 nM), a 32-fold increase in potency was observed compared to the PKI-displaying miniature protein (IC50 = 120 nM). Importantly, impressive selectivity was observed for PKA over closely related protein kinases. K252a and the bisubstrate inhibitor were assayed against AKT (PKB), PKCα, cGMP-dependent protein kinase (PKG), and Ca/calmodulin kinase II (CaMKII). While the small molecule K252a inhibits all of these kinases potently, the bivalent inhibitor was only observed to exhibit inhibition of PKA at concentrations up to 100 nM.
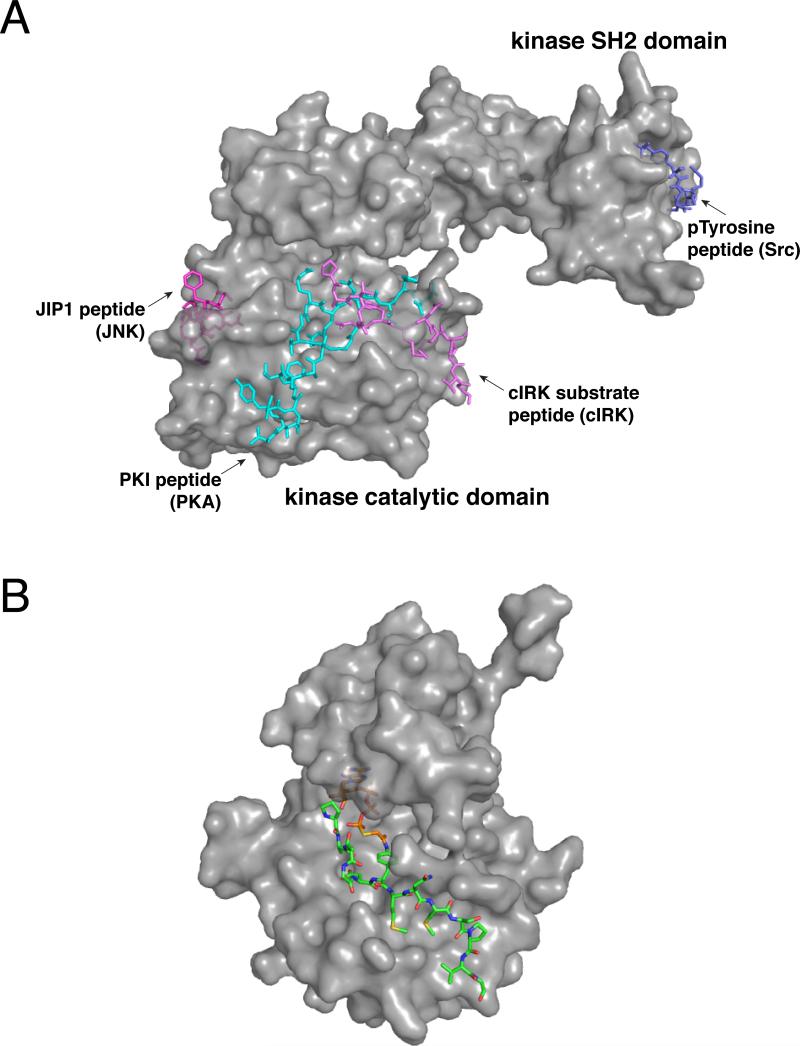
Figure 2.
Reported bisubstrate/bivalent inhibitors of protein kinases. (a) Crystallographic representation of secondary binding ligands used in previously described bisubstrate/bivalent inhibitors superimposed on a protein kinase. [PDB codes 1GAG, 2CPK, 1UKI, 1Y57] (b) Crystal structure of cIRK in complex with a bisubstrate inhibitor. [PDB code 1GAG]
Several earlier attempts at bisubstrate inhibition of PKA yielded modest inhibitors (Loog et al 1999, Medzihradszky et al 1994). One approach, similar to that reported by Ricouart and coworkers, linked an Arg6 peptide to adenosine-5'-carboxylic acid (AdoC) through a six carbon linker (Loog et al 1999). This construct inhibited PKA, PKC, and cyclin-dependent protein kinase-1 (CDK1) with IC50 values of 0.12, 0.27, 1.2 μM, respectively. As expected, the inhibitor was inactive against casein kinases 1 (CK1) and 2 (CK2), which are acidophilic protein kinases. In a subsequent report, AdoC was linked to a poly-arginine peptide in order to develop bisubstrate inhibitors capable of selectively enriching PKA from lysate (Loog et al 2000). Medzihradszky and coworkers generated a bisubstrate inhibitor by directly linking adenosine diphosphate (ADP), ATP or adenosine tetraphosphate (AP4) to the PKA substrate, kemptide (Medzihradszky et al 1994). It was seen that inhibitor potency, while modest, increased with the number of phosphates in the linker (IC50 = 935, 226, 68 μM for the ADP, ATP, and AP4 linkers, respectively). Inhibition was observed to be competitive with ATP but not with respect to kemptide, indicating that these inhibitors primarily interact with the ATP-binding site.
Bisubstrate inhibitors of tyrosine protein kinases
In an early report from Kruse and coworkers, a strategy for developing bisubstrate inhibitors of Abelson tyrosine kinase (Abl) was described (Table 1) (Kruse et al 1988). This method involves coupling a neutral ATP-mimetic, 5'-[4-(fluorosulfonyl)benzoyl]adenosine (5’-FSBA) to a tyrosine derivative. These assembled bisubstrate inhibitors were hypothesized to take advantage of the tyrosine substrate recognition preference exhibited by protein tyrosine kinases (PTKs). While modest inhibitors of Abl (IC50 = 19-120 μM) were developed by varying the tyrosine derivative and the linker to 5’-FSBA, the similarity of IC50 values displayed by the panel of compounds suggests that the inhibitors were binding only to the ATP-binding site. Further supporting this notion, these bisubstrate inhibitors possessed similar potencies against a non-tyrosine kinase, phosphorylase. These results highlight the importance of understanding substrate selectivity determinants for successful development of bisubstrate inhibitors. In a similar approach by Traxler and coworkers, sulfonylbenzoylnitrostyrene derivatives were synthesized as bisubstrate inhibitors of the epidermal growth factor receptor tyrosine kinase (EGFR) (Traxler et al 1991). The sulfonylbenzoyl moiety represents the ATP-competitive portion of these molecules, while the nitrostyrene was selected to mimic the natural product Erbstatin, a peptide substrate-competitive PTK inhibitor (Imoto et al 1987). With this method, a potent (IC50 = 0.054 μM) EGFR inhibitor was developed that showed high selectivity with respect to Abl (IC50 = 27 μM) and PKC (IC50 = 500 μM). While a highly potent and seemingly selective inhibitor of EGFR was developed, studies were not carried out to verify that this bisubstrate inhibitor occupied both substrate-binding sites. A subsequent report from the same group described bisubstrate inhibitors that are composed of EGFR consensus sequence substrate motifs tethered to an adenosine moiety through a 2-hydroxy-4-sulfonylbenzoyl linker (Rossé et al 1997). This strategy resulted in the generation of a less potent (IC50 = 33 μM) bisubstrate inhibitor that demonstrated similar potency to the 2-hydroxy-4-sulfonylbenzoyl linker group. The low activity of this compound was hypothesized to be a result of weak interactions between EGFR and the substrate peptide.
Parang and Cole generated a bisubstrate inhibitor of the insulin receptor protein tyrosine kinase (IRK) by tethering a well characterized pseudosubstrate of this kinase, IRS727, to ATPγS (Figure 2 a and b) (Parang et al 2001). This bisubstrate inhibitor was designed based on the principle that phosphate transfer in tyrosine kinases occurs through a dissociative mechanism. In a dissociative mechanism transition state, the bond between the attacked phosphorus and the leaving group (ADP) is largely broken before the bond between the nucleophile (the alcohol of Ser, Thr, or Tyr) and the phosphorus has fully formed (Hines et al 2005, Parang et al 2001). For a bisubstrate inhibitor to accommodate such a transition state, the tether between peptide and the nucleotide must be at least 5 Å in length. An acetyl linker, which provides a distance between the Tyr oxygen surrogate of the peptide and γ-phosphorus of ATP of 5.7 Å in an extended conformation, was envisioned as being optimal. Gratifyingly, the bisubstrate inhibitor was shown biochemically to have increased binding affinity compared to both ATPγS and the IRS727 peptide. Additionally, a crystal structure of IRK in complex with the bisubstrate inhibitor was solved. This confirmed that both the nucleotide and peptide binding sites of IRK were occupied by the bisubstrate inhibitor. Furthermore, the distance between the γ-phosphorus and the Tyr oxygen surrogate was found to be 5 Å, supporting the hypothesis of a dissociative mechanism of phosphate transfer. To further investigate the affinity contributions of this bisubstrate inhibitor, a series of changes were introduced into the linker, the amino acid sequence of the pseudosubstrate, and the identity of the Tyr oxygen surrogate (Hines et al 2005). The parent bisubstrate IRK inhibitor displayed an anilino group where the tyrosine hydroxyl would appear in the native peptide substrate. This allowed the peptide moiety to be linked to ATPγS, while still retaining an important hydrogen bond. A hydrogen bond donor at this position can potentially interact with the carboxylate side chain of the catalytic Asp of IRK and was proposed to be important for the potency of the interaction between substrate and enzyme. As expected, replacement of the nitrogen with an ether linkage, which cannot make the same hydrogen bond, resulted in 80-fold weaker inhibition of IRK. Furthermore, when the acetyl linker was substituted with a longer propionyl group (~1.2 Å longer than acetyl), an 18-fold less potent bisubstrate IRK inhibitor resulted. In addition, replacement of ATP with ADP, shortening the distance between adenosine and the anilino nitrogen by ~4 Å, resulted in a 200-fold less potent inhibitor of IRK. These results further support the notion of a dissociative transition state for phosphate transfer.
Shen and Cole also generated a bisubstrate inhibitor of the protein tyrosine kinase Csk (Shen and Cole 2003). Csk is responsible for the negative regulation of SFK activity through phosphorylation of their C-terminal tails (Levinson et al 2009). A bisubstrate inhibitor of Csk was assembled in vitro using expressed protein ligation to fuse a recombinant Src fragment (83-524) to a pseudosubstrate peptide/ATPγS conjugate. With this method, a bisubstrate inhibitor displaying an IC50 of 600 nM against Csk was generated, with the recombinant Src fragment alone showing no measureable inhibition at a concentration of 10 μM. The immobilized bisubstrate construct was also able to isolate Csk from Csk-enriched NIH3T3 cell lysate. The Src fragment alone showed no significant enrichment of Csk under the same conditions. This method presents a potential means to discover cellular actors of phosphorylation events through immobilization of a downstream protein.
Bivalent inhibitors of protein kinases
Several of the previously described bisubstrate kinase inhibitors represent excellent examples of potent kinase inhibition by targeting the ATP- and substrate-binding sites. The presence of domains and interaction sites distal to the site of phosphate transfer, however, presents an avenue for the development of numerous, potent bivalent inhibitors that are capable of selecting between closely-related kinases. Moving away from kinase active sites has given researchers the ability to target protein kinases of interest without dependence on the generally low-affinity substrate-binding site.
Bivalent inhibitors of serine/threonine protein kinases
In 2011, Stebbins and coworkers reported a bivalent inhibitor of the MAPK c-Jun N-terminal kinase 1 (JNK1) (Figure 2 a and Table 1) (Stebbins et al 2011). This bivalent inhibitor was generated by linking an ATP-competitive inhibitor of the JNK family, an analog of SP600125, to a peptide that targets JNK1's docking groove. The peptide selected for this study was a minimal sequence derived from the D-domain of the JNK1 scaffolding protein, JIP1. While the minimal peptide selected does not inhibit JNK1 activity (IC50 > 50 μM), it is capable of displacing the inhibitory parent peptide, pepJIP1, in a dissociation enhanced lanthanide fluoro-immuno assay (DELFIA) platform. The bivalent inhibitor was linked through a glycine-glycine-diaminopropyl linkage and displayed sub-nanomolar (IC50 = 0.7 nM) JNK1 inhibition in vitro, a 20000-fold increase in JNK1 inhibition compared to the ATP-competitive inhibitor alone (IC50 = 14 μM). To confirm that this inhibitor interacts with both targeted sites in vitro, JNK1 was pre-incubated with an excess of the promiscuous kinase inhibitor staurosporine to eliminate one binding site prior to treatment with the bivalent inhibitor. This resulted in a 600-fold loss in the ability of the bivalent inhibitor to compete with pepJIP for the docking site of JNK1. Thus, a potent bivalent inhibitor of JNK1 was developed by linking a minimal peptide derived from the native interactor JIP1 to an ATP-competitive small molecule inhibitor of modest potency.
Meyer and coworkers took a different approach in developing a potent and selective bivalent inhibitor of PKA (Meyer et al 2007). An in vitro bivalent selection strategy was envisioned where small molecule targeting could be combined with biological selection so that no previous structural or peptide substrate data would be necessary for inhibitor design. In this approach, a highly promiscuous staurosporine derivative of intermediate potency was used in a warhead-guided phage display selection strategy to yield cyclic peptide binders of PKA surfaces near the active site. The library and small molecule are tethered through a self-assembling heterodimer of Jun and Fos proteins. The small molecule tethered to Jun and cyclic peptide library displayed from Fos are brought within proximity of each other upon self-assembly of the Jun/Fos coiled coil. In this way, a PKA-binding cyclic peptide was selected and subsequently tethered to the staurosporine derivative to generate a bivalent inhibitor. The resulting inhibitor displayed an impressive increase in both potency and selectivity of inhibition. By introducing a bivalent interaction, potency increases of 93- and 21000-fold were observed compared to the small molecule and cyclic peptide, respectively. Additionally, five kinases that share a similar staurosporine inhibition profile with PKA (ASK1, CaMKIIβ, cSrc, EphA5 and Mnk2) were assayed for activity in the presence of the bivalent inhibitor. Only PKA displayed a significant reduction in kinase activity in response to the inhibitor at a concentration of 100 nM. A bivalent inhibitor with an optimized linker between both monovalent ligands demonstrated even greater potency against PKA and showed remarkable selectivity for this kinase over an extended panel of 90 kinases (Shomin et al 2009). These reports not only demonstrate the utility of bivalent inhibitors for enhancing potency and selectivity but also present a model for the generation of bivalent inhibitors of kinases without a need for prior understanding of protein-protein interactions made by the kinase of interest. The generality of this strategy was demonstrated by the application of the same selection methodology to the serine/threonine kinase Aurora A (Shomin et al 2011). These efforts resulted in the identification of cyclic peptides that target sites outside of the ATP- and substrate-binding clefts of Aurora A.
Bivalent inhibitors of tyrosine protein kinases
In 1999, Profit and Lawrence reported a bivalent inhibitor targeting two distinct domains of Src tyrosine kinase (Figure 2 a and Table 1) (Profit et al 1999). The phospho-recognition site of the Src SH2 domain was targeted with a peptidic derivative of the SH2 recognition sequence found in hamster polyomavirus middle-T antigen. This ligand was coupled to an active site-directed peptide displaying pentafluorophenylalanine as a non-phosphorylatable tyrosine analog. The optimal length for linking these two binding elements was determined empirically, with the most potent bivalent inhibitor exhibiting a 230-fold lower IC50 against Src (IC50 = 6.9 μM) than the active site-directed peptide alone (IC50 = 1590 μM). While this report validated the concept of generating bivalent inhibitors that target two different domains in multi-domain tyrosine kinases, the highest affinity inhibitor only demonstrated modest potency against Src (IC50 = 6.9 ± 0.4 μM). Furthermore, the selectivity of this inhibitor was not examined. In a later report from the same group, the active site-directed peptide inhibitor of Src was optimized and nanomolar inhibition of this kinase was achieved (IC50 = 36 ± 2 nM) (Hah et al 2006). Additionally, this inhibitor discriminated between the two SFK subgroups: Group A (Fyn, Fgr, Src, and Yes) and Group B (Blk, Hck, Lck, and Lyn). Inhibition selectivity from 13- to 1600-fold was observed between the two subgroups. The development of inhibitors that are selective for specific SFK members will prove useful in the quest to disentangle the overlapping roles of SFKs in the cell.
Cellular applications of bivalent and bisubstrate inhibitors
Due to their high molecular weights and overall polar natures, bisubstrate and bivalent inhibitors are usually not cell permeable. This makes using these reagents in a cellular setting very challenging. The ability to study protein kinase function in a cellular environment with these reagents would be exceptionally valuable because the roles of many members of this enzyme family are not well understood, and there is a dearth of pharmacological tools that possess the necessary selectivity. However, several groups have pursued strategies that allow intra-cellular application of their bivalent reagents. Stebbins and coworkers reported a strategy for the intra-cellular targeting of their bivalent, sub-nanomolar JNK1 inhibitor (Stebbins et al 2011). To do this, an all-D retroinverso version of their bivalent inhibitor was fused to the cell penetrating HIV-TAT peptide sequence (GRKKRRQRRR). While the cell penetrant bivalent inhibitor was less active than the parent conjugate, this inhibitor still exhibited an IC50 of 18 nM against JNK1 in vitro, which is 780-fold lower than the monovalent ATP-competitive inhibitor alone. The cell penetrant bivalent inhibitor effectively inhibited JNK1-dependent processes in mammalian cells, while processes dependent on the closely related MAPK, p38α, were unaltered. Additionally, intraperitoneal injection of the inhibitor into glucose intolerant mice increased the ability of mice to process glucose compared to vehicle.
Ameijde and coworkers pursued a similar approach for conferring cellular penetrance to their bisubstrate PKC inhibitor. Two cell-penetrating peptides (CPPs), HIV-TAT and antennapedia homeodomain derived penetratin, were conjugated to their bisubstrate PKC inhibitor, either through linear synthesis onto the pseudosubstrate or via a disulfide linkage (van Wandelen et al 2013). HeLa cells incubated with fluorescently labeled constructs showed cellular uptake by confocal microscopy and FACS for both CPPs and attachment methods. The intracellular activity of these bisubstrate inhibitors was assayed by ELISA using a phospho-specific antibody for the PKC substrate, MARCKS. The linear TAT-conjugate, linear penetratin-conjugate, and disulfide TAT-conjugate inhibited intra-cellular PKC phosphorylation of MARCKS, with IC50 values of 10, 19, and 17 μM, respectively. Interestingly, the disulfide penetratin-conjugate did not show significant inhibition in cells. This construct did exhibit a significantly shorter half-life compared to the other three constructs, perhaps explaining this result. Although these constructs showed decreased intra-cellular inhibition in comparison to the monovalent staurosporine analog (IC50 = 3.8 μM), microarray assays of HeLa lysate treated with either penetratin- or TAT-based bisubstrate inhibitors had diminished MARCKS phosphorylation, while the profile of other phosphorylation sites was similar to that of the control, which demonstrates selective on-target inhibition.
Bivalent inhibitors based on a SNAP-tag protein scaffold
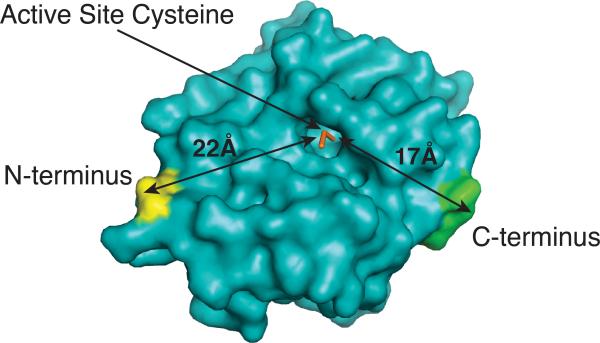
In order to provide a bivalent inhibition method that can be used in cells without the use of intra-cellular delivery methods, we developed a chemical genetic strategy based on a self-labeling protein called SNAP-tag (or AGT). This allows one component of a bivalent inhibitor to be genetically encoded as a SNAP-tag fusion protein and the other component to be assembled through a rapid and chemo-selective covalent reaction (Hill et al 2009, 2011, 2012). This overall strategy is shown in Figure 3 a. A SNAP-tag fusion protein, which contains a ligand that targets a region outside the ATP-binding site, is selectively labeled with an ATP-competitive small molecule inhibitor conjugate that contains a chemo-selective tag. This is a modular strategy that allows different combinations of monovalent reagents to be assembled rapidly. This approach in generating bivalent kinase inhibitors with SNAP-tag is unique from most reports in that, an entire, folded protein is used as a scaffold for ligand display. The labeled active site residue is located in a relatively shallow hydrophobic pocket, which allows favorable access to the ATP-competitive small molecule, while the secondary binding ligand can be displayed from either the N- or C-terminus of the protein scaffold. The N- and C-termini both sit on the same face of the active site of SNAP-tag, which allows for favorable ligand display from the scaffold to the kinase target (Figure 4). Furthermore, the active site is only ~22 Å from the N-terminus and ~17 Å from the C-terminus.
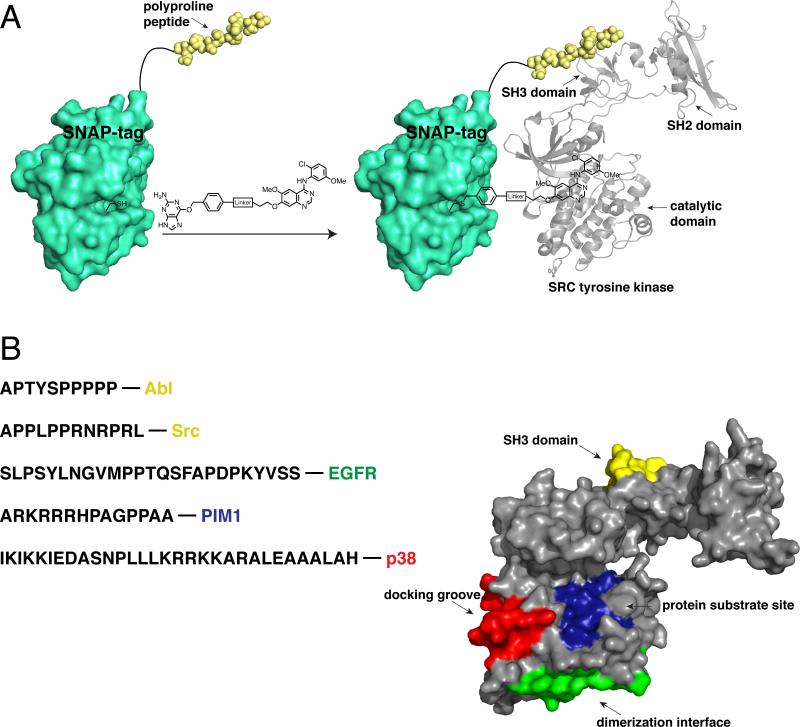
Figure 3.
A SNAP-tag-based bivalent inhibitor approach. (a) Schematic depiction of a representative SNAP-tag-based bivalent inhibitor. SNAP-tag self-labels an active site cysteine with a quinazoline-based kinase inhibitor conjugated to a benzylguanine (BG) group. The genetically encoded SNAP-tag fusion contains a polyproline peptide ligand, which recognizes the SH3 domain of Src. The two ligands of the bivalent inhibitor interact with distinct Src tyrosine kinase binding sites. [PDB codes 3L00 and1Y57] (b) The sequences of peptide ligands that have been successfully used in SNAP-tag-based bivalent/bisubstrate inhibitors, and the kinases they target, are shown. The binding sites that have been targeted with SNAP-tag-based bivalent inhibitors are shown. These sites are superimposed on a crystal structure of Src tyrosine kinase. [PDB code 1Y57]
Figure 4.
Crystallographic representation of the engineered protein SNAP-tag. The N- and C-termini, as well as the active site cysteine, are shown. Lengths (in Å) represent the distances between the N-terminus and the active site or the C-terminus and the active site. [PDB code 3L00]
The key to our bivalent inhibitor strategy is the unique reactivity of SNAP-tag. SNAP-tag is an engineered form of the DNA repair enzyme O6-alkylguanine-DNA alkyltransferase (AGT). The normal function of AGT in the cell is to repair O6-alkylated guanine bases in DNA by irreversibly transferring the alkyl group from the damaged base to its active site cysteine residue (Pegg 2000, Tubbs et al 2007). Johnsson and coworkers exploited the already relaxed substrate specificity of AGT to engineer variants that show significantly increased rates of reactivity with O6-benzylguanine (BG) conjugates (Gronemeyer et al 2006, Juillerat et al 2003, 2005, Keppler et al 2003). To do this, a library of hAGT mutants were displayed on filamentous phage M13 as g3p fusions and selected for labeling by BG conjugates (Juillerat et al 2003). After multiple rounds of enrichment, hAGT variants with greatly enhanced reactivity relative to the wild type protein were identified. Johnsson and coworkers have shown that optimized hAGT variants (referred to as SNAP-tag) can be rapidly and specifically labeled in vitro and in situ with BG derivatives conjugated to fluorophores or affinity handles (Corrêa et al 2013, Juillerat et al 2005, Keppler et al 2003). Furthermore, New England Biolabs sells cloning vectors and BG conjugates that can be used to label a SNAP-tag fusion protein of interest.
SNAP-tag-based bivalent inhibitors that target multi-domain protein kinases
Like Src, the tyrosine kinase Abl is a multi-domain tyrosine kinase that contains SH2 and SH3 domains. These domains modulate the catalytic activity and localization of Src and Abl. In addition, the catalytic domains of Src and Abl are very similar and most inhibitors have comparable affinities for their ATP-binding sites. Therefore, we felt that Src and Abl would be a good test for the ability of our bivalent inhibitor strategy to discriminate between two highly related kinases (Figure 3 b) (Hill et al 2009). In order to target Src and Abl, bivalent inhibitors that contain an ATP-competitive inhibitor and a SH3 domain-targeting ligand were generated. Two BG-derivatives based on a 4-anilinoquinazoline scaffold, and with differing linker lengths, were used as ATP-competitive small molecule inhibitors. In vitro activity assays showed that attaching BG through a linker had a minimal effect on the abilities of these conjugates to inhibit Abl and Src, relative to the parent 4-anilinoquinazoline compound. To target the SH3 domains of Src and Abl, SNAP-tag fusion proteins that contain polyproline (PP)-containing peptides linked through a flexible serine-glycine spacer to the N- or C-terminus were expressed and purified. As the SH3 domain interaction was designed to be the selectivity determinant for these bivalent inhibitors, peptides that have previously been shown to have a preference for Src or Abl were used.
The monovalent reagents described above were assembled into bivalent inhibitors and tested for their abilities to inhibit the catalytic activities of Src and Abl in vitro (Table 2). As expected, the 4-anilinoquinazoline ATP-competitive inhibitor was a less potent inhibitor of Src and Abl when displayed from the active site of wild type SNAP-tag (SNAP(wt)). However, when this same inhibitor was displayed from a SNAP-tag fusion containing either an Abl- or Src-selective PP-containing peptide, a significant increase in potency for the targeted kinase was observed. Interestingly, increased potency was observed whether the SH3 ligand was displayed from the N- or C-terminus of SNAP-tag, and the length of the serine-glycine linker had only a small effect on the IC50. The most potent bivalent Src inhibitor (IC50 = 13 nM) displayed a >23-fold lower IC50 relative to the monovalent BG-derivative ATP-competitive inhibitor and a >380-fold lower IC50 relative to the SNAP(wt)-ATP-competitive inhibitor conjugate. Similar increases in potency against Abl using bivalent constructs that contain the Abl-selective PP peptide were observed, with the most potent inhibitor (IC50 <6 nM) demonstrating >85 fold more potent inhibition than the monovalent BG-derivative ATP-competitive inhibitor and >200 fold potency increase relative to SNAP(wt)-ATP-competitive inhibitor conjugate. Src-selective and Abl-selective bivalent inhibitors were assayed against a panel of 13 different kinases. As expected, all protein kinases in the panel lacking a SH3 domain were not significantly inhibited by either bivalent construct. The Src-selective PP bivalent construct showed >150-fold selectivity for Src (IC50 = 0.013 μM) over Abl (IC50 = 2.5 μM) and the SFK Lck (IC50 = 2.4 μM). Similarly, the Abl-selective bivalent construct used in this study had an IC50 for Abl of 0.018 μM, which was >30-fold lower than Src (IC50 = 0.640 μM) and >130-fold lower than Lck (IC50 >2.5 μM). Thus, targeting the SH3 domains of Src and Abl allowed highly selective inhibition to be achieved.
Table 2.
Bisubstrate and bivalent inhibitors of protein kinases based on the SNAP-tag protein scaffold.
| Kinase | Specificity | Ligand A (active-site directed) | Fold Inc Over A | Ligand B | Fold Inc Over B | Linker | Reference |
|---|---|---|---|---|---|---|---|
| ABL | Tyr | 4-anilinoquinazoline | >85 | ABL-selective PP motif (SH3 domain) | N/A | (GS)2 | Hill et al. (2009) |
| SRC | Tyr | 4-anilinoquinazoline | 23 | SRC-selective PP motif (SH3 domain) | N/A | (SG)2 | Hill et al. (2009) |
| EGFR | Tyr | Gefitinib | 18 | MIG6 peptide (dimer interface) | 330 | (SG)5 | Hill et al. (2012) |
| PIM1 | Ser/Thr | SGI-1776 | 12 | Pimtide pseudosubstrate (substrate site) | 64 | (GS)2 | Hill et al. (2012) |
| p38α | Ser/Thr | 4-anilinoquinazoline | 150 | MAPKAPK2 (docking site) | >1600 | (SG)5 | Hill et al. (2012) |
Fold increase refers to the potency of kinase inhibition. (N/A = not applicable)
A more detailed analysis of the contributions of each monovalent component of the assembled bivalent inhibitors that target Src and Abl was undertaken (Hill et al 2011). It had already been determined that the length of the linker and the fusion orientation (C- or N-terminal) of the SH3 ligand had only a small effect on bivalent inhibitor potency. However, varying the length of the linker between the 4-anilinoquinazoline ATP-competitive inhibitor and the SNAP-tag fusion protein had a more pronounced effect on bivalent inhibitor potency. While tether length only had a small effect on the potencies of the Src-selective bivalent inhibitors, with the construct containing the longest linker possessing the lowest IC50, there was a greater than 15-fold difference between Abl-selective bivalent inhibitors with variable ATP-competitive inhibitor linker lengths. Surprisingly, the bivalent construct with an intermediate linker length possessed the lowest IC50 for Abl activity. These results suggest that Abl may have a different relative orientation between its SH3 and catalytic domains compared to Src, which could potentially be exploited to further differentiate between these two kinases. In the same study, the contributions of the ATP-competitive inhibitor and PP-containing SH3 domain ligand were also probed. A small panel of BG-linked ATP-competitive inhibitors that displayed varying affinities for the active site of Abl and Src were tested. In all cases, conjugating the BG-derivative inhibitors to a SNAP-tag SH3 domain ligand fusion protein resulted in at least a small increase in potency against the kinase target. Furthermore, small differences in affinity between these monovalent ATP-competitive inhibitors had a direct and predictable effect on the potency of the corresponding bivalent inhibitor. This same trend was observed for the PP-containing SH3 domain ligand, where relative differences in the affinities of the peptide ligands for the SH3 domain of the kinase being targeted led to a predictable corresponding change in the potency of the assembled bivalent inhibitor. These studies further highlighted that the high selectivity exhibited by the Abl-selective and Src-selective bivalent inhibitors is due to the relative specificities of their corresponding SH3 domain ligands.
SNAP-tag-based bivalent inhibitors that target diverse protein kinases
SNAP-tag-based bivalent inhibitors that are directed towards the diverse protein kinases PIM1, p38α, and EGFR have also been generated (Figure 3 b and Table 2) (Hill et al 2012). Importantly, these bivalent inhibitors all exploit diverse signaling interaction sites that are located in the catalytic domains of their kinase targets. To target PIM1, N- and C-terminal SNAP-tag fusion proteins that contain a modified pseudosubstrate called Pimtide (ARKRRRHPSGPPTA), which is the peptide ligand corresponding to the consensus substrate motif of this kinase, were generated. A linkable analog of an imidazo[1,2-b]pyridazine inhibitor was used to target the ATP-binding site of Pim1. While the imidazo[1,2-b]pyridazine inhibitor exhibited a >9-fold loss in potency when displayed from the active site of SNAP-tag(wt), its conjugation to SNAP-tag fusions that contain Pimtide ligands led to an increase in potency relative to both monovalent components. For the bivalent inhibition of the MAPK p38α, its docking groove, which interacts with a number of activators and substrates, was targeted using a 31-amino acid peptide derived from the mitogen activated protein kinase-activated protein kinase 2 (MAPKAPK2). This peptide is a ligand for a docking site on p38α and does not overlap with the kinase active site. Two different ATP-competitive inhibitors with variable affinities for the ATP-binding site of p38α were selected as the other monovalent components of the bivalent inhibitor. Consistent with the ability to simultaneously target p38α's ATP-binding site and docking groove, conjugation of either inhibitor to a SNAP-tag-MAPKAPK2-peptide fusion protein led to more potent inhibition of p38α than either monovalent component alone. In contrast to PIM1 and p38α, a slightly different approach was needed for selecting a non-ATP-binding site ligand for EGFR because this kinase does not possess substrate interaction sites of sufficient binding affinity. Therefore, a 25-amino acid peptide from the mitogen-induced gene 6 (MIG6) protein, which binds to the hydrophobic dimer interface on the C-lobe of EGFR, was chosen as the genetically encodable ligand that was displayed as a SNAP-tag fusion. Bivalent inhibitors were generated by conjugating the clinically approved drug gefitinib to SNAP-tag fusions displaying the MIG6 peptide from their N-termini. Similar to p38α and PIM1, the assembled bivalent inhibitors were significantly more potent against their target, EGFR, than either individual monovalent component. These three examples demonstrate that SNAP-tag-based bivalent inhibitors can target a number of different binding sites outside of the ATP-binding clefts of protein kinases.
Beyond showing increased potency for specific kinase targets, the SNAP-tag-based bivalent inhibitors described above demonstrated increased overall selectivity. The most potent bivalent inhibitors of PIM1, p38α, and EGFR were assayed against a panel of 27 kinases that were selected based on diversity and being known off-targets of the ATP-competitive inhibitor components used in the bivalent inhibitors. While the ATP-competitive inhibitor components of the EGFR- and p38α-targeting inhibitors were already fairly selective, the assembled bivalent conjugates were even more so. In both cases, the assembled bivalent inhibitors were >900-fold selective for their intended targets over the 26 other kinases tested. For the PIM1-targeting bivalent inhibitor, only PIM2 kinase showed any significant inhibition (IC50 = 1700 nM) compared to PIM1 (IC50 = 45 nM). However, the assembled bivalent inhibitor showed enhanced selectivity between these two kinases relative to the monovalent substituents.
Cellular applications of SNAP-tag-based bivalent inhibitors
Like the bisubstrate inhibitors described above, the first cellular experiments with SNAP-tag-based bivalent inhibitors involved the use of an intra-cellular delivery method (Gregersen et al 2010). Specifically, a fully assembled bivalent inhibitor of Abl was encapsulated in dye-sensitized lipid nanocapsules, which were taken up by BCR-Abl-dependent Ba/F3 cells through endocytosis. Nanosecond laser pulses in the far red (645 nm) region allowed photothermal release of the bivalent inhibitor into the cytosol. As Ba/F3 cells are dependent on the activity of BCR-Abl for survival in the absence of interleukin-3 (IL-3), intra-cellular release of the bivalent Abl inhibitor led to rapid cell death. However, when a similar experiment was performed with Ba/F3 cells in the presence of IL-3, no cell death occurred, confirming that the phenotypic effect observed upon bivalent inhibitor release is through BCR-Abl.
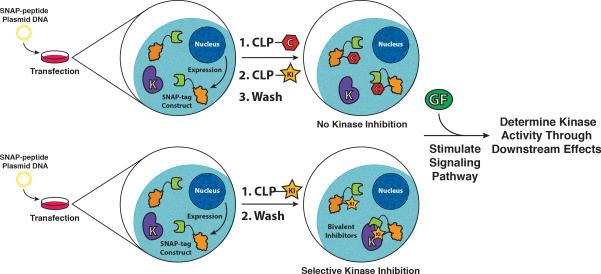
While intra-cellular delivery methods allow bivalent SNAP-tag-based inhibitors to be used in cells, the most general strategy for application of this methodology involves the assembly of bivalent inhibitors in cells that are expressing SNAP-tag fusion proteins (Figure 5). Johnsson and co-workers have demonstrated that BG derivatives conjugated to fluorophores or affinity handles are cell permeable and can undergo selective intra-cellular labeling of SNAP-tag (Keppler et al 2003). We have found that, in general, BG-derivatized ATP-competitive inhibitors have low cell permeability. However, ATP-competitive inhibitors that are conjugated to an O4-benzyl-2-chloro-6-aminopyrimidine (CLP) tag, which can also rapidly and selectivity label SNAP-tag, are cell permeable and efficiently label intra-cellular SNAP-tag. Four diverse CLP-conjugated ATP-competitive inhibitors of PIM1, p38α, and EGFR were able to label SNAP-tag in a variety of cell lines (COS-7, HeLa, and HEK293) (Hill et al 2012). These results set the stage for the intra-cellular assembly of potent and selective SNAP-tag-based bivalent inhibitors in cells and their use in a number of cellular studies.
Figure 5.
Schematic representation of the strategy for intra-cellular assembly and use of SNAP-tag-based bivalent inhibitors. Mammalian cells express a genetically encoded SNAP-tag fusion protein. A CLP-derivatized ATP-competitive kinase inhibitor (CLP-KI) passively diffuses into cells and labels the monovalent SNAP-tag fusion. Excess monovalent CLP-KI is removed through several wash steps. Upon stimulation of the signaling pathway, kinase activity can be determined by monitoring downstream effects. As a control, a CLP-derivatized small molecule lacking inhibitory activity is used to label (block) the SNAP-tag active site before addition of CLP-KI. Any effects observed are due to CLP-KI and not the SNAP-tag-based bivalent inhibitor.
Conclusions
Over the past two decades, there has been widespread interest in the use of bisubstrate and bivalent inhibitors to study protein kinases. Examples in the literature have shown that these tools, when built with appropriate linkage and ligand display, can be potent (nanomolar dissociation constants) inhibitors of target protein kinases. In order for bivalent inhibitors to be useful tools for the study of kinase function, they must also be highly selective. Early examples of bivalent inhibitors sought to differentiate between two or more closely related protein kinases, occasionally investigating selectivity over more diverse kinase family members (Hines and Cole 2004, Ricouart et al 1991, Traxler et al 1991). In certain instances, such as the work performed by Ricouart and coworkers, the observed selectivity was, regrettably, opposite of the authors’ intensions. In general, all of these earlier studies only profiled a small panel of potential kinase off targets, and it is difficult to judge the true kinome selectivity of these reagents. More recent examples have included profiling against larger panels of protein kinases, ranging from 27 to 90 family members (Hill et al 2012, Shomin et al 2009). In these larger screens, the developed bivalent inhibitors were shown to be highly selective for the protein kinase of interest over both closely related family members and more distantly related kinases. With the recent advent of whole-kinome screening techniques (Davis et al 2011, Fabian et al 2005, Karaman et al 2008, Urbaniak et al 2012), the ability to extensively profile bivalent reagents is now possible. With the application of these techniques to bisubstrate and bivalent inhibitors, it will be interesting to see if these reagents are truly as selective as theorized.
Another major challenge in the application of bisubstrate and bivalent inhibitors is in their use in a cellular environment. To date, there have been very few examples of the successful use of bivalent kinase inhibitors in cells. Where there has been success is in the use of transport mechanisms to deliver these high molecular weight reagents to cells. For example, the attachment of a HIV-TAT peptide to a potent JNK1 inhibitor by Stebbins and coworkers, allowed the authors to achieve JNK1 inhibition both in cultured cells and in glucose-intolerant mice (Stebbins et al 2011). Furthermore, we have shown that it is possible to photothermally release bivalent inhibitors that are encapsulated in dye-sensitized lipid nanocapsules (Gregersen et al 2010). Examples of this type point to the likelihood of increased use of bisubstrate and bivalent inhibitors in cellular studies. However, the routine delivery of biomolecules of this kind is still a challenge. An alternative strategy is to assemble bivalent inhibitors inside the cell using the diversity of highly chemoselective conjugation methods that are currently available. This approach is actively being pursued (Hill et al 2012, 2009, 2011).
Protein kinases represent a large and interesting family of proteins within the human proteome. Key players in diverse cellular signaling networks, these enzymes represent a large set of druggable targets. Despite this, the specific roles that many protein kinases play in both native signaling and disease states are not well understood. Likewise, selective and potent inhibitors of most protein kinases do not exist. The ability to develop powerful tools for the cellular study of a specific protein kinase is invaluable, and we feel that the development of cellular bivalent inhibitors is an excellent starting point in developing the necessary toolset for the cellular study of protein kinases.
Footnotes
Declaration of interest
The authors report no declarations of interest.
REFERENCES
- Barr FA, Silljé HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5(6):429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Benes CH, Wu N, Elia AEH, Dharia T, Cantley LC, Soltoff SP. The C2 Domain of PKCδ Is a Phosphotyrosine Binding Domain. Cell. 2005;121(2):271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. Annual Reviews. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- Biondi RM, Nebreda AR. Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem J. 2003;372(1):1–13. doi: 10.1042/BJ20021641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Carnegie GK, Means CK, Scott JD. A-kinase anchoring proteins: from protein complexes to physiology and disease. IUBMB life. 2009;61(4):394–406. doi: 10.1002/iub.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Turk BEB. Analysis of serine-threonine kinase specificity using arrayed positional scanning peptide libraries. Current protocols in molecular biology / edited by Frederick M. Ausubel ... [et al.] 2010 doi: 10.1002/0471142727.mb1814s91. Chapter 18: Unit–Un14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nature reviews. Drug discovery. 2002;1(4):309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- Corrêa IR, Baker B, Zhang A, Sun L, Provost CR, Lukinavičius G, Reymond L, Johnsson K, Xu M-Q. Substrates for improved live-cell fluorescence labeling of SNAP-tag. Current pharmaceutical design. 2013;19(30):5414–5420. doi: 10.2174/1381612811319300011. [DOI] [PubMed] [Google Scholar]
- Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nature biotechnology. 2011;29(11):1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- Durocher D, Jackson SP. The FHA domain. FEBS letters. Elsevier. 2002;513(1):58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lélias J-M, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nature biotechnology. 2005;23(3):329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Fedorov O, Marsden B, Pogacic V, Rellos P, Muller S, Bullock AN, Schwaller J, Sundstrom M, Knapp S. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proceedings of the National Academy of Sciences. 2007;104(51):20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen KAD, Hill ZB, Gadd JC, Fujimoto BS, Maly DJ, Chiu DT. Intracellular Delivery of Bioactive Molecules using Light-Addressable Nanocapsules. ACS Nano. 2010;4(12):7603–7611. doi: 10.1021/nn102345f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer T, Chidley C, Juillerat A, Heinis C, Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling. Protein Eng Des Sel. 2006;19(7):309–316. doi: 10.1093/protein/gzl014. [DOI] [PubMed] [Google Scholar]
- Hah J-M, Sharma V, Li H, Lawrence DS. Acquisition of a ‘Group A’-Selective Src Kinase Inhibitor via a Global Targeting Strategy. Journal of the American Chemical Society. 2006;128(18):5996–5997. doi: 10.1021/ja060136i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. The FASEB Journal. 1995;9(8):576–596. [PubMed] [Google Scholar]
- Hill ZB, Perera BGK, Maly DJ. A Chemical Genetic Method for Generating Bivalent Inhibitors of Protein Kinases. Journal of the American Chemical Society. 2009;131(19):6686–6688. doi: 10.1021/ja900871y. [DOI] [PubMed] [Google Scholar]
- Hill ZB, Perera BGK, Maly DJ. Bivalent inhibitors of the tyrosine kinases ABL and SRC: determinants of potency and selectivity. Molecular BioSystems. 2011;7(2):447–456. doi: 10.1039/c0mb00108b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill ZB, Perera BGK, Andrews SS, Maly DJ. Targeting Diverse Signaling Interaction Sites Allows the Rapid Generation of Bivalent Kinase Inhibitors. ACS Chemical Biology. 2012;7(3):487–495. doi: 10.1021/cb200387g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines AC, Cole PA. Design, synthesis, and characterization of an ATP-peptide conjugate inhibitor of protein kinase A. Bioorganic & Medicinal Chemistry Letters. 2004;14(11):2951–2954. doi: 10.1016/j.bmcl.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Hines AC, Parang K, Kohanski RA, Hubbard SR, Cole PA. Bisubstrate analog probes for the insulin receptor protein tyrosine kinase: Molecular yardsticks for analyzing catalytic mechanism and inhibitor design. Bioorganic Chemistry. 2005;33(4):285–297. doi: 10.1016/j.bioorg.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Hunter T. Signaling--2000 and beyond. Cell. 2000;100(1):113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nature Chemical Biology. 2004;1(1):27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- Imoto M, Umezawa K, Isshiki K, Kunimoto S, Sawa T, Takeuchi T, Umezawa H. Kinetic studies of tyrosine kinase inhibition by erbstatin. Journal of antibiotics. Nature Publishing Group. 1987;40(10):1471–1473. doi: 10.7164/antibiotics.40.1471. [DOI] [PubMed] [Google Scholar]
- Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chemistry & biology. 2003;10(4):313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- Juillerat A, Heinis C, Sielaff I, Barnikow J, Jaccard H, Kunz B, Terskikh A, Johnsson K. Engineering substrate specificity of O6-alkylguanine-DNA alkyltransferase for specific protein labeling in living cells. ChemBioChem. 2005;6(7):1263–1269. doi: 10.1002/cbic.200400431. [DOI] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nature biotechnology. Nature Publishing Group. 2008;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nature biotechnology. 2003;21(1):86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chemistry & biology. 2005;12(6):621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128(3):425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Kothe M, Kohls D, Low S, Coli R, Rennie GR, Feru F, Kuhn C, Ding Y-H. Selectivity-determining residues in Plk1. Chemical biology & drug design. 2007;70(6):540–546. doi: 10.1111/j.1747-0285.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- Krishnamurty R, Maly DJ. Chemical genomic and proteomic methods for determining kinase inhibitor selectivity. Combinatorial chemistry & high throughput screening. 2007;10(8):652–666. doi: 10.2174/138620707782507368. [DOI] [PubMed] [Google Scholar]
- Kruse CH, Holden KG, Pritchard ML, Feild JA, Rieman DJ, Greig RG, Poste G. Synthesis and evaluation of multisubstrate inhibitors of an oncogene-encoded tyrosine-specific protein kinase. Journal of Medicinal Chemistry. 1988;31(9):1762–1767. doi: 10.1021/jm00117a015. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J. 2000;350(1):1–18. [PMC free article] [PubMed] [Google Scholar]
- Levinson NM, Visperas PR, Kuriyan J. The tyrosine kinase Csk dimerizes through Its SH3 domain. PloS one. 2009;4(11):e7683. doi: 10.1371/journal.pone.0007683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loog M, Uri A, Järv J, Ek P. Bi-substrate analogue ligands for affinity chromatography of protein kinases. FEBS letters. 2000;480(2-3):244–248. doi: 10.1016/s0014-5793(00)01948-7. [DOI] [PubMed] [Google Scholar]
- Loog M, Uri A, Raidaru G, Järv J, Ek P. Adenosine-5′-carboxylic acid peptidyl derivatives as inhibitors of protein kinases. Bioorganic & Medicinal Chemistry Letters. 1999;9(10):1447–1452. doi: 10.1016/s0960-894x(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Lowery DM, Lim D, Yaffe MB. Structure and function of Polo-like kinases. Oncogene. 2005;24(2):248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Current biology. 1995;5(3):296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- Medzihradszky D, Chen SL, Kenyon GL, Gibson BW. Journal of the American Chemical Society. 21. Vol. 116. ACS Publications; 1994. Solid-phase synthesis of adenosine phosphopeptides as potential bisubstrate inhibitors of protein kinases. pp. 9413–9419. [Google Scholar]
- Meyer SC, Shomin CD, Gaj T, Ghosh I. Tethering Small Molecules to a Phage Display Library: Discovery of a Selective Bivalent Inhibitor of Protein Kinase A. Journal of the American Chemical Society. 2007;129(45):13812–13813. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
- Mok J, Kim PM, Lam HYK, Piccirillo S, Zhou X, Jeschke GR, Sheridan DL, Parker SA, Desai V, Jwa M, Cameroni E, Niu H, Good M, Reményi A, Ma J-LN, Sheu Y-J, Sassi HE, Sopko R, Chan CSM, De Virgilio C, Hollingsworth NM, Lim WA, Stern DF, Stillman B, Andrews BJ, Gerstein MB, Snyder M, Turk BE. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Science Signaling. 2010;3(109):ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA. Mechanism-based design of a protein kinase inhibitor. Nature structural biology. 2001;8(1):37–41. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutation research. 2000;462(2-3):83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- Poot AJ, van Ameijde J, Slijper M, van den Berg A, Hilhorst R, Ruijtenbeek R, Rijkers DTS, Liskamp RMJ. Development of Selective Bisubstrate-Based Inhibitors Against Protein Kinase C (PKC) Isozymes By Using Dynamic Peptide Microarrays. ChemBioChem. 2009;10(12):2042–2051. doi: 10.1002/cbic.200900199. [DOI] [PubMed] [Google Scholar]
- Profit AA, Lee TR, Lawrence DS. Bivalent Inhibitors of Protein Tyrosine Kinases. Journal of the American Chemical Society. 1999;121(2):280–283. [Google Scholar]
- Qiu H, Miller WT. Role of the Brk SH3 domain in substrate recognition. Oncogene. 2003;23(12):2216–2223. doi: 10.1038/sj.onc.1207339. [DOI] [PubMed] [Google Scholar]
- Ricouart A, Gesquiere JC, Tartar A, Sergheraert C. Design of potent protein kinase inhibitors using the bisubstrate approach. Journal of Medicinal Chemistry. 1991;34(1):73–78. doi: 10.1021/jm00105a012. [DOI] [PubMed] [Google Scholar]
- Rosse C, Linch M, Kermorgant S, Cameron AJM, Boeckeler K, Parker PJ. Nature Reviews Molecular Cell Biology. Vol. 11. Nature Publishing Group; 2010. PKC and the control of localized signal dynamics. pp. 103–112. [DOI] [PubMed] [Google Scholar]
- Rossé G, Séquin U, Mett H, Furet P, Traxler P, Fretz H. Synthesis of Modified Tripeptides and Tetrapeptides as potential bisubstrate inhibitors of the epidermal growth factor receptor protein tyrosine kinase. Helvetica Chimica Acta. 1997;80(3):653–670. [Google Scholar]
- Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Science's STKE : signal transduction knowledge environment. 2003;2003(191):RE12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- Schneider TL, Mathew RS, Rice KP, Tamaki K, Wood JL, Schepartz A. Increasing the Kinase Specificity of K252a by Protein Surface Recognition. Organic Letters. 2005;7(9):1695–1698. doi: 10.1021/ol050179o. [DOI] [PubMed] [Google Scholar]
- Shen K, Cole PA. Conversion of a Tyrosine Kinase Protein Substrate to a High Affinity Ligand by ATP Linkage. Journal of the American Chemical Society. 2003;125(52):16172–16173. doi: 10.1021/ja0380401. [DOI] [PubMed] [Google Scholar]
- Sheridan DL, Kong Y, Parker SA, Dalby KN, Turk BE. Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. The Journal of biological chemistry. 2008;283(28):19511–19520. doi: 10.1074/jbc.M801074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomin CD, Meyer SC, Ghosh I. Staurosporine tethered peptide ligands that target cAMP-dependent protein kinase (PKA): optimization and selectivity profiling. Bioorg Med Chem. 2009;17(17):6196–6202. doi: 10.1016/j.bmc.2009.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomin CD, Restituyo E, Cox KJ, Ghosh I. Selection of cyclic-peptide inhibitors targeting Aurora kinase A: problems and solutions. Bioorg Med Chem. 2011;19(22):6743–67499. doi: 10.1016/j.bmc.2011.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins JL, De SK, Pavlickova P, Chen V, Machleidt T, Chen L-H, Kuntzen C, Kitada S, Karin M, Pellecchia M. Design and Characterization of a Potent and Selective Dual ATP- and Substrate-Competitive Subnanomolar Bidentate c-Jun N-Terminal Kinase (JNK) Inhibitor. Journal of Medicinal Chemistry. 2011;54(18):6206–6214. doi: 10.1021/jm200479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Bubis J, Toner-Webb J, Saraswat LD, First EA, Buechler JA, Knighton DR, Sowadski J. CAMP-dependent protein kinase: prototype for a family of enzymes. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1988;2(11):2677–2685. doi: 10.1096/fasebj.2.11.3294077. [DOI] [PubMed] [Google Scholar]
- Traxler PM, Wacker O, Bach HL, Geissler JF, Kump W, Meyer T, Regenass U, Roesel JL, Lydon N. Sulfonylbenzoyl-nitrostyrenes: potential bisubstrate type inhibitors of the EGF-receptor tyrosine protein kinase. Journal of Medicinal Chemistry. 1991;34(8):2328–2337. doi: 10.1021/jm00112a003. [DOI] [PubMed] [Google Scholar]
- Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA repair. 2007;6(8):1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk BE. Understanding and exploiting substrate recognition by protein kinases. Current Opinion in Chemical Biology. 2008;12(1):4–10. doi: 10.1016/j.cbpa.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8(7):530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- Urbaniak MD, Mathieson T, Bantscheff M, Eberhard D, Grimaldi R, Miranda-Saavedra D, Wyatt P, Ferguson MAJ, Frearson J, Drewes G. Chemical proteomic analysis reveals the drugability of the kinome of Trypanosoma brucei. ACS Chemical Biology. 2012;7(11):1858–1865. doi: 10.1021/cb300326z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ameijde J, Poot AJ, van Wandelen LTM, Wammes AEM, Ruijtenbeek R, Rijkers DTS, Liskamp RMJ. Preparation of novel alkylated arginine derivatives suitable for click-cycloaddition chemistry and their incorporation into pseudosubstrate- and bisubstrate-based kinase inhibitors. Organic & biomolecular chemistry. 2010;8(7):1629–1639. doi: 10.1039/b922928k. [DOI] [PubMed] [Google Scholar]
- van Wandelen LTM, van Ameijde J, Ismail-Ali AF, van Ufford HCLQ, Vijftigschild LAW, Beekman JM, Martin NI, Ruijtenbeek R, Liskamp RMJ. Cell-Penetrating Bisubstrate-Based Protein Kinase C Inhibitors. ACS Chemical Biology. 2013;8:1479–1487. doi: 10.1021/cb300709g. [DOI] [PubMed] [Google Scholar]
- Yadav SS, Miller WT. The Evolutionarily Conserved Arrangement of Domains in Src Family Kinases Is Important for Substrate Recognition. Biochemistry. 2008;47(41):10871–10880. doi: 10.1021/bi800930e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Bhattacharyya RP, Lim WA. The structure and function of proline recognition domains. Science's STKE : signal transduction knowledge environment. 2003;2003(179):RE8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]