Abstract
A cross between the sweet cherry (Prunus avium) cultivars ‘Wanhongzhu’ and ‘Lapins’ was performed to create a mapping population suitable for the construction of a linkage map. The specific-locus amplified fragment (SLAF) sequencing technique used as a single nucleotide polymorphism (SNP) discovery platform and generated 701 informative genotypic assays; these, along with 16 microsatellites (SSRs) and the incompatibility (S) gene, were used to build a map which comprised 8 linkage groups (LGs) and covered a genetic distance of 849.0 cM. The mean inter-marker distance was 1.18 cM and there were few gaps > 5 cM in length. Marker collinearity was maintained with the established peach genomic sequence. The map was used to show that trunk diameter (TD) is under the control of 4 loci, mapping to 3 different LGs. Different locus influenced TD at a varying stage of the tree’s development. The high density ‘W×L’ genetic linkage map has the potential to enable high-resolution identification of QTLs of agronomically relevant traits, and accelerate sweet cherry breeding.
Introduction
The economically significant fruit tree species sweet cherry (Prunus avium) is self-incompatible, and therefore has a highly heterozygous genetic background. However, sweet cherry has a small genome size (338 M), just two times that of Arabidopsis [1]. It shares the same chromosome number (2n = 2x = 16) with its relatives peach (P. persica), almond (P. dulcis) and apricot (P. armeniaca). As with other woody perennials, sweet cherry has a long juvenile period (average of 6 years) and, thus requiring long-term breeding strategies. Genetic linkage maps provide opportunities for unlocking the complex genetics of quantitatively inherited traits through the localization of quantitative trait loci (QTL), and serve as a repository of markers useful in marker-assisted breeding (MAB) [2–4]. Several intact genetic linkage maps for sweet cherry have been assembled [5–8,9] (S1 Table). While, some of these have been based on gel-based DNA assasys, more recently a SNP approach has been exploited for mapping [10–14].
Basing marker discovery on a reduced representation libraries (RRL) has proven to be an efficient and cost-saving strategy across a wide range of animal, plant and microorganism species [15–20]. One such application, denoted “specific-locus amplified fragment sequencing” (SLAF-seq) [21], has been successfully developed for markers developing in at least three plant species [22–24]. Meanwhile, the acquisition of full genome sequence of peach [25, 26 and www.rosaceae.org/peach/genome] has allowed advantage to be taken of the extensive synteny which has been shown to exist in the genus Prunus [27].
Here, the SLAF-seq method was exploited to rapidly provide the large number of markers needed to generate a high density linkage map of sweet cherry. The map was based on a segregating population bred from a cross between two elite sweet cherry cultivars ‘Wanhongzhu’ (‘W’) and ‘Lapins’ (‘L’). ‘W’ fruit matures about 10 days later than those of ‘L’, and forms fruits which are large and of excellent quality. In three continuous years, a broad variety in tree habit and fruit characters were observed and scored in the progeny. The resulting linkage map was used to explore the genetic determination of the trunk diameter (TD), a trait which has been correlated with tree vigor, resistance and fruit yield [28]. Trees which have a large TD, along with a wide brunch angle and lower tree height are seen as advantageous in the context of crop management and so are favored by breeders. However, unlike tree height and brunch angle, TD cannot be manipulated by prune. Thus a genetic means of controlling this trait would aid the genetic advancement of the crop.
Materials and Methods
Plant materials
The cross between ‘W’ as female and ‘L’ as male was made in 2007. ‘W’ (S 6 S 9) was a seedling of ‘19–11’, which was a progeny of ‘Bing’ and ‘Sunburst’. ‘L’ (S 1 S 4’) was a self-compatible cultivar, widely used in cherry breeding. A population of 860 F1 progeny was planted in rows in which the intra-row spacing was 1.0 m and the inter-row spacing was 3.5 m at the Shangzhuang experiment station of institute of forestry and pomology’s in Beijing in the spring of 2008. Because these field experiments were done in the test bases of Institute of Forestry and Pomology. And, the field studies did not involve endangered or protected species. No specific permissions were required for field experiment locations/activities. A subset of 100 of the population was selected as the linkage mapping population, ensuring a balance of S allele representation: 16 progeny were of genotype S 4 ’S 6, 18 were S 4 ’S 9, 23 were S 1 S 6 and 22 were S 1 S 9.
Marker development
The methods used to generate SLAF markers (pre-design, library construction, Illumina sequencing, SNP discovery and genotyping) largely followed those described in [21]. Since the full genome sequence of sweet cherry has not yet been acquired, that of peach were used for in-silico analysis of restriction enzyme recognition sites. Following the construction of the library, pair-end sequencing targeting fragments in the size range 400~500 bp was performed by Beijing Biomarker Technologies Co. Ltd (www.biomarker.com.cn/english/) using a genome analyzer II instrument (Illumina, Inc; San Diego, CA, U.S.). The resulting raw sequence reads were processed by custom Perl scripts (E.A.J.) to optimize read number and to reduce artifacts (five bases with Q score < 20). After the SLAF reads were assigned into individual plants according to their given index, sequences sharing > 96% identity were considered as a single SLAF locus. Only sequences represented at least 107 times and harboring at most 4 genotypes were accepted as representing a high quality SLAFs; those harboring 2–4 genotypes were then carried forward for mapping. In addition, a set of 53 published SSR assays, along with 150 SSR assays designed from sequences represented in the peach genome (www.rosaceae.org/node/355) were used to fingerprint the two parental cultivals and the members of the mapping population. The PCR conditions were identical to those published [5].
Linkage map construction
Based on the parental genotypes, the informative set of SLAF loci formed 5 allelic classes (ab×cd, ef×eg, hk×hk, lm×ll and nn×np). Sixty-three aa×bb type markers were convertible into lm×ll, nn×np or ef×eg types based on the separation pattern produced by the F1 progeny. Homozygosity was confirmed only where a sequence depth of at least three was recorded; heterozygosity was confirmed when either the sequence depth of the least frequent allele was > 3 or the depth ratio of the two alleles was > 1:6 or 2:18, otherwise it would be considered homozygous or missing. For the informative SSR loci, the allelic status of the mapping population progeny fell into the 3 classes hk×hk, lm×ll and nn×np.
The full set of genotypic data (SLAF, SSR and S gene) were combined and then subjected to analysis using JoinMap®4.0 software [29], employing the “CP” model. A two-step strategy was implemented, in which a framework map was first generated based on a set of the highest quality marker (missing value rate < 30% and fitting the expected segregation ratio with P-value < 0.05), applying a stringent LOD thresholds (5.0) for grouping. Recombination frequencies were converted to cM using the Kosambi function regression mapping method [30]. The goodness-of-fit jump threshold for removal a loci was set to 5.0, the ‘suspect linkage’ and ‘genotype probabilities’ tools were applied to improve the reliability of the map. Linkages with a LOD = 3 were used for mapping and the second round maps were selected. Then, a strongest crosslink values (SCLs) of 1 were applied to assign ungrouped loci to their most likely LG [29]. By using similar parameters and procedures, a ‘W×L’ map was assembled. Synteny analysis was performed between the frame map and the ‘W×L’ map repeatedly to test how marker rearrangement was affected by addition of newly-added markers. In addition, Sequence alignment between SLAF sequences and the peach genome sequence was based on a similarity threshold of 80%. Each of the sweet cherry LGs was related by name to the eight peach LGs, according to their content of anchored SLAF markers. A Pearson correlation analysis and the synteny analysis of anchored marker positions between ‘W×L’ genetic linkage map and peach physical map were performed using SPSS software.
QTL analysis for TD
TDs were measured at a fixed point of 20 cm above ground in four continuous years. Anural trunk net growth (ATNG) was calculated from the difference between TD year(n+1) and TD year(n). The mapping population’s statistical parameters were obtained using the SPSS 16.0 software. QTL detection was carried out using the MapQTL 4.0 software, using the interval mapping (IM) procedure [29]. The genome-wide LOD score threshold for QTL significance was determined using the permutation test (PT, [31]), from which a LOD of 3.0 was set. QTL positions were drawn using MapChart [32].
Results
Informative markers
The intention was to construct a sweet cherry linkage map in which the mean interval between adjacent markers was about 1 cM. Since the estimated genetic length of sweet cherry genome was 600–800 cM (S1 Table), a target of 600–800 markers was chosen. Based on the known level of marker polymorphism between sweet cherry varieties (10–15%) [5–8, 9], about 6000–8000 SLAF fragments should be produced by digestion in SLAF-seq method. In peach, the restriction enzyme combination Bfa I+Mse I was predicted to produce 12,856 well distributed DNA fragments of size range 380–430 bp (S1 Fig) with the expected repetitive sequences might be controlled within 1.13%, so this was selected an appropriate combination for sweet cherry. A total of ~ 4.7 G of raw sequence data (NIH Short Read Archive accession number: SRP063722), derived from 31,048,840 pair-end (PE) reads was obtained, resulting in the identification of 14,634 high quality SLAFs. The coverage of these loci was 27×in ‘L’, 20× in ‘W’ and average 4.9× among the mapping population progeny. Of these, 1,838 (12.6%) were classified as polymorphic (S2 Table), in agreement with the predicted range of marker informativeness. Repeated sequences accounted for 4.97% of the high quality SLAFs, a slightly higher frequency than in peach. The final set of high quality SLAFs with full integrity in parents and 82% integrity in progenies was 953. With respect to SSRs, 28 (13.79%) were informative. The S gene segregated consistently with its inheritance from an ab×cd cross. Thus in all, segregation data at 982 loci (953 SLAFs, 28 SSRs and S) were available for the construction of the linkage map (S3 Table).
The sweet cherry linkage map
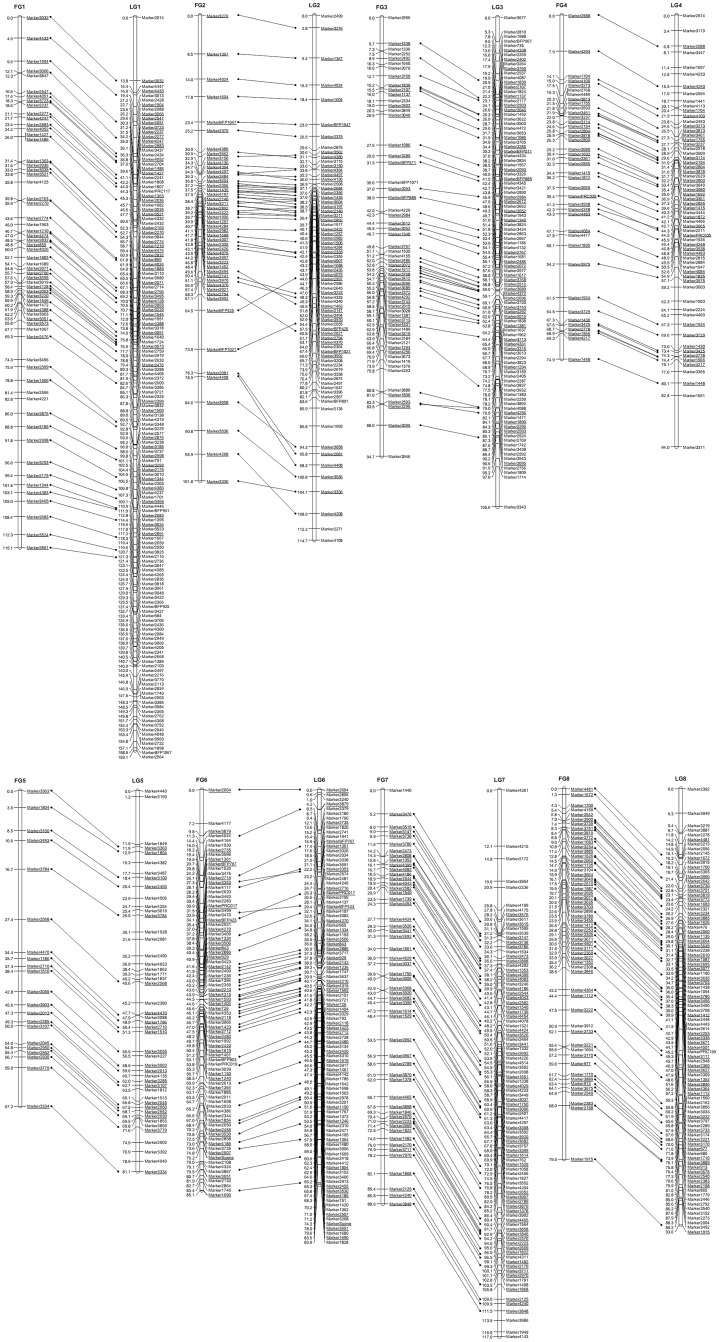
The frame map comprised 8 LGs (FG1-FG8) and was based on 409 markers was built (Fig 1). Its genetic length was 703.5 cM; the shortest LG was FG5 (67.2 cM) and the longest was FG1 (115.1 cM). The mean inter-marker interval was 1.86 cM (range from 1.18 cM on FG6 to 3.20 cM on FG5) (S4 Table). In all, 24 interval (5.87% of the total) were longer than 5.0 cM. The high density map included 309 additional markers, which increased the overall map length to 849.0 cM, while reducing the mean inter-marker distance to 1.18 cM and leaving just 10 (1.39%) marker intervals longer than 5.0 cM. Within each LG, the mean inter-marker interval ranged from 0.86 cM (LG6) to 1.73 cM (LG5) (Table 1). The incorporation of only a small number of the added markers perturbed the marker order predicted by the framework map, so that the extent of collinearity between the two maps was high (Fig 1). Once again, LG5 was the shortest LG and harbored the fewest loci, while LG1 was the longest and harbored the most loci. Segregation distortion affected 130 (18.57%) of the markers, and was noted on all LGs except for LG2; there were clusters of distorted markers on LG1 (123.5~159.1 cM), LG6 (1.1~24.4 cM and 54.0~58.0 cM) and LG7 (40.1~44.1 cM) (Table 1, S2 Fig). In total, 718 markers including 701 SLAFs, 16 SSRs and S gene, were mapped in the integrated ‘W×L’ map (Fig 1, S2 Fig).
Fig 1. Collinearity between sweet cherry LGs derived the framework map (FG1-8) and the high density map (LG1-8).
Sequence-anchored markers are indicated by connecting lines and are shown underlined. Inter-marker distance given in cM.
Table 1. LG by LG breakdown of the markers contributing to the high density ‘W×L’ linkage map.
| Linkage group | Marker number | Map length (cM) | Map density | > 5 cM marker distance | Sig < 0.05 marker | Ratio a | Added markers | Added map length |
|---|---|---|---|---|---|---|---|---|
| LG1 | 150 | 159.10 | 1.06 | 1 | 43 | 28.67% | 88 | 44 |
| LG2 | 74 | 114.70 | 1.55 | 3 | 0 | 0.00% | 26 | 12.9 |
| LG3 | 100 | 105.60 | 1.06 | 2 | 19 | 19.00% | 40 | 10.9 |
| LG4 | 63 | 94.00 | 1.49 | 1 | 10 | 15.87% | 23 | 19.1 |
| LG5 | 47 | 81.10 | 1.73 | 1 | 10 | 21.28% | 26 | 13.9 |
| LG6 | 98 | 83.90 | 0.86 | 0 | 25 | 25.51% | 29 | 2.2 |
| LG7 | 95 | 117.00 | 1.23 | 1 | 13 | 13.68% | 42 | 28.4 |
| LG8 | 91 | 93.60 | 1.03 | 1 | 10 | 10.99% | 35 | 14.1 |
| Total | 718 | 849.00 | 1.18 | 10 | 130 | 18.11% | 309 | 145.5 |
a The ratio is No. of sig < 0.05 markers to total markers in each LG.
Of 203 SSR assays attempted, 28 (13.8%) were informative between the parental cultivars, 16 were assigned a map location and 14 were also represented on both the Prunus reference map [33], and sweet cherry ‘EF×NY’ map [5]. Matching LG assignment applied to 11 of the 14 common SSR loci (Table 2). The 3 exceptions were Prc117 (LG4 in peach, LG1 in sweet cherry) and Prc139 (LG1 in peach, LG8 in sweet cherry) and BPPCT009 (LG6 in peach, but mapping to LG2, LG4 and LG6 in sweet cherry). LG5 and LG7 were devoid of SSR loci.
Table 2. Conservation of SSRs between the sweet cherry ‘W×L’ map, other sweet cherry maps and the peach genome sequence.
| ‘W×L’ linkage goup | Marker name | SSR name | Peach genome | Prunus reference map | Sweet cherry ‘EF×NY’ map |
|---|---|---|---|---|---|
| WL1 | BFP1067 | BPPCT028 | scaffold_1 | G1 | |
| WL1 | BFP925 | MA073a | scaffold_1 | G1 | - |
| WL1 | Prc117 | Prc117 | scaffold_4 | - | - |
| WL1 | BFP941 | UDAp-485 | scaffold_1 | G1 | - |
| WL2 | BFP1021 | BPPCT009 | scaffold_6 | G4 | NY6 |
| WL2 | BFP1047 | UCD-CH36 | - | - | - |
| WL2 | BFP425 | SC2 | - | - | - |
| WL2 | BFP801 | UDAp-461 | scaffold_2 | G2 | EF2 |
| WL3 | BFP967 | BPPCT007 | scaffold_3 | G3 | - |
| WL3 | BFP885 | EPDCU3083 | scaffold_3 | - | EF3 |
| WL3 | BFP511 | BPPCT039 | scaffold_3 | G3 | EF3 |
| WL4 | Prc035 | Prc035 | scaffold_4 | - | - |
| WL6 | BFP767 | BPPCT008 | scaffold_6 | G6 | EF6 |
| WL6 | Prc017 | Prc017 | scaffold_6 | - | - |
| WL6 | BFP423 | SC1 | scaffold_6 | - | - |
| WL8 | Prc139 | Prc139 | scaffold_1 | - | - |
The S locus mapped at 74.3 cM on LG6, flanked by the SLAF loci 3651 and 3268. The 3651 sequence shares homology with the region 27367312–27366944 bp of scaffold 6 in the peach genome, and the peach S locus in located between 26446961 and 26448303 bp on the same scaffold 6; thus the separation between the S locus and the sequence matching SLAF locus 3651 in peach was less 1 Mb.
Synteny between the sweet cherry and peach genomes
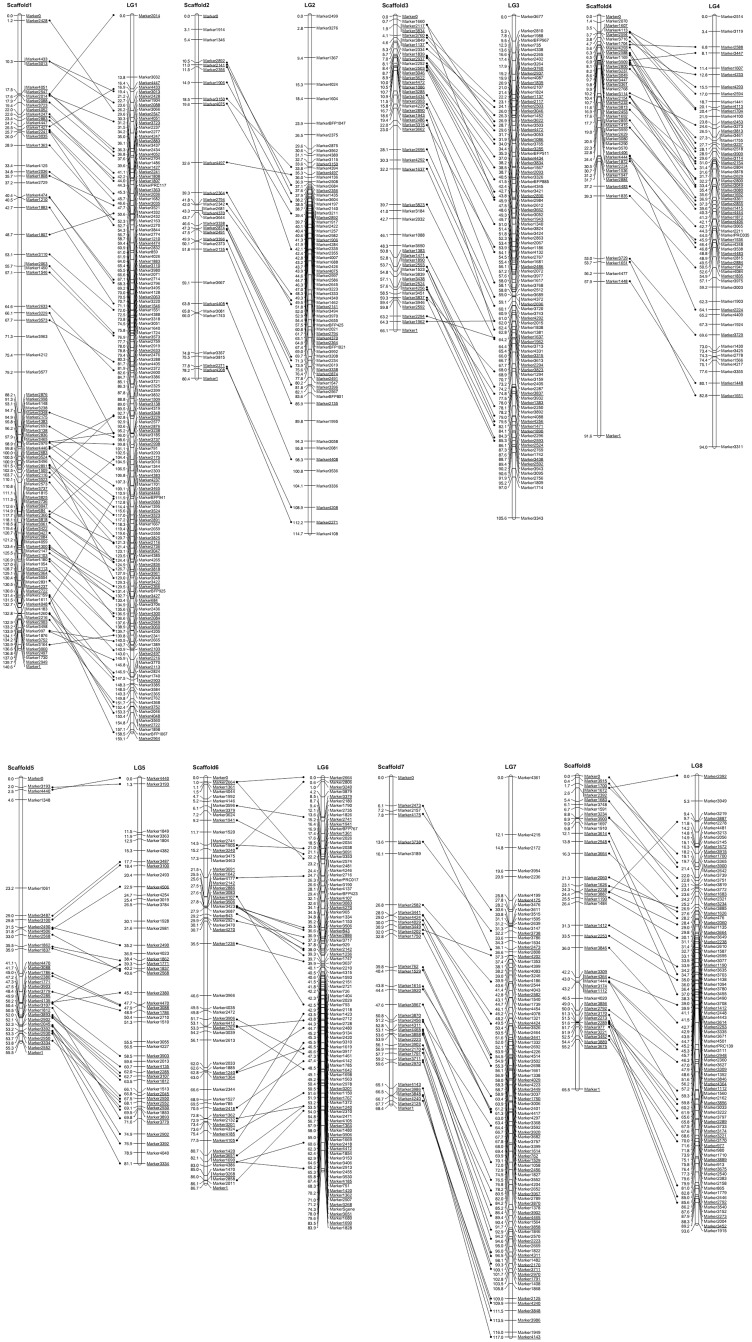
Among the 953 SLAF sequences, 326 (34.2%) SLAFs shared at least 80% homology with a sequence represented in the peach genome; these were designated as similar sequences (SSs). In which, 272 SLAFs were mapped in the sweet cherry linkage map. Pearson correlation coefficient between the mapped location of these 272 SLAF loci and the location of the matching sequence in the peach genome sequence ranged from 0.904 to 0.979, indicative of a high level of synteny obtaining between the two genomes (Table 3, Fig 2). Regions of imperfect collinearity were observed on each LG: 0.4~8.2 Mb and 31.6~46.5 Mb of LG1, 3.5~10.8 Mb of LG2, 0.6 and 21.4 Mb of LG3, 2.1~5.6 Mb of LG4, 15.7~17.9 Mb of LG5, 0.3~5.0 Mb and 17.2~25.8 Mb of LG6, 2.03~10.7 Mb of LG7, 4.0~8.1 Mb of LG8 (Fig 2).
Table 3. LG by LG breakdown of SLAF sequences matching sequences present in the peach genome.
| Linkage group | No. of similar sequence | No. of markers | Ratio of similar sequence to total marker | Pearson correlation (two-tails) |
|---|---|---|---|---|
| LG1 | 69 | 150 | 46.00% | 0.979 |
| LG2 | 18 | 74 | 24.32% | 0.944 |
| LG3 | 34 | 100 | 34.00% | 0.923 |
| LG4 | 34 | 63 | 53.97% | 0.923 |
| LG5 | 28 | 47 | 59.57% | 0.930 |
| LG6 | 25 | 98 | 25.51% | 0.931 |
| LG7 | 31 | 95 | 32.63% | 0.964 |
| LG8 | 32 | 91 | 35.16% | 0.904 |
| Total | 271 | 718 | 37.74% |
Fig 2. Collinearity between the sweet cherry high density map (LG1-8) and peach genome sequence (Scaffold1-8).
In the latter, markers ‘0’ and ‘1’ refer to the two ends of each scaffold. Positions of anchored markers in each peach scaffold = physical position×3×10–6. Sequence-anchored markers are indicated by connecting lines and are shown underlined. Inter-marker distance given in cM.
The detection of TD QTL
TD was continuous distributions, (S3 Fig) and increased year on year (S3 Fig). ATNG was highest between 2013 and 2014 (S3 Fig). Highly significant correlations were found among TDs from year to year over the period (r ranging from +0.60 to +0.93 across the full set of pairwise combinations). However, ATNG was not correlated in this way, presumably because this trait is so heavily determined by the environment. Correlations between TD and ATNG differed from year to year (Table 4). ATNG2012 was more correlated with TD2012~2014 (+0.62~0.65) than TD2011 (-0.16). ATNG2013 was more correlated with TD2013 and TD2014 (+0.58 and +0.52) than TD2012 (+0.36). ATNG2014 was most correlated with TD2014 (+0.50) and none correlated with TD2013.
Table 4. Year on year correlation for TD and ATNG.
| Traits | TD2012 | TD2013 | TD2014 | ATNG2012 | ATNG2013 | ATNG2014 |
|---|---|---|---|---|---|---|
| TD2011 | 0.77** | 0.75** | 0.61** | 0.16* | 0.21 | -0.16 * |
| TD2012 | 0.93** | 0.78** | 0.62** | 0.36** | 0.00 | |
| TD2013 | 0.83** | 0.62** | 0.58** | 0.01 | ||
| TD2014 | 0.65** | 0.52** | 0.50** | |||
| ATNG2012 | 0.34** | 0.26* | ||||
| ATNG2013 | 0.07 |
** Correlation is significant at the 0.01 level (2-tailed).
* Correlation is significant at the 0.05 level (2-tailed).
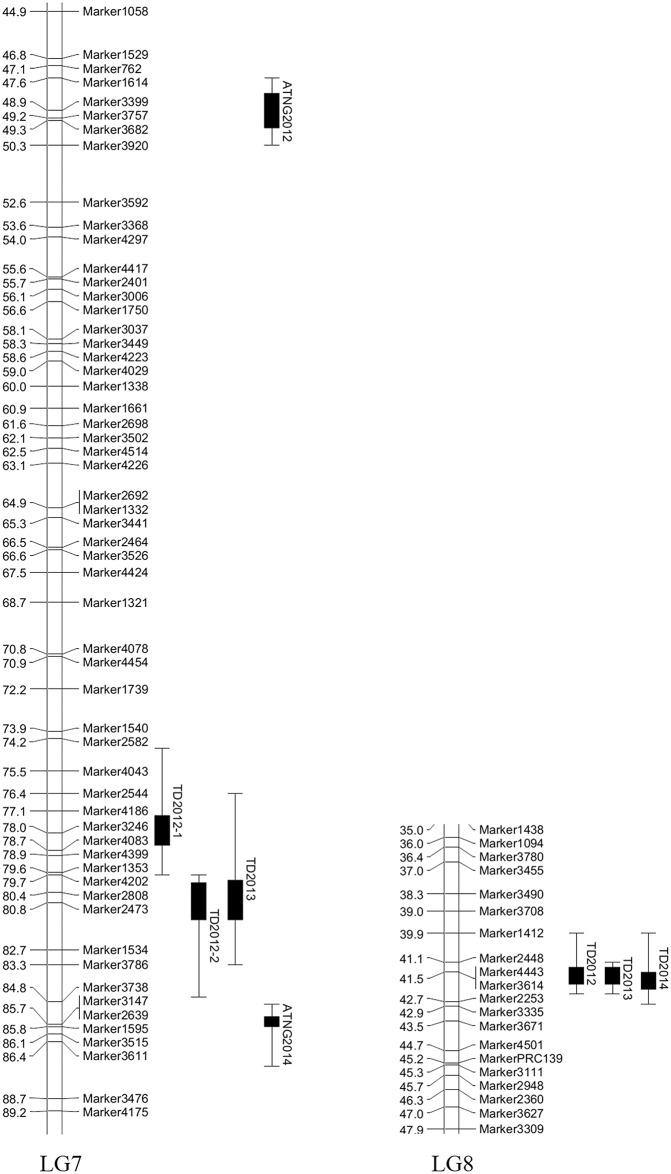
TD QTL was detected on 3 LGs. One mapped to LG6 but was only expressed in 2011. Two closely linked loci on LG7, one mapping in the region around 78.0 cM and the other around 80.4 cM (Table 5, Fig 3). The former explained 15.7% of the phenotypic variance for TD in 2012 and 15.1% in 2013; the latter explained 21.1% and 20.1%, respectively. The final locus mapped to LG8 in the region of 41.5 cM; this locus was expressed in 2012, 2013 and 2014, explaining, respectively, 21.5%, 21.5%, and 16.8% of the variance for TD. For the trait ATNG2012 and ATNG2014, QTL were mapped to different regions of LG7 (respectively, 49.2 cM and 85.7 cM) while for ATNG2013, no QTL was detected. The site of the ATNG2014 QTL was closed to a TD QTL. About 30% of the mapping population trees first bore fruit in 2012, while above 90% of them bore fruit in 2013. These results revealed that different QTLs control TD in different tree development stage, those on LG7 and LG8 mainly controlled TD after fruit.
Table 5. TD and ATNG QTL identified in the ‘W×L’ mapping population.
| Trait | Year | Linkage group | LOD | Position | Marker | Expl% |
|---|---|---|---|---|---|---|
| TD | 2011 | LG6 | 3.04 | 28.1 | Marker4137 | 18.10% |
| 2012 | LG7 | 3.24 | 78.0 | Marker3246 | 15.70% | |
| 3.61 | 80.4 | Marker2808 | 21.10% | |||
| 3.29 | 80.8 | Marker2473 | 17.30% | |||
| LG8 | 4.28 | 41.5 | Marker3614 | 21.50% | ||
| 2013 | LG7 | 3.61 | 80.4 | Marker2808 | 20.10% | |
| 3.29 | 80.8 | Marker2473 | 17.50% | |||
| 2.97 | 78.0 | Marker3246 | 15.10% | |||
| LG8 | 4.11 | 41.5 | Marker3614 | 21.50% | ||
| 2014 | LG8 | 3.01 | 41.5 | Marker3614 | 16.80% | |
| ATNG | 2012 | LG7 | 3.67 | 48.9 | Marker3399 | 23.50% |
| LG7 | 3.24 | 49.2 | Marker3757 | 21.50% | ||
| 2014 | LG7 | 3.02 | 85.7 | Marker2639 | 16.20% |
Fig 3. QTL for trunk diameter (TD) detected over the period 2012–2014 and annual trunk net growth (ATNG) in 2012 and 2014.
1-LOD and 2-LOD support intervals of each QTL are marked by thick and thin bars, respectively. The partial genetic map only includes the LGs harboring relevant QTL.
Determining the favorable QTL alleles, identified by their flanking markers, is critical for further QTL validation and eventual utilization in MAS. A haplotype analysis was conducted featuring the alleles present at the two LG7 SLAF marker2808 and marker2473 and the LG8 SLAF marker3614 (Table 5). Mapping population trees of genotype marker2808 ‘lm’, 2473 ‘nn’ and 3614 ‘lm’ produced the largest TD over the period 2012–2014 (Table 6). All three favorable alleles were inherited from ‘Lapins’. The only sequence hit obtained from a Blast-based search using the three marker sequences as query was for 3614, which almost completely matched a sequence present in an exon of GIGANTEA in almond (KJ502316.1).
Table 6. Mean TD values of mapping population progeny classified according to marker haplotype in loci linked to 3 TD QTL.
| Linkage group | Marker | Allele | SNP | Average trait value | ||
|---|---|---|---|---|---|---|
| 2012 | 2013 | 2014 | ||||
| LG7 | Marker2808 | ll | G | 49.40 | 56.44 | 70.23 |
| lm | G/C | 52.34 | 60.44 | 77.12 | ||
| LG7 | Marker2473 | nn | T | 53.31 | 60.46 | 73.98 |
| np | T/C | 48.65 | 55.99 | 73.78 | ||
| LG8 | Marker3614 | ll | C | 46.87 | 54.10 | 69.40 |
| lm | T/C | 59.62 | 68.51 | 82.41 | ||
Discussion
Linkage between a conveniently assayable marker and a gene of breeding value is the foundation of marker assisted breeding [34]. Establishing such linkages for one or more QTL requires the elaboration of a genome-wide linkage map, which in turn requires that a substantial number of informative markers be generated and that a suitable mapping population be constructed. Here, the aim was to rapidly generate a high density sweet cherry genetic linkage map, and the SLAF-seq strategy proved to be most effective in both producing the necessary markers and in performing the genotyping. Sequencing depth is an important consideration with respect to the quality of a SLAF-based linkage map. It has been suggested that sequencing depth over 4× had relatively little influence on sequencing error rates [21]. In this paper, the sequence depth was 5.38× for progenies and above 20× for parents. The 20× sequence depth for parents could ensure the sequence correction of SLAFs mapped in linkage map. The 5.38× sequence depth for progenies could ensure the correction in genotyping.
The current ‘W×L’ sweet cherry linkage map compares well with already established ones. Its length (849 cM) lies in the same ball park as that of both the ‘BT×K’ map (752.9 cM) [8] and a Prunus consensus map built from four populations (779.4 cM) [7]. With respect to the individual LGs, LG1 is consistently the longest and LG5 the shortest, while the S gene in each case has been mapped to one end of LG6 [5–8]. About 18.57% of the markers represented in the ‘W×L’ sweet cherry linkage map suffered from significant segregation distortion, and there was evidence of certain hot spots of distortion on 123.5~159.1 cM of LG1 and 1.1~24.4 cM of LG6, regions which also have been noted to be liable to this phenomenon in ‘PA×PN’ [6] and ‘Lapins’ [8] genetic background.
Creating a linkage map in a highly heterozygous species such as sweet cherry is less straight-forward than in a species where the mapping parents are highly homozygous; the heterozygosity of the parents means that a larger number of gametic types will be generated. A possible strategy used in some cases was first to construct two separate parental maps, and then to combine them using common markers to produce an integrated map [8,9]. However, a problem would appear if homozygous hk×hk pattern marker existed in one group. It would be difficult to combine the parental map and reduce the information into an integrated map. Here, an alternative strategy was pursued, in which a set of high-quality markers was initially used to build a framework map first, and subsequently the remaining markers were added; this latter step caused very little perturbation to mark order, so that the level of collinearity between the framework map and high-density map was very high.
DNA sequence information of the SLAF markers facilitated a cross-species comparison of the linkage map with the peach genome. About 34.2% of the SLAF markers were homologous with peach sequences, which defined a substantial number of sequence-anchored marker position and confirmed the suggestion made elsewhere [8, 27] that these two Prunus species are highly syntenous. Synteny extended to the S locus, since in sweet cherry, the SLAF marker (3165) most closely linked to the gene harbored a sequence which is highly homologous to a peach sequence lying within 1 Mb of the peach S locus.
TD is the two-way channel for nutrients transportation between root and blade, which reflects the overall condition of root biomass, foliage mass, root efficiency and leaf quantity [35]. TD has been documented to exert a direct influence on yield and fruit quality [35, 28]; unlike tree shape, it is not readily manipulable by pruning. Until now, the genetic determinism of TD has hardly been addressed in sweet cherry [36]; although, some effort in this direction has been invested both in apricot (Prunus armeniaca) [37] and mei (Prunus mume) [38]. In the former case, TD QTL have been mapped to both LG1 and LG2 [37], while in the latter, the major site was on LG8 [38]. Here, the indications were that in sweet cherry, the important TD QTL are sited on LG6, LG7 and LG8, which is a partly similar result with that of mei.
The trait proved to be highly correlated across years, which was not unexpected given that the TD measured in any particular year represents the tree’s accumulated growth over the preceding years. In contrast, the ATNG trait was poorly amenable to genetic analysis because it is strongly mediated by the growing environment. So, TD is a more stable tree character than ATNG. The results, QTLs of TD were stabled in 78~80 cM of LG7 and 41.5 cM of LG8, and QTLs of ATNG in 2012 and 2014 were located in different position of LG7, were consistent with traits relationship. ATNGs show more correlations with this year TD than last TD. In corresponding, QTLs for ATNG2012 and TD2012 were located in LG7 while QTL of TD2011 was in LG6. The various TD QTL was expressed at a different stage of the tree’s development. Thus, once the trees had started to produce fruit, TD was more strongly affected by the LG7 and LG8 QTL and not at all by the LG6 locus. A similar transition in genetic control has been noted for Populus sp. [39, 40].
The LG8 TD QTL was responsible for some 20% of the phenotypic variation once the trees were old enough to set fruit. Its most closely linked marker (SLAF 3614) harbored sequence present in almond GIGANTEA, a gene which in A. thaliana regulates a number of developmental processes [41, 42, 43]; its expression has also been correlated with fruit set [44] and the regulation of wall in-growth deposition in phloem parenchyma transfer cells [45]. Both these two latter activities are in keeping with the major function of the tree trunk, which provides the physical connection between the plant's root system and it photosynthetic apparatus.
The ‘W×L’ progeny varied not just for TD, but also for a number of significant fruit characters. The high density linkage map offers a straight-forward means of determining the genetic basis of this phenotypic variation, thereby opening the way to accelerating sweet cherry improvement by exploiting marker assisted selection.
Supporting Information
Eight scaffolds correspond to the eight peach chromosomes. The color (yellow to dark) indicated the number (0 to 10) of predicted SLAFs attributed in 8 peach chromosomes.
(TIF)
Inter-marker distance given in cM. Loci exhibiting skewed distribution are marked by asterisks to indicate distortion level (* for p<0.1; **p<0.05; ***p<0.01; **** p<0.005; ***** p<0.001; ****** p<0.0005; ******* p<0.0001).
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors thank Lijie Jiang for producing the hybrids. We would like also to thank Professor Xiaoquan Qi from Chinese academy of science and Dr. Richard Harrison from East Malling research for their assistance concerning linkage map construction. The authors would also like to thank Professor Kevin M. Folta from Florida University for his comments and advice during the revision of the manuscript.
Data Availability
Raw sequence data are available from the NIH Short Read Archive under accession number: SRP063722.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31401827); URL: (http://www.nsfc.gov.cn/). JW received the funding. The funder had a role in study design, data collection and analysis, and decision to publish. This work was supported by the National Natural Science Foundation of China (31272123); URL: (http://www.nsfc.gov.cn/). KCZ received the funding. The funder had a role in the decision to publish. This work was supported by the Special Fund for the Construction of Scientific and Technological Innovation Capability (KJCX201101009, KJCX201004003, KJCX20140110); URL: (http://www.baafs.net.cn/). KCZ received these funding. The funder had a role in the decision to publish. This work was supported by the National Plan for Science, Technology and Innovation (2013BAD02B03-1-04); URL: (http://program.most.gov.cn/). KCZ received these funding. The funder had role in decision to publish. This work was supported by the Key Project of Beijing Municipal Science and Technology Commission (D131100000113001); URL: (http://www.bjkw.gov.cn). KCZ received these funding. The funder had a role in the decision to publish. This work was supported by the National Crop Germplasm Resources Platform Project (2012-068); URL: (http://www.most.gov.cn/). GHY received these funding. The funder had a role in data collection and analysis.
References
- 1. Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9(3): 208–218. [Google Scholar]
- 2. Lambert P, Hagen LS, Arus P, Audergon JM. Genetic linkage maps of two apricot cultivars (Prunus armeniaca L.) compared with the almond Texas × peach Earlygold reference map for Prunus . Theor Appl Genet. 2004; 108:1120–1130. [DOI] [PubMed] [Google Scholar]
- 3. Emebiri LC, Moody DB, Panozzo JF, Read BJ. Mapping of QTL for malting quality attributes in barley based on a cross of parents with low grain protein concentration. Field Crop Res. 2004; 87:195–205. [Google Scholar]
- 4. Sooriyapathirana SS, Khan A, Sebolt AM, Wang D, Bushakra JM, Wang KL, et al. QTL analysis and candidate gene mapping for skin and flesh color in sweet cherry fruit (Prunus avium L.). Tree Genet Genomes. 2010;6: 821–832. [Google Scholar]
- 5. Olmstead JW, Sebolt AM, Cabrera A, Sooriyapathirana SS, Hammar S, Iriarte G, et al. Construction of an intra-specific sweet cherry (Prunus avium L.) genetic linkage map and synteny analysis with the Prunus reference map. Tree Genet Genomes. 2008;4: 897–910. [Google Scholar]
- 6. Clarke JB, Sargent DJ, Bošković RI, Belaj A, Tobutt KR. A cherry map from the inter-specific cross Prunus avium ‘Napoleon’ × P. nipponica based on microsatellite, gene-specific and isoenzyme markers. Tree Genet Genomes. 2009;5: 41–51. [Google Scholar]
- 7. Cabrera A, Rosyara UR, Franceschi PD, Sebolt A, Sooriyapathirana SS, Dirlewanger E, et al. Rosaceae conserved orthologous sequences marker polymorphism in sweet cherry germplasm and construction of a SNP-based map. Tree Genet Genomes. 2012;8: 237–247. [Google Scholar]
- 8. Klagges C, Campoy JA, Quero-García J, Guzma´n A, Mansur L, Grataco´ s E, et al. Construction and comparative analyses of highly dense linkage maps of two sweet cherry intra-specific progenies of commercial cultivars. PloS ONE. 2013;8(1):e54743 10.1371/journal.pone.0054743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guajardo V, Solis S, Sagredo B, Gainza F, Munoz C, Gasic K, et al. Construction of high density sweet cherry (Prunus avium L.) linkage maps using microsatellite markers and SNPs detected by genotyping-by-sequencing (GBS). PloS ONE. 2015. May 26;10(5):e0127750 10.1371/journal.pone.0127750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suh Y, Vijg J. SNP discovery in associating genetic variation with human disease phenotypes. Mutat Res. 2005;573(1–2): 41–53. [DOI] [PubMed] [Google Scholar]
- 11. Mammadov JA, Chen W, Ren R, Pai R, Marchione W, Yalçin F, et al. Development of highly polymorphic SNP markers from the complexity reduced portion of maize (Zea mays, L.) genome for use in marker-assisted breeding. Theor Appl Genet. 2012;121: 577–588. [DOI] [PubMed] [Google Scholar]
- 12. Wittenberg AH, Van der Lee T, Cayla C, Kilian A, Visser RG, Schouten HJ. Validation of the high-throughput maker technology DArT using the model plant Arabidopsis thaliana . Mol Genet and Genomics. 2005;274: 30–39. [DOI] [PubMed] [Google Scholar]
- 13. Pfender WF, Saha MC, Johnson EA, Slabaugh MB. Mapping with RAD (restriction-site associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne . Theor Appl Genet. 2011;122: 1467–1480. 10.1007/s00122-011-1546-3 [DOI] [PubMed] [Google Scholar]
- 14. Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011. May 4;6(5):e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, et al. A first generation haplotype map of maize. Science. 2009;326: 1115–1117. 10.1126/science.1177837 [DOI] [PubMed] [Google Scholar]
- 16. Hyten DL, Cannon SB, Song Q, Weeks N, Fickus EW, Shoemaker RC, et al. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics. 2010. January 15;11:38 10.1186/1471-2164-11-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3(10):e3376 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baxter SW, Davey JW, Johnston JS, Shelton AM, Heckel DG. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS ONE. 2011. April 26;6(4):e19315 10.1371/journal.pone.0019315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chutimanitsakun Y, Nipper RW, Cuesta-Marcos A, Cistue L, Corey A, Filichkina T, et al. Construction and application for QTL analysis of a Restriction Site Associated DNA (RAD) linkage map in barley. BMC Genomics. 2011. January 4;12:4 10.1186/1471-2164-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lewis ZA, Shiver AL, Stiffler N, Miller MR, Johnson EA, Selker EU. High density detection of restriction-site associated DNA markers for rapid mapping of mutated loci in Neurospora. Genetics. 2007;177: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, et al. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. Plos ONE. 2013;8(3): e58700 10.1371/journal.pone.0058700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen S, Huang Z, Dai Y, Qin S, Gao Y, Chen J. The development of 7E chromosome-specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLoS ONE. 2013;8(6): e65122 10.1371/journal.pone.0065122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang YX, Wang LH, Xin HG, Li DH, Ma CX, Ding X, et al. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013. September 24;13:141 10.1186/1471-2229-13-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xia C, Chen LL, Rong TZ, Li R, Xiang Y, Wang P, et al. Identification of a new maize inflorescence meristem mutant and association analysis using SLAF-seq method. Euphytica. 2015;202(1) 35–44. [Google Scholar]
- 25. Aruús P, Verde I, Sosinski B, Zhebentyayeva T, Abbott A. The peach genome. Tree Genet Genomes. 2012;8: 531–547. [Google Scholar]
- 26.Sosinski B, Verde I, Morgante M, Rokhsar D. The international peach genome initiative. A first draft of the peach genome sequence and its use for genetic diversity analysis in peach. In: 5th Intl Rosaceae Genomics Conf, Stellenbosch, South Africa. 2010.
- 27. Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere´ F, Cosson P, Howad W, et al. Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci. 2004;101: 9891–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller S, Sorza R. Response of two novel peach tree growth habits to in-row tree spacing, training system, and pruning: effect on growth and pruning. J Am Pomol Soc. 2010;64: 199–217. [Google Scholar]
- 29. Van Ooijen JW. JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, 33 2006. [Google Scholar]
- 30. Kosambi DD. The estimation of map distances from recombination values. Ann Eugenics. 1944;12: 172–175. [Google Scholar]
- 31. Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voorrips RE. Map Chart: software for the graphical presentation of linkagemaps and QTLs. J Hered. 2002;93(1):77–78. [DOI] [PubMed] [Google Scholar]
- 33. Aranzana MJ, Pineda A, Cosson P, Dirlewanger E, Ascasibar J, Cipriani G, et al. A set of simple-sequence repeat markers covering the Prunus genome. Theor Appl Genet. 2003;106: 819–825. [DOI] [PubMed] [Google Scholar]
- 34. Gupta PK, Varshney RK, Sharma PC, Ramesh B. Molecular markers and their applications in wheat breeding. Plant Breeding. 1999;118(5): 369–390. [Google Scholar]
- 35. Laurens F, Audergon J, Claverie J, Duval H, Germain E, Kervella J, et al. Integration of architectural types in French programmes of ligneous fruit species genetic improvement. Fruits. 2000;54: 441–449. [Google Scholar]
- 36. Alfonso Salazar J, Ruiz D, Campoy JA, Sanchez-Perez R, Crisosto CH, Martinez-Garcia PJ, et al. Quantitative Trait Loci (QTL) and Mendelian Trait Loci (MTL) analysis in Prunus: a breeding perspective and beyond. Plant Mol Biol Rep. 2014;32(1): 1–18. [Google Scholar]
- 37. Socquet-Juglard D, Christen D, Devenes G, Gessler C, Duffy B, Patocchi A. Mapping architectural, phenological, and fruit quality QTLs in apricot. Plant Mol Biol Rep. 2013;31(2): 387–397. [Google Scholar]
- 38. Sun L, Wang Y, Yan X, Cheng T, Ma K, Yang W, et al. Genetic control of juvenile growth and botanical architecture in an ornamental woody plant, Prunus mume Sieb. et Zucc. as revealed by a high-density linkage map. BMC Genetics. 2014;15 Suppl 1:S1 10.1186/1471-2156-15-S1-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu RL, Stettler RF. Quantitative genetics of growth, development in Populus. I. A three-generation comparison of tree architecture during the first two years of growth. Theor Appl Genet. 1994;89: 1046–1054. 10.1007/BF00224537 [DOI] [PubMed] [Google Scholar]
- 40. Wu R, Stettler RF. The genetic dissection of juvenile canopy structure and function in a three-generation pedigree of Populus . Trees Struc Fun. 1996;11: 99–108. [Google Scholar]
- 41. Cao S, Ye M, Jiang S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Report. 2005;24(11): 683–690. [DOI] [PubMed] [Google Scholar]
- 42. Han Y, Zhang X, Wang Y, Ming F. The Suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana . PloS ONE. 2013. November 6;8(11):e73541 10.1371/journal.pone.0073541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. LaFlamme B. The two sides of GIGANTEA. Nature genetics. 2015; 47(4): 311. [Google Scholar]
- 44. Brock MT, Tiffin P, Weinig C. Sequence diversity and haplotype associations with phenotypic responses to crowding: GIGANTEA affects fruit set in Arabidopsis thaliana . Mol Ecol. 2007;16(14): 3050–3062. [DOI] [PubMed] [Google Scholar]
- 45. Edwards J, Martin AP, Andriunas F, Offler CE, Patrick JW, McCurdy DW. GIGANTEA is a component of a regulatory pathway determining wall ingrowth deposition in phloem parenchyma transfer cells of Arabidopsis thaliana . Plant Journal. 2010;63(4):651–661. 10.1111/j.1365-313X.2010.04269.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Eight scaffolds correspond to the eight peach chromosomes. The color (yellow to dark) indicated the number (0 to 10) of predicted SLAFs attributed in 8 peach chromosomes.
(TIF)
Inter-marker distance given in cM. Loci exhibiting skewed distribution are marked by asterisks to indicate distortion level (* for p<0.1; **p<0.05; ***p<0.01; **** p<0.005; ***** p<0.001; ****** p<0.0005; ******* p<0.0001).
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Raw sequence data are available from the NIH Short Read Archive under accession number: SRP063722.