SUMMARY
Despite the continuing progress made towards mapping kinase signaling networks, there are still many phosphorylation events for which the responsible kinase has not yet been identified. We are interested in addressing this problem through forming covalent crosslinks between a peptide substrate and the corresponding phosphorylating kinase. Previously we reported a dialdehyde-based kinase binding probe capable of such a reaction with a peptide containing a cysteine substituted for the phosphorylatable ser/thr/tyr residue. Here, we examine the yield of a previously reported dialdehyde-based probe, and report that the dialdehyde based probes possesses a significant limitation in terms of crosslinked kinase-substrate product yield. To address this limitation, we develop a crosslinking scheme based on a kinase activity-based probe, and this new cross-linker provides an increase in efficiency and substrate specificity, including in the context of cell lysate.
INTRODUCTION
The protein kinase-catalyzed transfer of phosphate from ATP to protein substrates constitutes a major form of information transfer in eukaryotic cells. With 518 human kinases (Manning, 2002) and an estimated 20,000 or more phosphorylation sites (Goel et al., 2012), the phosphoproteome is a complex network of enzyme-substrate relationships. While robust methods exist for identifying downstream substrates of a particular protein kinase (Allen et al., 2005; Garber and Carlson, 2013; Garske et al., 2011), the discovery of new phosphorylation sites outpaces the identification of kinase-substrate pairs by these methods (Garber and Carlson, 2013). A method to match kinase-substrate pairs by the reverse approach, i.e., starting with a known phosphosite and discovering the kinase responsible for installing the phosphate group would provide a much needed tool for deconvoluting signaling networks. Due to the weak affinity between kinases and their substrates, a method to covalently crosslink a known substrate to its upstream kinase would facilitate unbiased approaches to identify the kinase(s) responsible for a particular phosphorylation event (Eyrich et al., 2011; Suwal and Pflum, 2010). However, development of a suitable chemical reaction to crosslink and identify new kinase-substrate pairs has remained elusive (Parang et al., 2002; Suwal and Pflum, 2010), while the need for such a tool has increased as more phosphosites are discovered (Lemeer and Heck, 2009).
We have previously reported a three-component chemical reaction capable of covalently linking an engineered “bait” quasi-substrate peptide to a kinase (Maly et al., 2004). The quasi-substrate contains a cysteine residue in place of the target serine, threonine or tyrosine residue, creating a traceable reactant in a bio-orthogonal reaction. These crosslinkers are comprised of a promiscuous kinase binding group and an aromatic-dialdehyde, which is able to covalently link the cysteine residue from the quasi-substrate to the conserved lysine residue on a kinase via a three-component cascade reaction as shown in Figure 1A (Statsuk et al., 2008). In this report we investigate the step-wise yield of the dialdehyde based crosslinker and found that the initial reaction between the target kinase and the crosslinker is robust, however, the subsequent reaction with the cysteine peptide is very inefficient. Although the reaction produces sufficient crosslinked product for detection by western blot, the yield is too low to allow for unbiased identification of the kinase by mass spectrometry. Thus, the poor yield of our previously described crosslinking reaction limits our ability to use this technique for the discovery of up-stream kinases.
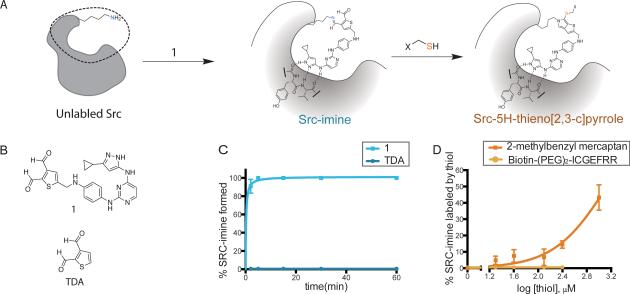
Figure 1.
Reactions of thiophene dialdehyde based crosslinkers with c-Src. (A) Reaction scheme of crosslinker 1 with c-Src. (B) Structures of crosslinker 1 and thiophene dialdehyde. (C) Time course of imine formation with 20 μM crosslinker and 4 μM c-Src as quantified by LC-MS. Error bars represent the standard error of the mean (SEM) of duplicate data points. Data is representative of three separate experiments. (D) Dose response curve of thiol reaction with Src-imine. Indicated thiol was added to 4 μM c-Srcimine and allowed to react for 25 minutes at room temperature. Results were analyzed as in (C).
To develop a crosslinker suitable for unbiased kinase-substrate detection, we designed a new ATP based crosslinker which proceeds through a two step mechanism as opposed to a three component cyclization. The new crosslinker is based on the well-validated acyl-phosphate activity probe (ATP-biotin) for biotinylation of lysine residues in the kinase active site (Patricelli et al., 2011; 2007). Replacement of the biotin with an acrylate resulted in efficient tethering of an acrylamide to an active site lysine residue, which is then primed for reaction with the quasi-substrate cysteine containing peptide. We demonstrate that this new crosslinking approach significantly improves the yield of the crosslinking reaction while retaining kinase substrate selectivity.
RESULTS AND DISCUSSION
LC/MS investigation of thiophene dialdehyde based crosslinker
The tyrosine kinase c-Src was chosen as a model as it is readily expressed in e. Coli (Seeliger et al., 2005), well-behaved in vitro, and amenable to analysis by mass spectrometry. Reaction of c-Src with aromatic dialdehydes in the absence of cysteine containing peptides yielded the intermediate imine which was trapped by reduction with sodium borohydride (Means and Feeney, 1995) (Figure 1A-Src-imine). Incubation of Src with 1 followed by treatment with NaBH4 revealed quantitative adduct formation in five minutes (Figure 1C). The extent of modification was assessed by ultra pressure liquid chromatography electrospray mass spectrometry (UPLC-ESI-TQD) with the intact protein. The technique relies on the assumption that a covalent addition of a small molecule is modest modification to a protein and therefore does not alter the protein's ionization efficiently (Adamczyk et al., 2000; Bateman et al., 2004; Green et al., 1996; Weber et al., 1995). We have confirmed experimentally that the signal intensities measured for c-Src are linear with concentration and not altered by a small molecule adduct (Figure S1). To test whether the dialdehyde reactivity alone was sufficient for reaction, or whether targeting to the ATP site contributed to adduct formation, we treated c-Src with thiophene dialdehyde (TDA) (Figure 1B), which contains no adenine mimetic. As predicted, the product formed in the absence of the kinase-binding element is almost undetectable, suggesting the reaction occurs in the active site and is greatly enhanced by non-covalent positioning of the dialdehyde near a lysine residue in the active site.
The final step of the reaction requires nucleophilic attack of the kinase-imine intermediate by the thiol of the quasi-substrate cysteine residue, followed by cyclization to yield the final crosslinked product. We used an optimized Src substrate peptide sequence (Srctide) with a cysteine in place of the target tyrosine residue as our quasi-substrate (Biotin-ZZICGEFRRR, Cys-Srctide, Z = ethylene glycol). However, no product formation was detected upon addition of Cys-Srctide to the c-Src-1 complex despite using concentrations exceeding 200 μM, significantly above the Km for the peptide substrate (Till et al., 1994). Two possible explanations for the failure to detect the expected product were considered: either the reaction is taking place, but the cyclized product is not stable enough to withstand LC/MS detection, or there are steric constrains keeping the cysteine from accessing the imine. To rule out the first possibility, we utilized a small molecule thiol, 2-methylbenzylmercaptan, in place of the quasi-substrate peptide. At high concentrations 2-methylbenzylmercaptan shows significant product formation (Figure 1D) suggesting the assay was suitable for LC-MS detection of the product. Based on this result our hypothesis is that the Cys-Srctide is not appropriately positioned for attack of the imine once the peptide binds to the kinase. Both the crosslinker and the peptide contain kinase-binding elements, which we postulated to impose geometric constraints limiting the variety of orientations the imine moiety and cysteine residue can adopt, which is not the case for the benzyl mercaptan.
Design of a new crosslinker
In considering how to optimize thiol capture of an electrophile in the kinase active site we were inspired by acyl carrier protein domains in which a phosphopantetheinyl prosthestic group is attached through a phosphodiester bond to the hydroxyl group of a conserved serine reside (Walsh et al., 1997). The 13-atom phosphopantetheinyl addition effectively converts the serine residue into a flexibly tethered thiol which is available to perform additional chemical reactions at the enzyme active site. With this in mind we envisioned a prosthetic electrophile attached to the catalytic lysine, thus converting the conserved lysine residue into a cysteine-reactive electrophile capable of trapping the bound Cys-Srctide.
To deliver a cysteine reactive electrophile we took advantage of the chemoselective nature of acyl-phosphate based activity probes pioneered by AcitvX Biosciences, such as ATP-biotin. These probes contains a phospho-anhydride group which targets one of the two conserved active site lysine residues in the vast majority of kinases (Patricelli et al., 2007; 2011). Our selection of electrophile was largely based on two factors. First, our previous crosslinker designs based on the dialdehydes strongly suggested that more reactive thiol-electrophiles were subject to side reactions and second, the crosslinker must be stable in solution. Methacrylamides are less reactive in comparison to other Michael acceptors (Tsou et al., 2001), and the methylacrylate anhydride is more stable than corresponding acrylate, so we proceeded with a methylacrylate based crosslinker. In contrast to the dialdehyde-based crosslinker which remains covalently attached throughout the reaction sequence, ATP is released in the acrylamide formation step. We thus anticipated that the methacrylamide being attached by a flexible tether to the active site lysine might provide for more efficient kinase substrate crosslinking (Figure 2A).
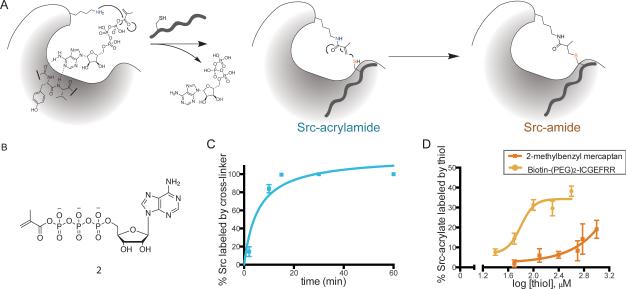
Figure 2.
Reactions of crosslinker 2 with c-Src. (A) Reaction scheme of crosslinker 2 with c-Src. (B) Structure of crosslinker 2. (C) Time course of acrylamide formation with 20 μM crosslinker added to 4 μM c-Src and quantified by LC-MS. Error bars represent the SEM of duplicate data points. Data is representative of three separate experiments. (D) Dose response curve of thiol reaction with Src-acrylamide. Indicated thiol was added to 4 μM c-Src-acrylamide and allowed to react for 25 minutes at room temperature. Results were analyzed as in (C).
LC/MS investigation of ATP-acrylate crosslinker
Consistent with data from ATP-biotin (Figure S2), crosslinker 2 showed complete modification of c-Src after 20 minutes (Figure 2C). This modification can be completely inhibited by the addition of a covalent Src inhibitor (Gushwa et al., 2012) that targets an active site catalytic lysine (Figure S3). In contrast to the previous dialdehyde-based strategy, addition of Cys-Srctide to the acrylamide adduct showed significant crosslinked product. The Cys-Srctide product formation shows a sigmoidal dose dependence, with half-maximal activity at 63 μM near the published Km for the non cysteine containing version of this peptide (Till et al., 1994) (Figure 2D). Again, in contrast to the corresponding reaction with crosslinker 1, 2-methylbenzylmercaptan is much less efficient, which is consistent with the lack of binding interaction with the kinase. Taken together, these results show improvement in the overall yield of the reaction from below the limit of detection for LC/MS to 40% conversion in 25 minutes.
Substrate selectivity
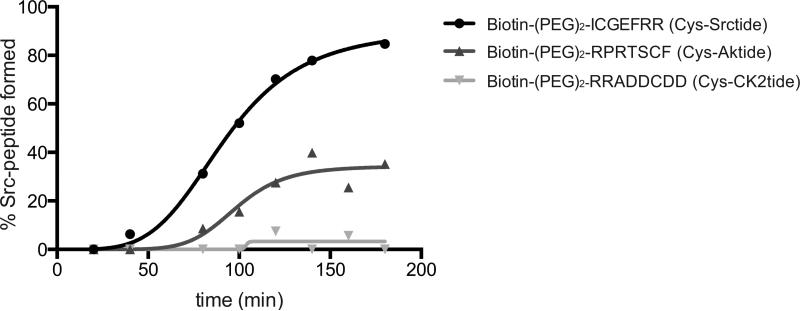
To trap bona fide kinase-substrate pairs, the crosslinking must be selective; meaning the crosslinking of kinases to their substrates must be more favorable than the crosslinking of a kinase to a quasi-substrate derived from another kinase phosphorylation sequence. To test the selectivity of the reaction between c-Src and Cys-Srctide we compared this reaction to that of c-Src with cysteine containing peptides lacking the c-Src consensus motif over a three hour time course. First, we incubated c-Src with 2 and Cys-Srctide and allowed the reaction to proceed to completion. Next, we performed the same experiment with two other peptides, Cys-Aktide (Biotin-ZZRPRTSCF) and Cys-CK2tide (Biotin-ZZRRADDCDDDD), which both contain a cysteine residue but are designed as quasi-substrates for two other kinases, AKT and CK2, respectively. Cys-Aktide reacted more slowly than Cys-Srctide, and did not reach completion during the time course of the experiment, and Cys-CK2tide resulted in less than 10% crosslinked product (Figure 3). These data suggests our system retains the native specificity of kinase-substrate pairs.
Figure 3.
Product formation as a function of time. The indicated peptide (74 μM) was added to 4 μM c-Src with 20 μM crosslinker 2. Percentage final product was calculated as a percentage of total c-Src present as quantified by LC-MS. Also see figure S4.
Streptavidin precipitation of recombinant c-Src from cell lysate
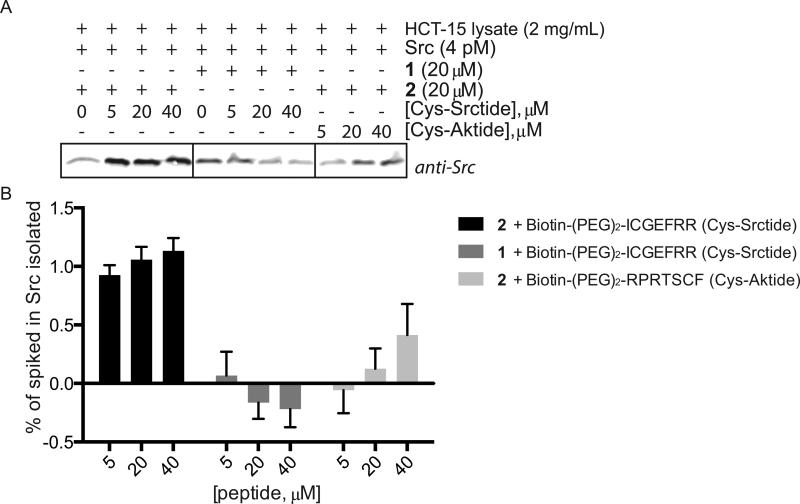
To determine if the strengths of the acrylamide based kinase-substrate crosslinking strategy are maintained in biologically relevant systems, we tested the reaction in cell lysates. Recombinant c-Src kinase was added to HCT-15 cell lysate with crosslinker 2, followed by the addition of varying concentrations of biotinylated quasi-substrates. After 25 minutes the reaction was stopped by addition of 8M Urea, incubated with Streptavidin beads and the isolated proteins were analyzed by Western blot (Figure 4A). To compare the effect of the dialdehyde vs. acrylamide based crosslinkers we compared the efficiency of c-Src isolation between crosslinker 1 and crosslinker 2, respectively. Based on our previous LC/MS experiments we expected very little capture of c-Src by crosslinker 1. In the experiments with crosslinker 2 and Cys-Srctide, 1% of the total recombinant c-Src added to the cell lysate was recovered by precipitation (Figure 4B). In contrast, the yield of c-Src from the experiments using the previous crosslinker, 1 was not higher than the background signal even at the highest concentration of Cys-Srctide.
Figure 4.
Streptavidin-agarose precipitation of recombinant c-Src added to HCT-15 cell lysate. (A) Representative western blot. 4 pmol recombinant c-Src was added to 500 μL of 2 mg/mL HCT-15 cell lysate followed by 20 μM crosslinker. After 10 minutes the indicated quasi-substrate was added and the reaction proceeded for 25 minutes. The proteins were captured using High Capacity Streptavidin beads. Bands were quantified using Odyssey Imaging System Imager. (B) Quantification of c-Src obtained from Streptavidin precipitation experiments as a percentage of total c-Src added to the HCT-15 cell lysate. Error bars represent the SEM of duplicate data points. Data are representative of two separate experiments.
To test peptide selectively in the same cell lysate, we compared Cys-Srctide and Cys-Aktide in reactions using crosslinker 2. As expected, the yield of c-Src using Cys-Aktide as the bait quasi-substrate was less than that of Cys-Srctide, reaching only 0.5% of recombinant c-Src with the highest concentration of peptide. However, unlike the results with crosslinker 1, the percentage of c-Src recovered with 40 μM Cys-Aktide and crosslinker 2 is significantly above the negative control, indicating the reaction is occurring in a manner consistent with our experiments performed using purified components. At the lowest concentration of peptide, 5 μM, Cys-Srctide and crosslinker 2 still capture roughly 1% c-Src, while c-Src is not captured at a detectable level by Cys-Aktide at the same concentration. These data show that low concentrations of peptide may be optimal for capturing the desired kinase while minimizing background from off target reactions.
In conclusion, mass spectrometry quantification of kinase-substrate crosslinking reactions led us to explore a new crosslinking strategy based on a tethered acrylamide rather than previously reported dialdehyde based cyclization reactions. The new acrylamide based method produces kinase-substrate crosslinking with higher yield while maintaining selectivity for correct kinase-substrate pairs. This approach is inspired by biological examples of nucleotides delivering chemical prosthetic groups to proteins both in nature, such as coenzyme A transferring phosphopantetheine to acyl carrier proteins, and previous work demonstrating the power of activity-based probes (Cravatt et al., 2008), such as ATP-biotin (Patricelli et al., 2007; 2011). The synthetic route to crosslinker 2 allows for derivatization at both the acrylate arm and the kinase-binding element. These structural modifications of 2 may further improve the yield and specificity of the reaction for other kinases. In particular, the development of ADP-methacrylate may position the acrylate to attack a distinct lysine residue in kinases, providing different tethering sites for different kinases. Many kinases have two conserved active site lysine residues, both of which are capable of being biotinylated by ATP and ADP based biotin probes, and it is possible that only one of the lysine adducts will be appropriately positioned for crosslinking to cysteine containing peptides or proteins. In addition to kinase identification, 2 could prove useful in obtaining co-crystal structures of kinase-substrate pairs with low binding affinity. Using full-length proteins as the quasi-substrate should further increase selectivity of the reaction, due to the greater binding affinity between the kinase and substrate as compared to peptides. Future experiments will include using full-length proteins to continue to optimize the kinase precipitation protocol.
SIGNIFICANCE
In order to gain a better understanding of kinase signaling networks, matching kinases to their substrate(s) is essential. Although there are tools to discover the substrates of known kinases, there is currently no robust discovery method to unambiguously discover the upstream kinases of known phosphoproteins in complex cellular mixtures. A selective crosslinking method with high yield will aid in mapping kinase substrate networks as new phosphosites continue to be identified. We have demonstrated both high yield and selectivity for our new ATP based crosslinker, and the yield is high enough to isolate purified kinase from cell lysate via streptavidin precipitation at a significantly higher concentration than has been previously reported. This represents a large improvement towards the goal of discovering new kinase substrate connections in signaling pathways.
EXPERIMENTAL PROCEDURES
General procedure for crosslinking reactions with Src
Kinase labeling experiments were performed by incubating 10 μL of Src (final concentration 4 μM) in kinase reaction buffer (25 mM HEPES pH=6.5 for thiophene based compounds, PBS pH=8 for acrylates) with 20 μM crosslinker (1 μL of 220 μM stock, final DMSO concentration 0.025%) for a final volume of 11 μL. Reaction was quenched with 3 μL NaBH4 freshly dissolved in cold MeOH (66 mM) and analyzed by LC mass spectrometry. In all cases, the extent of modification was assessed by electrospray mass spectrometry using a Waters Acquity UPLC/ESI-TQD with a 2.1 x 50 mm Acquity UPLC BEH300 C4 column. MaxEnt 1 software within Waters MassLynx version 4.1 was used for deconvolution of the multiple charged ion.
Thiol reactions
The protein was labeled with a crosslinker in conditions described above for 15 minutes but no quench step was performed. The product was incubated with 2-methylbenzylmercaptan (50 μM-1 mM) or peptide (25-400 μM) for 40 minutes. Reaction was analyzed as described above.
Streptavidin precipitation of Src from cell lysate
This procedure was adapted from Pierce Kinase Enrichment Kits for ActivX Probes. Lysate was diluted with Reaction Buffer (Pierce) to 2mg/mL and 500 μL were transferred to a micocentrifuge tube, to which 10 μL of 1M MgCl2 and 4 pmol of recombinant c-Src was added to each sample. 1 μL of 10 mM stock solution of crosslinker was added to the samples and incubated for 10 minutes at room temperature. Peptide was added at final concentration ranging from 5 μM to 40 μM and incubated for 25 minutes. 500 uL of 8M urea/IP Lysis Buffer (Pierce) was added to each reaction followed by 50 uL of 50% High Capacity Streptavidin Agrose resin slurry (Pierce) and incubated for 1 hour and room temperature with shaking. Samples were centrifuged at 1000 x g for 1 minute and supernatant was removed. Pellets were washed with 4M Urea/Lysis Buffer (Pierce) and centigrade at 1000 x g for 1 minute 4 times. Proteins were eluted in 50 μL 2X Laemmli reducing sample buffer (4% w/v SDS, 100 mM Tris, pH 6.5, 0.2 mg/mL bromophenol blue, 0.2 M DTT, 20% w/v Glycerol) by boiling for 5 minute. Proteins were analyzed by SDS-PAGE and Western blot using anti-Src monoclonal antibody (Cell Signaling). Signal intensities from each Western blot were quantified by using the LI-COR/Odyssey infrared image system. A sample corresponding to the concentration of c-Src added to cell lysate was used to determine percentage of total c-Src recovered.
Supplementary Material
HIGHLIGHTS.
A kinase-substrate crosslinker derivatizes kinase active sites with an electrophile
The crosslinker enables complex formation between Src kinase and a pseudo-substrate
The reaction occurs preferentially with true kinase-substrate pairs
Kinase-substrate crosslinks can be formed in cell lysates, enabling precipitation
ACKNOWLEDGMENTS
We thank members of the Shokat lab for helpful discussions, in particular, Greg Hamilton and Jon Ostrem for contributing to the design of our chemical crosslinkers. Rebecca Lavine provided HTC-15 cell lysate. We thank Nathan Gushwa and Jack Taunton for supplying the small molecule covalent Src inhibitor. We thank Matthew Patricelli for a careful reading of this manuscript and helpful suggestions. This work was supported by the NIH 2R01EB001987.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes four figures, supplemental experimental procedures and chemical characterization, and can be found with this article online.
REFERENCES
- Adamczyk M, Gebler J, Shreder K, Wu J. Region-Selective Labeling of Antibodies as Determined by Electrospray Ionization-Mass Spectrometry (ESI-MS). Bioconjugate Chem. 2000;11:557–563. doi: 10.1021/bc990181y. [DOI] [PubMed] [Google Scholar]
- Allen JJ, Lazerwith SE, Shokat KM. Bio-orthogonal Affinity Purification of Direct Kinase Substrates. J. Am. Chem. Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman KP, Baker J, Wilke M, Lee J, LeRiche T, Seto C, Day S, Chauret N, Ouellet M, Nicoll-Griffith DA. Detection of Covalent Adducts to Cytochrome P450 3A4 Using Liquid Chromatography Mass Spectrometry. Chem. Res. Toxicol. 2004;17:1356–1361. doi: 10.1021/tx0498861. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Eyrich B, Sickmann A, Zahedi RP. Catch me if you can: Mass spectrometry-based phosphoproteomics and quantification strategies. Proteomics. 2011;11:554–570. doi: 10.1002/pmic.201000489. [DOI] [PubMed] [Google Scholar]
- Garber KCA, Carlson EE. Thiol-ene Enabled Detection of Thiophosphorylated Kinase Substrates. ACS Chem. Biol. 2013;8:1671–1676. doi: 10.1021/cb400184v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garske AL, Peters U, Cortesi AT, Perez JL, Shokat KM. Chemical genetic strategy for targeting protein kinases based on covalent complementarity. Proceedings of the National Academy of Sciences. 2011;108:15046–15052. doi: 10.1073/pnas.1111239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R, Harsha HC, Pandey A, Prasad TSK. Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis. Mol. BioSyst. 2012;8:453–463. doi: 10.1039/c1mb05340j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BN, Hutton T, Vinogradov SN. Analysis by Mass Spectrometry. Humana Press; New Jersey: 1996. Analysis of Complex Protein and Glycoprotein Mixtures by Electrospray Ionization Mass Spectrometry with Maximum Entropy Processing. pp. 279–294. [DOI] [PubMed] [Google Scholar]
- Green KD, Pflum MKH. Kinase-Catalyzed Biotinylation for Phosphoprotein Detection. J. Am. Chem. Soc. 2007;129:10–11. doi: 10.1021/ja066828o. [DOI] [PubMed] [Google Scholar]
- Gushwa NN, Kang S, Chen J, Taunton J. Selective Targeting of Distinct Active Site Nucleophiles by Irreversible Src-Family Kinase Inhibitors. J. Am. Chem. Soc. 2012;134:20214–20217. doi: 10.1021/ja310659j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeer S, Heck AJ. The phosphoproteomics data explosion. Current Opinion in Chemical Biology. 2009;13:414–420. doi: 10.1016/j.cbpa.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Maly DJ, Allen JA, Shokat KM. A Mechanism-Based Cross-Linker for the Identification of Kinase–Substrate Pairs. J. Am. Chem. Soc. 2004;126:9160–9161. doi: 10.1021/ja048659i. [DOI] [PubMed] [Google Scholar]
- Manning G. The Protein Kinase Complement of the Human Genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Means GE, Feeney RE. Reductive alkylation of proteins. Analytical Biochemistry. 1995;224:1–16. doi: 10.1006/abio.1995.1001. [DOI] [PubMed] [Google Scholar]
- Parang K, Kohn JA, Saldanha SA, Cole PA. Development of photo-crosslinking reagents for protein kinase–substrate interactions. FEBS Letters. 2002;520:156–160. doi: 10.1016/s0014-5793(02)02778-3. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, et al. In Situ Kinase Profiling Reveals Functionally Relevant Properties of Native Kinases. Chemistry & Biology. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional Interrogation of the Kinome Using Nucleotide Acyl Phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statsuk AV, Maly DJ, Seeliger MA, Fabian MA, Biggs WH, Lockhart DJ, Zarrinkar PP, Kuriyan J, Shokat KM. Tuning a Three-Component Reaction For Trapping Kinase Substrate Complexes. J. Am. Chem. Soc. 2008;130:17568–17574. doi: 10.1021/ja807066f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwal S, Pflum MKH. Phosphorylation-Dependent Kinase–Substrate Cross-Linking. Angewandte Chemie. 2010;122:1671–1674. doi: 10.1002/anie.200905244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till JH, Annan RS, Carr SA, Miller WT. Use of synthetic peptide libraries and phosphopeptide-selective mass spectrometry to probe protein kinase substrate specificity. The Journal of Biological Chemistry. 1994;269:7423–7428. [PubMed] [Google Scholar]
- Tsou H-R, Mamuya N, Johnson BD, Reich MF, Gruber BC, Ye F, Nilakantan R, Shen R, Discafani C, DeBlanc R, et al. 6-Substituted-4-(3-bromophenylamino)quinazolines as Putative Irreversible Inhibitors of the Epidermal Growth Factor Receptor (EGFR) and Human Epidermal Growth Factor Receptor (HER- 2) Tyrosine Kinases with Enhanced Antitumor Activity. J. Med. Chem. 2001;44:2719–2734. doi: 10.1021/jm0005555. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Gehring AM, Weinreb PH, Quadri LE, Flugel RS. Post-translational modification of polyketide and nonribosomal peptide synthases. Current Opinion in Chemical Biology. 1997;1:309–315. doi: 10.1016/s1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- Weber RE, Malte H, Braswell EH, Oliver RWA, Green BN, Sharma PK, Kuchumov A, Vinogradov SN. Mass Spectrometic Composition, Molecular Mass and Oxygen Binding of Macrobdella decora Hemoglobin and its Tetramer and Monomer Subunits. Journal of Molecular Biology. 1995;251:703–720. doi: 10.1006/jmbi.1995.0466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.