Abstract
Cases of H7N9 human infection were caused by a novel, avian-origin H7N9 influenza A virus that emerged in eastern China in 2013. Clusters of human disease were identified in many cities in China, with mortality rates approaching 30%. Pandemic concerns were raised, as historically, influenza pandemics were caused by introduction of novel influenza A viruses into immunologically naïve human population. Currently, there are no approved human vaccines for H7N9 viruses. Recombinant protein vaccine approaches have advantages in safety and manufacturing. In this review, we focused on evaluation of the expression of recombinant hemagglutinin (rHA) proteins as candidate vaccines for H7N9 influenza, with the emphasis on the role of oligomeric and particulate structures in immunogenicity and protection. Challenges in preparation of broadly protective influenza vaccines are discussed, and examples of broadly protective vaccines are presented including rHA stem epitope vaccines, as well as recently introduced experimental multi-HA VLP vaccines.
Introduction
The H7N9 virus belongs to influenza A viruses (family Orthomyxoviridae). Cases of H7N9 human infection caused by an avian-origin H7N9 virus emerged in eastern China in 2013 [1,2]. This raised pandemic concerns as historically, previous pandemics were caused by introduction of new influenza A viruses into immunologically naïve human population [3]. Chicken H9N2 viruses with similarity to A/brambling/Beijing/16/2012-like viruses provided all six of the novel reassortant’s internal genes [1,3]. The gene encoding hemagglutinin (HA) shared the highest identity with A/duck/Zhejiang/12/2011 (H7N3, subtype ZJ12), while the gene encoding neuraminidase (NA) protein was most closely related to A/wild bird/Korea/A14/2011 (H7N9, subtype KO14). Thus, phylogenetic results indicated that novel H7N9 virus was a triple reassortant [1].
Human H7N9 cases were identified in many cities in China, with two epidemiological peaks of infection in 2013–2014 [4]. Since 2013, influenza surveillance at live poultry markets routinely detected H7N9 virus [5]. Human infection with avian influenza A (H7N9) virus is associated mainly with the exposure to infected poultry [5,6]. The mode of subtype H7N9 virus transmission between avian species remains largely unknown, with various wild birds suggested as a source of transmission [5].
Influenza A virus particles are pleomorphic, mostly spherical in shape and approximately 80–120 nm in diameter. Ultrastructural changes in the human lung cells infected with H7N9 virus have been studied by electron microscopy [7]. One characteristic of H7N9 viral infection was the formation of numerous tubular structures within the nucleus and the cytoplasm, which have previously been observed for some influenza A viruses including the 2009 pandemic (H1N1) virus [7]. In general, the envelope of the influenza A virus is made of a lipid bilayer derived from the host cell and contains glycoprotein spikes of two types, hemagglutinin (HA), a ~14 nm long trimer, and neuraminidase (NA), a ~16 nm long tetramer [8,9] (Figure 1).
Figure 1.
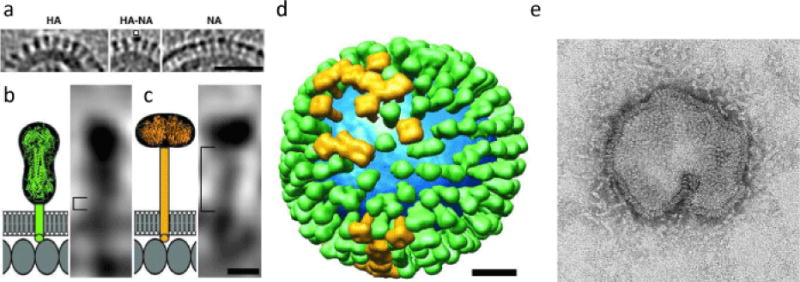
Distributions and shape-based differentiation of HA and NA spikes. (a) HA cluster (Left); single NA (marked) in a cluster of HAs (Center); and cluster of mainly NA spikes (Right). (Scale bar, 50 nm) (b and c) The stem lengths of HA and NA (square brackets in b and c, respectively). The structures of the stems, trans-membrane domains, and C-terminal endodomain tails are shown schematically. Molecules in the matrix layer are shown as oval shapes in a monolayer with a spacing of ≈ 4 nm (Scale bar, 5 nm.) (d) Model of the distribution of glycoprotein HA (green) and NA (gold) on a single influenza virion. The lipid bilayer is blue. (Scale bar 20 nm.) From [9], with publisher’s permission (Copyright (2006) National Academy of Sciences, U.S.A.). (e) Spherical/pleomorphic influenza A H7N9 virus particle, by electron microscopy. From http://www.cdc.gov/flu/avianflu/h7n9-images.htm.
It has been estimated that a spherical virion of average diameter 120 nm has ~375 spikes [9]. The HA protein is responsible for virus attachment to the cell receptors and subsequent fusion with the host cell membrane. It is made in the infected cells as HA0 precursor that is cleaved posttranslationally by cellular proteases into HA1 and HA2 subunits. Cleavage exposes the hydrophobic N-terminus of HA2, which mediates fusion. There is a direct link between cleavage and virulence of avian influenza viruses [10]. Highly pathogenic H5 and H7 viruses contain multiple basic amino acid cleavage sites between HA1 and HA2, which can be cleaved by furin and PC6 proteases [11] in many host cells and organs that can lead to efficient spread of the virus and severe disease in humans. In contrast, HA of low pathogenicity viruses does not have furin cleavage site (Figure 2). In this sense, the 2013 H7N9 viruses are considered “low pathogenicity” and they are often asymptomatic in birds although they can cause severe disease in humans.
Figure 2.

Alignment of H5 protein of the high pathogenicity A/Viet Nam/1203/2004 (H5N1) virus (designated h5) to the H7 protein of the low pathogenicity A/Shanghai/2/2013 or A/Anhui/1/2013 (H7N9) virus (designated h7). Alignment was done using ClustalW2 software with default parameters. Furin protease cleavage site between HA1 and HA2 in the H5 is indicated in grey colour.
After infection, influenza A virus induces both innate and adaptive immune responses. The virus uses various strategies to evade innate immune responses and recognition by components of the humoral and cellular immunity, which may result in reduced clearing of the virus and virus-infected cells [12,13]. Three families of pattern recognition receptors, toll-like receptors (TLRs), retinoic acid-inducible gene 1 protein like helicases (RLRs) and nucleotide-binding domain and leucine-rich-repeat-containing proteins (NLRs), are involved in recognition of influenza virus and they cooperate to respond to the virus in cell culture or in vivo [14].
Vaccines are also designed to induce broad spectrum of immune response. However, although the role of the innate and adaptive immune responses including CD4+ and CD8+ T cells in recovery from influenza infection including H7N9 are important and well documented [15], we will focus in this review mostly on the antibody-mediated protective immunity. Despite influenza vaccines were approved over 60 years ago, the hemagglutination-inhibition serum antibody titer is currently the only universally accepted immune correlate of protection against influenza [16].
The surface envelope proteins generally represent the best targets for vaccines. HA represents the main target for vaccine development. Induction of immune responses against the surface envelope proteins has advantages, because virus-neutralizing response to the viral envelope proteins would prevent early steps of viral infection. Human monoclonal antibodies targeting the HA glycoprotein can neutralize H7N9 influenza virus [17]. The neuraminidase (NA) and M2 also localize in the viral envelope. The NA protein has a sialidase activity that facilitates release of the progeny virions from infected cells. In addition, NA can facilitate virus attachment to the epithelial cells by removal of sialic acid from the mucin layer [18,19]. The M2 protein is a minor component of the envelope that has been implicated in the ion channel activity and efficient uncoating of incoming viruses during the infection cycle.
Electron microphotograph of H7N9 particle is shown (Figure 1, e). As of February 23, 2015, the World Health Organization (WHO) had reported 571 infections and 212 deaths from H7N9 virus, mostly in eastern China [20].
Cases of H7N9 infection in travellers have also been reported [4,21,22]. However, no considerable human-to-human transmission has been described. The factors that allow interspecies transmission but limit human-to-human transmission are not understood. Recent study has shown that A/Anhui/1/2013 (H7N9) influenza virus infection in chickens (natural hosts) is asymptomatic and that it generates a high genetic diversity. In contrast, virus diversity was tightly restricted in infected ferrets, thus limiting further adaptation to a transmissible form. Experimental airborne transmission in ferrets was accompanied by the mutations in PB1, NP and NA genes [6].
Genetic diversity of the 2013–14 human isolates of influenza H7N9 in China has been studied [23]. Two distinct phylogenetic groups of influenza H7N9 currently circulate in China and cause infections in humans as a consequence of cross-species transfer from the avian species. The expansion to different geographic areas and persistence of H7N9 viruses in live poultry market demands effective vaccines for protection against H7N9 viruses.
Prophylactic vaccination is the best way to reduce impact of influenza. However, there are currently no approved human vaccines for H7N9 viruses. Experimental vaccines have been developed for H7N9 and other H7 subtype viruses including live attenuated H7N7 and H7N3 vaccines [24–26], inactivated H7N7 vaccine [27], cell-based H7N1 split virus vaccine [28], and viral vector vaccines [29,30]. Recently, immunization with a live attenuated H7N9 influenza experimental vaccine protected mice against lethal challenge [31]. Furthermore, immunization with modified vaccinia virus vector resulted in protection against H7N9 virus challenge in a ferret model [32]. Recombinant H7N9 protein vaccines based on expression of HA in heterologous expression systems have been also reported (see below).
Recombinant protein vaccine approaches have advantages in safety and manufacturing. Currently, the majority of influenza vaccines are manufactured in fertilized eggs; and during the process of virus production, egg-adaptive mutations can occur in the vaccine antigen resulting in loss in vaccine efficacy. Furthermore, in the case of an outbreak of avian influenza or other agricultural disease affecting chickens, the supply of eggs can be diminished.
The recombinant technology does not have the same vulnerabilities to mutations during production or to supply of eggs for vaccine manufacturing. Furthermore, recombinant HA-based vaccines have already been shown to be efficacious against influenza including H7N9 [33–41]. Due to significant variability of the HA proteins, the immune response to HA is expected to be strain-specific and not capable of efficient cross-neutralizing many strains of influenza. Conserved stem domain of HA has been considered as a candidate for the development of broadly protective influenza vaccines capable of protecting against many isolates of the virus [42]. M2 protein is also highly conserved among influenza A viruses and has been considered for the development of a universal vaccine. Conserved inner proteins of the virion such as NP has been also considered as candidates for universal vaccine along with conserved epitopes within HA, NA, and M2 proteins [42–44].
In this review, we will focus on the expression of recombinant hemagglutinin (rHA) as experimental vaccines for H7N9 influenza, with the emphasis on the role of oligomeric and particulate structures on immunogenicity and protection. Challenges in preparation of broadly protective influenza vaccines will be described, and examples of broadly protective vaccines such as HA stem epitope and recently introduced experimental multi-HA VLPs will be reviewed and discussed.
Subunit rHA Vaccines
Epitope rHA vaccines
Early research suggested that subunit vaccines comprising purified influenza proteins are well tolerated clinically [45]. Several approaches for recombinant subunit H7N9 vaccines are being developed including epitope vaccines, full-length HA vaccines, and virus-like particles (VLPs). B- and T-cell epitopes for many influenza viruses have been reported in the literature, including a number of protective epitopes, and the antigenic architecture of the HA for some influenza viruses has been determined at high resolution [46,47]. Methods for fine mapping of viral epitopes have also been developed [48–53].
Potentially, information regarding protective epitopes can be used for the development of epitope-based vaccines including recombinant proteins and even synthetic peptides. However, despite their high safety profile, attempts to develop vaccines based on short epitopes need to take into account limitations in the immunogenicity of epitope-based immunogens, HLA polymorphism and the high mutation rate of influenza viruses. The most promising application for epitope vaccines is the development of broadly protective influenza vaccines based on highly conserved epitopes.
Recent efforts for the development of such broadly protective influenza epitope vaccine have focused on the highly conserved HA stem domain [42,54] (Figure 3). For example, H3-stem based chimeric HA vaccines were used to protect against H7N9 virus challenge in mice. Chimeric HA constructs protected from viral challenge in the context of different administration routes and a generic oil-in-water adjuvant [54]. In another study, influenza HA stem-fragment also elicited broadly neutralizing antibodies and heterologous protection [55]. The trimeric HA stem-fragment immunogen mimics the native, pre-fusion conformation of HA and binds conformation specific broadly neutralizing antibodies with high affinity.
Figure 3.
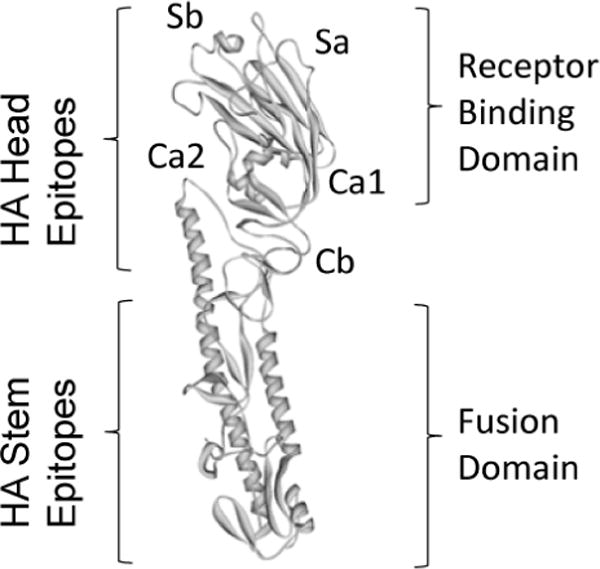
Comparison of influenza HA globular head epitopes and HA stem region epitopes, adapted from [91]. A structural view of the H1 hemagglutinin molecule monomer showing the location of the different globular head epitopes and their localization in the vicinity of the receptor binding domain. The receptor binding domain and fusion domain are also indicated.
This immunogen elicited antibodies that neutralized highly divergent H1, H5 and H3 subtype influenza virus strains in vitro. Stem-derived immunogens conferred protection against a lethal heterologous A/Puerto Rico/8/34 virus challenge in vivo. However, protection against H7N9 virus has not been tested. An advantage of bacterial expression of such designed immunogens allows for rapid scale-up during pandemic outbreaks [55].
Because most epitopes are conformation-dependent, the increased length of the epitope can be advantageous for vaccination. For example, the HA1 neutralizing domain of H5N1 influenza in an optimal conformation induces strong cross-protection [56]. Recombinant proteins covering regions of HA1 of H5N1 virus were prepared that differed in their ability to oligomerize. Modified HA-13-263 construct, which formed an oligomeric conformation, induced the strongest neutralizing antibody response and cross-protection against challenges of two tested H5N1 virus strains. The dimeric and trimeric HA-13-263 recombinant proteins elicited higher neutralizing antibody response and protection than their monomeric counterpart. These results suggested that the oligomeric form of the protein containing the receptor binding domain can be further developed as an effective and safe vaccine for cross-protection against divergent strains of influenza [56].
Immunogenicity and efficacy of recombinant HA2 subunit derived from H7N9 virus was prepared in Escherichia coli and tested in mice [57]. The HA2 subunit was chosen as the vaccine antigen because it is highly conserved among the human H7N9 virus strains. Moreover, in silico analysis predicted two immunogenic regions within the HA2 subunit that may contain potential human B-cell epitopes [57]. In BALB/c mice, intraperitoneal immunization with one or two doses of HA2 with or without imiquimod adjuvant resulted in induction of immune response. In a viral challenge experiment, the 2-dose-adjuvant group had the best survival rate (100%), followed by the 2-dose-no-adjuvant group (90%), the 1-dose-adjuvant group (70%) and the 1-dose-no-adjuvant group (40%). Thus, rHA HA2 subunit protected mice against influenza A H7N9 virus infection [57].
Various expression systems are actively explored for manufacturing of recombinant subunit HA vaccines. For example, recombinant HA was produced in Leishmania tarentolae and its immunogenicity was studied in mice [58]. Soluble HA proteins were secreted into the cell culture medium and were easily purified via a His-Tag domain fused to the proteins. The recombinant HA proteins expressed by L. tarentolae were characterized by dynamic light scattering and were observed to be mostly monomeric. The recombinant HA proteins were immunogenic in mice at a dose of 10 μg when administered twice with an oil-in-water emulsion-based adjuvant. The authors suggest that the L. tarentolae expression system may be an alternative to the current egg-based vaccine production [58]. However it is not clear if immune response is highly protective against live virus challenge.
Full-length rHA vaccines
In general, most of the epitope vaccines were found to be relatively weak immunogens. Efficacy of epitope vaccines could be considerably improved by using adjuvants such as squalene (MF59), which represents an unsaturated aliphatic hydrocarbon, or by using liposomes [59,60]. The full-length HA contains virus-neutralizing epitopes and is included in all currently approved human influenza vaccines, as well as in the majority of experimental vaccines. A trivalent, baculovirus-generated, recombinant subunit HA0 (rHA0) vaccine was safe and immunogenic in a healthy adult population [61]. Thus, recombinant full-length HA alone is capable of protecting against infection with influenza viruses [62,63] including in animal models and in humans [61,64]. Recombinant subunit vaccine under trade name FluBlok recently has been approved by the FDA as the first recombinant human influenza vaccine [64]. Studies have also shown that immune response can depend upon the molecular structure of recombinant HA (rHA) protein [65].
Recombinant HA H7 vaccine against H7N9 virus has been prepared [40]. A previous study has shown that purified rH3 HA forms rosette-shaped subviral particles (SVPs) of approximately 30–50 nm in diameter [66]. We recently prepared the full-length H7 rHA from A/Anhui/1/2013 (H7N9) using a modified purification conditions for rH7 HA [67]. Similarly to a previous study [40], we found that purified rHA is arranged into particulate structures, or subviral particles (SVP), with approximately 20 nm in diameter (Figure 4). The size of rH7 SVP resembled the 22 nm particles of hepatitis B virus (HBV) surface antigen (HBsAg), an effective recombinant vaccine for HBV [68,69]. Thus, it is likely that formation of SVP mimics to some extent the conformational epitopes of the virus, which can explain the high immunogenicity of influenza rH7 HA. Recent data demonstrated that SVPs comprised of the full-length rHA from A/Anhui/1/2013 (H7N9) are immunogenic and protective in animal models [67].
Figure 4.
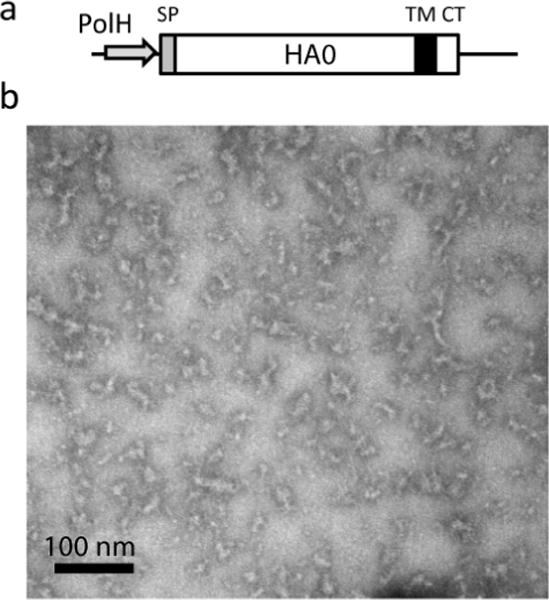
Preparation of the full-length rH7 HA0. (a) Baculovirus expression cassette for rH7 derived from A/Anhui/1/2013 (H7N9) influenza virus. Indicated within rH7 HA0 are locations of the baculovirus polyhedrin promoter (PolH), HA0 signal peptide (SP), trans-membrane domain (TM), and C-terminus (CT). (b) Transmission electron microscopy of purified rH7. The rH7 preparation was negatively stained with 1% phosphotungstic acid and visualized on a Hitachi H-7600 transmission electron microscope. Bar, 100nm.
In general, the advantages of rH7 as a vaccine include high level of safety, rapid manufacturing and the ability to generate highly-purified, well characterized vaccine. HA provides subtype-specific immunity and is effective against infection with a homologous virus. However, one of the limitations of rH7 vaccine is that by definition, it lacks important immunogenic epitopes from other influenza proteins including NA. It has been shown that high level of anti-NA immunity results in cross-protection to viruses expressing divergent neuraminidases [70]. Therefore, inclusion of NA into recombinant vaccine formulation would be justified.
Taken together, previous studies have shown that oligomeric form may be critically important for immunogenicity of rHA. It has been known that for a wide range of pathogens, presentation of vaccine antigens in particulate form has clear advantages over the presentation of soluble antigen alone [71,72]. Therefore, attempts have been made to improve immunogenicity of subunit proteins by oligomerization using adjuvants and virosomes. For example, cylindrical (diameter 80 nm, height 320 nm) poly (lactide-co-glycolide) (PLGA) based PRINT particles were designed to electrostatically bind commercial trivalent injectable influenza vaccine. In a variety of blended PLGA formulations, these particles were safe and showed enhanced immune responses to influenza HA in murine models [73].
Furthermore, liposome formulations were used to improve vaccine immunogenicity. The combination of subunits of the influenza virus with a liposome is called a virosome. The viral antigens HA and NA can be anchored into the nanoparticle or liposome lipid layer, thus resembling natural influenza virus particle [74]. Commercial virosomal influenza vaccines have been available in Europe, such as Inflexal V, licensed for all age groups. Inflexal V represents an inactivated vaccine which has virosomes in its formulation. To the best of our knowledge, there were no published reports regarding preparation of virosomal formulations as H7N9 vaccine.
Influenza VLP Vaccines
Influenza VLP vaccines have been demonstrated as a promising approach to co-express multiple influenza antigens in the native conformation and to elicit highly protective immune response capable of protecting from influenza infections [34,69]. During the course of infection in virus-infected host cells, structural proteins and genomic nucleic acids of viruses including influenza virus assemble into progeny virion particles, which are released from the infected cells. During past decade, considerable progress has been achieved in expression of various types of VLPs for many viruses, as reviewed elsewhere [69]. For example, expression of L1 protein of human papilloma virus (HPV) in cells results in production of HPV VLPs [75,76]. Influenza structural proteins also maintain the intrinsic ability to self-assemble into VLPs following expression of the structural genes in various cell culture systems [37,69,77,78]. The size and morphology of such self-assembled influenza VLPs resemble those of influenza virions. However, VLPs are non-infectious, because in the cell culture expression systems, viral proteins self-assemble in the absence of the viral genetic material. This ensures intrinsic safety of recombinant VLPs.
Recombinant virus-like particles (VLPs) have been previously shown to be safe and effective vaccines for influenza. In many cases, recombinant VLP were comprised of HA and additional influenza proteins such as NA and/or M1 [34,35,37,78,79]. VLPs comprised of only HA, NA, and M1 proteins [37] have been shown to elicit highly efficient protective immune responses, which in some cases exceeded immune responses elicited by rHA [36,80]. In influenza VLPs, HA and other proteins are arranged into ~120 nm pleomorphic particles that morphologically resemble influenza virions, which is viewed as advantageous feature for immunogenicity and efficacy of VLP vaccines [34,69,81].The observed high immunogenicity of influenza VLP vaccines has been attributed to the organization of HA into regular, highly repeated patterns in the virus resembling structures that favor activation of immune effector functions [34,69].
The early influenza VLP studies have utilized cloning of three strain-specific genes, HA, NA, and M1, into a single baculovirus vector in order to generate VLPs [35,37,80,82]. Infection with such baculovirus vector resulted in a uniform co-expression of all three genes in Sf9 cells. The need to clone several strain-specific genes can delay cloning, vector preparation and production process of the VLP vaccine, especially, if sequences of all three genes are not available. Therefore, in some studies, a “generic” M1 was used for VLP production process, while HA and NA were derived from the vaccine-relevant strain of interest. Furthermore, in order to speed up preparation of VLPs, co-infection with several baculovirus vectors, each expressing only one influenza gene, was used for H7N9 influenza VLP preparation.
For example, H7N9 VLPs were prepared using baculovirus expression and immunogenicity and protective efficacy of a H7N9 VLP vaccine was evaluated in the ferret challenge model [41]. In VLPs, the full-length HA and NA were derived from A/Anhui/1/2013 (H7N9) virus, while M1 was from A/Indonesia/5/2005 (H5N1) virus. VLPs were made by co-infection of Sf9 cells with three distinct baculovirus vectors, each expressing one influenza gene (H7, N9 or M1). VLPs comprised of H7, N9 and M1 proteins were harvested from Sf9 growth medium. Purified recombinant H7N9 VLPs morphologically resembled influenza virions and elicited high-titer serum hemagglutination inhibition (HI) and neutralizing antibodies specific for A/Anhui/1/2013 (H7N9) virus. H7N9 VLP-immunized ferrets subsequently were challenged with homologous virus. Vaccinated animals displayed considerable reductions in fever, weight loss, and virus shedding as compared to these parameters in unimmunized control ferrets. H7N9 VLPs were also effective in protecting against lung and tracheal infection. The addition of either ISCOMATRIX or Matrix-M1 adjuvant improved immunogenicity and protection of the VLP vaccine against challenge with H7N9 virus. These results provide support for the development of a safe and effective human VLP vaccine against avian influenza H7N9 virus with pandemic potential [41].
Furthermore, we have prepared H7N9 VLPs, in which HA was derived from A/Shanghai/2/2013 (H7N9) virus, while both NA and M1 proteins were derived from A/Puerto Rico/8/1934 (H1N1) virus, a standard virus that is often used for preparation of influenza reassortant viruses used in human influenza vaccine production (Figure 5). By electron microscopy, VLP envelope was observed, similarly to the native H7N9 virus (Figure 1e). For VLP preparation, in these cases all three genes were expressed from a single baculovirus vector. The advantage of this approach is that it achieves a uniform co-expression of three genes in Sf9 cells, and that it allows cloning of a single vaccine-relevant antigen (rHA) into a prefabricated baculovirus transfer vector containing “standard” NA and M1 genes. This considerably facilitates gene cloning and baculovirus vector preparation thus accelerating influenza VLP vaccine production process.
Figure 5.
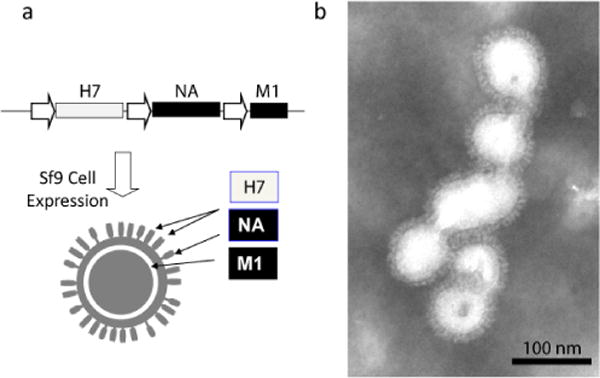
Preparation of influenza VLPs containing the full-length rH7 HA0, as well as NA and M1 proteins. (a) Baculovirus expression cassette and schematic composition of VLP comprised of rH7 protein derived from A/Shanghai/2/2013 (H7N9) virus, as well as NA and M1 proteins derived from A/Puerto Rico/8/1934 (H1N1) virus. Indicated within the baculovirus expression cassette are rH7 HA0 (open box), NA and M1 (solid boxes), as well as baculovirus polyhedrin promoters (arrows). (b) Transmission electron microscopy of purified rH7 VLPs. The VLP preparation was negatively stained with 1% phosphotungstic acid and visualized on a Hitachi H-7600 transmission electron microscope. Bar, 100nm.
In a recent study, a recombinant influenza vaccine was also developed by expressing HA of H7N9 (A/Shanghai/2/2013) on the surface of recombinant baculovirus. Although this approach would not result in a typical influenza VLP, the spatial conformation of H7 in the resulting vaccine is expected to be similar to that in an influenza virion or in a VLP. Mice were immunized twice either intranasally or subcutaneously with the vaccine. Immunogenicity and cross-protective efficacy of the vaccine was assessed against H7N9 or H7N7 subtype challenges. The authors concluded that intranasal administration of H7 protein expressed on the baculovirus envelope can be an alternative way to prime the immune system against influenza infection during a pandemic situation [83].
Potential pandemic concerns were raised for H7N9 virus [1,3]. However, in addition to H7N9 virus, multiple other viruses with pandemic potential including H5N1 and H9N2 subtypes continue to circulate in birds and other animals. Furthermore, additionally to the potential of causing lethal human disease and pandemics, avian influenza can cause devastating epizootics in poultry that can threaten food supply and safety, and also result in reduced capability to manufacture egg-derived vaccines worldwide including those against influenza, yellow fever, mumps and measles viruses. Therefore, vaccines capable of protecting against multiple potentially pandemic influenza strains are needed. Candidate vaccines against H5, H7, and H9 viruses have been prepared in the past [84]. In order to enhance pandemic preparedness, inactivated H5N1 vaccines have been also developed and approved by regulatory agencies [85] including recently prepared cell culture-derived Flucelvax vaccine. In recent studies, we described novel, multi-subtype VLPs containing three subtypes of the full-length rHA within the envelope [79,86]. Recombinant multi-HA VLPs represent a conceptually novel broadly protective influenza vaccine, which is designed to elicit specific immunity to multiple influenza strains and does not require blending of individual vaccines to prepare a trivalent formulation. Initially, a baculovirus vector was configured to co-express the H2, H7, and H5 genes derived from potentially pandemic A/Swine/Missouri/4296424/2006 (H2N3), A/New York/107/2003 (H7N2) and A/Viet Nam/1203/2004 (H5N1) viruses [86]. NA and M1 gene sequences were derived from A/Indonesia/05/2005 (H5N1). VLPs that contained H2, H7 and H5 genes were made in Sf9 cells. We have also prepared VLPs that contained the full-length rHA genes from seasonal influenza strains A/New Caledonia/20/1999 (H1N1), A/New York/55/2004 (H3N2) and B/Shanghai/361/2002 [86]. Again, NA and M1 gene sequences were derived from A/Indonesia/05/2005 (H5N1). By immunoelectron microscopy, we showed that multiple subtypes of HA can co-localize within VLP structure. Finally, such multi-subtype VLPs induced highly protective immune responses against pandemic and seasonal strains of influenza when administered to ferrets intramuscularly [86].
In another study, we prepared a triple-subtype VLP to contain the H5, H7, and H9 proteins derived from potentially pandemic influenza viruses [79]. A baculovirus vector was prepared to co-express the H5, H7, and H9 genes derived from A/Viet Nam/1203/2004 (H5N1), A/New York/107/2003 (H7N2) and A/Hong Kong/33982/2009 (H9N2) viruses, respectively, as well as neuraminidase (NA) and matrix (M1) genes from A/Puerto Rico/8/1934 (H1N1) virus. VLPs were prepared in Sf9 cells (Figure 6), and immunogenicity and efficacy of such H5/H7/H9 VLPs were evaluated in ferrets following intranasal (i.n) vaccination. We showed that i.n vaccination with the H5/H7/H9 triple-subtype VLP induced immune responses and protected ferrets from experimental challenges with three avian influenza viruses [79].
Figure 6.
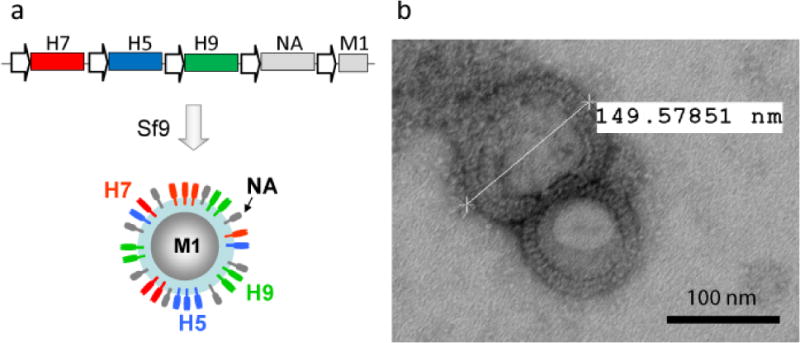
Preparation of influenza multi-HA VLPs. (a) Baculovirus expression cassette and schematic composition of multi-HA VLPs [79]. The H5, H7, and H9 genes were derived from A/Viet Nam/1203/2004 (H5N1), A/New York/107/2003 (H7N2) and A/Hong Kong/33982/2009 (H9N2) viruses, respectively. Neuraminidase (NA) and matrix (M1) proteins were derived from A/Puerto Rico/8/1934 (H1N1) virus (shown in gray). (b) Transmission electron microscopy of purified multi-HA VLPs. The VLP preparation was negatively stained with 1% phosphotungstic acid and visualized on a Hitachi H-7600 transmission electron microscope. Size of VLP is indicated. Bar, 100nm.
We have recently made another multi-HA VLPs that co-localize A/Vietnam/1203/2004 (H5N1), A/Hong Kong/33982/2009 (H9N2) and A/Shanghai/2/2013 (H7N9) rHA proteins. Animal studies are being conducted.
Thus, multi-subtype VLP that co-localize multiple subtypes of the full-length rHA within the same particle can be used to protect against multiple influenza viruses following intramuscular (i.m) or i.n. vaccination. Since this multi-HA VLP is a novel approach, additional research and pre-clinical testing is needed. For example, additional data are needed regarding immunogenicity and protective effects of triple-subtype VLPs given i.m or i.n against heterologous virus challenges. It has been previously shown that mucosally vaccinated ferrets had broader protection against challenge with unrelated strains than parenterally vaccinated animals [35].
In summary, multi-subtype VLPs can be a promising approach for pre-pandemic vaccine stockpiling purposes in addition to, or instead of, current H5N1-only vaccine. In the case of a pandemic involving influenza viruses, such multi-HA VLPs can be used as a first line of defence during the time period that is required to make a specialized vaccine against a specific pandemic virus strain.
Conclusion
Novel H7N9 virus causes severe disease in people and represents a pandemic threat. Efforts are being made to develop effective vaccines against H7N9 virus. These efforts included live attenuated viruses, inactivated vaccines, virus vectors, and recombinant proteins. Recombinant rHA vaccines included epitopes derived from H7, the full-length rH7, as well as VLPs containing rH7. The tendency was observed that oligomeric and particulated rH7 displayed higher immunogenicity. Therefore, efforts have been made to increase the oligomeric state of rHA by using genetic engineering methods, advanced adjuvants and VLPs. Two types of recombinant H7 rHA vaccines for H7N9 viruses have been made in Sf9 insect cells using baculovirus expression system. The first type is recombinant rHA including Flublok technology that has recently been approved by the Food and Drug Administration (FDA) for human use [64]. Another type of recombinant influenza vaccine is recombinant VLP comprised of H7 rHA and additional influenza proteins such as NA and/or M1 [34,35,37,78,79]. VLPs have been shown to elicit highly efficient protective immune responses, which in some cases exceeded immune responses elicited by rHA [36,80]. In the influenza VLPs, HA and other proteins are arranged into ~120 nm pleomorphic particles that morphologically resemble influenza virions, which is viewed as advantageous for immunogenicity and efficacy of VLP vaccines [34,69,81]. However, rHA is also immunogenic and protective in animal models and in humans [61,64]. Immunogenicity of rHA at least in part can be explained by the observed arrangement of rHA into subviral particles, SVP, with up to 30–50 nm in diameter and containing multiple rHA molecules [66]. Thus, it appears that successful recombinant vaccines including L1-based HPV, HBsAg-based HBV, and even rHA-based influenza represent particulate structures such as VLPs or SVP, with oligomeric/particulate structures likely responsible for high immunogenicity of these vaccines.
For preparation of influenza vaccine in Sf9 cells using baculovirus expression system, rapid manufacturing is important including production of recombinant baculovirus vector stocks. Rapid manufacturing is especially important in the case of novel influenza virus outbreaks with a pandemic potential such as H7N9 virus. Recent introduction of automated virus counters allowed determination of baculovirus titer in minutes, instead of days if using traditional plaque assay. For example, ViroCyt® virus counter that has been used for quantitation of various viruses [87] has been configured for quantitation of recombinant baculoviruses. Furthermore, methods have been described to improve the turnaround time for manufacturing by designing VLPs using HA and NA [41] or HA only (Figure 5) from the vaccine-relevant strain, while using the same generic M1 (or NA and M1) proteins for various VLP preparations.
In the future H7N9 vaccine developments, the challenge of genetic diversity of H7N9 viruses needs to be addressed [88]. It will be important to test the efficacy of vaccines against different genotypes of H7N9 viruses, the spread of which may follow the same pattern as H5N1 and H9N2 cases in China [3,89,90]. Potentially, the challenge of genetic diversity of H7N9 viruses can be addressed by using multi-HA VLP vaccine approach. In addition, multi-HA VLPs can also be a useful tool to prepare multi-subtype VLP vaccines to protect against H7, H5 and H9 subtypes, as well as against other potentially pandemic and epidemic subtypes of influenza viruses [79,86].
Acknowledgments
This work was supported in part by grants 1R01AI111532 (NIH NIAID) and 2013-33610-21041 (USDA NIFA). The opinions and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agencies.
References
- 1.Gao R, Cao B, Hu Y, Feng Z, Wang D, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 2.Chen F, Li J, Sun B, Zhang H, Zhang R, et al. Isolation and characteristic analysis of a novel strain H7N9 of avian influenza virus A from a patient with influenza-like symptoms in China. Int J Infect Dis. 2015;33:130–131. doi: 10.1016/j.ijid.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Pu J, Wang S, Yin Y, Zhang G, Carter RA, et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A. 2015;112:548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang JR, Kuo CY, Huang HY, Wu FT, Huang YL, et al. Characterization of influenza A (H7N9) viruses isolated from human cases imported into Taiwan. PLoS One. 2015;10:e0119792. doi: 10.1371/journal.pone.0119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones JC, Sonnberg S, Webby RJ, Webster RG. Influenza A(H7N9) virus transmission between finches and poultry. Emerg Infect Dis. 2015;21:619–628. doi: 10.3201/eid2104.141703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaraket H, Baranovich T, Kaplan BS, Carter R, Song MS, et al. Mammalian adaptation of influenza A(H7N9) virus is limited by a narrow genetic bottleneck. Nat Commun. 2015;6:6553. doi: 10.1038/ncomms7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terrier O, Carron C, Cartet G, Traversier A, Julien T, et al. Ultrastructural fingerprints of avian influenza A (H7N9) virus in infected human lung cells. Virology. 2014:456–457. 39–42. doi: 10.1016/j.virol.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, et al. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci U S A. 2013;110:4592–4597. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horimoto T, Kawaoka Y. Pandemic threat posed by avian influenza A viruses. Clin Microbiol Rev. 2001;14:129–149. doi: 10.1128/CMR.14.1.129-149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horimoto T, Nakayama K, Smeekens SP, Kawaoka Y. Proprotein-processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J Virol. 1994;68:6074–6078. doi: 10.1128/jvi.68.9.6074-6078.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Sandt CE, Kreijtz JH, Rimmelzwaan GF. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses. 2012;4:1438–1476. doi: 10.3390/v4091438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale BG, Albrecht RA, García-Sastre A. Innate immune evasion strategies of influenza viruses. Future Microbiol. 2010;5:23–41. doi: 10.2217/fmb.09.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S, Metcalf JP, Wu W. Innate immune response to influenza virus. Curr Opin Infect Dis. 2011;24:235–240. doi: 10.1097/QCO.0b013e328344c0e3. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Wan Y, Qiu C, Quiñones-Parra S, Zhu Z, et al. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat Commun. 2015;6:6833. doi: 10.1038/ncomms7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reber A, Katz J. Immunological assessment of influenza vaccines and immune correlates of protection. Expert Rev Vaccines. 2013;12:519–536. doi: 10.1586/erv.13.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Wang J, Bao L, Guo L, Zhang W, et al. Human monoclonal antibodies targeting the haemagglutinin glycoprotein can neutralize H7N9 influenza virus. Nat Commun. 2015;6:6714. doi: 10.1038/ncomms7714. [DOI] [PubMed] [Google Scholar]
- 18.Palese P, Tobita K, Ueda M, Compans RW. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61:397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 19.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO. Summary of surveillance and investigation findings. Human cases of avian influenza A (H7N9) virus infection 2015 [Google Scholar]
- 21.William T, Thevarajah B, Lee SF, Suleiman M, Jeffree MS, et al. Avian influenza (H7N9) virus infection in Chinese tourist in Malaysia, 2014. Emerg Infect Dis. 2015;21:142–145. doi: 10.3201/eid2101.141092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin PH, Chao TL, Kuo SW, Wang JT, Hung CC, et al. Virological, serological, and antiviral studies in an imported human case of avian influenza A(H7N9) virus in Taiwan. Clin Infect Dis. 2014;58:242–246. doi: 10.1093/cid/cit638. [DOI] [PubMed] [Google Scholar]
- 23.Farooqui A, Leon AJ, Huang L, Wu S, et al. Genetic diversity of the 2013–14 human isolates of influenza H7N9 in China. BMC Infect Dis. 2015;15:109. doi: 10.1186/s12879-015-0829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min JY, Vogel L, Matsuoka Y, Lu B, Swayne D, et al. A live attenuated H7N7 candidate vaccine virus induces neutralizing antibody that confers protection from challenge in mice, ferrets, and monkeys. J Virol. 2010;84:11950–11960. doi: 10.1128/JVI.01305-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talaat KR, Karron RA, Callahan KA, Luke CJ, DiLorenzo SC, et al. A live attenuated H7N3 influenza virus vaccine is well tolerated and immunogenic in a Phase I trial in healthy adults. Vaccine. 2009;27:3744–3753. doi: 10.1016/j.vaccine.2009.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babu TM, Levine M, Fitzgerald T, Luke C, Sangster MY, et al. Live attenuated H7N7 influenza vaccine primes for a vigorous antibody response to inactivated H7N7 influenza vaccine. Vaccine. 2014;32:6798–6804. doi: 10.1016/j.vaccine.2014.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Couch RB, Patel SM, Wade-Bowers CL, Niño D. A randomized clinical trial of an inactivated avian influenza A (H7N7) vaccine. PLoS One. 2012;7:e49704. doi: 10.1371/journal.pone.0049704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox RJ, Major D, Hauge S, Madhun AS, Brokstad KA, et al. A cell-based H7N1 split influenza virion vaccine confers protection in mouse and ferret challenge models. Influenza and other respiratory viruses. 2009;3:107–117. doi: 10.1111/j.1750-2659.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goff PH, Krammer F, Hai R, Seibert CW, Margine I, et al. Induction of cross-reactive antibodies to novel H7N9 influenza virus by recombinant Newcastle disease virus expressing a North American lineage H7 subtype hemagglutinin. J Virol. 2013;87:8235–8240. doi: 10.1128/JVI.01085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreijtz JH, Wiersma LC, De Gruyter HL, Vogelzang-van Trierum SE, van Amerongen G, et al. A Single Immunization With Modified Vaccinia Virus Ankara-Based Influenza Virus H7 Vaccine Affords Protection in the Influenza A(H7N9) Pneumonia Ferret Model. J Infect Dis. 2014;211:791–800. doi: 10.1093/infdis/jiu528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Zhao J, Wang C, Duan Y, Zhao Z, et al. Immunization with a live attenuated H7N9 influenza vaccine protects mice against lethal challenge. PLoS One. 2015;10:e0123659. doi: 10.1371/journal.pone.0123659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreijtz JH, Wiersma LC, De Gruyter HL, Vogelzang-van Trierum SE, van Amerongen G, et al. A single immunization with modified vaccinia virus Ankara-based influenza virus H7 vaccine affords protection in the influenza A(H7N9) pneumonia ferret model. J Infect Dis. 2015;211:791–800. doi: 10.1093/infdis/jiu528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galarza JM, Latham T, Cupo A. Virus-like particle (VLP) vaccine conferred complete protection against a lethal influenza virus challenge. Viral Immunol. 2005;18:244–251. doi: 10.1089/vim.2005.18.244. [DOI] [PubMed] [Google Scholar]
- 34.Kang SM, Pushko P, Bright RA, Smith G, Compans RW. Influenza virus-like particles as pandemic vaccines. Curr Top Microbiol Immunol. 2009;333:269–289. doi: 10.1007/978-3-540-92165-3_14. [DOI] [PubMed] [Google Scholar]
- 35.Perrone LA, Ahmad A, Veguilla V, Lu X, Smith G, et al. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. J Virol. 2009;83:5726–5734. doi: 10.1128/JVI.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bright RA, Carter DM, Daniluk S, Toapanta FR, Ahmad A, et al. Influenza virus-like particles elicit broader immune responses than whole virion inactivated influenza virus or recombinant hemagglutinin. Vaccine. 2007;25:3871–3878. doi: 10.1016/j.vaccine.2007.01.106. [DOI] [PubMed] [Google Scholar]
- 37.Pushko P, Tumpey TM, Bu F, Knell J, Robinson R, et al. Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine. 2005;23:5751–5759. doi: 10.1016/j.vaccine.2005.07.098. [DOI] [PubMed] [Google Scholar]
- 38.Quan FS, Vunnava A, Compans RW, Kang SM. Virus-like particle vaccine protects against 2009 H1N1 pandemic influenza virus in mice. PLoS One. 2010;5:e9161. doi: 10.1371/journal.pone.0009161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross TM, Mahmood K, Crevar CJ, Schneider-Ohrum K, Heaton PM, et al. A trivalent virus-like particle vaccine elicits protective immune responses against seasonal influenza strains in mice and ferrets. PLoS One. 2009;4:e6032. doi: 10.1371/journal.pone.0006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buckland B, Boulanger R, Fino M, Srivastava I, Holtz K, et al. Technology transfer and scale-up of the Flublok recombinant hemagglutinin (HA) influenza vaccine manufacturing process. Vaccine. 2014;32:5496–5502. doi: 10.1016/j.vaccine.2014.07.074. [DOI] [PubMed] [Google Scholar]
- 41.Liu YV, Massare MJ, Pearce MB, Sun X, Belser JA, et al. Recombinant virus-like particles elicit protective immunity against avian influenza A(H7N9) virus infection in ferrets. Vaccine. 2015;33:2152–2158. doi: 10.1016/j.vaccine.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol. 2013;3:521–530. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res. 2013;100:567–574. doi: 10.1016/j.antiviral.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 44.Zheng M, Luo J, Chen Z. Development of universal influenza vaccines based on influenza virus M and NP genes. Infection. 2014;42:251–262. doi: 10.1007/s15010-013-0546-4. [DOI] [PubMed] [Google Scholar]
- 45.Nicholson KG, Tyrrell DA, Harrison P, Potter CW, Jennings R, et al. Clinical studies of monovalent inactivated whole virus and subunit A/USSR/77 (H1N1) vaccine: serological responses and clinical reactions. J Biol Stand. 1979;7:123–136. doi: 10.1016/s0092-1157(79)80044-x. [DOI] [PubMed] [Google Scholar]
- 46.Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci U S A. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velkov T, Ong C, Baker MA, Kim H, Li J, et al. The antigenic architecture of the hemagglutinin of influenza H5N1 viruses. Mol Immunol. 2013;56:705–719. doi: 10.1016/j.molimm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Meisel H, Sominskaya I, Pumpens P, Pushko P, Borisova G, et al. Fine mapping and functional characterization of two immuno-dominant regions from the preS2 sequence of hepatitis B virus. Intervirology. 1994;37:330–339. doi: 10.1159/000150397. [DOI] [PubMed] [Google Scholar]
- 49.Bichko V, Schödel F, Nassal M, Gren E, Berzinsh I, et al. Epitopes recognized by antibodies to denatured core protein of hepatitis B virus. Mol Immunol. 1993;30:221–231. doi: 10.1016/0161-5890(93)90051-c. [DOI] [PubMed] [Google Scholar]
- 50.Sominskaya I, Bichko V, Pushko P, Dreimane A, Snikere D, et al. Tetrapeptide QDPR is a minimal immunodominant epitope within the preS2 domain of hepatitis B virus. Immunol Lett. 1992;33:169–172. doi: 10.1016/0165-2478(92)90043-n. [DOI] [PubMed] [Google Scholar]
- 51.Sominskaya I, Pushko P, Dreilina D, Kozlovskaya T, Pumpen P. Determination of the minimal length of preS1 epitope recognized by a monoclonal antibody which inhibits attachment of hepatitis B virus to hepatocytes. Med Microbiol Immunol. 1992;181:215–226. doi: 10.1007/BF00215767. [DOI] [PubMed] [Google Scholar]
- 52.Ulrich R, Pushko P, Kozlovskaya T, Siakkou H, Pumpen P, et al. Precise localization of the epitope of major BLV envelope protein. Acta Virol. 1991;35:302. [PubMed] [Google Scholar]
- 53.Levy R, Forsyth CM, LaPorte SL, Geren IN, Smith LA, et al. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J Mol Biol. 2007;365:196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krammer F, Margine I, Hai R, Flood A, Hirsh A, et al. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol. 2014;88:2340–2343. doi: 10.1128/JVI.03183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mallajosyula VV, Citron M, Ferrara F, Lu X, Callahan C, et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci. 2014;111:2514–2523. doi: 10.1073/pnas.1402766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du L, Zhao G, Sun S, Zhang X, Zhou X, et al. A critical HA1 neutralizing domain of H5N1 influenza in an optimal conformation induces strong cross-protection. PLoS One. 2013;8:e53568. doi: 10.1371/journal.pone.0053568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.To KK, Zhang AJ, Chan AS, Li C, Cai JP, et al. Recombinant influenza A virus hemagglutinin HA2 subunit protects mice against influenza A(H7N9) virus infection. Arch Virol. 2015;160:777–786. doi: 10.1007/s00705-014-2314-x. [DOI] [PubMed] [Google Scholar]
- 58.Pion C, Courtois V, Husson S, Bernard MC, Nicolai MC, et al. Characterization and immunogenicity in mice of recombinant influenza haemagglutinins produced in Leishmania tarentolae. Vaccine. 2014;32:5570–5576. doi: 10.1016/j.vaccine.2014.07.092. [DOI] [PubMed] [Google Scholar]
- 59.Podda A. The adjuvanted influenza vaccines with novel adjuvants: experience with the MF59-adjuvanted vaccine. Vaccine. 2001;19:2673–2680. doi: 10.1016/s0264-410x(00)00499-0. [DOI] [PubMed] [Google Scholar]
- 60.Pushko P, Tumpey TM, Van Hoeven N, Belser JA, Robinson R, et al. Evaluation of influenza virus-like particles and Novasome adjuvant as candidate vaccine for avian influenza. Vaccine. 2007;25:4283–4290. doi: 10.1016/j.vaccine.2007.02.059. [DOI] [PubMed] [Google Scholar]
- 61.Treanor JJ, Schiff GM, Hayden FG, Brady RC, Hay CM, et al. Safety and immunogenicity of a baculovirus-expressed hemagglutinin influenza vaccine: a randomized controlled trial. JAMA. 2007;297:1577–1582. doi: 10.1001/jama.297.14.1577. [DOI] [PubMed] [Google Scholar]
- 62.Treanor JJ, El Sahly H, King J, Graham I, Izikson R, et al. Protective efficacy of a trivalent recombinant hemagglutinin protein vaccine (FluBlok(R)) against influenza in healthy adults: a randomized, placebo-controlled trial. Vaccine. 2011;29:7733–7739. doi: 10.1016/j.vaccine.2011.07.128. [DOI] [PubMed] [Google Scholar]
- 63.King JC, Cox MM, Reisinger K, Hedrick J, Graham I. Evaluation of the safety, reactogenicity and immunogenicity of FluBlok trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy children aged 6–59 months. Vaccine. 2009;27:6589–6594. doi: 10.1016/j.vaccine.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 64.Traynor K. First recombinant flu vaccine approved. Am J Health Syst Pharm. 2013;70:382. doi: 10.2146/news130016. [DOI] [PubMed] [Google Scholar]
- 65.Wei CJ, Xu L, Kong WP, Shi W, Canis K, et al. Comparative efficacy of neutralizing antibodies elicited by recombinant hemagglutinin proteins from avian H5N1 influenza virus. J Virol. 2008;82:6200–6208. doi: 10.1128/JVI.00187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holtz KM, Robinson PS, Matthews EE, Hashimoto Y, McPherson CE, et al. Modifications of cysteine residues in the transmembrane and cytoplasmic domains of a recombinant hemagglutinin protein prevents cross-linked multimer formation and potency loss. BMC Biotechnol. 2014;14:1. doi: 10.1186/s12896-014-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pushko P, Pujanauski L, Sun X, Pearce M, Hidajat R, et al. Recombinant H7 hemagglutinin forms subviral particles that protect mice and ferrets from challenge with H7N9 influenza virus. Vaccine. 2015 doi: 10.1016/j.vaccine.2015.07.026. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu R, Lin Q, Sun Y, Lu X, Qiu Y, et al. Expression, purification, and characterization of hepatitis B virus surface antigens (HBsAg) in yeast Pichia Pastoris. Appl Biochem Biotechnol. 2009;158:432–444. doi: 10.1007/s12010-009-8527-x. [DOI] [PubMed] [Google Scholar]
- 69.Pushko P, Pumpens P, Grens E. Development of virus-like particle technology from small highly symmetric to large complex virus-like particle structures. Intervirology. 2013;56:141–165. doi: 10.1159/000346773. [DOI] [PubMed] [Google Scholar]
- 70.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, et al. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. MBio. 2015;6:e02556. doi: 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aguilar JC, Rodríguez EG. Vaccine adjuvants revisited. Vaccine. 2007;25:3752–3762. doi: 10.1016/j.vaccine.2007.01.111. [DOI] [PubMed] [Google Scholar]
- 72.Singh M, O’Hagan D. Advances in vaccine adjuvants. Nat Biotechnol. 1999;17:1075–1081. doi: 10.1038/15058. [DOI] [PubMed] [Google Scholar]
- 73.Galloway AL, Murphy A, DeSimone JM, Di J, Herrmann JP, et al. Development of a nanoparticle-based influenza vaccine using the PRINT technology. Nanomedicine. 2013;9:523–531. doi: 10.1016/j.nano.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 74.Moser C, Müller M, Kaeser MD, Weydemann U, Amacker M. Influenza virosomes as vaccine adjuvant and carrier system. Expert Rev Vaccines. 2013;12:779–791. doi: 10.1586/14760584.2013.811195. [DOI] [PubMed] [Google Scholar]
- 75.Sasagawa T, Pushko P, Steers G, Gschmeissner SE, Hajibagheri MA, et al. Synthesis and assembly of virus-like particles of human papillomaviruses type 6 and type 16 in fission yeast Schizosaccharomyces pombe. Virology. 1995;206:126–135. doi: 10.1016/s0042-6822(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 76.Kirnbauer R1. Papillomavirus-like particles for serology and vaccine development. Intervirology. 1996;39:54–61. doi: 10.1159/000150475. [DOI] [PubMed] [Google Scholar]
- 77.Latham T, Galarza JM. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J Virol. 2001;75:6154–6165. doi: 10.1128/JVI.75.13.6154-6165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe T, Watanabe S, Neumann G, Kida H, Kawaoka Y. Immunogenicity and protective efficacy of replication-incompetent influenza virus-like particles. J Virol. 2002;76:767–773. doi: 10.1128/JVI.76.2.767-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tretyakova I, Pearce MB, Florese R, Tumpey TM, Pushko P. Intranasal vaccination with H5, H7 and H9 hemagglutinins co-localized in a virus-like particle protects ferrets from multiple avian influenza viruses. Virology. 2013;442:67–73. doi: 10.1016/j.virol.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 80.Pushko P, Tumpey TM, Van Hoeven N, Belser JA, Robinson R, et al. Evaluation of influenza virus-like particles and Novasome adjuvant as candidate vaccine for avian influenza. Vaccine. 2007;25:4283–4290. doi: 10.1016/j.vaccine.2007.02.059. [DOI] [PubMed] [Google Scholar]
- 81.Zabel F, Mohanan D, Bessa J, Link A, Fettelschoss A, et al. Viral particles drive rapid differentiation of memory B cells into secondary plasma cells producing increased levels of antibodies. J Immunol. 2014;192:5499–5508. doi: 10.4049/jimmunol.1400065. [DOI] [PubMed] [Google Scholar]
- 82.Pushko P, Kort T, Nathan M, Pearce MB, Smith G, et al. Recombinant H1N1 virus-like particle vaccine elicits protective immunity in ferrets against the 2009 pandemic H1N1 influenza virus. Vaccine. 2010;28:4771–4776. doi: 10.1016/j.vaccine.2010.04.093. [DOI] [PubMed] [Google Scholar]
- 83.Prabakaran M, Kumar SR, Raj KV, Wu X, He F, et al. Cross-protective efficacy of baculovirus displayed hemagglutinin against highly pathogenic influenza H7 subtypes. Antiviral Res. 2014;109:149–159. doi: 10.1016/j.antiviral.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 84.Couch RB, Decker WK, Utama B, Atmar RL, Niño D, et al. Evaluations for in vitro correlates of immunogenicity of inactivated influenza a H5, H7 and H9 vaccines in humans. PLoS One. 2012;7:e50830. doi: 10.1371/journal.pone.0050830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Neill E, Donis RO. Generation and characterization of candidate vaccine viruses for prepandemic influenza vaccines. Curr Top Microbiol Immunol. 2009;333:83–108. doi: 10.1007/978-3-540-92165-3_4. [DOI] [PubMed] [Google Scholar]
- 86.Pushko P, Pearce MB, Ahmad A, Tretyakova I, Smith G, et al. Influenza virus-like particle can accommodate multiple subtypes of hemagglutinin and protect from multiple influenza types and subtypes. Vaccine. 2011;29:5911–5918. doi: 10.1016/j.vaccine.2011.06.068. [DOI] [PubMed] [Google Scholar]
- 87.Rossi CA, Kearney BJ, Olschner SP, Williams PL, Robinson CG, et al. Evaluation of ViroCyt® Virus Counter for rapid filovirus quantitation. Viruses. 2015;7:857–872. doi: 10.3390/v7030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lam TT, Zhou B, Wang J, Chai Y, Shen Y, et al. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature. 2015;522:102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 89.Guan Y, Smith GJ. The emergence and diversification of panzootic H5N1 influenza viruses. Virus Res. 2013;178:35–43. doi: 10.1016/j.virusres.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu KM, Smith GJ, Bahl J, Duan L, Tai H, et al. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol. 2007;81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ellebedy AH, Ahmed R. Re-engaging cross-reactive memory B cells: the influenza puzzle. Front Immunol. 2012;3:53. doi: 10.3389/fimmu.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]


