Abstract
Background
The human retinal pigment epithelium (RPE) plays an important role in the pathogenesis of age related macular degeneration (AMD). AMD is the leading cause of blindness worldwide. There is currently no effective treatment available. Preclinical studies in AMD mouse models are essential to develop new therapeutics. This requires further in-depth knowledge of the similarities and differences between mouse and human RPE.
Methods
We performed a microarray study to identify and functionally annotate RPE specific gene expression in mouse and human RPE. We used a meticulous method to determine C57BL/6J mouse RPE signature genes, correcting for possible RNA contamination from its adjacent layers: the choroid and the photoreceptors. We compared the signature genes, gene expression profiles and functional annotations of the mouse and human RPE.
Results
We defined sets of mouse (64), human (171) and mouse–human interspecies (22) RPE signature genes. Not unexpectedly, our gene expression analysis and comparative functional annotation suggested that, in general, the mouse and human RPE are very similar. For example, we found similarities for general features, like “organ development” and “disorders related to neurological tissue”. However, detailed analysis of the molecular pathways and networks associated with RPE functions, suggested also multiple species-specific differences, some of which may be relevant for the development of AMD. For example, CFHR1, most likely the main complement regulator in AMD pathogenesis was highly expressed in human RPE, but almost absent in mouse RPE. Furthermore, functions assigned to mouse and human RPE expression profiles indicate (patho-) biological differences related to AMD, such as oxidative stress, Bruch’s membrane, immune-regulation and outer blood retina barrier.
Conclusion
These differences may be important for the development of new therapeutic strategies and translational studies in age-related macular degeneration.
Introduction
Age related macular degeneration (AMD) is the leading cause of blindness worldwide. The disease affects 4% of the population over age 60. With the increase of the aging population, AMD is becoming an even more important public health issue. The etiology of AMD remains largely unknown. The first clinical manifestations of the disease include the appearance of sub-retinal drusen and pigmentary or degenerative changes of the RPE. Ultimately, the disease affects the RPE, Bruch’s membrane (BM), photoreceptors (PR) and choriocapillaries (CH). We focused this study on the RPE.
The RPE is a monolayer of pigmented neuro-epithelial cells, which forms part of the outer blood-retina barrier. It closely interacts with the PR to maintain visual function. The apical membrane of the RPE faces the photoreceptor outer segments and its basolateral membrane faces the BM. The BM separates the RPE from CH, which nourishes the RPE and outer layers of the retina [1]. In healthy eyes, BM functions as a structural support that is permeable to fluid and small molecules. Additionally it acts as a physical barrier, containing anti-angiogenic molecules, which protect the retina against neovascularization [2,3].
A healthy RPE is essential for visual function. It supplies the PR with nutrients, absorbs the excess light energy focused by the lens on the retina, recycles retinal from the PR, regulates the ion balance in the sub retinal space and maintains the function and survival of the PR by phagocytosis of the shed photoreceptor outer segments [1]. Failure of any of these functions can lead to degeneration of the retina, loss of visual function and, eventually, blindness in retinal diseases such as AMD or retinitis pigmentosa.
In AMD, RPE dysfunction or degeneration leads to a dystrophy of the PR and thereby vision loss [4]. The early stage of AMD is characterized by the presence of drusen and vision loss is relatively mild. Later stages of the disease involve two forms: the dry form (geographic atrophy) and the wet form (choroidal neovascularization). Both forms affect about half of the late stage AMD patients. AMD has a multifactorial etiology [5], and is caused by a variety of environmental and genetic risk factors [4]. There is evidence that positive life style changes (quit smoking; healthy food) and dietary supplements (Zn2+) may postpone the onset or progression of the disease [6]. Patient-unfriendly, repeated intra-ocular injections with anti-VEGF may temporarily halt the progression of the wet form of AMD. However, it does not prevent the atrophy of RPE and PR [7,8]. Once vision is lost, a possible (future) cure for AMD may be cell replacement therapy. Pre-clinical experiments indicate that transplantation of stem cell derived RPE cells can successfully be used to rescue PR and vision [9–11]. However, these preclinical studies are predominantly performed in mice. To translate results and start clinical studies in man further knowledge of the similarities and differences between mouse and human RPE is essential.
In this study we compared the gene expression profiles and functional annotation of mouse and human RPE on a single microarray platform to further improve translational studies.
Results
First, we determined the gene expression profiles of the mouse RPE, CH and PR (raw data available in the Gene Expression Omnibus database with the accession number GSE66916). We confirmed our microarray methodology by checking the expression of (well established) RPE genes using semi-quantitative RT-PCR (sqRT-PCR) (Fig 1 and S1 Fig). Subsequently, we determined mouse and human RPE signature genes, we defined the functionalities of the gene expression profiles of mouse and man, and we analyzed the most extreme differences in RPE gene expression between the two species. Also these results were partly confirmed using sqRT-PCR (S2 Fig).
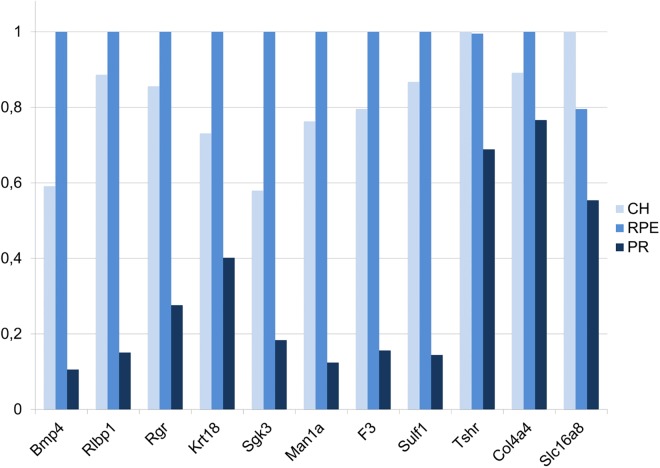
Fig 1. Confirmation of microarray results by sqRT-PCR.
Beta-actin (Bact), a housekeeping gene, was used to normalize gene expression in mouse CH, RPE and PR. The light blue bars indicate expression levels in CH, the blue bars expression levels in the RPE and the dark blue bars indicate expression levels in PR. Similar to the microarray data the expression level is highest in the RPE and lowest in the PR. The sqRT-PCR results confirm our findings; however Tshr and Slc16a8 show expression lower in RPE compared to choroid. Overall, the sqRT-PCR confirmation rate in this, and in all our previous studies (combined), using exactly the same methodology and platform to investigate neuroepithelia from human donor eyes and brains was 87% [12–14].
Mouse, human and inter-species RPE signature genes
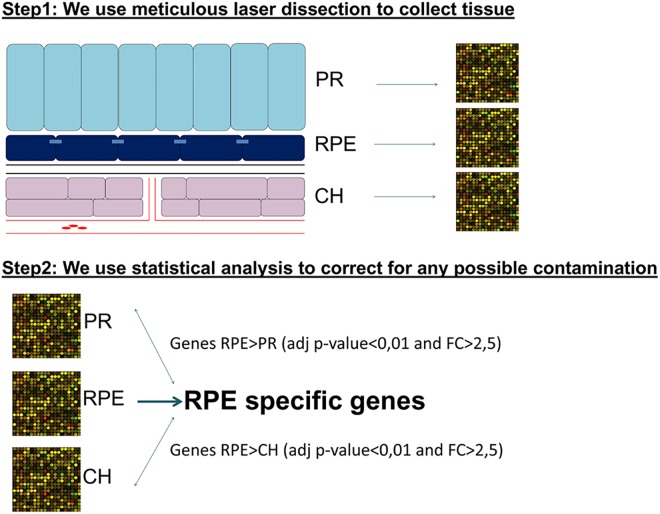
In our lab, we previously designed a new strategy to select RPE signature genes (Fig 2). RPE specificity was determined by comparison of the gene expression levels between the RPE and its adjacent layers: the CH and PR [12]. In the current study, we applied this strategy to the mouse retina in order to select mouse RPE signature genes.
Fig 2. Strategy to select RPE signature genes.
In the first step of this strategy we laser dissect the RPE (and its adjacent layers, the CH and PR) for specific tissue collection. In the second step we statistically correct for possible contamination by adjacent layers.
We selected the genes that have a significant higher expression level in the mouse RPE compared to their expression in the CH and the PR, with a fold change (FC) higher than 2.5 and a B-H adjusted p-value<0.01. This resulted in a list of 64 genes that are specifically expressed in the RPE relative to both its adjacent layers; the CH and PR. We annotated this set the “Mouse RPE signature genes” dataset (see Table 1).
Table 1. Our “Mouse RPE signature genes” dataset: 64 mouse RPE genes with an average expression of at least 2.5 fold higher in the mouse RPE than in both the PR and the CH with an adjusted p-value smaller than 0.01.
| CH<RPE>PR genes | RPE compared to CH | RPE compared to PR | |||
|---|---|---|---|---|---|
| GeneName | SystematicName | adj.P.Val | FC RPE-CH | adj P value | FC RPE-PR |
| Rgr | ENSMUST00000022338 | 5,93E-03 | 4,9 | 5,90E-06 | 306,1 |
| LOC100045988 | XM_001475309 | 6,03E-03 | 4,6 | 3,81E-03 | 3,5 |
| Pon1 | NM_011134 | 1,01E-03 | 4,2 | 2,10E-07 | 95,8 |
| Rdh10 | NM_133832 | 1,46E-03 | 4,1 | 9,38E-08 | 75,1 |
| Arl6ip1 | NM_019419 | 4,41E-03 | 3,2 | 2,73E-06 | 24,6 |
| Rlbp1 | NM_020599 | 5,78E-03 | 3,2 | 4,29E-07 | 42,4 |
| Tbx5 | NM_011537 | 2,65E-03 | 3,2 | 3,61E-04 | 3,5 |
| Bmp4 | NM_007554 | 3,03E-03 | 3,2 | 2,31E-05 | 47,9 |
| F3 | NM_010171 | 1,42E-03 | 3,1 | 4,09E-07 | 102,2 |
| 5730469M10Rik | NM_027464 | 2,28E-03 | 3,1 | 1,03E-06 | 14,0 |
| Rrh | NM_009102 | 2,40E-03 | 3,1 | 8,94E-08 | 47,2 |
| Man1a | NM_008548 | 2,07E-04 | 3,0 | 6,11E-08 | 10,3 |
| Sema3c | NM_013657 | 5,52E-04 | 2,9 | 9,66E-08 | 504,3 |
| Vldlr | NM_013703 | 1,35E-03 | 2,9 | 1,79E-05 | 4,6 |
| Atp1b1 | NM_009721 | 2,54E-03 | 2,9 | 1,29E-07 | 34,1 |
| Ctsd | NM_009983 | 6,72E-03 | 2,9 | 6,81E-05 | 5,7 |
| Cspg5 | NM_001166273 | 5,34E-03 | 2,9 | 1,94E-06 | 15,1 |
| Cldn2 | NM_016675 | 6,79E-04 | 2,9 | 6,75E-07 | 7,8 |
| Sulf1 | NM_172294 | 4,05E-04 | 2,9 | 2,38E-07 | 22,9 |
| BC048943 | NM_001127685 | 1,48E-03 | 2,9 | 8,23E-05 | 3,4 |
| Slc39a12 | NM_001012305 | 6,97E-04 | 2,9 | 8,03E-08 | 123,3 |
| Loxl4 | NM_001164311 | 4,47E-04 | 2,8 | 7,52E-07 | 13,2 |
| NAP114398-1 | NAP114398-1 | 5,88E-04 | 2,8 | 2,70E-07 | 9,4 |
| Slc1a1 | NM_009199 | 6,31E-03 | 2,8 | 1,30E-07 | 27,2 |
| Slc6a13 | NM_144512 | 3,86E-03 | 2,8 | 9,05E-08 | 49,1 |
| Car12 | NM_178396 | 5,92E-03 | 2,8 | 2,86E-07 | 34,5 |
| Iqgap2 | NM_027711 | 3,55E-04 | 2,8 | 5,11E-08 | 13,4 |
| Hist2h2aa1 | NM_013549 | 2,53E-04 | 2,8 | 2,57E-07 | 5,7 |
| Tgfa | NM_031199 | 1,07E-03 | 2,8 | 2,66E-07 | 11,9 |
| Spon1 | NM_145584 | 3,68E-04 | 2,7 | 2,54E-07 | 7,4 |
| Flot2 | NM_008028 | 4,72E-03 | 2,7 | 1,50E-05 | 7,1 |
| Tmem27 | NM_020626 | 1,64E-03 | 2,7 | 3,15E-05 | 108,8 |
| Trhde | NM_146241 | 1,06E-03 | 2,7 | 7,15E-08 | 19,8 |
| Hist2h4 | NM_033596 | 8,85E-03 | 2,7 | 5,29E-05 | 7,0 |
| Itgb8 | NM_177290 | 2,57E-03 | 2,7 | 4,33E-07 | 14,9 |
| Cbfa2t3 | NM_001109873 | 6,18E-03 | 2,7 | 2,75E-06 | 11,8 |
| Tcfl5 | NM_178254 | 1,92E-03 | 2,7 | 4,74E-07 | 12,2 |
| Adora2b | NM_007413 | 3,61E-04 | 2,7 | 1,30E-07 | 37,0 |
| Spock1 | NM_009262 | 2,29E-03 | 2,7 | 1,28E-06 | 9,3 |
| Gpam | ENSMUST00000086868 | 8,06E-03 | 2,7 | 1,09E-03 | 3,0 |
| Acsl6 | NM_001033599 | 2,80E-03 | 2,7 | 2,96E-04 | 2,8 |
| Lrp2 | NM_001081088 | 6,73E-03 | 2,7 | 3,74E-06 | 10,7 |
| Slc6a20a | NM_139142 | 5,91E-04 | 2,6 | 3,38E-07 | 7,6 |
| Nt5dc2 | NM_027289 | 1,30E-03 | 2,6 | 9,65E-07 | 7,7 |
| Krt18 | NM_010664 | 2,04E-03 | 2,6 | 1,73E-06 | 7,2 |
| Slc16a8 | NM_020516 | 1,87E-03 | 2,6 | 2,46E-07 | 14,9 |
| Gabrb3 | NM_008071 | 3,18E-04 | 2,6 | 4,83E-06 | 3,3 |
| Mogat1 | NM_026713 | 2,79E-03 | 2,6 | 5,42E-07 | 12,9 |
| Hkdc1 | NM_145419 | 2,81E-03 | 2,6 | 7,31E-06 | 5,9 |
| Tmem56 | NM_178936 | 4,12E-03 | 2,6 | 1,71E-08 | 148,2 |
| Col4a4 | NM_007735 | 2,96E-03 | 2,6 | 3,44E-06 | 4,0 |
| Pebp4 | NM_028560 | 4,39E-04 | 2,6 | 4,15E-08 | 13,5 |
| Trpm3 | NM_001035246 | 1,61E-04 | 2,6 | 5,35E-09 | 23,2 |
| Hist1h4i | NM_175656 | 1,08E-03 | 2,6 | 4,09E-06 | 4,7 |
| A2m | NM_175628 | 1,55E-03 | 2,5 | 8,00E-05 | 3,0 |
| Bphl | NM_026512 | 8,30E-04 | 2,5 | 2,48E-06 | 3,1 |
| Slc7a10 | NM_017394 | 1,64E-03 | 2,5 | 3,27E-08 | 33,6 |
| Tshr | NM_011648 | 4,22E-04 | 2,5 | 1,22E-08 | 58,3 |
| Car14 | NM_011797 | 3,58E-03 | 2,5 | 2,60E-07 | 18,4 |
| Adra2c | NM_007418 | 4,16E-03 | 2,5 | 4,30E-08 | 54,8 |
| Fam13a | NM_153574 | 5,08E-04 | 2,5 | 2,20E-08 | 19,6 |
| Sgk3 | NM_133220 | 3,38E-03 | 2,5 | 3,87E-06 | 7,0 |
| Pde4b | NM_019840 | 5,03E-04 | 2,5 | 8,98E-07 | 5,0 |
| Slco1a4 | NM_030687 | 3,07E-04 | 2,5 | 6,65E-09 | 32,7 |
Using the same cut-off criteria; we determined a set of genes that is specific for the mouse CH compared to the RPE and a set of genes mouse PR specific when compared to the RPE (S1 Table).
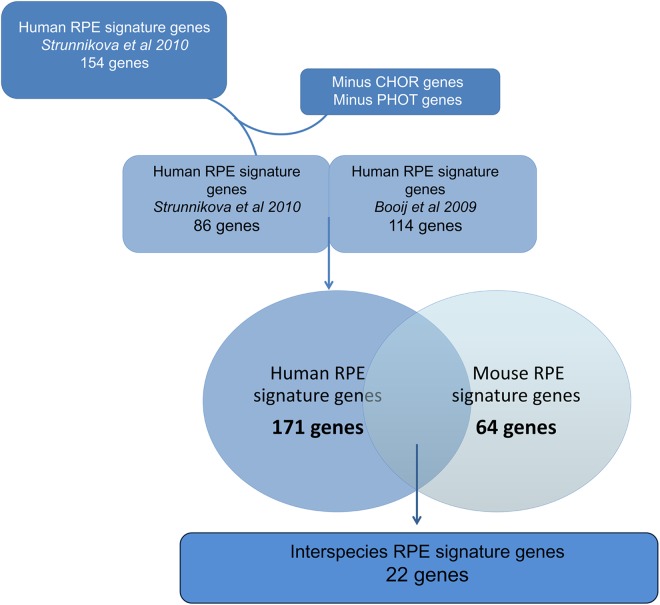
We next defined a new “Human RPE signature genes” dataset. We carefully selected two previously published human RPE specific gene expression datasets for a comprehensive comparison between mouse and human RPE (Fig 3) [12,15]. The first study was conducted in our lab using a similar methodology for determining RPE specific gene expression resulting in identification of 114 RPE specifically expressed genes [12]. The second microarray study included multiple RPE types but the investigators did not correct for possible contamination of adjacent tissues [15]. For the latter dataset, we removed possible CH and PR RNA contamination (see Methods), and generated a list of 86 human RPE specifically expressed genes. We subsequently merged the two human RPE specific gene expression datasets, resulting in 171 human RPE signature genes (S2 Table).
Fig 3. Strategy to determine “Interspecies RPE signature genes”.
Schematic overview of our comparison strategy: our “Mouse RPE signature genes” dataset and “Human RPE signature genes” dataset, which contains (a modification of) two human RPE transcriptome datasets [12,15]. This resulted in a new dataset, “Interspecies RPE signature genes”.
Finally, in order to facilitate comparative retinal studies between mouse and human, we aimed to develop a list of interspecies RPE signature genes. We determined the overlap between the mouse (64) and the human (171) RPE signature gene lists, resulting in an interspecies RPE signature gene list of 22 genes (Table 2).
Table 2. The 22 signature genes that are specifically expressed in both RPE in mouse and in human.
| Gene Symbol | Genbank ID Mus Musculus | Genbank Homo Sapiens |
|---|---|---|
| ADORA2B | NM_007413 | NM_000676 |
| G-protein coupled adenosine receptor. This integral membrane protein stimulates adenylate cyclase activity in the presence of adenosine. | ||
| BMP4 | NM_007554 | NM_001202 |
| A member of the bone morphogenic protein family which is part of the transforming growth factor-beta superfamily. The superfamily includes large families of growth and differentiation factors. | ||
| CA14 | NM_011797 | NM_012113 |
| Carbonic anhydrases are a large family of zinc metalloenzymes that catalyze the reversible hydratyion of carbon dioxide. | ||
| CSPG5 | NM_001166273 | NM_001206942.1 |
| A proteoglycan that may function as a neural growth and differentiation factor. | ||
| CTSD | NM_009983 | NM_001909.4 |
| An aspartic protease resident in endosomal and lysosomal compartments of all eukaryotic cells. | ||
| GPAM | ENSMUST00000086868 | |
| A mitochondrial enzyme which prefers saturated fatty acids as its substrate for the synthesis of glycerolipids. | ||
| ITGB8 | NM_177290 | NM_002214.2 |
| Cell surface adhesion receptor mediating cell-adhesion to extra cellular matrix or to other cells, through hetero dimerization and connecting to the cytoskeleton and various signaling molecules within cells. | ||
| KRT18 | NM_010664 | NM_000224 |
| Keratin 18 is a type I cytokeratin (this type constitutes the type I intermediate filaments of the intracytoplasmatic cytoskeleton, which is present in all mammalian epithelial cells), together with Krt8 is the most common found product of the intermediate filament gene family. They are expressed in single layer epithelial tissues of the body. | ||
| RDH10 | NM_133832 | [NM_172037 |
| A retinol dehydrogenase, which converts all-trans-retinol to all-trans-retinal, with preference for NADP as a cofactor. | ||
| RGR | ENSMUST00000022338 | NM_002921 |
| A putative retinal G-protein coupled receptor and acts as a photoisomerase to catalyze the conversion of all-trans-retinal to 11-cis-retinal. | ||
| RLBP1 | NM_020599 | NM_000326 |
| Retinaldehyde binding protein 1. carries 11-cis-retinaldehyde or 11-cis-retinal as physiological ligands. It may be a functional component of the visual cycle. | ||
| SEMA3C | NM_013657 | NM_006379.3 |
| Binds to plexin family members and plays an important role in the regulation of developmental processes. | ||
| SLC16A8 [MCT3] | NM_020516 | NM_013356 |
| Belongs to a family of monocarboxylate transporters. It is expressed in the basolateral membrane of the RPE. | ||
| SLC39A12 | NM_001012305 | NM_152725 |
| Zinc transporter, which is a cofactor for hundreds of enzymes and therefore normal cell function. | ||
| SLC6A13 | NM_144512 | NM_016615 |
| Encodes a sodium- and chloride-dependent GABA transporter [GAT2] | ||
| SLC6A20 | NM_139142 | NM_020208 |
| Encodes an amino acid transmembrane transporter that mediates the transport of small hydrophilic substances across cell membranes. | ||
| SLC7A10 | NM_017394 | NM_019849 |
| Encodes an amino acid transmembrane transporter that mediates high-affinity transport of D-serine and several other neutral amino acids. | ||
| SPOCK1 | NM_009262 | NM_004598 |
| Encodes he protein core of a seminal plasma proteoglycan containing chondroitin- and heparin-sulfate chains. | ||
| SULF1 | NM_172294 | NM_015170 |
| Enzyme which can modulate the activity if heparan sulfate, thereby influencing the regulation of cell growth, proliferation, differentiation and migration. | ||
| TMEM27 | NM_020626 | NM_020665 |
| binds to amino acid transporters and regulates their expression on the plasma membrane | ||
| TMEM56 | NM_178936 | NM_152487 |
| function unknown | ||
| TRPM3 | NM_001035246 | NM_206948 |
| Belongs to the family of transient receptor potential channels. TRP channels are cation-selective channels important for cellular calcium signaling and homeostasis. | ||
Gene expression profiles and functions of the mouse and human RPE
Using our previously published methodology [12–14], we analyzed the highest expressed genes (highest 10th percentile: >90thP) of our mouse gene expression dataset (designated “Mouse high RPE gene expression”) to determine the most important functionalities of the mouse RPE. Subsequently, we compared the gene expression pathways and functional annotations of the mouse and human RPE. The latter dataset was available from our previous studies (“Human high RPE specific gene expression dataset” [12]. We used the Ingenuity Knowledge Database to determine biological functions, canonical pathways and molecular networks specific for mouse and human RPE in vivo.
Functional annotation yielded statistically significant biological functions that were the same for mouse and human (Table 3). We also found that many important canonical pathways for mouse and human RPE were similar. A summary of these findings is presented in Fig 4.
Table 3. Overview of the major biological functions found in a functional annotation by Ingenuity of the “Mouse High RPE gene expression” and “Human High RPE gene expression” datasets.
| Mouse High RPE expression | Human High RPE expression | ||
|---|---|---|---|
| Disease and Disorders | p-value | Disease and Disorders | p-value |
| Neurological Disease | 7,31E-51-9,20E-05 | Neurological Disease | 7,54E-57-2,18E-05 |
| Psychological Disorders | 8,09E-44-9,20E-05 | Psychological Disorders | 2,04E-49-2,26E-07 |
| Skeletal and Muscular Disorders | 2,84E-41-2,62E-05 | Skeletal and Muscular Disorders | 5,84E-47-2,18E-05 |
| Infectious Disease | 5,89E-36-9,33E-05 | Hereditary Disorder | 1,77E-39-2,18E-05 |
| Hereditary Disorder | 6,10E-34-4,77E-05 | Infectious Disease | 2,29E-32-1,08E-05 |
| Molecular and Cellular Functions | Molecular and Cellular Functions | ||
| Cellular Growth and Proliferation | 7,20E-39-8,57E-05 | Cell Death and Survival | 3,55E-42-2,06E-05 |
| Cell Death and Survival | 3,43E-38-5,57E-05 | Cellular Growth and Proliferation | 6,25E-37-1,64E-05 |
| Cell Morphology | 5,70E-25-9,33E-05 | Protein Synthesis | 1,77E-22-2,14E-05 |
| Protein Synthesis | 5,77E-22-9,28E-06 | Cell Morphology | 9,15E-21-1,66E-05 |
| Cellular Development | 3,25E-17-8,57E-05 | Gene Expression | 2,01E-20-2,15E-05 |
| Physiological System Development & Function | Physiological System Development & Function | ||
| Organismal Survival | 7,97E-23-7,93E-06 | Organismal Survival | 3,18E-23-3,18E-23 |
| Embryonic Development | 9,01E-18-8,15E-05 | Organismal Development | 2,63E-16-2,01E-05 |
| Organ Development | 9,01E-18-8,15E-05 | Nervous System Development and Function | 3,17E-16-1,66E-05 |
| Organ Morphology | 9,01E-18-9,33E-05 | Embryonic Development | 2,01E-14-1,65E-05 |
| Organismal Development | 9,01E-18-8,15E-05 | Organ Development | 3,73E-13-1,45E-05 |
The p-value for these categories are indicated as a range because each category contains sub-functions that have their own p-value.
Fig 4. Most significant canonical pathways identified by Ingenuity for the “Mouse High RPE gene expression” and “Human High RPE expression gene expression” datasets.
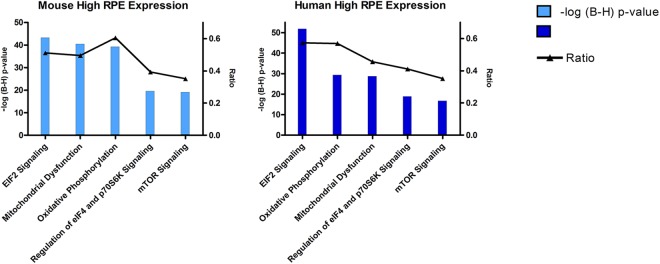
The left y-axis displays the–log of Benjamini-Hochberg corrected p-value. The right y-axis displays the ratio of the number of genes derived from our dataset, divided by the total number of genes in the pathway. The blue line indicates the threshold of the BH corrected p-value of 0.1.
In addition, we studied the molecular networks that were assigned to both the “Mouse” and “Human” “high RPE gene expression” datasets. Functions annotated to these datasets on a network level were more or less comparable (~75% overlap). The annotated functions included developmental disorders, hereditary disorders, small molecule/drug metabolism and cellular movement and maintenance. For an overview of the 10 most important networks for the “Mouse high RPE gene expression” dataset and the “Human high RPE gene expression” dataset, and to see which networks overlap, see S3 Table and S4 Table. For illustrative purpose we included an example of such a network (S3 Fig). For additional support of our findings in Ingenuity we also included a functional enrichment pathway analysis (KEGG analysis) in Webgestalt [16]. This gives approximately the same results (S5 Table).
Genes highly expressed in mouse RPE but hardly in human RPE
To investigate the largest gene expression and functional differences between the mouse and human RPE we subsequently compared the most extreme gene expression datasets of the two species, namely the very high (highest 10th percentile, >90thP; high expression) and very low (lowest 10th percentile <10thP; leaky expression) RPE expression datasets (GSE 66916).
Unexpectedly, the “Mouse high RPE gene expression” dataset (>90th P, 2663 genes) and the “Human very low RPE gene expression”dataset (0-10th P, 1770 genes) had 101 genes in common (S6 Table). Functional annotation of these genes yielded 31 canonical pathways in Ingenuity, whose activity or metabolic route may be differentially affected in mouse and human RPE. An overview is presented in Table 4.
Table 4. Overview of significant canonical pathways assigned by the Ingenuity knowledge database to the 101 genes that are the result of comparing the “Mouse high RPE gene expression” and the “Human very low RPE gene expression” datasets.
| Endocrine Signaling & Metabolic Function |
| Ephrin Receptor Signaling |
| PEDF Signaling |
| Protein Kinase A Signaling |
| Gαq Signaling |
| FGF Signaling |
| Phospholipase C Signaling |
| NGF Signaling |
| GNRH Signaling |
| PXR/RXR Activation |
| Ephrin B Signaling |
| Immunological Function |
| iCOS-iCOSL Signaling in T Helper Cells |
| Role of NFAT in Regulation of the Immune Response |
| Dendritic Cell Maturation |
| B Cell Receptor Signaling |
| IL-8 Signaling |
| Thrombin Signaling |
| PKCθ Signaling in T Lymphocytes |
| CD28 Signaling in T Helper Cells |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis |
| GM-CSF Signaling |
| PI3K Signaling in B Lymphocytes |
| Basic pathways of cellular (dys)function |
| Prostate Cancer Signaling |
| Regulation of the Epithelial-Mesenchymal Transition Pathway |
| Wnt/Ca+ pathway |
| P2Y Purigenic Receptor Signaling Pathway |
| Estrogen-Dependent Breast Cancer Signaling |
| Colorectal Cancer Metastasis Signaling |
| Epithelial junctions |
| Tight Junction Signaling |
| Epithelial Adherens Junction Signaling |
| Vesicle mediated transport |
| Clathrin-mediated Endocytosis Signaling |
| Oxidative stress |
| Hypoxia Signaling in the Cardiovascular System |
The core analysis of the 101 differentially expressed genes resulted in 7 molecular networks. The associated representative functions include developmental disorders, connective tissue disorders, ophthalmic disease, neurological disease, drug metabolism and cancer. These networks are presented in S7 Table. For illustrative purpose we included an example of one of these networks, see S4 Fig. For additional support of our findings in Ingenuity we also included a functional enrichment pathway analysis (KEGG analysis) in Webgestalt. This gives approximately the same results (S8 Table).
Genes highly expressed in human RPE but hardly in mouse RPE
In order to identify additional differences between mouse and human RPE, we also compared the “Human high RPE gene expression” dataset (>90th P, 2399 genes) and the “Mouse very low RPE gene expression”dataset (10th P, 3374 genes). This analysis yielded 54 genes (S9 Table). We also functionally annotated this set of genes using the Ingenuity knowledge database. The significant canonical pathways assigned to this dataset included PXR/RXR activation, nicotine degradation and bupropion degradation. Ingenuity analysis yielded four networks. The functional annotations of these networks include drug metabolism, nucleic acid metabolism, small molecule biochemistry, cardiovascular disease and humoral immune response. The molecular pathways are presented in S10 Table. We also included an illustrative example of such a network, see S5 Fig.
Among the major biological functions and disease that came out of this analysis were hereditary hearing loss and Usher syndrome. Major differences in molecular cellular functions identified by Ingenuity were drug metabolism, nucleic acid metabolism, small molecule biochemistry and lipid metabolism. For additional support of our findings in Ingenuity we included a functional enrichment pathway analysis (KEGG analysis) in Webgestalt. This gives approximately the same results (see S11 Table).
Discussion
In this study, we aimed to find similarities and differences between mouse and human RPE using RPE specific gene expression profiles and functional annotation on the same experimental platform. Our current data may be important for translational studies in age related macular degeneration, for creating and use of a representative AMD mouse model. Thus, we discuss here those aspects of our analyses of human and mouse RPE that are relevant for AMD.
Similarities and differences between mouse and human RPE transcriptomes in relation to AMD
Apart from the obvious similarities, there are a number of well-known differences between human and mouse RPE and adjacent tissues. These include the absence of a macula in the mouse, the difference in rod and cone number and distribution, and a thinner Bruch’s membrane in the mouse. Mouse models are available for wet and dry AMD, mimicking several of the pathological features seen in AMD, but no model recreates all of the AMD characteristics [17–20].
We were interested in the potential usefulness of our entire comparative human and mouse gene expression dataset for the investigation of AMD (mouse models). Interestingly we did find similarities and differences in relation to a number of previously published (patho-) biological aspects related to AMD, namely oxidative stress, zinc homeostasis, presence of proteins of the complement system that are found in drusen, proteins in Bruch´s membrane, involvement in neovascularization and tight junctions. These differences and similarities are important to develop and use representative mouse models for AMD, and they may be partly responsible for (the observed) discrepancies between mouse model and human patients.
Age related macular degeneration: Oxidative Stress
The RPE suffers from chronic oxidative stress due to the exposure to light, relatively low oxygen levels, and daily phagocytosis and digestion of photoreceptor outer segments [21]. The mainstream hypothesis in AMD is that prolonged oxidative stress harms the vitality of the RPE and oxidatively modified drusen-bound fatty acids and proteins. These are subsequently recognized by the body as non-self, and invoke a chronic, complement mediated, immune response [22,23].
Oxidative stress in the RPE is, among others, mediated by the manganese superoxidase dismutase protein family, consisting of SOD1, SOD2 and SOD3; Respectively, these SODs exert their antioxidant effect in the cytosol, mitochondria and extracellular matrix [24,25]. We found similar expression of SOD1 (very high) and SOD3 (moderate) in human and mouse RPE. In contrast, we found that the SOD2 gene was highly expressed in the mouse RPE but only at low levels in the human RPE (Table 5). Reactive Oxygen species (ROS)-associated mitochondrial DNA damage was previously correlated with the progression of AMD [26–28]. But association studies between genetic variants in the SOD2 gene and AMD pathogenesis yielded conflicting results [27,29].
Table 5. SOD1, SOD2, SOD3 gene expression in human and mouse RPE.
| Mouse | Human | |||
|---|---|---|---|---|
| isoenzyme | Reporter | Percentile | Reporter | Percentile |
| SOD1 | NM_011434 | High | NM_000454 | High |
| SOD2 | NM_013671 | High | NM_000636, BM994509, AL050388 | Low |
| SOD3 | NM_011435 | Intermediate | NM_003102 | Intermediate |
Sod1 and Sod3 are highly and moderately expressed respectively, in both species. Sod2 gene expression has a low expression in human RPE. In contrast it has a high expression in mouse RPE.
For both SOD1 and SOD2 mouse models were developed. Sod1-/- mice and Sod2-/- mice both show a thickened Bruch’s membrane, photoreceptor atrophy and reduced electroretinographic response [30]. Sod2-/- mice lacked drusen like deposits but have RPE atrophy [31,32]. In the Sod1-/- mice, 10% of the older animals showed choroidal neovascularization and 86% showed drusen-like deposits that contained several markers of drusen [33].
The previous studies on SOD family members and the different expression we find between mouse and human, indicate that all three SOD family members may be critically involved in the local defense against oxidative stress, although the mitochondrial SOD2 may play a more important role in the mouse RPE than in the human RPE.
In addition to SODs, zinc has also been implicated in mediating oxidative stress. The retina and especially drusen contain high amounts of zinc [34]. There is an age related decrease in systemic and cellular zinc levels in human RPE cells, that correlates with several age related pathologies like AMD [35]. In 1988 the first clinical trial favoring zinc supplementation in AMD was published [36]. Since that time, multiple studies suggested that zinc reduces the oxidative burden on the retina although the underlying molecular mechanism(s) is unknown [7,37–39]. Zinc ions reach the retina by specific transporters. We determined which zinc transporters are highly expressed in mouse and human RPE. In the highest 10th percentile of the mouse RPE transcriptome we found expression of Slc39a1, Slc39a4, Slc39a7 and Slc39a12. In the highest 10th percentile of the human RPE transcriptome we observed expression of SLC39A8, SLC39A12 and SLC39A13 (Table 6). Our data are largely in agreement with those of Leung and coworkers [40], who determined the expression of a large number of zinc transporters in cultured human RPE cells.
Table 6. Overview of zinc transporters that are highly expressed in human and mouse RPE.
Interestingly, we found that Slc39a4 was highly expressed in mouse RPE, but not in the human RPE. Indeed, Dufner-Beattie et al demonstrated the importance of this transporter in a Slc39a4 knockout mouse, which develops severe abnormalities of the nervous system, such as anophthalmia, exencephaly and hydrocephalus [41]. Our finding (in older human donor eyes) may be explained by an age-related effect, since Leung et al [40] found that Slc39a2 and Slc39a4 expression and corresponding zinc uptake are reduced in RPE from older individuals. We determined the Slc39a4 expression in RPE of five-month-old mice.
Age-related macular degeneration: drusen and complement system
Chronic inflammatory and immune mediated events at the level of the Bruch’s membrane and drusen play critical roles in AMD pathogenesis [42]. Initially, complement system related factors were immune-localized to drusen, a hallmark of AMD. Subsequently, genetic studies showed an association between polymorphisms of several complement pathway genes, such as CFH, CFB, C3, CFHRs, and AMD [2]. In our dataset we found high expression of several complement factors in the human and mouse RPE (Fig 5), which may be of interest for studies of the complement system and AMD pathogenesis in a mouse model.
Fig 5. Overview of highly expressed complement factors in the human and mouse RPE.
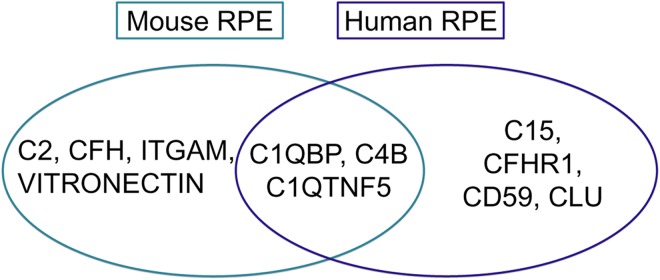
Complement factors in the overlay of the circles are highly expressed in RPE of both species.
Interestingly, C1QTNF5 is highly expressed in the both mouse and human RPE. Mutations in C1QTNF5 have been associated with late-onset retinal degeneration. A C1qtnf5 S163R knock-in mouse model developed by Chavali et al showed many pathological features of AMD, such as RPE abnormalities, photoreceptor loss, retinal vascular leakage [43]. Contrary to this, Shu et al. developed a C1qtnf5 Ser163Arg knock-in mouse model that lacked any phenotypic abnormality [44]. The reason for this discrepancy is currently not clear.
We observed that the complement factor H related 1 gene (CFHR1) is highly expressed in human RPE, but not in mouse RPE. Our data corroborate, in part, the data of Luo et al (2011) who found absence of Cfhr1 expression in mouse retina, RPE and choroid [45]. The regulation of the complement system (in AMD) is extremely complex, multiple regulators and feedback loops exist and the detailed mechanisms underlying the complement regulation at the RPE interface and macular area is not known. Nonetheless, several studies suggested that CFHR1, together with CFHR3, plays a central role in complement regulation of AMD. Several studies suggest that the absence of CFHR1 and/or its family member CFHR3 are highly protective against AMD in humans [46–48].
Age-related macular degeneration: Bruch’s membrane and neovascularization
Our data reveal differences in mouse and human RPE gene expression related to two other essential aspects of age-related macular degeneration: The build-up and turnover of Bruch’s membrane and neovascularization. Bruch’s membrane is a sheet of extracellular matrix that lies in between the RPE and the choroid. The extracellular matrix components that form the BM are made by RPE and CH. The BM has a major clinical significance because of its critical role in the pathogenesis of AMD [2].
We found that Timp2 and Col3a, genes involved in extracellular matrix formation or turnover, are highly expressed in the mouse RPE, but not in human RPE. Vice versa, COL16A1 is highly expressed in the human RPE, but not in mouse RPE.
We also found large inter-specifics gene expression differences annotated with the term “Angiogenesis”. “Angiogenesis” refers to the process whereby new blood vessels are formed. In the context of our RPE/AMD analysis, choroidal blood vessels usually penetrate the BM and from new (leaky) vessels underneath the RPE. Our data specifically suggest expression and functional differences for the angiogenic factors Fgf23 and Fgfr1 (highly expressed in mouse RPE), as well as the prostaglandin synthase PTGES and HS6ST1 (highly expressed in human RPE). In summary, our results suggest specific differences between mouse and man in terms of BM buildup or turnover, as well as related to neovascularization.
Age-related macular degeneration: tight junctions of the outer blood-retina barrier
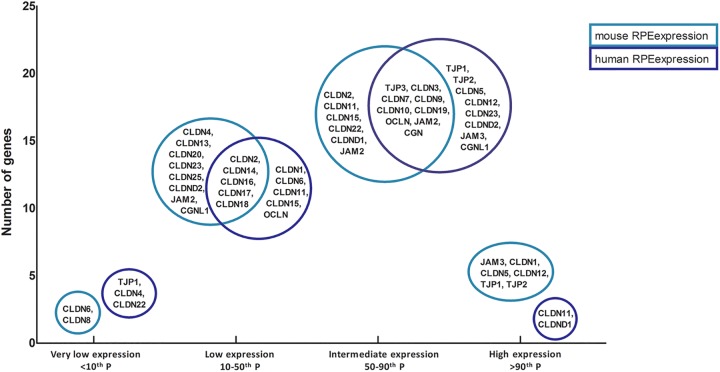
The RPE constitutes the outer blood-retina barrier (oBRB). The tight junctions between neighboring RPE cells bind the monolayer and separates the outer layer of the neural retina from the choriocapillaris [49]. The RPE maintains the integrity of the oBRB through the tight junctions, which is important for control of fluid leakage, solute transport and immune reactions. oBRB supports the functional homeostasis of the retina. Disruptions of RPE cell junction and barrier integrity are associated with AMD [50,51]. We compared the tight junction gene expression of mouse and human RPE by investigating the distribution of these genes in four categories: high expression (>90th percentile), moderate (50-90th percentile), low (10-50th percentile) and very low (<10th percentile) (Fig 6).
Fig 6. This diagram depicts the tight junction gene expression of mouse and human RPE, divided in four categories: high expression (>90th percentile), moderate (50-90th percentile), low (10-50th percentile) and very low (<10th percentile).
On the x-axis the four categories are displayed and on the y-axis the amount of genes found in a category is depicted. Light blue circles contain genes expressed in mouse RPE, Dark blue circles genes expressed in human RPE. Genes inside the overlapping parts of the circles are expressed in the RPE in both species in that category.
Overall, we find limited overlap of tight junction gene expression between mouse and human RPE. Our data suggest that the composition of the outer blood retina barrier differs between mouse and human. More investigation is necessary to determine the possible physiological or pathobiological effect of these differences.
Conclusion
In summary, in this study we determined 64 signature genes for mouse RPE, 171 signature genes for human RPE. We also deduced 22 mouse-human interspecies signature genes. We next analyzed the general mouse and human RPE gene expression profiles, and we found that (patho-) biological functions and canonical pathways assigned to the RPE of both species were highly similar. Nonetheless, more detailed studies, including analysis of specific molecular networks as well as extreme gene expression differences between mouse and human (expression of 155 genes), suggests substantial biological differences.
These similarities and differences may be important for the development of new therapeutic strategies and translational studies in age-related macular degeneration.
Methods
Strengths and limitations of the study design
The technical and methodological strengths and limitations of this approach have been extensively discussed elsewhere [12,13]. Our lab has more than 10 years’ experience in cellular microarray studies. In short, the strengths of this study include the use of selected healthy and freshly frozen samples with short post-mortem times. Sample preparation is characterized by minimal technical handling (such as mechanical or enzymatic dissociation, scraping, heating etc). In this way, the native “in vivo” gene expression profile is preserved. Next, we use laser dissection microscopy (LDM) of cryosections of the relevant cell-type. The use of the LDM ensures highly specific and homogeneous cell-type collection. After RNA isolation, we check RNA integrity and the quantity using the Agilent Bio-analyzer and Nanodrop during the procedure multiple times. Samples are labeled with Cy3 and Cy5 from the 3’-prime end to minimize effects from possible RNA degradation. RNA/samples that do not meet our quality criteria at any point in the procedure are discarded of. We use a common reference design, which also serves as an internal technical control, and a large-scale 44k microarray.
There are also a number of limitations of our studies. Given the lengthy procedure of sample selection, procedure and extensive quality controls, we usually include a limited number of “the very best samples” in our final microarray analysis. The consequence is that we only can detect consistent similarities and differences (and not all, strongly variable or transient ones) in gene expression between samples.
Another limitation is that some degree of cellular contamination of adjacent cell layers in samples is unavoidable, even when we use meticulous laser dissection microscopy in the nicely structurally stacked retina. To overcome this problem we included the gene expression of the adjacent layers in our analysis: the so-called “double selection procedure” (Fig 2) [12].
There are two limitations which are specific to this mouse-human study: The first is that there may be an oligo design difference for the comparative orthologous human and mouse genes on the Agilent whole Mouse and whole Human microarray. While, frequently, multiple different oligo’s for a single gene and reference genes may be present on the micro-array, this may hamper direct comparison between mouse and human gene expression data. To overcome this problem, we ranked the gene expression data of each sample/species according to percentiles, and divided it into four expression groups: high, moderate, low and very low expression [13,52]. By comparing the most extreme datasets, the high expressed genes with the very low expressed genes, between the two species; we could identify physiological relevant differences between the mouse and human RPE gene expressions, since these major differences could not be caused by different affinities alone. For further confirmation, we identified the genes that we described in our paper also by sqRT-PCR.
Finally, the mouse and human tissue used in this study had different post-mortem times: The mouse eyes were enucleated and embedded immediately after death, while for the donor eyes the post-mortem delay was between 16 and 22 hours. On the other hand, it has been shown that this has minimal effect on the RNA integrity of brain tissue. Up to 30 hours postmortem delay did not affect the mRNA [53]. During our experiments, we thoroughly checked the RNA integrity using BioAnalyzer, multiple times. In addition, since we designed labeling primers on the 3 prime end of the genes, potential starting degradation (first occurring at the 5 prime end) did not affect our gene expression results. A full description of the methodological (dis) advantages is beyond the scope of this paper. However, our approach enables us to determine highly specific RPE gene expression with a very limited amount of contamination, which is also corrected for in the analysis.
Mouse eyes, tissue processing and cell sampling
The study on mouse material was carried out in strict accordance with the recommendation in the Guide for the Care and Use of Laboratory Animals under the Dutch law, which is in accordance with the international declaration of Helsinki. The protocol was approved by the Committee on Ethical of Animal Experiments of the Netherlands Institute for Neuroscience (NIN), Royal Dutch Academy for Science (KNAW), the Netherlands (DEC protocol NIN 09.45). Mouse choroid, RPE and photoreceptors were obtained from eyes of healthy 5 months (-/+2 weeks) old C57BL/6 mice (J strain). We confirmed by sqRT-PCR that the mice of this sub strain (C57BL/6JOlaHsd) did not carry the rd8 mutation in the Crb1 gene that has been found in C57BL/6N strains [54]. For each tissue we used 3 mouse eyes (and 3–6 selected human donor eyes). Mice were raised in a room with a temperature around 21°C, on a 12:12-h light-dark cycle, and fed with standard pellet laboratory chow and water ad libitum. By the age of 5 months (-/+2 weeks) they were anesthetized with CO2/O2 and killed by cervical dislocation. The eyes were enucleated, embedded in OCT and snap frozen in liquid nitrogen. Until further use the eyes were stored in an -80°C freezer. We selectively cut out the CH, RPE and PR with a laser dissection microscope (PALM Carl Zeiss, MicroImaging GmbH, Munich, Germany). For this the eyes were cut in 20uM cryosections for the photoreceptors and 12uM for the RPE and the choroid. For every sample one whole eye was used. We used Cresyl Violet staining to identify photoreceptor cells. Before the dissection of the choroid the RPE was removed to prevent as much contamination as possible. After processing the tissue and running the microarray, we determined the (low) variability of the samples using a multidimensional scaling plot (S6 Fig).
RNA isolation and amplification
RNA isolation, amplification and labelling procedures were carried out essentially as described elsewhere [14]. Quality of tRNA was checked with a Bioanalyzer assay (RNA 6000 Pico Kit, Agilent Technologies, Amstelveen, The Netherlands). RNA integrity numbers of tRNA of mouse CH ranged from 4.9 to 6.9, of the mouse RPE ranged from 4.9 to 7.2 and of mouse PR ranged from 5.5 to 7.6. In our microarray study we used a common reference design. The common reference was prepared from mouse RPE/choroid that was isolated, amplified using the same methodology as our experimental samples, and labelled with Cy3 (Cy3 mono-reactive dye pack, GE Healthcare UK, Little Chalfont, Buckinghamshire, UK).
See Janssen et al. [14] for a more detailed description of the laser dissection procedures, RNA processing and microarray procedures.
Microarray data analysis
The microarray files were analysed and processed using Agilent Feature Extraction Software (Agilent Technologies, version 9.5.3.1). We included examples of our strict quality control assessment of the hybridizations in the supplementary file S7 Fig and S8 Fig. Data were imported into R (version 2.14.0 for Windows, R Development Core Team, 2009) using LIMMA in the Bioconductor package. We studied the differences between the RPE and the photoreceptors making a statistical comparison using LIMMA for determining significantly changed genes (R package LIMMA, including Bayesian statistics). We did the same for RPE and the choroid:
Using a common reference design, in LIMMA we first estimated the difference between the sample (either CH / RPE / PR) and the common reference (hybridized against each other on a two channel array). Next, the differences between both RPE and CH or between RPE and PR were estimated. Subsequently, LIMMA fitted a linear model to the expression data for each gene. LIMMA uses empirical Bayes statistics to moderate the standard error of the estimated log-fold changes which results in more stable inference and improved power [55]. A fully detailed description of the script that was used in LIMMA is available upon request.
We selected the genes that had a positive fold change, meaning that they are higher expressed in the RPE than in either the photoreceptors or the choroid, of more than 2.5. Cut-off value for statistical significant difference was an adjusted p-value of less than 0.01 after Benjamini-Hochberg correction for multiple testing. Two data subsets were created that either contained all genes that show a significant higher expression in the RPE compared to the photoreceptors (RPE>PR), or a significant higher expression in the RPE compared to the choroid (RPE>CH). The volcano plots that visualize the symmetrical spread of the differentially expressed genes are included in S9 Fig. Next we compared these two subsets to determine the genes that are present in both lists using a comparison analysis in IPA (Ingenuity Systems). These represent RPE specifically expressed genes. To visualize these significant differences in gene expression levels we included a figure depicting the mean and (low) standard deviations (S10 Fig) and we included a figure of the mean and (the low) standard deviations of the genes mentioned in the discussions section (S11 Fig).
Data analysis of two microarray studies on the human RPE transcriptome
To detect possible contamination of the choroid and the photoreceptors in the list of Strunnikova et al we used the expression data of the human choroid and the human photoreceptors as determined within our group [12]. We assumed that the main source of contamination of RPE sample(s) using this methodology comes primarily from the set of highest expressed genes in either the PR or CH. Consequently, we determined the highest 10th percentile of the average gene expression for both the photoreceptors and the choroid in Microsoft Excel. We ran a comparison analysis in IPA (Ingenuity Systems) to subtract the genes in the highest 10th percentile of photoreceptors and choroid from the 154 genes determined as RPE specific by Strunnikova et al [15]. We merged the two human RPE specific gene expression lists in Ingenuity using a comparison analysis.
Interspecies RPE signature genes
We compared the human RPE signature gene expression list, that we determined using the Booij et al list and the corrected Strunnikova et al list (Fig 3), with our new mouse RPE signature gene expression. We ran a comparison analysis in Ingenuity, which uses the Entrez Gene identifier, to investigate which genes are found in both datasets and thus are interspecies specific.
Confirmation of gene expression data by sqRT-PCR
We confirmed our microarray data with sqRT-PCR. For a detailed description of the sqRT-PCR, see Janssen et al. 2013 [13]. In short, sqRT-PCR was carried out using intron-spanning primers on cDNA from laser dissection microscopy derived samples, using three biological replicates. To minimize effects of RNA degradation artefacts, we generated primers near the 3’end of the gene. We quantified the gene expression in ImageJ and normalized expression by comparing it to the measured expression of housekeeping gene Bact.
Previously, we confirmed the human gene microarrays [12]. In the current study study, we selected a total of 27 genes to confirm our mouse microarray data. First, we selected randomly 11 highly expressed genes from our “Mouse RPE signature genes” dataset (Bmp4, Rlbp1, Rgr, Krt18, Sgk3, Man1a, F3, Sulf1, Thsr, Col4a4, SLc16a8). For 9 out of the 11 “Mouse RPE signature genes” we found the highest expression levels in the mouse RPE and the lowest in the CH and PR (Fig 1). Only Thsr and Slc16a8 showed highest expression in CH.
We next selected 8 well-established RPE specifically expressed genes (Mertk, Rrh, Tyr, Rpe65, Rdh5, Lrat, Tjp1 and Trpm3). For 7 out of 8 of the well-established RPE specific genes, we found highest expression in the mouse RPE and lower in the CH and PR (S1 Fig). Only Tyr showed highest expression in CH in our RT-PCR.
We also included sqRT-PCR for genes that were mentioned in the Discussion section to further technically validate our microarray. We selected 8 genes, 6 genes that are highly expressed (found in the highest 10th percentile of the mouse RPE microarray; Sod1, Sod2, Slc39a4, Timp2, Col3a, Cldn1) and 2 genes that are low expressed (found in the lowest 10th percentile of the mouse RPE microarray; Cldn8, Hs6st1). We compared the expression levels of these genes with the expression level of Bact. For all genes we found the expected confirmatory result (S2 Fig). Overall, in this study, we confirmed the expression levels for 24 out of 27 genes (89%), which is in line with the cumulative RT-PCR confirmation rate (87%) of all previous microarray studies (using similar tissue and methodology).
Supporting Information
Bact was used to normalize gene expression in mouse CH, RPE and PR. The light blue bars indicate the CH, the blue bars indicate RPE and the dark blue bars indicate PR. Similar to the microarray data the expression level is highest in the RPE and lowest in the PR.
(TIF)
Dark blue bar indicate Bact expression level, the blue bars show the expression level of genes that were highly expressed on the microarray en light blue bars indicate expression of genes that showed very low expression on the microarray.
(TIF)
(TIF)
(TIF)
(TIF)
We analyzed the normalized expression intensities of the nine samples that were used in this study, with a multidimensional scaling plot. It shows that the samples within a group are highly similar.
(TIF)
Plot of the log of the red background-corrected signal versus the log of the green background-corrected signal for non-control inlier features for all nine samples after hybridization. The linearity of the plots indicate the appropriateness of background method choices.
(TIF)
The spatial distribution of up- and down-regulated features is evenly spread throughout the array for all nine samples.
(TIF)
Shown is the spread of the differentially expressed genes derived from the t-tests. The positive (higher expression in RPE) and negative (higher expression in the CH or in the PR) fold changes are approximately symmetrically divided.
(TIF)
Shown are the means and standard deviations in the mouse CH, RPE and PR.
(TIF)
Shown are the means and the standard deviation in the mouse RPE. The upper line indicates the cut-off value for the highest 10th percentile, the middle line the cut-off value for the 50th percentile and the lowest line the cut-off value for the lowest 10th percentile.
(TIF)
(XLSX)
(XLS)
(XLS)
(XLS)
(XLSX)
(XLS)
(XLS)
(XLSX)
(XLS)
(XLS)
(XLSX)
Data Availability
All microarray files are available from the Gene Expression Omnibus database (accession number GSE66916).
Funding Statement
This study was supported by grants from the General Dutch Foundation Preventing Blindness, the foundation Blinden-Penning, the National Foundation for Blindness and Low Vision (LSBS); The National Foundation of Macular Degeneration (MD); The Netherlands Eye Foundation (Oogvereniging); The Gelderse Foundation for the Blind; Retina Netherlands Foundation; The Foundation Winckel-Sweep; The Rotterdam Foundation for the Blind (RvB); and The Hague Foundation “Care for the Blind” (all coordinated through the UitZicht platform, project 2011-6). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85: 845–881. 10.1152/physrev.00021.2004 [DOI] [PubMed] [Google Scholar]
- 2. Booij JC, Baas DC, Beisekeeva J, Gorgels TGMF, Bergen A a. B. The dynamic nature of Bruch’s membrane. Prog Retin Eye Res. 2010;29: 1–18. 10.1016/j.preteyeres.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 3. Schlingemann RO. Role of growth factors and the wound healing response in age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol Albrecht Von Graefes Arch Für Klin Exp Ophthalmol. 2004;242: 91–101. 10.1007/s00417-003-0828-0 [DOI] [PubMed] [Google Scholar]
- 4. de Jong PTVM. Age-related macular degeneration. N Engl J Med. 2006;355: 1474–1485. 10.1056/NEJMra062326 [DOI] [PubMed] [Google Scholar]
- 5. Hawkins BS, Bird A, Klein R, West SK. Epidemiology of age-related macular degeneration. Mol Vis. 1999;5: 26 [PubMed] [Google Scholar]
- 6. Age-Related Eye Disease Study 2 Research Group. Lutein + zeaxanthin and omega-3 fatty acids for age-related macular degeneration: the Age-Related Eye Disease Study 2 (AREDS2) randomized clinical trial. JAMA. 2013;309: 2005–2015. 10.1001/jama.2013.4997 [DOI] [PubMed] [Google Scholar]
- 7. Schmidl D, Garhöfer G, Schmetterer L. Nutritional supplements in age-related macular degeneration. Acta Ophthalmol (Copenh). 2015;93: 105–121. 10.1111/aos.12650 [DOI] [PubMed] [Google Scholar]
- 8. Campa C, Harding SP. Anti-VEGF compounds in the treatment of neovascular age related macular degeneration. Curr Drug Targets. 2011;12: 173–181. [DOI] [PubMed] [Google Scholar]
- 9. Stanzel BV, Liu Z, Somboonthanakij S, Wongsawad W, Brinken R, Eter N, et al. Human RPE stem cells grown into polarized RPE monolayers on a polyester matrix are maintained after grafting into rabbit subretinal space. Stem Cell Rep. 2014;2: 64–77. 10.1016/j.stemcr.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maeda T, Lee MJ, Palczewska G, Marsili S, Tesar PJ, Palczewski K, et al. Retinal pigmented epithelial cells obtained from human induced pluripotent stem cells possess functional visual cycle enzymes in vitro and in vivo. J Biol Chem. 2013;288: 34484–34493. 10.1074/jbc.M113.518571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwartz SD, Hubschman J-P, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379: 713–720. 10.1016/S0140-6736(12)60028-2 [DOI] [PubMed] [Google Scholar]
- 12. Booij JC, Brink JB ten, Swagemakers SMA, Verkerk AJMH, Essing AHW, van der Spek PJ, et al. A new strategy to identify and annotate human RPE-specific gene expression. PloS One. 2010;5: e9341 10.1371/journal.pone.0009341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janssen SF, van der Spek SJF, Brink JB Ten, Essing AHW, Gorgels TGMF, van der Spek PJ, et al. Gene expression and functional annotation of the human and mouse choroid plexus epithelium. PloS One. 2013;8: e83345 10.1371/journal.pone.0083345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Janssen SF, Gorgels TGMF, Bossers K, Brink JB Ten, Essing AHW, Nagtegaal M, et al. Gene expression and functional annotation of the human ciliary body epithelia. PloS One. 2012;7: e44973 10.1371/journal.pone.0044973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Strunnikova NV, Maminishkis A, Barb JJ, Wang F, Zhi C, Sergeev Y, et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum Mol Genet. 2010;19: 2468–2486. 10.1093/hmg/ddq129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41: W77–W83. 10.1093/nar/gkt439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coffey PJ, Gias C, McDermott CJ, Lundh P, Pickering MC, Sethi C, et al. Complement factor H deficiency in aged mice causes retinal abnormalities and visual dysfunction. Proc Natl Acad Sci U S A. 2007;104: 16651–16656. 10.1073/pnas.0705079104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nozaki M, Raisler BJ, Sakurai E, Sarma JV, Barnum SR, Lambris JD, et al. Drusen complement components C3a and C5a promote choroidal neovascularization. Proc Natl Acad Sci U S A. 2006;103: 2328–2333. 10.1073/pnas.0408835103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hollyfield JG, Bonilha VL, Rayborn ME, Yang X, Shadrach KG, Lu L, et al. Oxidative damage-induced inflammation initiates age-related macular degeneration. Nat Med. 2008;14: 194–198. 10.1038/nm1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ambati J, Anand A, Fernandez S, Sakurai E, Lynn BC, Kuziel WA, et al. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nat Med. 2003;9: 1390–1397. 10.1038/nm950 [DOI] [PubMed] [Google Scholar]
- 21. Beatty S, Koh H-H, Phil M, Henson D, Boulton M. The Role of Oxidative Stress in the Pathogenesis of Age-Related Macular Degeneration. Surv Ophthalmol. 2000;45: 115–134. 10.1016/S0039-6257(00)00140-5 [DOI] [PubMed] [Google Scholar]
- 22. Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134: 411–431. [DOI] [PubMed] [Google Scholar]
- 23. Nita M, Grzybowski A, Ascaso FJ, Huerva V. Age-related macular degeneration in the aspect of chronic low-grade inflammation (pathophysiological parainflammation). Mediators Inflamm. 2014;2014: 930671 10.1155/2014/930671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Behndig A, Svensson B, Marklund SL, Karlsson K. Superoxide dismutase isoenzymes in the human eye. Invest Ophthalmol Vis Sci. 1998;39: 471–475. [PubMed] [Google Scholar]
- 25. Newsome DA, Dobard EP, Liles MR, Oliver PD. Human retinal pigment epithelium contains two distinct species of superoxide dismutase. Invest Ophthalmol Vis Sci. 1990;31: 2508–2513. [PubMed] [Google Scholar]
- 26. Karunadharma PP, Nordgaard CL, Olsen TW, Ferrington DA. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51: 5470–5479. 10.1167/iovs.10-5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kondo N, Bessho H, Honda S, Negi A. SOD2 gene polymorphisms in neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Mol Vis. 2009;15: 1819–1826. [PMC free article] [PubMed] [Google Scholar]
- 28. Tokarz P, Kaarniranta K, Blasiak J. Role of antioxidant enzymes and small molecular weight antioxidants in the pathogenesis of age-related macular degeneration (AMD). Biogerontology. 2013;14: 461–482. 10.1007/s10522-013-9463-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kimura K, Isashiki Y, Sonoda S, Kakiuchi-Matsumoto T, Ohba N. Genetic association of manganese superoxide dismutase with exudative age-related macular degeneration. Am J Ophthalmol. 2000;130: 769–773. [DOI] [PubMed] [Google Scholar]
- 30. Pennesi ME, Neuringer M, Courtney RJ. Animal models of age related macular degeneration. Mol Aspects Med. 2012;33: 487–509. 10.1016/j.mam.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Justilien V, Pang J-J, Renganathan K, Zhan X, Crabb JW, Kim SR, et al. SOD2 knockdown mouse model of early AMD. Invest Ophthalmol Vis Sci. 2007;48: 4407–4420. 10.1167/iovs.07-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seo S, Krebs MP, Mao H, Jones K, Conners M, Lewin AS. Pathological consequences of long-term mitochondrial oxidative stress in the mouse retinal pigment epithelium. Exp Eye Res. 2012;101: 60–71. 10.1016/j.exer.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Imamura Y, Noda S, Hashizume K, Shinoda K, Yamaguchi M, Uchiyama S, et al. Drusen, choroidal neovascularization, and retinal pigment epithelium dysfunction in SOD1-deficient mice: a model of age-related macular degeneration. Proc Natl Acad Sci U S A. 2006;103: 11282–11287. 10.1073/pnas.0602131103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lengyel I, Flinn JM, Peto T, Linkous DH, Cano K, Bird AC, et al. High concentration of zinc in sub-retinal pigment epithelial deposits. Exp Eye Res. 2007;84: 772–780. 10.1016/j.exer.2006.12.015 [DOI] [PubMed] [Google Scholar]
- 35. Flinn JM, Kakalec P, Tappero R, Jones B, Lengyel I. Correlations in distribution and concentration of calcium, copper and iron with zinc in isolated extracellular deposits associated with age-related macular degeneration. Met Integr Biometal Sci. 2014;6: 1223–1228. 10.1039/c4mt00058g [DOI] [PubMed] [Google Scholar]
- 36. Newsome DA, Swartz M, Leone NC, Elston RC, Miller E. Oral zinc in macular degeneration. Arch Ophthalmol. 1988;106: 192–198. [DOI] [PubMed] [Google Scholar]
- 37. Smailhodzic D, van Asten F, Blom AM, Mohlin FC, Hollander AI den, van de Ven JPH, et al. Zinc supplementation inhibits complement activation in age-related macular degeneration. PloS One. 2014;9: e112682 10.1371/journal.pone.0112682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buschini E, Fea AM, Lavia CA, Nassisi M, Pignata G, Zola M, et al. Recent developments in the management of dry age-related macular degeneration. Clin Ophthalmol Auckl NZ. 2015;9: 563–574. 10.2147/OPTH.S59724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chew EY, Clemons T, SanGiovanni JP, Danis R, Domalpally A, McBee W, et al. The Age-related Eye Disease Study 2 (AREDS2): Study Design and Baseline Characteristics (AREDS2 Report Number 1). Ophthalmology. 2012;119: 2282–2289. 10.1016/j.ophtha.2012.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leung KW, Liu M, Xu X, Seiler MJ, Barnstable CJ, Tombran-Tink J. Expression of ZnT and ZIP Zinc Transporters in the Human RPE and Their Regulation by Neurotrophic Factors. Invest Ophthalmol Vis Sci. 2008;49: 1221–1231. 10.1167/iovs.07-0781 [DOI] [PubMed] [Google Scholar]
- 41. Dufner-Beattie J, Weaver BP, Geiser J, Bilgen M, Larson M, Xu W, et al. The mouse acrodermatitis enteropathica gene Slc39a4 (Zip4) is essential for early development and heterozygosity causes hypersensitivity to zinc deficiency. Hum Mol Genet. 2007;16: 1391–1399. 10.1093/hmg/ddm088 [DOI] [PubMed] [Google Scholar]
- 42. Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, Curletti CR, et al. The Pivotal Role of the Complement System in Aging and Age-related Macular Degeneration: Hypothesis Re-visited. Prog Retin Eye Res. 2010;29: 95–112. 10.1016/j.preteyeres.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chavali VRM, Khan NW, Cukras CA, Bartsch D-U, Jablonski MM, Ayyagari R. A CTRP5 gene S163R mutation knock-in mouse model for late-onset retinal degeneration. Hum Mol Genet. 2011;20: 2000–2014. 10.1093/hmg/ddr080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shu X, Luhmann UFO, Aleman TS, Barker SE, Lennon A, Tulloch B, et al. Characterisation of a C1qtnf5 Ser163Arg Knock-In Mouse Model of Late-Onset Retinal Macular Degeneration. PLoS ONE. 2011;6: e27433 10.1371/journal.pone.0027433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luo C, Chen M, Xu H. Complement gene expression and regulation in mouse retina and retinal pigment epithelium/choroid. Mol Vis. 2011;17: 1588–1597. [PMC free article] [PubMed] [Google Scholar]
- 46. Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38: 1173–1177. 10.1038/ng1890 [DOI] [PubMed] [Google Scholar]
- 47. Hageman GS, Hancox LS, Taiber AJ, Gehrs KM, Anderson DH, Johnson LV, et al. Extended haplotypes in the complement factor H (CFH) and CFH‐related (CFHR) family of genes protect against age‐related macular degeneration: Characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38: 592–604. 10.1080/07853890601097030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fritsche LG, Lauer N, Hartmann A, Stippa S, Keilhauer CN, Oppermann M, et al. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD). Hum Mol Genet. 2010;19: 4694–4704. 10.1093/hmg/ddq399 [DOI] [PubMed] [Google Scholar]
- 49. Rizzolo LJ. Barrier properties of cultured retinal pigment epithelium. Exp Eye Res. 2014;126: 16–26. 10.1016/j.exer.2013.12.018 [DOI] [PubMed] [Google Scholar]
- 50. Kaur C, Foulds WS, Ling EA. Blood-retinal barrier in hypoxic ischaemic conditions: basic concepts, clinical features and management. Prog Retin Eye Res. 2008;27: 622–647. 10.1016/j.preteyeres.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 51. Bailey TA, Kanuga N, Romero IA, Greenwood J, Luthert PJ, Cheetham ME. Oxidative stress affects the junctional integrity of retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2004;45: 675–684. [DOI] [PubMed] [Google Scholar]
- 52. Booij JC, van Soest S, Swagemakers SMA, Essing AHW, Verkerk AJMH, van der Spek PJ, et al. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics. 2009;10: 164 10.1186/1471-2164-10-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ervin JF, Heinzen EL, Cronin KD, Goldstein D, Szymanski MH, Burke JR, et al. Postmortem delay has minimal effect on brain RNA integrity. J Neuropathol Exp Neurol. 2007;66: 1093–1099. 10.1097/nen.0b013e31815c196a [DOI] [PubMed] [Google Scholar]
- 54. Mattapallil MJ, Wawrousek EF, Chan C-C, Zhao H, Roychoudhury J, Ferguson TA, et al. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci. 2012;53: 2921–2927. 10.1167/iovs.12-9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3: Article3. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bact was used to normalize gene expression in mouse CH, RPE and PR. The light blue bars indicate the CH, the blue bars indicate RPE and the dark blue bars indicate PR. Similar to the microarray data the expression level is highest in the RPE and lowest in the PR.
(TIF)
Dark blue bar indicate Bact expression level, the blue bars show the expression level of genes that were highly expressed on the microarray en light blue bars indicate expression of genes that showed very low expression on the microarray.
(TIF)
(TIF)
(TIF)
(TIF)
We analyzed the normalized expression intensities of the nine samples that were used in this study, with a multidimensional scaling plot. It shows that the samples within a group are highly similar.
(TIF)
Plot of the log of the red background-corrected signal versus the log of the green background-corrected signal for non-control inlier features for all nine samples after hybridization. The linearity of the plots indicate the appropriateness of background method choices.
(TIF)
The spatial distribution of up- and down-regulated features is evenly spread throughout the array for all nine samples.
(TIF)
Shown is the spread of the differentially expressed genes derived from the t-tests. The positive (higher expression in RPE) and negative (higher expression in the CH or in the PR) fold changes are approximately symmetrically divided.
(TIF)
Shown are the means and standard deviations in the mouse CH, RPE and PR.
(TIF)
Shown are the means and the standard deviation in the mouse RPE. The upper line indicates the cut-off value for the highest 10th percentile, the middle line the cut-off value for the 50th percentile and the lowest line the cut-off value for the lowest 10th percentile.
(TIF)
(XLSX)
(XLS)
(XLS)
(XLS)
(XLSX)
(XLS)
(XLS)
(XLSX)
(XLS)
(XLS)
(XLSX)
Data Availability Statement
All microarray files are available from the Gene Expression Omnibus database (accession number GSE66916).