Abstract
Scale up of Long Lasting Insecticide Nets (LLINs) has massively contributed to reduce malaria mortality across Africa. However, resistance to pyrethroid insecticides in malaria vectors threatens its continued effectiveness. Deciphering the detailed molecular basis of such resistance and designing diagnostic tools is critical to implement suitable resistance management strategies. Here, we demonstrated that allelic variation in two cytochrome P450 genes is the most important driver of pyrethroid resistance in the major African malaria vector Anopheles funestus and detected key mutations controlling this resistance. An Africa-wide polymorphism analysis of the duplicated genes CYP6P9a and CYP6P9b revealed that both genes are directionally selected with alleles segregating according to resistance phenotypes. Modelling and docking simulations predicted that resistant alleles were better metabolizers of pyrethroids than susceptible alleles. Metabolism assays performed with recombinant enzymes of various alleles confirmed that alleles from resistant mosquitoes had significantly higher activities toward pyrethroids. Additionally, transgenic expression in Drosophila showed that flies expressing resistant alleles of both genes were significantly more resistant to pyrethroids compared with those expressing the susceptible alleles, indicating that allelic variation is the key resistance mechanism. Furthermore, site-directed mutagenesis and functional analyses demonstrated that three amino acid changes (Val109Ile, Asp335Glu and Asn384Ser) from the resistant allele of CYP6P9b were key pyrethroid resistance mutations inducing high metabolic efficiency. The detection of these first DNA markers of metabolic resistance to pyrethroids allows the design of DNA-based diagnostic tools to detect and track resistance associated with bednets scale up, which will improve the design of evidence-based resistance management strategies.
Author Summary
Scale up of Long Lasting Insecticide Nets has massively reduced malaria mortality across Africa. However, resistance to pyrethroid insecticides in malaria vectors threatens its continued effectiveness. Here, we established that allelic variation in two CYP450s is the most important driver of pyrethroid resistance in the major African vector Anopheles funestus and detected key mutations controlling this resistance. The duplicated P450s CYP6P9a and CYP6P9b are directionally selected across Africa with alleles segregating according to resistance phenotypes. Alleles from resistant mosquitoes present significantly higher metabolic activities toward pyrethroids compared with alleles from susceptible mosquitoes. Furthermore, transgenic flies over-expressing resistant alleles of both genes were significantly more resistant to pyrethroids. Three amino acid changes from the resistant CYP6P9b allele are the key pyrethroid resistance mutations which induce high metabolic efficiency. The detection of these first DNA markers of metabolic resistance to pyrethroids allows the design of diagnostic tools to detect and track resistance.
Introduction
Despite the recent decrease in malaria mortality (47%) [1], the disease remains a serious public health burden in the tropical world, with 584,000 deaths globally in 2013, of which 90% occurred in WHO African region, and mostly in children under the age of 5. Malaria control relies heavily on the use of insecticide-impregnated LLINs and indoor residual spraying (IRS) [2]. Unfortunately, resistance to insecticides, especially pyrethroids (the only class approved by WHO for LLINs [3]), in major malaria vectors such as and An. funestus [4–6] and An. gambiae [7, 8] is threatening to derail these intervention tools [9]. An. funestus is widely, geographically distributed across Sub-Saharan Africa [10], and it has high vectorial capacity in some places surpassing even that of An. gambiae [11]. It reaches maximal abundance in the dry season when the density of An. gambiae and An. arabiensis have declined, thereby extending the period of malaria transmission [12]. Cases of resistance to pyrethroid, carbamate and organochlorine insecticides are increasingly reported in An. funestus populations across Africa [4, 5, 13–15]. It is imperative to design and implement suitable resistance management strategies to limit the impact of such resistance (WHO, 2012). One prerequisite is the development of appropriate diagnostic tools to facilitate the monitoring of insecticide resistance at an early stage, in order to inform control programs of the best course of action to take. However, the design of DNA-based diagnostic tools requires a thorough understanding of the molecular basis of the resistance.
To date, efforts to characterise mechanisms of resistance in malaria vectors have implicated knockdown resistance (kdr) and metabolic resistance through elevated expression of resistance genes, especially cytochrome P450s [6, 8] as the two major mechanisms conferring resistance to pyrethroids. No kdr mutation has been reported in the voltage-gated sodium channel of An. funestus [2]; pyrethroid resistance is therefore mainly metabolic. However, despite the numerous reports of implications of over-expressed P450s in pyrethroid resistance, the detailed molecular mechanisms through which they confer pyrethroid resistance in mosquitoes remain largely uncharacterised. It also remains unclear whether mechanisms other than P450 over-expression are also involved in resistance, for example, allelic variation with changes in the coding sequences through mutations of key amino acid residues or cis and/or trans mutations which could impact gene regulation [16]. Recent observation of polymorphism variations for the two most important pyrethroid resistance genes, CYP6P9a and CYP6P9b, in the malaria vector An. funestus [17, 18] suggests that this mosquito species is an excellent candidate to assess the impact of allelic variations of resistance genes on pyrethroid resistance. Recently, for the glutathione S-transferase gene, GSTe2, in An. funestus, a single point mutation (Leu119Phe) was detected and established to confer DDT resistance [15]. If such causative mutation(s) could be identified for pyrethroid resistance mediated by P450 genes, it will facilitate the design of DNA-based diagnostic tools to easily detect and track such resistance in field populations.
Knowledge of insect P450s’ preferential sites of metabolism of insecticides can facilitate the design of highly specific synergist inhibitors against the detoxification genes. For example, in insects and other organisms the 4´ spot of the phenoxybenzyl ring have been shown to be the preferential sites of hydroxylation [19, 20], which is followed by hydrolysis to generate intermediates including alcohols and acids which could easily be conjugated. On the other hand fluorogenic probes are increasingly used to identify inhibitors of P450s, and as diagnostic compounds to establish the degree of binding of insecticide substrates to P450s [21, 22].
In this study, sequence characterisation of CYP6P9a and CYP6P9b across Africa detected important polymorphism variations. In silico predictions, in vitro and in vivo functional characterisation tools were then applied to demonstrate that allelic variation in CYP6P9a and CYP6P9b is the key molecular change through which An. funestus mosquitoes acquire high resistance to pyrethroid insecticides. Furthermore, site-directed mutagenesis coupled with in vitro functional characterisation of the mutant recombinant proteins detected three major amino acid changes responsible for high pyrethroid metabolising efficiency of CYP6P9b from resistant populations of An. funestus s.s., compared to susceptible allele. This will allow the design of DNA-based diagnostic assay to detect and track this resistance in field populations in Africa.
Results
Africa-wide patterns of genetic variability of CYP6P9a and CYP6P9b
Analysis of cDNA polymorphism
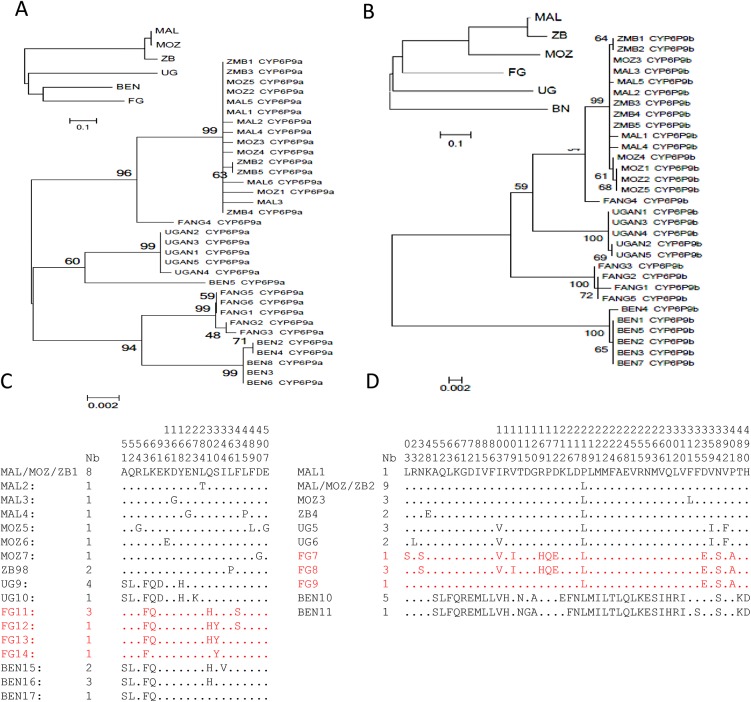
Analysis of the polymorphisms patterns of full-length cDNA sequences of CYP6P9a and CYP6P9b (1527bp for both) from different regions of Africa revealed a relative homogeneity within each geographical region but significant variations between haplotypes from different regions of Africa, and also the laboratory susceptible population (FANG). CYP6P9a has 17 haplotypes and 74 polymorphic sites of which 22 were non-synonymous, with the highest polymorphism observed in Benin and FANG (S1 Table). CYP6P9b has 11 haplotypes across Africa and 138 polymorphic sites the bulk of which were contributed from larger variations in the Benin and FANG alleles compared with Uganda and southern African alleles (Malawi, Mozambique and Zambia). Overall, the relative genetic homogeneity in each region is shown by (i) the fact that haplotypes from each country and region cluster together on the maximum likelihood phylogenetic tree (Fig 1A and 1B), notably those from the three southern African countries; (ii) the fact that only few amino acid changes (5 to 7) are observed within regions in contrast to the higher number of replacements between them (22 to 51) (S1 Table). A predominant haplotype is also observed for each gene in each region, notably across southern Africa (Fig 1C and 1D), where the predominant allele for each gene is the one previously associated with pyrethroid resistance when comparing alive (resistant) and dead (susceptible) mosquitoes after permethrin exposure in [4, 17]. Interestingly, the predominant haplotype in FANG was found exclusively in permethrin susceptible mosquitoes in southern Africa, suggesting that this allele is associated with susceptibility to pyrethroids [17]. The majority of the amino acid replacements are observed between the resistant haplotypes from southern Africa and those from FANG (Fig 1C and 1D) with the exception of CYP6P9b haplotype from Benin. These significant allelic variations are further supported by the neighbour-joining tree based on the Nst genetic distances showing high genetic distances between samples apart from those of southern Africa which are very close genetically (Fig 1A and 1B).
Fig 1. Schematic representation of haplotypes of An. funestus CYP6P9a and CYP6P9b from resistant populations and the susceptible strain FANG, across Africa.
(A) and (B) shows maximum likelihood tree of CYP6P9a and CYP6P9b cDNA sequences which forms clades specific to each phenotype. The small trees on the upper left of the maximum likelihood trees indicate the Nst genetic distances; (C) and (D) shows the polymorphic positions for both CYP6P9a and CYP6P9b amino acid sequences, respectively. A number is given to each haplotype preceded by country of origin (BEN, UGAN, MAL, MOZ, ZMB or FANG, for Benin, Uganda, Malawi, Mozambique, Zambia and FANG strains, respectively). The column Nb stands for the number of individual mosquitoes sharing a haplotype.
To determine if the allelic variations between the haplotypes could significantly impact the ability of these two genes in conferring pyrethroid resistance, a comparative analysis of the metabolic activity of all these alleles was carried out.
Allelic variations and in silico analysis of CYP6P9a and CYP6P9b alleles
Functional mapping of key amino acid changes
In comparison to FANG, for both CYP6P9a and CYP6P9b sequences, several amino acid changes observed in field resistant alleles were mapped to important domains of these P450s (S1 and S2 Figs).
Based on the haplotype differences, a single predominant allele in each region of Africa or in the FANG was chosen for further characterisation. For CYP6P9a, the selected alleles were: MAL/MOZ/ZB1 (Fig 1C) for southern Africa (hereafter MALCYP6P9a), UG9 for East Africa (UGANCYP6P9a), BEN16 for West Africa (BENCYP6P9a), and FG11 for FANG (FANGCYP6P9a). For CYP6P9b, the selected alleles were: MAL/MOZ/ZB2 (Fig 1D) for southern Africa (hereafter MALCYP6P9b), UG5 for East Africa (UGANCYP6P9b), BEN10 for West Africa (BENCYP6P9b) and FG8 for the FANG (FANGCYP6P9b). Both FANGCYP6P9a and FANGCYP6P9b were previously shown to be present only in the pyrethroid susceptible mosquitoes in southern Africa [17] and thus, are representative of the field pyrethroid susceptible mosquitoes.
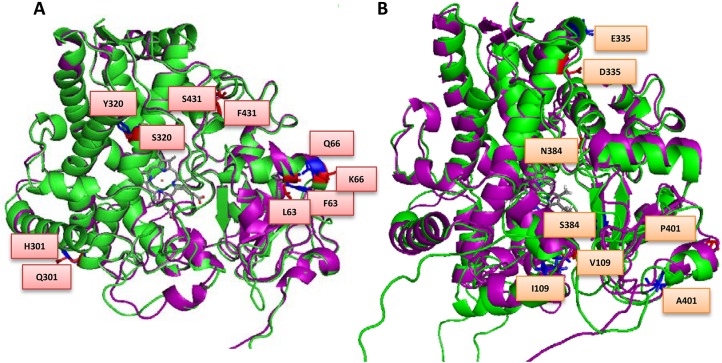
The predicted impact of the amino acids substitutions with relevant references used for the analysis are outlined in S2 Table, and the key mutations predicted to be important for catalysis are presented in Fig 2A and 2B, for both MALCYP6P9a and MALCYP6P9b in comparison to FANG.
Fig 2. Overlay of (A) MALCYP6P9a (green helices) and FANGCYP6P9a (purple helices) and (B) MALCYP6P9b (green helices) and FANGCYP6P9b (purple helices) showing amino acid residues changes.
Key residues are in stick format and those belonging to MALCYP6P9a or MALCYP6P9b are highlighted in red and annotated, while corresponding residues in FANGCYP6P9a and FANGCYP6P9b are in blue colour. Heme atoms are shown in stick format and grey.
Prediction of activity of allelic variants using molecular docking
Docking simulations predicted differences in the binding conformations and affinities of pyrethroids within the active sites of the different CYP6P9a and CYP6P9b models from resistant alleles compared with models from the susceptible alleles. Productive poses were defined as docking solutions with the 4´ spot of phenoxy ring approaching the heme iron (the preferred site of hydroxylation described for insect P450s [19, 23]), or the solutions with cis/trans-methyl group approaching the heme iron (minor route of hydroxylation) if the 4´ spot is away for optimal interaction to take place. Distances of between 3-6Å were considered productive to allow for optimal van der Waals contacts with minimum overlaps [24]. Higher ChemScore values and/or lower free energy of binding predicted by the GOLD software were considered to reflect higher catalytic activity.
Permethrin (Type I pyrethroid) docking
For CYP6P9a, permethrin docked into the active sites of models from resistant alleles (e.g. MALCYP6P9a) productively with trans methyl group oriented for hydroxylation at a distance of 4.2Å from the heme iron (S3A Fig), while in FANGCYP6P9a the insecticide docked with trans methyl group away (6.5Å) for optimal metabolism to occur (S3B Fig). The FANGCYP6P9a produced lowest ChemScore and highest free energy of binding suggesting that the susceptible allele may possess the lowest activity toward permethrin (S3 Table).
For CYP6P9b, permethrin docked productively in the model of the resistant allele (e.g. MALCYP6P9b) with the 4ʹ spot of the phenoxy ring oriented above the heme at a distance of 3.2Å (S3C Fig). The insecticide docked unproductively in the active site of FANGCYP6P9b with dichlorovinyl groups approaching the heme, and the possible sites of attack away from the catalytic centre: trans-methyl group located 10.4Å from heme iron (Figs 3D and S3D). Binding parameters for permethrin in the models of the resistant alleles and FANG are provided in S3 Table.
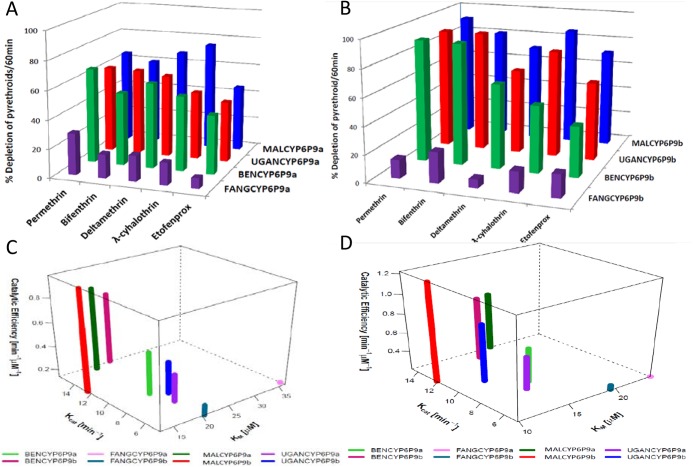
Fig 3. Substrates depletion assay results and kinetic profiles of recombinant CYP6P9a and CYP6P9b.
Percentage depletion of 20μM pyrethroids by (A) CYP6P9a and (B) CYP6P9b proteins. Results are average of three replicates (n = 3) compared with negative control (-NADPH); (C and D) 4D plot of the kinetic constants and catalytic efficiencies of CYP6P9a and CYP6P9b metabolism of permethrin and deltamethrin, respectively.
Deltamethrin (Type II pyrethroid) docking
Deltamethrin docked productively in the active site of MALCYP6P9a and MALCYP6P9b with trans methyl group within 3.5Å distance of the heme iron in MALCYP6P9a (S4A Fig) and the 4ʹ spot of phenoxy ring exposed for hydroxylation at reasonable distance of 3.7Å from the heme iron, in MALCYP6P9b (S4C Fig). The insecticide did not bind productively in the corresponding models from FANGCYP6P9a and FANGCYP6P9b. In FANGCYP6P9a, the insecticide docked with trans methyl group approaching the heme (6.0Å), but the dibromovinyl group projects toward the heme (S4B Fig). In FANGCYP6P9b the 4ʹ spot of phenoxy ring is the closest site of metabolism, but is located 8.9Å from heme iron for optimal interaction (S4D Fig). Binding parameters for deltamethrin in the models of the resistant alleles and FANG are provided in S3 Table.
Moreover, MALCYP6P9b allele was predicted to possess highest activity towards both permethrin and deltamethrin, compared with all the alleles of CYP6P9a and CYP6P9b analysed. In addition, the docking software predicted cis/trans-methyl hydroxylation to occur as well from MALCYP6P9b-mediated metabolism of deltamethrin, leading to multiple primary metabolites.
Comparison of substrate access channels between CYP6P9a and CYP6P9b models
Identification of pw2a substrates access channel
Because substrate binding and product release by P450s may impact their role in drug metabolism [25], a molecular simulation of substrate access and/or product egress routes was performed for all CYP6P9a and CYP6P9b models. The algorithm predicted that the models from the resistant and susceptible alleles of CYP6P9a alleles did not differ in their substrate access and/or product egress channels. In contrast, differences were observed in the channels composition and parameters between the models of resistant (S5A Fig) and susceptible (S5B Fig) alleles from CYP6P9b. For example, for MALCYP6P9b model, a total of 30 tunnels were predicted from the interior active site of the protein to the bulk solvent. Of these channels the first ranked with highest throughput (0.76), lowest energy cost (0.27) and which is longest (21.42Å) has its gorge lined with about 41 residues (S5A Fig). One of these residues is the Val109 from BʹC loop which was established to be a member of channel pw2a proposed to be the most common route of access and egress for most CYP450s [26, 27]. Channel pw2a has been established as the most energetically favourable of all the pathways observed in CYP101 [27] and was described as a product egress route for CYP101 and CYP2B4. This later property was possible because the pw2a is a holey channel (wide-open); a property made possible due to the potential motion in the FG loop and/or the BC loop. Val109 of SRS-1, a substitution common to all resistant alleles of CYP6P9b is located within contact distance (2.9Å) of this pw2a tunnel. For CYP6P9b the mutation Ile109Val possibly modifies the substrates accessing machinery in the resistant allele with impact on substrate recognition and/or affinity.
CAVER computed 23 tunnels for FANGCYP6P9b of which the first ranked with lower throughput (0.74), has higher cost (0.31) and is almost half short (13.91Å) (S5B Fig) compared with the first-ranked tunnel from MALCYP6P9b model. The corresponding Ile109 of FANGCYP6P9b is located 7.7Å from the tunnel and is therefore not a tunnel lining residue in all of the 23 predicted channels. This further strengthened the importance of the mutation Val109 in the resistant alleles of CYP6P9b. However, this simulation was conducted with deltamethrin only, although other substrates could access or egress differently in CYP6P9b or other P450s.
Comparative assessment of metabolic activity of CYP6P9a and CYP6P9b alleles using in vitro functional characterisation
Having established that the allelic variations observed between CYP6P9a and CYP6P9b haplotypes induced significant differences in their 3D structures and possibly their ability to interact and metabolise pyrethroid insecticides, we next validated these observations through in vitro and in vivo characterisation experiments.
In vitro expression and metabolism assays with recombinant enzymes
Recombinant proteins of the various CYP6P9a and CYP6P9b alleles were produced with optimal expression between 36–56 hours as described previously [4]. No significant differences were observed in P450 content and cytochrome P450 reductase activity between the recombinant proteins expressed from the various alleles of CYP6P9a and CYP6P9b (hereafter named resistant alleles) compared to proteins obtained from the FANG alleles (hereafter referred to as susceptible alleles) (S6A and S6B Fig). Thus, reductase content may not induce any allelic variation in metabolic activities.
Pyrethroid metabolism assays
Recombinant enzymes from resistant alleles of both CYP6P9a and CYP6P9b metabolize Type I (permethrin and bifenthrin) and Type II (deltamethrin and λ-cyhalothrin) pyrethroids, as well as etofenprox, with statistically significant depletion (+NADPH vs–NADPH) (S4 Table and Fig 3A and 3B). In contrast, only very low and non-significant depletions (not more than 20%) were obtained with proteins from susceptible alleles, for both genes. These results further support the observation that allelic variation in both genes is significantly impacting their ability to metabolise pyrethroid insecticides. In agreement with the docking results no major differences in pyrethroid metabolism were observed between the different field resistant alleles, although on average, proteins from the southern African alleles had higher depletion compared to corresponding proteins from other regions of Africa. For example, with CYP6P9a alleles, MALCYP6P9a exhibited highest activity against λ-cyhalothrin with a depletion of 75.41±1.72 significantly higher than for UGANCYP6P9a (47.74±1.5) (p<0.05). The same pattern was observed when depletion of λ-cyhalothrin and etofenprox from MALCYP6P9b was compared with values from BENCYP6P9b (p<0.05) (S4 Table). Overall, highest metabolic activities were observed from recombinant CYP6P9b proteins especially MALCYP6P9b which deplete more than 90% of permethrin and more than 80% of bifenthrin, deltamethrin and λ-cyhalothrin. CYP6P9b alleles consistently exhibited higher activities toward permethrin, bifenthrin and deltamethrin compared with CYP6P9a counterparts.
To further assess the impact of allelic variation on activity towards pyrethroids, steady-state kinetic parameters were established with permethrin and deltamethrin, for each CYP6P9a and CYP6P9b membrane. Reactions proceed via Michaelis-Menten mechanism (S7 Fig), with K m values within ranges (1–50μM) previously described for binding and metabolism of pyrethroid by insect P450s [28]. For CYP6P9a with permethrin, the K cat value obtained with MALCYP6P9a was the highest and 3-fold higher than the amount obtained with the FANGCYP6P9a from the susceptible allele (p<0.05) (Fig 3C and S5 Table). The K cat from Benin and Uganda recombinant proteins with permethrin, were also 2-fold higher than values from FANG (BENCYP6P9a vs FANGCYP6P9a, p<0.05). In terms of affinity, the susceptible FANGCYP6P9a proteins exhibited significantly higher K m, two-fold the values obtained with proteins from the resistant alleles (p<0.05). These differences translated into variation in catalytic efficiencies with MALCYP6P9a exhibiting the highest efficiency, 6-fold more efficient than FANGCYP6P9a (p<0.05), while BENCYP6P9a and UGANCYP6P9a were also 3-fold higher in activity compared with FANGCYP6P9a (p<0.05).
The same pattern of allelic differences in activities was also observed with respect to CYP6P9b (Fig 3). BENCYP6P9b exhibited the highest activity for permethrin compared with all the other alleles, with its K cat and that from MALCYP6P9b and UGANCYP6P9b 4-fold, 3-fold and 2-fold respectively higher compared to the K cat obtained from FANGCYP6P9b (p<0.05) (Fig 3C and S5 Table). No major differences were observed in terms of affinity toward permethrin (K m) between FANGCYP6P9b and the resistant alleles from East (UGANCYP6P9b) and West Africa (BENCYP6P9b), whereas, the southern African allele (MALCYP6P9b) significantly differs by producing low, reproducible K m (on average half the values obtained from FANGCYP6P9b as well as the other resistant alleles of CYP6P9b, p<0.05). These differences in kinetic parameters between resistant and susceptible alleles were reflected in the catalytic efficiency, with MALCYP6P9b 5-fold more efficient in permethrin metabolism than FANGCYP6P9b (p<0.05).
For deltamethrin, similar higher activities and catalytic efficiencies were obtained from the recombinant proteins expressed from the resistant alleles compared with the FANG (Fig 3D and S5 Table). Further analysis of kinetic parameters obtained with deltamethrin is discussed in more detail in S1 Text.
Consistent with the in silico predictions, the catalytic efficiencies for both pyrethroids metabolism by the alleles of CYP6P9b were higher than values from CYP6P9a. The K m of the southern African allele of CYP6P9b was also significantly lower compared with southern African CYP6P9a allele (p<0.05) (S5 Table), reflecting the pre-eminence of CYP6P9b over CYP6P9a in terms of pyrethroid metabolism and conferment of resistance.
Fluorogenic probes assays
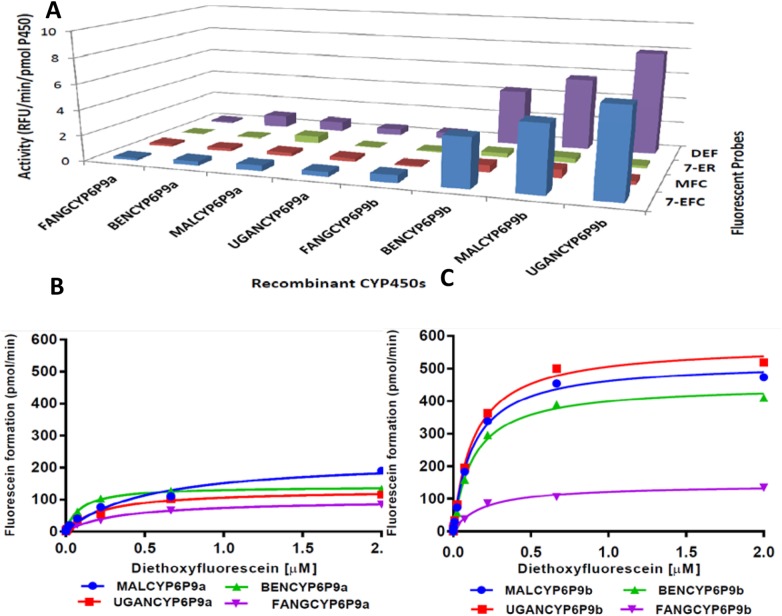
Fluorescent probes were screened to determine whether the different recombinant CYP6P9a and CYP6P9b proteins exhibit variations in O-dealkylation. Highest activities were observed with diethoxyfluorescein (DEF) and 7-ethoxy-4-trifluoromethylcuomarin (7-EFC) and low activities with 7-ethoxyresorufin (7-ER) and 7-methoxyfluoromethylcuomarin (MFC) (Fig 4A). Higher activities were consistently recorded for CYP6P9b than CYP6P9a, for example in Uganda a 17.9- and 15.9-fold activity was observed respectively toward 7-EFC and DEF for UGANCYP6P9b than UGANCYP6P9a. A comparison of the activity between recombinant proteins shows that for both genes, more evidently for CYP6P9b, the recombinant protein from FANG presented a significantly lower activity than proteins from all the field resistant alleles. For example, a 2.85- and 11.3-fold more activity was observed toward DEF, respectively for southern African CYP6P9a and CYP6P9b compared with corresponding proteins from FANG reflecting the higher O-dealkylation property of the resistant alleles. For both genes, no significant difference is observed between alleles from southern Africa and those from East and West Africa suggesting that the nature of the many amino acid changes observed between these alleles and their location do not critically affect their dealkylation activity.
Fig 4. Metabolism of probe substrates by various recombinant CYP6P9a and CYP69b proteins.
(A) Initial testing of O-dealkylation. The solid bars indicate average of significant turnovers of three experimental replicates compared to negative controls (-NADPH). DEF, diethoxyfluorescein; 7-ER, 7-ethoxyresorufin; MFC, 7-methoxyfluoromethylcuomarin; and 7-EFC, 7-ethoxy-4-trifluoromethylcuomarin. (B and C) Michaelis-Menten plots for recombinant (B) CYP6P9a and (C) CYP6P9b metabolism of diethoxyfluorescein.
The dealkylation of DEF follows Michaelis-Menten fashion (Fig 4B and 4C respectively), with proteins from resistant alleles exhibiting high activity and affinity compared with those from susceptible FANG (S6 Table). For CYP6P9a, turnover of DEF with MALCYP6P9a was 2-fold higher than obtained with FANGCYP6P9a, leading to catalytic efficiency more than 5-fold higher (p<0.05) (S8A Fig). The catalytic efficiency with UGANCYP6P9a and BENCYP6P9a were also respectively 2- and 6-folds higher than values obtained from FANGCYP6P9a (p<0.05) (S6 Table), reflecting the higher activity in the resistant alleles. For recombinant CYP6P9b, FANGCYP6P9b exhibited lower affinity (K m roughly two-fold the values obtained with proteins from the resistant alleles), and lower K cat (S8A Fig and S6 Table). These profound differences in the affinity and maximal activities are reflected in the catalytic efficiencies of CYP6P9b from resistant alleles toward DEF, which were several orders of magnitude (~20 fold) higher than the FANGCYP6P9b activity (p<0.05).
Overall, recombinant proteins from CYP6P9b alleles exhibited higher O-dealkylation activity than CYP6P9a alleles with K cat and catalytic efficiencies on average more than 10-fold the values obtained from CYP6P9a proteins suggesting a higher enzymatic activity in the former compared to the latter.
Inhibition assays
To determine whether the different field resistant alleles may exhibit variations in degree of binding to different insecticides, the southern African allele was compared to the Benin allele for CYP6P9a and to the Ugandan allele for CYP6P9b in an inhibition assay with insecticides from various classes, including Type I pyrethroids (permethrin and deltamethrin), Type II pyrethroids (deltamethrin, λ-cyhalothrin and cypermethrin), the pseudo-pyrethroid etofenprox, organochlorine (DDT), an organophosphate (chlorpyrifos), and two carbamate insecticides (bendiocarb and propoxur). Overall, the alleles exhibited a similar binding pattern to all the insecticides (inhibitors) screened (S8B Fig). Type II pyrethroids showed the most potent inhibitory activity against CYP6P9a/CYP6P9b-mediated dealkylation of DEF, with IC50s of less than 0.5μM indicating tighter binding compared with Type I pyrethroids, the pseudo-pyrethroid etofenprox and the organophosphate, chlorpyrifos. With DDT, the IC50 was averagely 10μM for all proteins excepting UGANCYP6P9b with a much higher value, suggestive of moderate binding of these P450s to this organochlorine, though the low affinity of DDT might be caused by its feeble solubility (low partitioning in water) compared with pyrethroids [29]. IC50 values higher than 25μM were obtained with bendiocarb and propoxur suggesting that these insecticides bind poorly to the P450s compared to other inhibitors tested. Tight-binding inhibitors have been defined as compounds with IC50 values of <10μM [30].
To establish the robustness of the docking analyses performed with the GOLD software, log IC50s of the pyrethroids permethrin, deltamethrin and the pseudo-pyrethroid etofenprox from assays with MALCYP6P9b were compared with the ChemScore values obtained from in silico docking of the same insecticides into MALCYP6P9b model. For all three pyrethroids, significantly good correlation was obtained between the experimental IC50s and the predicted ChemScores (S8C Fig).
Assessment of the importance of allelic variation in pyrethroid resistance using transgenic expression in Drosophila melanogaster
The extent of the role played by allelic variation of these P450s in pyrethroid resistance was determined in order to establish whether it is alone sufficient to confer resistance in vivo, even more than gene over-expression. For this purpose, the most resistant (MAL) and the susceptible alleles (FANG) of each gene were over-expressed in vivo through a transgenic expression using the GAL4/UAS system, and their ability to confer resistance compared using contact bioassays with permethrin and deltamethrin. qRT-PCR confirmed that both CYP6P9a and CYP6P9b were expressed only in the transgenic F1 progenies from the crosses between the Actin5C-GAL4 driver line and the different UAS-CYP6P9 lines, and not expressed in the control flies (S9A and S9B Fig) (see S1 Text). Control flies are generated by crossing flies with the same background as the experimental group (but devoid of the UAS element and the candidate gene), with the driver (Actin5C-GAL4) lines to generate Actin5C-GAL4-null lines without candidate P450s.
Pyrethroids contact bioassays
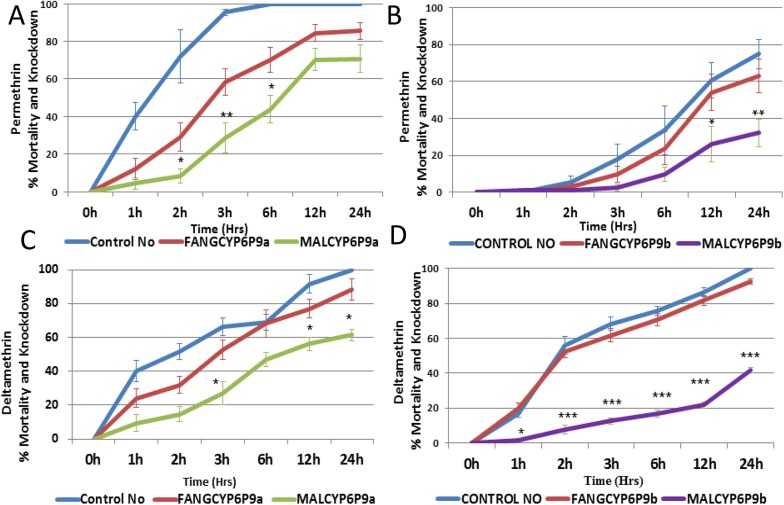
Bioassays with permethrin and deltamethrin revealed that the flies over-expressing the resistant MALCYP6P9a and MALCYP6P9b alleles were resistant to pyrethroids, with a very low mortality and knockdown compared with the flies over-expressing the susceptible FANGCYP6P9a and FANGCYP6P9b alleles, or the control group. For permethrin, progenies of crosses between the Actin5C-GAL4 and UAS-MALCYP6P9a (transgenic flies over-expressing MALCYP6P9a) showed significantly lower mortality (less than 10%, <30% and <45% in the first 2 hrs, 3 hrs and 6 hrs, respectively) compared with progenies of crosses between Actin5C-GAL4 and UAS-FANGCYP6P9a (30%, ~60% and ~70% mortality respectively in 2 hrs, 3 hrs and 6 hrs) (Fig 5). The same pattern was observed for CYP6P9b with transgenic flies over-expressing MALCYP6P9b showing significantly lower mortalities compared with the transgenic flies over-expressing the susceptible allele (FANGCYP6P9b) at the two exposure times of 12 hrs (<30% vs 55%) and 24 hrs (~30% vs ~60%).
Fig 5. Bioassay results with transgenic flies.
(A) Progenies of crosses between Actin5C-GAL4 and UAS-CYP6P9a (transgenic flies over-expressing CYP6P9a) with permethrin; (B) transgenic flies over-expressing CYP6P9b with permethrin; (C) transgenic flies over-expressing CYP6P9a with deltamethrin; and (D) transgenic flies over-expressing CYP6P9b with deltamethrin. Data shown as mean ±S.E.M. significantly different: * p<0.05, ** p<0.01 and *** p<0.001.
With deltamethrin, transgenic flies over-expressing MALCYP6P9a showed lower mortalities compared with transgenic flies over-expressing susceptible allele (FANGCYP6P9a), although these mortalities were only significant at the third (<30% vs 52%), twelfth and twenty fourth hours of exposure (Fig 5). For CYP6P9b, transgenic flies over-expressing the resistant allele MALCYP6P9b consistently showed significantly lower mortalities than flies over-expressing susceptible allele (FANGCYP6P9b). Statistically significant differences were obtained at every hour of exposure: 1 hr (1.5% vs 20%), 2 hrs (7.5% vs 53%), 3 hrs (12.5% vs 62%), 6 hrs (16% vs 70%), 12 hrs (21% vs 81%) and 24 hrs (41% vs 93%).
Similar to results from in vitro assays, for both pyrethroids especially deltamethrin, flies over-expressing MALCYP6P9b were significantly more resistant than those over-expressing MALCYP6P9a reflecting the pre-eminence of CYP6P9b with respective to pyrethroids metabolism and conferring of resistance.
Detection of causative mutations using site-directed mutagenesis
Expression patterns of CYP6P9b mutant recombinant proteins
Mutant recombinant proteins for CYP6P9b were successfully expressed along with the wild type MALCYP6P9b at 21°C and 150 rpm using E. coli cells JM109 (see S1 Text).
Pyrethroid metabolism assays
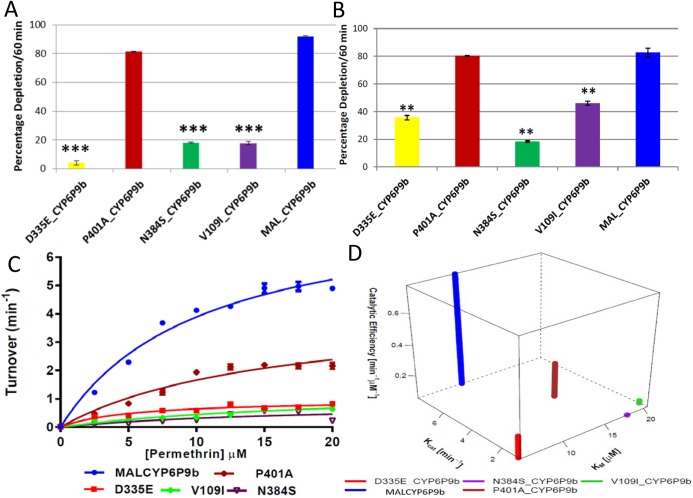
Initial depletion assays with permethrin and deltamethrin consistently revealed striking differences in the metabolic profiles of wild type resistant MALCYP6P9b (wrMALCYP6P9b) compared with membranes expressing the mutant CYP6P9b proteins. The mutants Asp335Glu and Asn384Ser, as well as Val109Ile exhibited significantly low activity against these pyrethroid insecticides, especially deltamethrin (Fig 6A). Indeed, a 23-, 5.1 and 5.0-fold less depletion is observed for deltamethrin when the resistant allele was mutated to the susceptible allele in positions 335, 109 and 384, respectively. The Pro401Ala replacement did not induce a significant change in deltamethrin depletion compared to wrMALCYP6P9b. The mutations showed a similar trend against permethrin (Fig 6B) but for Asp335Glu and Val109Ile a lower impact was observed than against deltamethrin with reduced depletion of 2.15- and 2-fold respectively, compared with amount of permethrin depleted by the wrMALCYP6P9b protein. However, for Asn384Ser mutant activities toward both permethrin and deltamethrin were similar suggesting that the 384 replacement confer the same impact on both permethrin and deltamethrin.
Fig 6. Substrate depletion and kinetic profiles of mutant CYP6P9b recombinant proteins.
(A and B) are percentages of depletion of 20μM deltamethrin and permethrin by CYP6P9b mutants, respectively. Results are average of three replicates (n = 3) compared with negative control (-NADPH). (C) Michaelis-Menten plots of mutant CYP6P9b proteins metabolism of permethrin; (D) 4D plot of the kinetic constants and catalytic efficiencies of mutant CYP6P9b proteins for metabolism of permethrin.
Metabolism of permethrin by the mutant membranes follow Michaelis-Menten mechanism (Fig 6C) with wrMALCYP6P9b having the highest catalytic activity (K cat), values 8-fold than that of Asp335Glu, 9-fold the K cat from Asn384Ser and 6-fold as obtained with Val109Ile mutant) (S7 Table). Val109Ile and Asn384Ser mutants exhibited the lowest affinity toward permethrin, with a higher K m, on average 2-fold as obtained with wrMALCYP6P9b and 4-fold the K m of Asp335Glu mutant. Pro401Ala mutant also exhibited lower K cat, half the value obtained with wrMALCYP6P9b but with K m values on average one-third higher than the wild type enzyme. Thus, the catalytic efficiency wrMALCYP6P9b was calculated on average as 3-fold the efficiency of Pro401Ala mutant, 4-fold the value obtained with Asp335Glu mutant, 13-fold higher than that of Val109Ile mutant and 20-fold higher than the efficiency of Asn384Ser mutant, showing that each of these amino acid change significantly affect the efficiency of CYP6P9b to metabolise permethrin efficiently.
The differences in the metabolic profiles of the wrMALCYP6P9b compared to the mutants can also be rationalised from the spatial positioning of the two residues Asp335 and Asn384 in MALCYP6P9b model compared with the corresponding residues Glu335 and Ser384 in FANGCYP6P9b. Overlay of MALCYP6P9b and FANGCYP6P9b models revealed striking differences in the overall 3D folding and backbone of the proteins (S10 Fig). These differences may result in differences in the manner through which these residues interact with the substrates or the redox partners with impact on catalysis (see S1 Text).
Fluorogenic probes assays
Differences in activities toward probe substrates were also observed between MALCYP6P9b and the mutants (S11A Fig). The wild type membrane exhibited higher activity towards diethoxyfluorescein (DEF) with catalytic efficiencies more than 8-fold, 10-fold and 15-fold greater than values from Asp335Glu, Val109Ile and Asn384Ser mutants, respectively (see S1 Text for further details).
Discussion
Detecting insecticide resistance at early stage is one of the prerequisite for the design and implementation of effective insecticide resistance management strategies. DNA-based diagnostic tools are essential for this purpose, but the diagnostics require a thorough understanding of the molecular basis of insecticide resistance. In this study, we dissected the molecular basis of a monooxygenase-mediated resistance to pyrethroid in one of the major African malaria vectors An. funestus demonstrating that (i) allelic variation in cytochrome P450 genes is a key molecular mechanism conferring pyrethroid resistance in field mosquitoes, and (ii) detecting key amino acid changes in resistant P450 alleles responsible for pyrethroids-metabolising efficiency. The finding of these resistance markers could help in the design of DNA-based diagnostic tools that will facilitate the detection and tracking of such resistance markers in the field.
Allelic variation is a key mechanism conferring pyrethroid resistance
This study has provided several evidences supporting the key role of allelic variation in conferring pyrethroid resistance.
Evidences from Africa-wide patterns of genetic variability of CYP6P9a and CYP6P9b alleles and in silico prediction of activity
Analysis of nucleotide sequences of different alleles of CYP6P9a and CYP6P9b across Africa revealed that the alleles from resistant populations are undergoing directional selection, with reduced genetic diversity and beneficial mutations selected, compared with the alleles from the susceptible population, FANG. Such signature of selection is similar to that reported in CYP6G1 P450 from D. melanogaster [31]. In silico prediction suggested that the allelic variants differ in their activity towards pyrethroids. Pyrethroids docked into the models of susceptible alleles in unproductive mode or away from the heme catalytic centre for catalysis to occur. In silico predictions have been applied in several studies to predict the ability of insect P450s to metabolise pyrethroids and other substrates [16, 19, 22].
Evidences from variation in metabolic activity towards pyrethroid and probe substrates
Metabolism assays established that the resistant alleles of CYP6P9a and CYP6P9b metabolise pyrethroid insecticides with higher turnover compared to the susceptible alleles. Highest activities were obtained from southern African alleles consistent with the highest pyrethroid resistance recorded in this region [4, 17, 32]. Kinetic parameters also differ significantly between resistant alleles of CYP6P9a and CYP6P9b and the susceptible ones. The resistant alleles exhibited high turnover (high K cat) and high affinity (lower K m) compared with the susceptible alleles, which translates into a very high efficiency of pyrethroid metabolism several-fold in the former compared with the latter. These findings established that allelic variation is impacting the pyrethroid metabolising efficiencies of resistant alleles of CYP6P9a and CYP6P9b. The kinetic parameters are compared with results established from several other studies with pyrethroids, as well as the probes assays in more detail in S1 Text.
Evidence from transgenic expression
Expression of alleles of CYP6P9a and CYP6P9b from resistant populations alone confers resistance to pyrethroids permethrin and deltamethrin in vivo, showing that allelic variation could even be more important in conferring resistance than gene over-expression. However, because resistant CYP6P9a and CYP6P9b alleles are always found to be also over-expressed in the field, it is likely that allelic variation combines with increased expression in this resistance mechanism as previously reported in other species notably in humans [33]. The ability of the transgenic expression to reveal phenotypic difference caused by gene polymorphism in CYP6P9a and CYP6P9b further highlights the reliability of this approach in characterising the functional role of genes in relation to phenotypes as done in other species such as Tribolium castaneum (CYP6BQ9) [34] and D. melanogaster (CYP6G1 and CYP12D1) [35].
Impact of allelic variation has previously been reported in the CYP6A2 P450 gene in D. melanogaster with regard to DDT resistance [36] suggesting that allelic variation of key resistance genes could be common in insect species (see S1 Text for further details).
Key amino acid changes control pyrethroid resistance and could lead to DNA-based diagnostic tools
Detection of DNA-based markers of metabolic resistance has so far proved challenging because of the redundancy of genes involved but also because of the multitude of mechanisms through which these mechanisms can operate. Here, using site-directed mutagenesis and comparative recombinant enzyme characterisation we established, for the first time in our knowledge in a mosquito species, that three amino acid replacements in the P450 CYP6P9b are responsible for high metabolic activity toward pyrethroids as their replacement in MALCYP6P9b with variants from the susceptible FANGCYP6P9b correlated with significant loss of catalytic activity and metabolic efficiency towards permethrin. We suggest that the three residues (Val109, Asn384 and Asp335) in the protein from the resistant alleles of CYP6P9b contribute in unique ways to create ensemble complementarity groups on the P450 active sites, a potent pharmacophore which recognise key features on the pyrethroid substrates, allowing optimal inter-molecular interactions and enhancing affinity and/or catalysis.
Impact of amino acid changes in metabolic activity of CYP450 genes have been described previously in relation to metabolisms of other chemicals but not against pyrethroids. Indeed, the Glu318Asp substitution in human CYP1A2 has been shown to increase the K cat of O-dealkylation of 7-ethoxycoumarin 13-fold [37] (see S1 Text).
As these are the first markers of metabolic resistance to pyrethroids involving P450s in An. funestus and/or even other mosquito species, these mutations should facilitate the design of DNA-based diagnostic tests for tracking resistance in the field. However, unlike the case of DDT-resistance gene GSTe2 (Leu119Phe substitution) in An. funestus, the CYP6P9a and CYP6P9b are duplicated genes with high sequence similarity which will require particular attention in the design of diagnostic assays. The detection of these causative mutations in CYP6P9b also set a pace for studies of this kind to be carried out in other insect vectors for which allelic variation in key resistance genes could also play an important role.
Conclusion
This study presents a detailed dissection of the genetic and molecular basis of metabolic resistance to insecticide in a major malaria vector demonstrating that allelic variation of key P450 genes is responsible for the resistance to pyrethroids, the bed nets insecticides. Key amino acid changes between resistant and susceptible alleles specifically, three residues, Val109, Asp335 and Asn384 in the resistant CYP6P9b alleles accounted for these metabolic differences. The finding and characterisation of these resistance markers paves a way to design a DNA-based diagnostic test that can allow tracking of these resistant alleles across Africa, enabling the design of evidenced-based resistance management strategies to mitigate the impact of this resistance on the success of ongoing and future control interventions.
Methods
Sequence characterisation and in silico analysis of CYP6P9a and CYP6P9b alleles
Polymorphism analysis of CYP6P9a and CYP6P9b alleles
To establish the presence of allelic variation in both CYP6P9a and CYP6P9b, the full-length cDNA of both genes was amplified from permethrin resistant mosquitoes from three regions of Africa with different resistance profiles, as well as from the susceptible laboratory strain FANG, originally from Angola. The resistant populations were An. funestus s.s. from southern Africa: Chikwawa in Malawi, Chokwe in Mozambique and Katete in Zambia [4, 17, 32], East-Africa: Tororo in Uganda [38], and West-Africa: Pahou in Benin [39]. Pyrethroid-resistance profiles of these mosquito populations as well as their relevant transcriptional analysis have been previously published [17, 38, 39]. The fully susceptible FANG from Calueque, southern Angola [40] was utilised as a susceptible population.
Full-length sequences of CYP6P9a and CYP6P9b were amplified using cDNA synthesised from total RNA extracted from three batches of 10 mosquitoes from Benin, Uganda, Malawi, Mozambique and Zambia, and the FANG, as previously described [17] (primers given in S8 Table). Polymorphisms were detected through manual examination of sequence traces using BioEdit version 7.2.3.0 [41] and sequence differences in multiple alignments using CLC Sequence Viewer 6.9 (http://www.clcbio.com/). Different haplotypes were compared by constructing a phylogenetic maximum likelihood tree using MEGA 6.0 [42]. Genetic parameters of polymorphism of both genes for each sample were determined using DnaSP 5.10 [43]. Locations of amino acids differences within the sequences of CYP6P9a and CYP6P9b were mapped by identifying helices A-L and substrate recognition sites (SRS1-6) using crystal structures [44, 45] and structurally conserved regions using CYPED database [46].
Comparison of various CYP6P9a and CYP6P9b alleles using molecular docking simulation
To predict the impact of observed amino acid changes on the structure of CYP6P9a and CYP6P9b models of both genes were created using MODELLER 9v2 [47] and CYP3A4 (PDB: 1TQN) [48] as a template, with 33% and 32% identity, respectively for CYP6P9a and CYP6P9b sequences. Ligand structures were retrieved from ZINC12 library (https://zinc.docking.org/). 3D protein models and ligands were prepared for docking using Molegro Molecular Viewer 2.5 (http://www.clcbio.com/). To predict the pattern of interaction between the genes and insecticides, docking was carried out with GOLD 5v2 [49], with ChemScore scoring function [50] and active site defined as a cavity of 20Å radius centred above the heme iron. 50 binding poses were obtained for each ligand (permethrin, deltamethrin and etofenprox) and scored according to binding parameters and the conformation of ligands in the active site of the respective P450s. Figures were prepared using the PyMOL 1.7 [51].
In silico prediction of substrate access/product egress channels
To identify potential substrate access and/or product egress channels between the binding sites of pyrethroids to the bulk solvent, comparative channels search was conducted using models generated from sequences of resistant alleles and those from the susceptible (FANG) alleles. Channel searches was conducted using the algorithm tool CAVER 3.1 [52], with settlings for tunnels calculations as described in the PyMOL plugin [53].
In vitro functional characterisation of metabolic activity of various recombinant CYP6P9a and CYP6P9b proteins
Cloning and heterologous expression of recombinant CYP6P9a and CYP6P9b in E. coli
Recombinant enzymes of both CYP6P9a and CYP6P9b genes were expressed for the different alleles. Expression plasmids pB13::ompA+2-CYP6P9a and pB13::ompA+2-CYP6P9b for the alleles: Benin-CYP6P9a and–b (hereby after BENCYP6P9a and BENCYP6P9b), Uganda (UGANCYP6P9a and UGANCYP6P9b), Malawi (MALCYP6P9a and MALCYP6P9b) and FANG (FANGCYP6P9a and FANGCYP6P9b) were constructed by fusing cDNA fragment from a bacterial ompA+2 leader sequence with its downstream ala-pro linker to the NH2-terminus of the P450 cDNA, in frame with the P450 initiation codon, as described [54]; and then cloned into NdeI- and XbaI-linearised pCW-ori+ vector [55]. Details of PCR conditions used to create this type of expression plasmid cassettes have already been described [4, 17] and list of primers are provided in S8 Table.
For all alleles, E. coli JM109 cells were co-transformed with the P450 expression cassettes and a plasmid containing the An. gambiae cytochrome P450 reductase (pACYC-AgCPR) fused to pelB leader sequence [56]. Membrane expression and preparations, measurement of P450 content, measurement of cytochrome c reductase activity, cytochrome b5 expression and measurement of its content were carried out as previously described [19, 57, 58].
Comparative assessment of pyrethroids metabolic activity of various recombinant CYP6P9a and CYP69b alleles using metabolism assays
To establish whether allelic variation in CYP6P9a and CYP6P9b impact their metabolic activities toward insecticides, pyrethroids were screened with the recombinant enzymes from these genes. As previously described [17, 28], 0.2M Tris-HCl and NADPH-regeneration components (1mM glucose-6-phosphate, 0.25mM MgCl2, 0.1mM NADP and 1U/ml glucose-6-phosphate dehydrogenase) were added to the bottom of 1.5ml tube chilled on ice. Membrane expressing recombinant CYP6P9a/-b and AgCPR, and reconstituted cytochrome b5 were added to the side of the tube and pre-incubated for 5 minutes at 30°C, with shaking at 1,200 rpm to activate the membrane. 20μM of test insecticide was then added into the final volume of 0.2ml (~2.5% v/v methanol), and reaction started by vortexing at 1,200 rpm and 30°C for 1 hour. Reactions were quenched with 0.1ml ice-cold methanol and incubated for 5 more minutes. Tubes were then centrifuged at 16,000 rpm and 4°C for 15 minutes, and 150μl of supernatant transferred into HPLC vials for analysis. Reactions were carried out in triplicates with experimental samples (+NADPH) and negative controls (-NADPH). 100μl of sample was loaded onto an isocratic mobile phase (90:10 v/v methanol to water) with a flow rate of 1ml/min, monitoring wavelength of 226nm and peaks separated with a 250mm C18 column (Acclaim 120, Dionex) on Agilent 1260 Infinity at 23°C. Enzyme activity was calculated as percentage depletion (the difference in the amount of insecticide(s) remaining in the +NADPH tubes compared with the–NADPH) and a t-test used for statistical analysis.
Steady-state kinetic parameters for each allele were obtained with permethrin and deltamethrin by measuring the rate of reaction under linear conditions for 10 minutes while varying the substrates concentrations (2.5μM-20μM). Reactions were performed in triplicates both for +NADPH (experimental tubes) and–NADPH (negative control). K m and V max were established from the plot of substrate concentrations against the initial velocities and fitting of the data to the Michaelis-Menten module using the least squares non-linear regression, as described in the GraphPad Prism 6.03 Software (GraphPad Inc., La Jolla, CA, USA).
Comparative assessment of metabolic activities of recombinant CYP6P9a and CYP6P9b using fluorescent probe assays
To detect potential differences in substrate preferences, recombinant proteins from the various alleles of CYP6P9a and CYP6P9b were screened with fluorescent probes, kinetic analysis carried out, and inhibition assays conducted. Probes such as diethoxyfluorescein undergo P450-mediated O-dealkylation with large Stokes shift which can be measured fluorometrically [59].
Four probes: 7-ethoxyresurofin (7-ER), 7-ethoxy-4-trifluoromethylcoumarin (7-EFC), 7-methoxy-4-trifluoromethylcoumarin (MFC) and diethoxyfluorescein (DEF) were initially tested with the various recombinant CYP6P9a and CYP6P9b proteins. In a total volume of 225μl containing ~10pmol P450, buffered with 50mM potassium phosphate buffer (KPi at pH 7.4 with 5mM MgCl2), 1μM probe substrate was added. Membranes were activated for 5 minutes at 37°C in fluorescence spectrophotometer Infinite M200 (TECAN) before 25μl NADPH regeneration buffer (1mM glucose-6-phosphate (G6P), 0.25mM MgCl2, 0.1mM NADP and 1U/ml glucose-6-phosphate dehydrogenase (G6PDH) prepared in KPi pH 7.4) was added. All reactions were conducted in three replicates with negative control (–NADPH) devoid of the regeneration buffer. Rate of fluorescent product formation for 7-ER (ʎexc = 544nm, ʎemi = 590nm), 7-EFC and MFC (ʎexc = 410nm, ʎemi = 535nm) and DEF (ʎexc = 485nm, ʎemi = 530nm) was determined by linear regression of measurement between 2 and 10 minutes after start of the reaction. Results were analysed using Magellan v6.2 software.
For kinetics, 0 to 2μM diethoxyfluorescein was assayed with amount of membranes which produced optimal activities (10pmol for CYP6P9a and 3.33pmol for CYP6P9b) in a total volume of 250μl. The protocol was as outlined above, only that the substrate concentration varies and incubation was done under conditions established to be linear with respect to time. Steady-state kinetic parameters were obtained by measuring the rate of reaction for 10 minutes while varying the substrate concentration from 0 to 2μM. K m and V max were established from the plot of substrate concentrations against the initial velocities through a non-linear regression, by fitting the data to the Michaelis-Menten equation using GraphPad Prism 6.03 (GraphPad Software).
To further assess differences in the degree of binding of the various recombinant proteins to insecticides from different classes, inhibition assay was conducted with DEF and ten insecticides (inhibitors). The potent P450s inhibitor miconazole [60] was utilised as a positive control inhibitor. IC50 determination was conducted as described in previous studies [21, 61]. In a total volume of 225μl buffered with 50mM KPi (pH 7.4), containing 0.1–0.13μM DEF (~K m values), 10pmol CYP6P9a or 3.33pmol CYP6P9b, 5μl (25μM) test inhibitors or miconazole was spiked in the top wells and serially diluted into eight-fold concentrations (25–0.011μM). The 96 well-plate was pre-incubated for 5 minutes at 37°C and then 25μl regeneration buffer (7.8mg glucose-6-phosphate (G6P), 0.25mM MgCl2, 1.7mg NADP, 6U/ml glucose-6-phosphate dehydrogenase and 2% w/v NaHCO3) incorporated. Fluorescence was monitored for 21 cycles at interval of 1 minute with shaking at every step. Results were analysed using Magellan v6.2 software and inhibition at each inhibitor concentration and incubation time calculated as residual control activity towards DEF.
Comparative transgenic expression of resistant and susceptible alleles of CYP6P9a and CYP6P9b in D. melanogaster
To assess whether allelic variation in CYP6P9a and CYP6P9b was the most important factor conferring pyrethroid resistance even more than gene over-expression, a resistant (Malawi) and a susceptible (FANG) alleles for each gene were independently expressed in D. melanogaster, using the GAL4-UAS system. The hypothesis being that if sequence variation in these two genes is the key determinant of pyrethroid resistance, then over-expression of resistant alleles of both genes will confer more resistance to pyrethroids than observed by the over-expression of the susceptible alleles.
Cloning and construction of transgenic plasmids
Four transgenic lines, UAS-MALCYP6P9a, UAS-FANGCYP6P9a, UAS-MALCYP6P9b and UAS-FANGCYP6P9b were constructed and crossed with Actin5C-GAL4 lines to drive ubiquitous expression. Primers used for this PCR are given in S8 Table. PCR conditions, cloning techniques, construction of transgenic flies and expression of candidate genes was conducted as described in previous studies [4], with more details provided in S1 Text. For control group, flies with the same background as the UAS transgenic lines but devoid of the UAS construct with the candidate gene were crossed with the driver Actin5C-GAL4 line to generate Actin5C-GAL4-null flies.
Insecticides contact bioassays
Insecticide (2% permethrin and 0.15% deltamethrin-impregnated) filter papers were prepared in acetone and Dow Corning 556 Silicone Fluid (BDH/Merk, Germany) and kept at 4°C prior to bioassay. These papers were then rolled and introduced into 45 cc plastic vials. The vials were then plugged with cotton wool soaked in 5% sucrose. 20–25 (2–4 days old, post-eclosion F1 females) were selected for the bioassays and introduced into the vials. Mortality plus knockdown was scored after 1 hr, 2 hrs, 3 hrs, 6 hrs, 12 hrs and 24 hrs post-exposure to the discriminating dose of the insecticides. For each assay, at least six replicates were performed for both experimental flies and control, and t-test used to carry out statistical analysis of mortality plus knockdown observed between experimental groups and control.
qRT-PCR validation of over-expression of transgenes
To confirm expression of the candidate genes in the experimental flies and its absence in the control groups, qRT-PCR was carried out as described previously [4, 17]. RNA was extracted from three pools of 5 F1 experimental and control flies separately and cDNA synthesized as described. qRT-PCR for both CYP6P9a and CYP6P9b mRNA was conducted using the primers given in S8 Table, with normalization using the housekeeping gene RPL11.
Detection of causative mutations in CYP6P9a and CYP6P9b using site-directed mutagenesis
To detect the key amino acid changes conferring high pyrethroid-metabolising efficiency in resistant CYP6P9a and CYP6P9b alleles, a hypothesis-driven approach was used to select candidate amino acids from resistant alleles which were mutated into variants existing in the susceptible alleles, FANGCYP6P9a and FANGCYP6P9b. The hypothesis being that if the target residue is linked with high pyrethroid metabolising-activity in the resistant allele then a significant loss of activity will be obtained when mutated into a variant from the susceptible allele. For CYP6P9a the substitution aimed at were Ser320Tyr, Phe431Ser, Gln301His and a double mutant Leu63Phe_Lys66Gln. For CYP6P9b the following replacements were effected: Val109Ile, Asp335Glu, Asn384Ser and Pro401Ala. For both genes, the first amino acid in each case represents residue from the resistant allele and the second amino acids being variants present in the susceptible allele. Amplification was achieved using mutagenic primers (S8 Table) in a whole plasmid primer extension PCR followed by self-ligation at transformation step [62, 63]. The PCR was carried out using plasmidic pJET1.2::ompA+2-MALCYP6P9a and pJET1.2::ompA+2-MALCYP6P9b respectively as templates, with Phusion HotStart II High-Fidelity DNA Polymerase (Thermo SCIENTIFIC, USA). In a reaction mix containing 1X Buffer (with 7.5mM MgCl2), 80μM dNTP mixes, 0.3μM each of forward and reverse mutagenic primers, 2-4ng plasmidic DNA template, 0.04U/μl of Phusion HotStart Taq and sterile water was added to give 50μl. The reaction was started by preheating the mixture to 98°C for 10 minutes; and then followed by 35 cycles each of 94°C for 30 seconds; annealing (at 65°C) for 30 seconds and extension at 72°C for 2.5 minutes. This is then followed with final extension at 72°C for 10 minutes, and 4°C (hold). The PCR product was incubated for 1 hr with 2μl of 1X FastDigest Buffer and 1μl of DpnI (Thermo SCIENTIFIC), at 37°C to digest the Dam +-methylated parental template [62]. 4μl of the digest was then transformed into E. coli DH5α. Positive colonies were mini-prepped and sequenced on both strands to confirm presence of desired mutations. Mutagenic plasmids of MALCYP6P9b were successfully digested with NdeI and XbaI restriction enzymes, gel extracted with QIAquick Gel Extraction Kit (QIAGEN) and ligated into pCWOri+ already linearized with the same restriction enzymes, to construct the mutagenic cassettes pB13::ompA+2-MALCYP6P9b; mutagenic plasmids of MALCYP6P9a could not be cut out from the pJET1.2 construct and consequently the P450 insert could not be introduced into the expression vector pCWOri+. The mutant plasmids of MALCYP6P9b were then co-transformed together with AgCPR into JM109 and functional membranes expressed as described previously. Efforts to introduce mutations into the constructs pB13::ompA+2-CYP6P9a and pB13::ompA+2-CYP6P9b directly were not successful due possibly to the larger size of pCWori+ plasmid and as such amplification of mutants from MALCYP6P9a plasmids were not successful.
Recombinant MALCYP6P9b membranes expressed from mutant plasmids were used alongside the wild type recombinant MALCYP6P9b (wrMALCYP6P9b) to screen for metabolic activities against Type I and Type II pyrethroids (permethrin and deltamethrin, respectively), in order to determine the functional impact of each amino acid changed on metabolism of insecticides. In addition, fluorescent probes were also screened to establish differences in the O-dealkylation activity of the mutants compared with the wild type enzyme. All in vitro and kinetic analyses were conducted as described above. However, as different mutants expressed with varying concentrations all assays were conducted within linear range, with the amount of enzyme and time which produced highest activities.
Accession numbers
The DNA sequences reported in this paper have been deposited in the GenBank database (accession numbers: GenBank KR866022-KR866069).
Supporting Information
Red, solid boxes represent helices A-L; blue, dashed boxes represent the predicted substrate recognition sites 1–6 (SRS1-6). Amino acid substitutions are highlighted in pink.
(TIF)
Red, solid boxes represent motifs and blue, solid boxes correspond with the protein β-sheets. Amino acids substitutions are highlighted in red.
(TIF)
Permethrin is in stick format and orange. Heme atoms are in stick format and grey. Distance between possible sites of metabolism and heme iron are annotated in Angstrom.
(TIF)
Deltamethrin is in stick format and cyan. Heme atoms are in stick format and grey. Distance between possible sites of metabolism and heme iron are annotated in Angstrom.
(TIF)
Helix I and BʹC loop are annotated; pw2a is in blue colour; deltamethrin is in stick format and pink; heme atoms are in stick format and grey. Key residues Val109 and Ile109 are highlighted in stick format and yellow colour, and distance to the channel annotated.
(TIF)
(TIF)
Each point is a mean ± S.E.M. of three independent replicates.
(TIF)
Insecticides tested include Type I (permethrin and bifenthrin) and Type II (deltamethrin, λ-cyhalothrin and cypermethrin) pyrethroids, etofenprox and non-pyrethroid insecticides; (B) Mean IC50 of the test insecticide inhibitors against CYP6P9a and CYP6P9b metabolism of diethoxyfluorescein. Data represent mean IC50 at eight concentrations of each insecticide plus or minus standard deviation. Error bars represent variation in the values of the IC50 between different concentrations. For this assay, miconazole was used a positive control inhibitor; (C) Correlation between the IC50 of test insecticides inhibitors on MALCYP6P9b metabolism of diethoxyfluorescein with ChemScore values from docking with GOLD: Numerals I, II and III represents permethrin, deltamethrin and etofenprox, respectively. r2 = regression coefficient.
(TIF)
(TIF)
(A) FANGCYP6P9b-Ser384: yellow stick; FANGCYP6P9b-Arg385: green stick with superscript F; MALCYP6P9b-Asn384: blue stick; MALCYP6P9b-Arg385: red stick with superscript M; heme atoms: stick format and grey; deltamethrin in stick format and pink; (B) MALCYP6P9b-Asp335 is in stick form and blue colour, and FANGCYP6P9b-Glu335 is in red colour.
(TIF)
(A) O-dealkylation of seven fluorescent probes by the recombinant CYP6P9b mutants. The solid bars indicate average of significant activity in three experimental replicates compared to negative controls (-NADPH). RBE, resorufin benzylether; 7-ER, 7-ethoxyresorufin; RME, resorufin methylether; RPE, resorufin pentylether; DEF, diethoxyfluorescein; 7-EFC, 7-ethoxy-4-trifluoromethylcuomarin; MFC, 7-methoxy-4-trifluoromethylcuomarin. (B) Michaelis-Menten plots of mutant CYP6P9b proteins metabolism of diethoxyfluorescein; (C) 4D plot of the kinetic constants and catalytic efficiencies of mutant CYP6P9b proteins’ metabolism of diethoxyfluorescein.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Dr Michael Voice, of Cypex BioDundee for his help with fluorescent probes assay and Dr Hanafy M. Ismail for advice on metabolism assay experiments.
Data Availability
The DNA sequences reported in this paper have been deposited in the GenBank database (accession numbers: GenBank KR866022–KR866069).
Funding Statement
This work was supported by Wellcome Trust Fellowship grants to CSW (083515/Z/07/Z and 101893/Z/13/Z). SSI was supported by the Nigerian Government TETFund PhD scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. WHO. World Malaria Report 2014: World Health Organization; 2014. [Google Scholar]
- 2. Hemingway J. The role of vector control in stopping the transmission of malaria: threats and opportunities. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014;369(1645):20130431 10.1098/rstb.2013.0431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Report of the sixteenth WHOPES working group meeting: WHO/HQ, Geneva, 22–30 July 2013: review of Pirimiphos-methyl 300 CS, Chlorfenapyr 240 SC, Deltamethrin 62.5 SC-PE, Duranet LN, Netprotect LN, Yahe LN, Spinosad 83.3 Monolayer DT, Spinosad 25 Extended release GR. Geneva, Switzerland: World Health Organization, 2013 924150630X.
- 4. Riveron JM, Ibrahim SS, Chanda E, Mzilahowa T, Cuamba N, Irving H, et al. The highly polymorphic CYP6M7 cytochrome P450 gene partners with the directionally selected CYP6P9a and CYP6P9b genes to expand the pyrethroid resistance front in the malaria vector Anopheles funestus in Africa. BMC Genomics. 2014;15(1):817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mulamba C, Riveron JM, Ibrahim SS, Irving H, Barnes KG, Mukwaya LG, et al. Widespread pyrethroid and DDT resistance in the major malaria vector Anopheles funestus in East Africa is driven by metabolic resistance mechanisms. PLoS One. 2014;9(10):e110058 10.1371/journal.pone.0110058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coetzee M, Koekemoer LL. Molecular systematics and insecticide resistance in the major African malaria vector Anopheles funestus. Annual review of entomology. 2013;58:393–412. 10.1146/annurev-ento-120811-153628 [DOI] [PubMed] [Google Scholar]
- 7. Edi CV, Djogbenou L, Jenkins AM, Regna K, Muskavitch MA, Poupardin R, et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10(3):e1004236 10.1371/journal.pgen.1004236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ranson H, N’Guessan R, Lines J, Moiroux N, Nkuni Z, Corbel V. Pyrethroid resistance in African anopheline mosquitoes: what are the implications for malaria control? Trends in parasitology. 2011;27(2):91–8. 10.1016/j.pt.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 9. WHO. Global Plan for Insecticide Resistance Management (GPIRM). Geneva, Switzerland: World Health Organization, 2012. [Google Scholar]
- 10. Dia I, Guelbeogo MW, Ayala D. Advances and Perspectives in the Study of the Malaria Mosquito Anopheles funestus INTECH, 2013. [Google Scholar]
- 11. Dia I, Diop T, Rakotoarivony I, Kengne P, Fontenille D. Bionomics of Anopheles gambiae Giles, An. arabiensis Patton, An. funestus Giles and An. nili (Theobald) (Diptera: Culicidae) and transmission of Plasmodium falciparum in a Sudano-Guinean zone (Ngari, Senegal). Journal of medical entomology. 2003;40(3):279–83 [DOI] [PubMed] [Google Scholar]
- 12. Fontenille D, Lochouarn L, Diagne N, Sokhna C, Lemasson JJ, Diatta M, et al. High annual and seasonal variations in malaria transmission by anophelines and vector species composition in Dielmo, a holoendemic area in Senegal. The American journal of tropical medicine and hygiene. 1997;56(3):247–53. [DOI] [PubMed] [Google Scholar]
- 13. Brooke BD, Kloke G, Hunt RH, Koekemoer LL, Temu EA, Taylor ME, et al. Bioassay and biochemical analyses of insecticide resistance in southern African Anopheles funestus (Diptera: Culicidae). Bulletin of entomological research. 2001;91(4):265–72. . [DOI] [PubMed] [Google Scholar]
- 14. Casimiro SL, Hemingway J, Sharp BL, Coleman M. Monitoring the operational impact of insecticide usage for malaria control on Anopheles funestus from Mozambique. Malaria journal. 2007;6:142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, et al. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014;15(2):R27 10.1186/gb-2014-15-2-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schuler MA, Berenbaum MR. Structure and function of cytochrome P450S in insect adaptation to natural and synthetic toxins: insights gained from molecular modeling. Journal of chemical ecology. 2013;39(9):1232–45. 10.1007/s10886-013-0335-7 [DOI] [PubMed] [Google Scholar]
- 17. Riveron JM, Irving H, Ndula M, Barnes KG, Ibrahim SS, Paine MJ, et al. Directionally selected cytochrome P450 alleles are driving the spread of pyrethroid resistance in the major malaria vector Anopheles funestus. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(1):252–7. 10.1073/pnas.1216705110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wondji CS, Irving H, Morgan J, Lobo NF, Collins FH, Hunt RH, et al. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome research. 2009;19(3):452–9. 10.1101/gr.087916.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stevenson BJ, Bibby J, Pignatelli P, Muangnoicharoen S, O'Neill PM, Lian LY, et al. Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: Sequential metabolism of deltamethrin revealed. Insect Biochem Mol Biol. 2011;41(7):492–502. Epub 2011/02/18. 10.1016/j.ibmb.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 20. Gilbert LI, Gill SS. Insect control: biological and synthetic agents: Academic Press; 2010. [Google Scholar]
- 21.Bambal RB, Bloomer JC. Screening assay for inhibitors of human cytochrome P-450. Google Patents; 2006.
- 22. Chandor-Proust A, Bibby J, Regent-Kloeckner M, Roux J, Guittard-Crilat E, Poupardin R, et al. The central role of mosquito cytochrome P450 CYP6Zs in insecticide detoxification revealed by functional expression and structural modelling. The Biochemical journal. 2013;455(1):75–85. 10.1042/BJ20130577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilbert LI, Iatrou K, Gill SS. Comprehensive molecular insect science: Elsevier; 2005. [Google Scholar]
- 24. BIOTOL. Molecular Fabric of Cells: Elsevier Science; 1991. [Google Scholar]
- 25. Schleinkofer K, Sudarko, Winn PJ, Ludemann SK, Wade RC. Do mammalian cytochrome P450s show multiple ligand access pathways and ligand channelling? EMBO Rep. 2005;6(6):584–9. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochimica et biophysica acta. 2007;1770(3):390–401. [DOI] [PubMed] [Google Scholar]
- 27. Wade RC, Winn PJ, Schlichting I, Sudarko. A survey of active site access channels in cytochromes P450. Journal of inorganic biochemistry. 2004;98(7):1175–82. [DOI] [PubMed] [Google Scholar]
- 28. Stevenson BJ, Pignatelli P, Nikou D, Paine MJ. Pinpointing P450s associated with pyrethroid metabolism in the dengue vector, Aedes aegypti: developing new tools to combat insecticide resistance. PLoS Negl Trop Dis. 2012;6(3):e1595 Epub 2012/04/06. 10.1371/journal.pntd.0001595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laskowski DA. Physical and chemical properties of pyrethroids. Reviews of environmental contamination and toxicology. 2002;174:49–170. [DOI] [PubMed] [Google Scholar]
- 30. Kemp CA, Flanagan JU, van Eldik AJ, Marechal JD, Wolf CR, Roberts GC, et al. Validation of model of cytochrome P450 2D6: an in silico tool for predicting metabolism and inhibition. Journal of medicinal chemistry. 2004;47(22):5340–6. [DOI] [PubMed] [Google Scholar]
- 31. Schlenke TA, Begun DJ. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc Natl Acad Sci U S A. 2004;101(6):1626–31. Epub 2004/01/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cuamba N, Morgan JC, Irving H, Steven A, Wondji CS. High level of pyrethroid resistance in an Anopheles funestus population of the Chokwe District in Mozambique. PloS one. 2010;5(6):e11010 10.1371/journal.pone.0011010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pastinen T, Ge B, Hudson TJ. Influence of human genome polymorphism on gene expression. Hum Mol Genet. 2006;15 Spec No 1:R9–16. [DOI] [PubMed] [Google Scholar]
- 34. Zhu F, Parthasarathy R, Bai H, Woithe K, Kaussmann M, Nauen R, et al. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(19):8557–62. 10.1073/pnas.1000059107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Daborn PJ, Lumb C, Boey A, Wong W, Ffrench-Constant RH, Batterham P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect biochemistry and molecular biology. 2007;37(5):512–9. [DOI] [PubMed] [Google Scholar]
- 36. Amichot M, Tares S, Brun-Barale A, Arthaud L, Bride JM, Berge JB. Point mutations associated with insecticide resistance in the Drosophila cytochrome P450 Cyp6a2 enable DDT metabolism. European journal of biochemistry / FEBS. 2004;271(7):1250–7. [DOI] [PubMed] [Google Scholar]
- 37. Hiroya K, Murakami Y, Shimizu T, Hatano M, Ortiz de Montellano PR. Differential roles of Glu318 and Thr319 in cytochrome P450 1A2 catalysis supported by NADPH-cytochrome P450 reductase and tert-butyl hydroperoxide. Archives of biochemistry and biophysics. 1994;310(2):397–401. [DOI] [PubMed] [Google Scholar]
- 38. Morgan JC, Irving H, Okedi LM, Steven A, Wondji CS. Pyrethroid resistance in an Anopheles funestus population from Uganda. PLoS One. 2010;5(7):e11872 Epub 2010/08/06. 10.1371/journal.pone.0011872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Djouaka R, Irving H, Tukur Z, Wondji CS. Exploring Mechanisms of Multiple Insecticide Resistance in a Population of the Malaria Vector Anopheles funestus in Benin. PLoS One. 2011;6(11):e27760 Epub 2011/11/24. 10.1371/journal.pone.0027760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hunt RH, Brooke BD, Pillay C, Koekemoer LL, Coetzee M. Laboratory selection for and characteristics of pyrethroid resistance in the malaria vector Anopheles funestus. Medical and veterinary entomology. 2005;19(3):271–5. [DOI] [PubMed] [Google Scholar]
- 41.Hall TA, editor BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series; 1999.
- 42. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30(12):2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 44. Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. The Journal of biological chemistry. 1992;267(1):83–90. [PubMed] [Google Scholar]
- 45. Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. The Journal of biological chemistry. 1985;260(30):16122–30. [PubMed] [Google Scholar]
- 46. Sirim D, Widmann M, Wagner F, Pleiss J. Prediction and analysis of the modular structure of cytochrome P450 monooxygenases. BMC structural biology. 2010;10:34 10.1186/1472-6807-10-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods in enzymology. 2003;374:461–91. [DOI] [PubMed] [Google Scholar]
- 48. Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. The Journal of biological chemistry. 2004;279(37):38091–4. [DOI] [PubMed] [Google Scholar]
- 49. Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. Journal of molecular biology. 1997;267(3):727–48. [DOI] [PubMed] [Google Scholar]
- 50. Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. Journal of computer-aided molecular design. 1997;11(5):425–45. [DOI] [PubMed] [Google Scholar]
- 51.PyMOL User's Guide [Internet]. The PyMOL Molecular Graphics System 2004
- 52. Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC bioinformatics. 2006;7:316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS computational biology. 2012;8(10):e1002708 10.1371/journal.pcbi.1002708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pritchard MP, Ossetian R, Li DN, Henderson CJ, Burchell B, Wolf CR, et al. A general strategy for the expression of recombinant human cytochrome P450s in Escherichia coli using bacterial signal peptides: expression of CYP3A4, CYP2A6, and CYP2E1. Archives of biochemistry and biophysics. 1997;345(2):342–54. [DOI] [PubMed] [Google Scholar]
- 55. McLaughlin LA, Niazi U, Bibby J, David JP, Vontas J, Hemingway J, et al. Characterization of inhibitors and substrates of Anopheles gambiae CYP6Z2. Insect molecular biology. 2008;17(2):125–35. 10.1111/j.1365-2583.2007.00788.x [DOI] [PubMed] [Google Scholar]
- 56. Pritchard MP, McLaughlin L, Friedberg T. Establishment of functional human cytochrome P450 monooxygenase systems in Escherichia coli Cytochrome P450 Protocols: Springer; 2006. p. 19–29. [DOI] [PubMed] [Google Scholar]
- 57. Omura T, Sato R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. I. Evidence for Its Hemoprotein Nature. The Journal of biological chemistry. 1964;239:2370–8. [PubMed] [Google Scholar]
- 58. Strobel HW, Dignam JD. Purification and properties of NADPH-cytochrome P-450 reductase. Methods in enzymology. 1978;52:89–96. [DOI] [PubMed] [Google Scholar]
- 59. Zhang R, Kang K-D, Shan G, Hammock BD. Design, synthesis and evaluation of novel P450 fluorescent probes bearing α-cyanoether. Tetrahedron letters. 2003;44(23):4331–4. [Google Scholar]
- 60. Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. Molecular basis of resistance to azole antifungals. Trends in molecular medicine. 2002;8(2):76–81. [DOI] [PubMed] [Google Scholar]
- 61. Kajbaf M, Longhi R, Montanari D, Vinco F, Rigo M, Fontana S, et al. A comparative study of the CYP450 inhibition potential of marketed drugs using two fluorescence based assay platforms routinely used in the pharmaceutical industry. Drug metabolism letters. 2011;5(1):30–9. [DOI] [PubMed] [Google Scholar]
- 62. Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic acids research. 2004;32(14):e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li S, Wilkinson M. Site-directed mutagenesis: a two-step method using PCR and DpnI. Biotechniques. 1997;23(4):588–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Red, solid boxes represent helices A-L; blue, dashed boxes represent the predicted substrate recognition sites 1–6 (SRS1-6). Amino acid substitutions are highlighted in pink.
(TIF)
Red, solid boxes represent motifs and blue, solid boxes correspond with the protein β-sheets. Amino acids substitutions are highlighted in red.
(TIF)
Permethrin is in stick format and orange. Heme atoms are in stick format and grey. Distance between possible sites of metabolism and heme iron are annotated in Angstrom.
(TIF)
Deltamethrin is in stick format and cyan. Heme atoms are in stick format and grey. Distance between possible sites of metabolism and heme iron are annotated in Angstrom.
(TIF)
Helix I and BʹC loop are annotated; pw2a is in blue colour; deltamethrin is in stick format and pink; heme atoms are in stick format and grey. Key residues Val109 and Ile109 are highlighted in stick format and yellow colour, and distance to the channel annotated.
(TIF)
(TIF)
Each point is a mean ± S.E.M. of three independent replicates.
(TIF)
Insecticides tested include Type I (permethrin and bifenthrin) and Type II (deltamethrin, λ-cyhalothrin and cypermethrin) pyrethroids, etofenprox and non-pyrethroid insecticides; (B) Mean IC50 of the test insecticide inhibitors against CYP6P9a and CYP6P9b metabolism of diethoxyfluorescein. Data represent mean IC50 at eight concentrations of each insecticide plus or minus standard deviation. Error bars represent variation in the values of the IC50 between different concentrations. For this assay, miconazole was used a positive control inhibitor; (C) Correlation between the IC50 of test insecticides inhibitors on MALCYP6P9b metabolism of diethoxyfluorescein with ChemScore values from docking with GOLD: Numerals I, II and III represents permethrin, deltamethrin and etofenprox, respectively. r2 = regression coefficient.
(TIF)
(TIF)
(A) FANGCYP6P9b-Ser384: yellow stick; FANGCYP6P9b-Arg385: green stick with superscript F; MALCYP6P9b-Asn384: blue stick; MALCYP6P9b-Arg385: red stick with superscript M; heme atoms: stick format and grey; deltamethrin in stick format and pink; (B) MALCYP6P9b-Asp335 is in stick form and blue colour, and FANGCYP6P9b-Glu335 is in red colour.
(TIF)
(A) O-dealkylation of seven fluorescent probes by the recombinant CYP6P9b mutants. The solid bars indicate average of significant activity in three experimental replicates compared to negative controls (-NADPH). RBE, resorufin benzylether; 7-ER, 7-ethoxyresorufin; RME, resorufin methylether; RPE, resorufin pentylether; DEF, diethoxyfluorescein; 7-EFC, 7-ethoxy-4-trifluoromethylcuomarin; MFC, 7-methoxy-4-trifluoromethylcuomarin. (B) Michaelis-Menten plots of mutant CYP6P9b proteins metabolism of diethoxyfluorescein; (C) 4D plot of the kinetic constants and catalytic efficiencies of mutant CYP6P9b proteins’ metabolism of diethoxyfluorescein.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The DNA sequences reported in this paper have been deposited in the GenBank database (accession numbers: GenBank KR866022–KR866069).