Abstract
The neuromuscular junction (NMJ) is the most extensively studied model of neuronal synaptogenesis. Acetylcholine receptor (AChR) clustering on the postsynaptic membrane is a cardinal event in the differentiation of NMJs. AChR clustering and postsynaptic differentiation is orchestrated by sophisticated interactions among three proteins: the neuron-secreted proteoglycan agrin, the co-receptor LRP4, and the muscle-specific receptor tyrosine kinase MuSK. LRP4 and MuSK act as scaffolds for multiple binding partners, resulting in a complex and dynamic network of interacting proteins that is required for AChR clustering. In this review, we discuss the structural basis for NMJ postsynaptic differentiation mediated by the agrin–LRP4–MuSK signaling pathway.
Keywords: Neuromuscular junction, NMJ development, Acetylcholine receptor, Postsynaptic differentiation, Protein–protein interaction, Signaling pathway, Receptor tyrosine kinase, Wnt signaling
Introduction
In vertebrates, innervation of muscle fibers by motor neurons leads to the formation of neuromuscular junctions (NMJs) composed of neuronal presynaptic and muscle postsynaptic membranes. Owing to their size and accessibility, NMJs serve as excellent models to study the differentiation, stabilization, and maintenance of synapses. Once established, NMJs efficiently convert the electrical impulses of the motor neuron into action potentials in the juxtaposed muscle fiber, a process that depends on the release of large quantities of acetylcholine (ACh) molecules by the presynaptic motor neuron, as well as high-density clustering of nicotinic ACh receptors (AChRs) on the postsynaptic muscle membrane [1, 2]. During postnatal development in mammals, the NMJ postsynaptic membrane forms a “pretzel-like” structure and AChRs are concentrated at the crests of the numerous folds [3]. The estimated density of the clustered AChRs is ~10,000–20,000/μm2, which compares with only ~10/μm2 in membranes in extrasynaptic regions [4, 5]. The process by which AChRs form high-density clusters during NMJ development involves an intricate network of signaling molecules and communication between presynaptic motor neurons and postsynaptic muscle fibers [6, 7]. Despite intense study, the molecular mechanisms that regulate and stabilize AChR clustering remain unclear.
In the past three decades, a number of nerve-derived and muscle-derived factors and signaling pathways that drive differentiation of NMJs [7] have been identified. These include transmembrane and cytoplasmic proteins, as well as components of the extracellular basal lamina, which co-cluster with AChRs during postsynaptic differentiation. The synaptic basal lamina contains two major signaling proteins, agrin and neuregulin, which are secreted by motor neurons and play essential roles in NMJ formation and maintenance [8, 9]. Other extracellular matrix proteins are also implicated, including laminin, collagen IV/XIII, perlecan, collagen Q, and biglycan [10–12]. On the muscle membrane, muscle-specific kinase (MuSK) is the key organizer and scaffold protein for recruitment of other extracellular and intracellular factors [5, 13]. The integral membrane protein LRP4 (low-density lipoprotein receptor-related protein 4) forms a complex with MuSK and is also critical to NMJ function [14–16]. Inside the muscle cells, many signaling molecules, such as the adaptor proteins Dok7, Tid1, and rapsyn, directly interact with the cytoplasmic domain of MuSK to form a multiprotein complex required for AChR clustering [7, 17, 18]. Collectively, the agrin–LRP4–MuSK signaling pathway plays a crucial role in NMJ development. Our understanding of the molecular interactions by which this complex functions has increased over the last decade, due in part to invaluable information on the structures of the individual components, as well as several protein–protein complexes. In this review, we briefly summarize recent studies that have shed light on the structures of the component proteins, and the roles played by the signaling molecules in the formation and stabilization of NMJs.
Agrin orchestrates AChR clustering
Agrin was first identified from the electric organ of the torpedo ray by McMahan and colleagues [19, 20]. As a large heparan sulfate proteoglycan, agrin has an estimated molecular weight of ~400 kDa, almost twice the molecular weight predicted from its amino acid composition [21]. Agrin is composed of a number of structural modules common to other basal lamina proteins, including nine follistatin-like (FS), two serine/threonine-rich, one laminin B-like, four EGF-like, and three laminin G-like (LG) domains. The N-terminus of agrin has alternative start sites that generate two isoforms of different length and tissue specificity. The short form of agrin has an N-terminal transmembrane region and is mainly expressed in the brain, whereas the long form, which lacks the transmembrane region, is expressed at NMJs and in other tissues. In addition, the C-terminus of agrin contains three conserved alternative splicing sites, termed X, Y, and Z in mammals. In chickens, the mammalian Y and Z sites are termed A and B, respectively. The A/Y site binds heparin and the B/Z site is essential for the AChR-clustering activity of agrin. The function of the X site is not yet known [22].
Agrin is expressed in various tissues including muscle, brain, and cells of the immune system [21]. In mammals, only the neuron-specific agrin contains an eight amino acid insert (ELTNEIPA, Z8) at the Z site. The Z8 insert lies within the C-terminal LG3 domain and is absolutely required for agrin’s AChR-clustering activity [21–24], although the exact mechanism for its involvement is unclear. Both the Z8+ and Z8− isoforms are expressed in brain [25, 26], where agrin expression is highest during the synaptogenic period of development. Agrin is also thought to be involved in excitatory synapse development [27], but its precise function in brain remains to be clarified.
The N-terminal domain
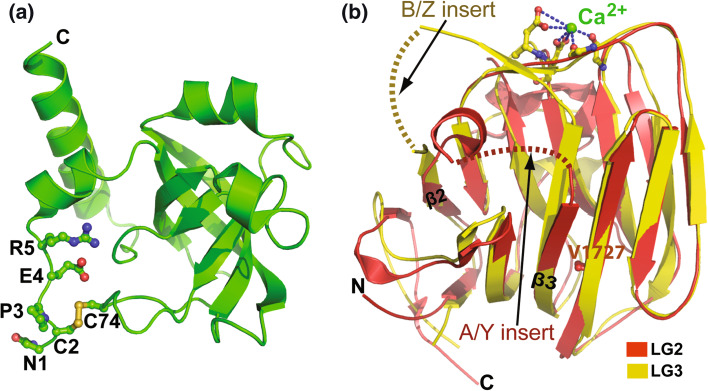
The N-terminal domain (NtA) of the secreted form of agrin (SS-NtA) interacts with the coiled-coil region of laminin, which localizes agrin to the synaptic basal lamina. Several crystal structures of chicken agrin NtA have been published [28–30], which all show identical core structures, but different conformations of the extreme N-terminal region.
The first crystal structure of chicken agrin NtA was determined with protein expressed in E. coli [30]. The NtA domain exhibits an OB (oligosaccharide/oligonucleotide binding) fold with a central β-barrel core flanked by N- and C-terminal α helices [30]. Interestingly, agrin NtA has striking structural similarity to tissue inhibitor of metalloproteinases-1 (TIMP-1), despite their low sequence identity. The first five N-terminal amino acids were disordered in the apo NtA. However, clear electron densities were observed for residues Cys2–Arg5, as well as a disulfide bond between Cys2 and Cys74, when NtA was co-crystallized with a synthetic 20-residue peptide corresponding to the NtA-binding site in the laminin γ1 chain. Although the functional role of this disulfide bond in NtA is still unknown, disruption of the homologous disulfide bond in TIMP-1 abolishes its function [31, 32]. In this structure, no electron density was visible for the peptide used in the co-crystallization, making it unclear whether it was bound to NtA. Nevertheless, deletion of the N-terminal residues of NtA did not affect its binding to laminin, raising the possibility that another laminin-binding region on NtA may compensate for the loss of the N-terminal residues.
Several years after these studies, the same chicken NtA fragment was expressed in HEK293 cells and crystallized [29]. The most remarkable feature of the new structure was the presence of highly structured N-terminal residues (Asn1–Arg5) in the absence of the laminin peptide (Fig. 1a), in contrast to the structure of the E. coli-expressed protein. Moreover, the disulfide bridge between Cys2 and Cys74 was also present in the HEK293-expressed protein (Fig. 1a). Solid-phase binding assays of NtA with its neighboring FS domain (NtA-FS) confirmed that NtA binds to the γ1 chain of laminin [29]. Furthermore, mutagenesis-based mapping experiments revealed that Leu117 and Val124 of NtA are critical for laminin binding [33]. A more recent study using soft X-ray diffraction showed that the most C-terminal cysteine residue (Cys123) of NtA forms an interdomain disulfide bridge with the adjacent FS module, which may be critical for stabilization of the overall structure of the full-length agrin [28].
Fig. 1.
Crystal structures of agrin domains. a The structure of the NtA domain is similar to tissue inhibitor of metalloproteinases-1 (TIMP-1). The five N-terminal residues, Asn1 (N1), Cys2 (C2), Pro3 (P3), Glu4 (E4) and Arg5 (R5), are ordered in the recombinant protein expressed in eukaryotic cells. All the amino acids in the figures are labeled with single-letter code and shown in ball-and-stick format, in which oxygen atoms are in red, nitrogen in blue, sulfur in yellow and carbon in the same color as the peptide main chain. The disulfide bond between C2 and C74, which was not observed in the protein expressed in E. coli, may stabilize the N-terminal region of the protein. The C-terminus of the domain is indicated by a single letter C. b The LG2 domain (in red) containing a 4-amino acid splice insert at the A/Y site is superimposed with the LG3 domain (in yellow) containing an 8-amino acid splice insert at the B/Z site. LG2 and LG3 both adopt a 13-stranded β-jellyroll fold structure. The insert at the A/Y site of the LG2 domain (A/Y insert, red dash line) and the insert at the B/Z site of the LG3 domain (B/Z insert, yellow dash line) are disordered in the absence of a binding partner. Genetic mutation of Val1727 (V1727) on the LG2 domain decreases the AChR clustering activity of agrin. The Ca2+ (green sphere) bound in the LG3 domain is also important for agrin function. The N- and C-termini of the LG3 domain are indicated by N and C, respectively
The LG2 and LG3 domains
The C-terminal domain (LG1–3) of agrin is the most critical region for its AChR clustering function. In fact, the 95 kDa LG1–3 region is almost as potent as the full-length agrin molecule in inducing AChR clustering in vitro [34]. Moreover, the 22 kDa C-terminal LG3 domain containing the B/Z insert is sufficient to induce AChR clustering, albeit with lower efficiency than the whole LG1–3 region [34, 35]. In addition to the B/Z insert, extracellular calcium is required for agrin function, which is believed to bind in the LG3 domain. Extensive structural studies have been performed on the LG3 domain to understand the function of the B/Z insert and Ca2+ in regulating AChR clustering [36–39]. The structure of LG3 domain B0 (lacking the B/Z insert) was determined by NMR in the presence of Ca2+ [39], and the structures of the B8+ and B11+ (11 residues insert) isoforms of LG3 were determined by X-ray crystallography in the presence of Ca2+ at 2.3 and 1.4 Å, respectively. For comparison, the LG3 B8+ structure was also determined in the absence of Ca2+ [39]. All forms of the LG3 domain share an almost identical core structure, a 13-stranded β jellyroll fold, which is structurally homologous to the LG/LNS domains found in laminin, neurexin, and sex hormone binding globulin protein. These LG/LNS domains have a similar ligand-binding site that is reminiscent of the antigen binding sites in immunoglobulin [40].
The neuron-specific B inserts lie between the L2 and 3 loops of agrin LG3. A recent circular dichroism study of agrin constructs containing mutated Z8 residues showed the Z8 asparagine residue to be crucial for functional activity [36]. However, none of the B insert amino acids were visible in the agrin crystal structures, suggesting the inserts may adopt flexible conformations that require stabilization by a binding partner [39]. The Ca2+-binding site is located on the rim of the β jellyroll, close to the B insert (Fig. 1b). The Ca2+ ion is coordinated by the side chains of Asp1817 and Asp1886 together with the backbone carbonyl oxygens of Leu1834 and Gln1884, which are structurally conserved in agrin and laminin [39]. The importance of Ca2+ for agrin function is revealed by the fact that mutation of either of the two Ca2+ coordinating Asp residues in rat agrin inhibits its AChR clustering activity [37, 39]. The proximity of the B insert and the bound Ca2+ also suggests the potential for crosstalk.
The role of the LG2 domain of agrin in synaptogenesis may be related to its association with α-dystroglycan, heparin, and integrins [34, 41]. The A/Y site in the LG2 domain is known to be important for heparin binding and contains a four amino acid splice insert. However, the KSRK peptide of the mouse agrin Y insert does not itself bind heparin. Interestingly, neural agrin isoforms that contain the insert at the B/Z site also contain the insert at the A/Y site, suggesting there may be a coordinated splicing mechanism in vivo [22, 42]. A crystal structure of the mouse agrin LG2 domain has recently been deposited in PDB by New York SGX Research Center for Structural Genomics (PDB code: 3PVE, unpublished). The structure reveals a 13-stranded β jellyroll fold that is highly similar to the LG3 domain (Fig. 1b) [40, 43]. The KSRK Y insert connecting the β2–3 strands is not observed in the LG2 structure, which is reminiscent of the LG3 structure in which the B/Z inserts were also invisible. These observations suggest that the A/Y insert loop is involved in binding to another molecule(s), heparin for example, which stabilizes the loop [44, 45]. Remarkably, a single V1727F mutation within the LG2 domain of human agrin causes severe congenital myasthenic syndrome in humans, attesting to the importance of this domain [46]. This was confirmed by functional analyses showing that agrin-V1727F had reduced AChR clustering activity compared with the wild-type molecule. When modeled on the structure of mouse LG2 domain, Val1727 is located close to the A/Y site. Thus, replacement of this valine with the bulky phenylalanine may disrupt the β sandwich structure of LG2 and impair the native conformation of agrin (Fig. 1b) [46].
MuSK is the key organizer at the postsynaptic membrane
Muscle specific kinase (MuSK) is member of the receptor tyrosine kinase (RTK) superfamily. RTK proteins are composed of an extracellular ligand-binding region, a single transmembrane-spanning domain, and an intracellular region that harbors the kinase domain. There are four major RTK activation mechanisms, as summarized by Lemmon and Schlessinger [47]. All four require direct binding of a ligand to RTK and dimerization of the RTK, which induces trans-autophosphorylation of the intracellular kinase domains. The activated RTKs then recruit other proteins to initiate a signaling cascade [47].
Autophosphorylation of MuSK and subsequent clustering of AChR on the postsynaptic membrane can be activated by antibodies that induce dimerization of MuSK, which suggests that the molecular mechanism of MuSK activation is similar to other RTKs [48, 49]. However, although there is considerable evidence that agrin is a ligand for MuSK, the two molecules do not directly interact [50, 51]. These observations implicated an intermediate protein that physically linked agrin and MuSK, which was later identified to be LRP4 [14–16]. Therefore, activation of MuSK requires not only a soluble agrin ligand, but also the involvement of the obligate co-receptor LPR4, making MuSK a unique member of the RTK family [47].
The kinase domain
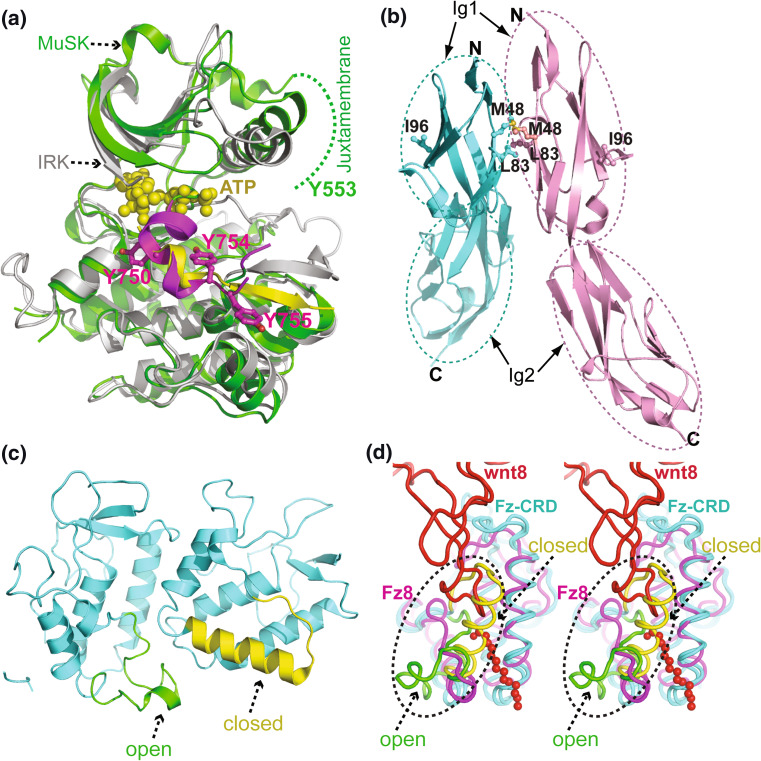
The MuSK kinase domain shares homology with serine/threonine kinases [52, 53]. The domain comprises an N-terminal lobe with a five-stranded β-sheet and a C-terminal lobe mainly composed of α helices (Fig. 2a). The active site is located in the cleft between the N-terminal and C-terminal lobes, and substrates bind mainly at the C-terminal lobe. The activity of the MuSK kinase domain appears to be regulated both by the activation loop (A-loop) and the juxtamembrane region. Prior to substrate binding, the A-loop partially occupies the active site cleft, and Tyr754 on this loop is positioned as a pseudo-substrate to suppress spontaneous autophosphorylation (Fig. 2a). Therefore, the A-loop simultaneously obstructs ATP and substrate binding. This dual inhibitory role of the A-loop is also observed in the kinase domain of insulin receptor kinase (IRK), which shows high structural similarity with MuSK (Fig. 2a).
Fig. 2.
Crystal structures of MuSK domains. a The apo MuSK kinase domain (in green and magenta) is superimposed with the ATP (yellow spheres)-bound insulin receptor kinase domain (in gray and yellow). The A-loops of MuSK (in magenta) and IRK (in yellow) are located in a similar position. Tyr754 (Y754) on the MuSK A-loop partially occupies the active site of the enzyme, suggesting this residue has an autoinhibitory function. The other two tyrosine residues, Y750 and Y755, are phosphorylated upon the activation of the protein. The N-terminal juxtamembrane region of MuSK is disordered (indicated by the green dash line) in the crystal structure. Tyr553 (Y553) within the juxtamembrane is positioned close to the active site of the kinase domain. b The two N-terminal Ig-like domains (Ig1–Ig2) of MuSK form a linear rod, which dimerizes in the crystal asymmetric unit at an angle of ~60° (the two protomers are colored in blue and magenta). The hydrophobic residues Met48 (M48) and Leu83 (L83), on the dimer interface, and Ile96 (I96) on the opposite side of the dimer interface (all shown in spheres), are important for agrin-induced AChR clustering. The N- and C-termini are labeled as N and C, respectively. c The Fz-CRD of MuSK was crystallized as an asymmetric dimer with either a “closed” (in yellow) or “open” (in green) conformation near the C-terminus. d The two protomers of the dimerized MuSK Fz-CRD (in cyan) are superimposed well with Fz8 (in magenta) of the Wnt8–Fz8 complex, shown in walleye stereo. Wnt8 and its palmitoleic acid (spheres) are shown in red. The variable C-terminal region of the MuSK Fz-CRD is shown with the same color scheme as in panel c and highlighted by a black dashed ellipse. In contrast to the open conformation, the closed conformation of the α helix clashes with the superimposed Wnt8, indicating that only the open conformation favors Wnt interaction
The juxtamembrane region of MuSK contains an NPXY (X represents any amino acid) motif, which is bound by a PTB (phosphotyrosine binding) domain in the substrate [54, 55]. Residue Tyr553 within the NPXY motif, which is disordered in the crystal structure, is also important for regulation of MuSK kinase activity. Upon agrin-induced activation of MuSK, Tyr754 and Tyr553 are thought to be autophosphorylated first [52], followed by the remaining two tyrosines in the A-loop (Tyr750 and Tyr755), which dramatically increases the kinase catalytic efficiency. Phosphorylation of Tyr553 in the juxtamembrane region allows binding of PTB domain-containing proteins, for example, the adaptor protein Dok7 [52].
Ligand-induced activation of MuSK creates a positive-feedback loop that induces further MuSK clustering [56]. This finding is consistent with the observation that overexpression of ectopic MuSK induces agrin-independent autophosphorylation and non-synaptic AChR clustering [57, 58]. Thus, the combined inhibition of MuSK kinase activity by the A-loop and juxtamembrane region is crucial to control ligand-independent activation and to maintain a basal activation state.
The Ig-like 1 and 2 domains
The ectodomain of MuSK is composed of three consecutive immunoglobulin-like (Ig-like) domains followed by a cysteine-rich domain (CRD) that is homologous to the CRD of Wnt-interacting Frizzled proteins. Ig-like domains are common in other RTKs and generally serve as ligand-binding sites [59]. The crystal structure of the first two Ig-like domains of MuSK (Ig1–2), which are mainly responsible for agrin-induced AChR clustering, has been determined at 2.2 Å resolution [60–62]. Ig1 and Ig2 are highly similar and aligned in a linear manner (Fig. 2b); there is no obvious linker region between the domains and consequently they form a semi-rigid rod-like structure. Interestingly, the domain–domain connections between Ig2 and Ig3 are similar to those between Ig1 and Ig2. Thus, the third Ig-like domain of MuSK, although not included in the crystal structure, may be connected to Ig1 and Ig2 in a similarly rigid manner.
Compared with Ig-like domains in other RTKs, the Ig1 domain of MuSK contains an additional disulfide bond that is critical for correct protein folding [62]. As shown in Fig. 2b, MuSK Ig1–2 was crystallized as a dimer in an asymmetric unit. The dimer interface is solely formed by the Ig1 domains and the two protomers are aligned at an angle of ~60°. The dimer interface is primarily hydrophobic, and significantly, the two hydrophobic residues in the interface, Met48 and Leu83 (shown in sphere in Fig. 2b), were shown to be critical for agrin-induced MuSK activation [62]. Mutation of Met48 or Leu83 did not affect expression of the mutant proteins, but severely reduced agrin-induced MuSK activation. The formation of this dimer interface is thus crucial to MuSK function. One possibility is that Ig1 homodimerization may be involved in dimerization of MuSK in vivo. Alternatively, the dimer interface may be involved in critical interactions of MuSK with other proteins. Now that LRP4 has been identified as an obligate MuSK binding partner; it will be interesting to determine whether this region of Ig1 is involved in the binding of MuSK and LRP4.
Interestingly, another hydrophobic residue, Ile96 (in sphere, Fig. 2b), which is located on the opposite site of the Ig1 dimer interface, was found to be functionally required, as mutation of this residue completely abolished agrin-induced MuSK activation [62]. In the homologous Trk receptor, ligand (NGF) binding is mediated by hydrophobic residues around the equivalent position of MuSK Ile96 [63]. Therefore, it is possible that the Ile96-containing region in the MuSK Ig1 domain might be involved in LRP4 binding.
The Frizzled-like CRD
The region following the third Ig-like domain of MuSK was initially thought to be a C6 box plus a fourth Ig-like domain [64]. However, it was later identified as a Frizzled-like CRD (Fz-CRD), which is homologous to the CRD of the Frizzled proteins that serve as receptors in the Wnt signaling pathway [65, 66]. Because Wnt signaling plays an important role in NMJ synaptogenesis, this finding raised the possibility that MuSK might bind Wnt proteins through the CRD. Notably, the Fz-CRD is also found in other RTKs engaged in Wnt signaling, including Ror1 and Ror2 [66].
The MuSK CRD appears to be dispensable for agrin-induced MuSK activation and AChR clustering, but may be involved in agrin-independent associations with other postsynaptic components. For example, Wnt11r has been shown to interact with MuSK Fz-CRD and to regulate the neuron-independent pre-patterning of AChRs on postsynaptic muscle membranes [67, 68]. Wnt4 also interacts with MuSK Fz-CRD and increases MuSK phosphorylation [69]. Importantly, Wnt9a, Wnt9b, Wnt10b, Wnt11, and Wnt16 all stimulate agrin-independent AChR clustering by directly interacting with MuSK Fz-CRD in a LRP4-dependent manner, suggesting the existence of a novel Wnt signaling pathway involving both MuSK and LRP4 [70].
The crystal structure of the MuSK Fz-CRD was determined at 2.1 Å resolution and found to be highly similar to the CRDs of Wnt receptor Frizzled-8 (Fz8) and secreted Frizzled-related protein-3 protein (sFRP3) [71, 72]. Interestingly, the structure shows two MuSK CRD molecules in a crystallographic asymmetric unit that adopt different conformations at their C-termini (residues 410–430), resulting in asymmetric dimeric packing (Fig. 2c). The interaction between the two CRD copies is mainly hydrophobic. In the case of Fz8 and sFRP3, their CRDs were monomers in solution, but they crystallized as dimers [72]. Other functional studies indicated that Xenopus laevis Frizzled-3 formed CRD-dependent homodimers [73], and that Ror2 CRD formed heterodimers with Frizzled-2 and Frizzled-5 in response to different stimuli [74]. Thus, the Fz-CRD might be involved in dimerization of MuSK on the cell surface in a manner similar to that observed in the crystal structure.
The MuSK CRD structure deviates from that of Fz8 CRD mainly at the C-terminal end, where the two copies of MuSK CRD in the asymmetric unit are structurally different (Fig. 2c). In one protomer, residues 410–431 form a well-ordered α helix that folds tightly against the core structure of the molecule (the “closed” conformation in Fig. 2c), while in the second protomer, the same sequence element extends into a much looser loop-like structure (“open” conformation). The Wnt8 binding site on Fz8 CRD was proposed to include the loop connecting the β1–β2 strands, the loop connecting the α3–α4 helices, and the C-terminal tail [72]. Indeed, a recent structural study revealed that Wnt8 clamps the two sides of Fz8 CRD simultaneously [75]. A structural comparison shows that the overall structure of the MuSK Fz-CRD superimposes well with Fz8 CRD in the Wnt8-Fz8 complex (Fig. 2d). The two potential Wnt binding sites on the MuSK CRD are structurally conserved with those of Fz8 CRD. Close examination of the Wnt binding sites of Fz8 CRD indicates that only the MuSK Fz-CRD with the “open” conformation is compatible with Wnt binding. By contrast, the “closed” conformation of MuSK CRD does not allow binding of Wnt8 and its palmitoleic acid (Fig. 2d). Further studies are needed to verify whether the observed conformations of MuSK CRD might represent physiological conformational changes induced by Wnt or other regulatory proteins.
LRP4 mediates the agrin–MuSK interaction
LRP4 (also known as MEGF7) is a member of the LDLR family of single-spanning transmembrane proteins composed of similar structural modules [76, 77]. LRP4 has a short intracellular C-terminal domain and a large and complex ectodomain composed of eight LDLa (LDL class A) repeats at the N-terminus followed by four homologous YWTD motif-containing β-propeller domains, which are separated by EGF-like modules. Although LDLR proteins were originally thought only to transport lipids or lipoproteins during cell metabolism [78, 79], it is now clear that these proteins play significant roles in cell signaling and are critically involved in a number of cellular functions. LRP4 was known to be important in limb development and Wnt signaling [80, 81], but its role in NMJ development was first suggested by the absence of AChR clustering in LRP4 null mice [16]. Subsequently, the critical involvement of LRP4 in NMJ development was shown to be mediated through its interactions with both agrin and MuSK.
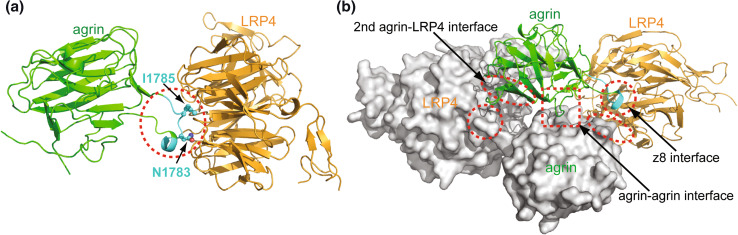
The binding of agrin and LRP4 is a key step in MuSK activation and initiation of downstream signaling. A systematic truncation study mapped the minimum agrin-binding domain of LRP4 to its first β-propeller domain [82]. The crystal structure of a complex between neural agrin LG3 (containing the Z8 insert) and the LRP4 β1-propeller domain flanked by EGF modules has been determined at 2.85 Å resolution [82]. The complex structure clearly shows that the agrin Z8 insert forms a loop projecting to the binding surface of the LRP4 β1 propeller (Fig. 3a). Because the same loop was disordered in the chicken apo agrin LG3 domain (Fig. 1b), it seems clear that the rigidification of the Z8 loop results from an induced-fit interaction between agrin and LRP4. Strikingly, only two residues, Asn1783 and Ile1785, on the tip of the Z8 insert loop are directly engaged in the interaction with LRP4 (Fig. 3a). Agrin Asn1783 forms multiple hydrogen bonds with LRP4, and Ile1785 is buried in a hydrophobic pocket. Notably, mutation of either residue abolished the agrin–LRP4 interaction and subsequent AChR clustering [82].
Fig. 3.
Structure of the agrin-LRP4 complex. a The binary complex of agrin–LRP4 is mediated by the neuron-specific Z8 alternative splice insert (in cyan and in red dashed circle). Asn1783 (N1783) and Ile1785 (I1785) on the tip of the Z8 insert loop are directly engaged in binding to LRP4. b The binary complex that is held together by the Z8 interface (red dashed circles) forms a homodimer mediated by an agrin–agrin interface (red dashed square) and two additional agrin–LRP4 interfaces (red dashed ellipses). The two agrin–LRP4 binary complexes are shown in ribbons and surface representations, respectively
We compared the agrin–LRP4 structure with other homologous structures composed of a YWTD motif-containing β-propeller domain and its ligand. Unexpectedly, we found a conserved NXI/V motif (where X is any amino acid) located in the receptor-interacting loop, which may represent a universal binding motif for such ligands [83]. Examples of the ligands are laminin, SOST (Sclerostin), Wise, and Dkk1 (Dickkopf 1) [83, 84]. Intriguingly, SOST, Wise, and Dkk1 were found to be ligands of LRP4, in addition to their known binding to LRP5/6 [85, 86]. Moreover, these proteins may bind to the third β-propeller domain of LRP4 [86], suggesting that the agrin pathway and Wnt pathway may converge at LRP4 during NMJ development. It will be of interest to determine how the different β-propeller domains (i.e. β1 and β3) of LRP4 are involved in binding distinct ligands through a similar mechanism, and to uncover how these protein interactions integrate the two signaling pathways that are essential for NMJ development and maintenance.
Agrin and LRP4 formed a 2:2 hetero-tetramer in a non-crystallographic asymmetric unit (Fig. 3b), and this 2:2 stoichiometry was also observed in solution [82]. In addition to the Z8 loop-mediated binary complex interface (termed the Z8 interface), the hetero-tetrameric complex is held together by two additional interfaces. One is between agrin and LRP4 (the agrin–LRP4 interface) and the other is between two agrin molecules (the agrin–agrin interface) (Fig. 3b). Interestingly, the two LRP4 molecules do not directly interact. The dimerization of the agrin–LRP4 heterodimer suggests a mechanism for MuSK activation through agrin–LRP4-induced dimerization. Structure-based mutagenesis and complementary functional studies confirmed that the agrin–LRP4 binary complex is necessary but not sufficient for the physiological function of agrin. Rather, dimerization of the binary complex is required for activation of MuSK and agrin-induced AChR clustering, suggesting that the tetrameric architecture of agrin and LRP4 is physiologically relevant.
On the postsynaptic membrane of the NMJ, LRP4 is thought to bind constitutively to MuSK, and the interaction is allosterically strengthened in the presence of agrin [14, 87]. Only the ectodomains of LRP4 and MuSK mediate this interaction [87, 88]. Recent studies using protein deletion mutants have indicated that the C-terminal moiety of LDLa repeats and the third β-propeller domain of LRP4 might be critical for binding of MuSK, whereas the fourth β-propeller may be dispensable [87]. Further studies are needed to fine map the interacting domains in LRP4 and MuSK, as well as to characterize the structural interplay among agrin, LRP4, and MuSK.
Dok7 is a MuSK adaptor protein
Downstream-of-kinase or docking-protein 7 (Dok7) belongs to a family of cytoplasmic signaling adaptor proteins that includes Dok1–7 and insulin receptor substrates 1–4 (IRS1–4) [89, 90]. Dok7 has an N-terminal pleckstrin homology (PH) domain, a PTB domain, and an extended C-terminal region with multiple sites of tyrosine phosphorylation. Dok7 is required for agrin-induced MuSK activation and AChR clustering. Agrin fails to induce MuSK autophosphorylation or AChR clustering in Dok7-deficient mice, which could be rescued by exogenous expression of Dok7 [91, 92]. Genetic mutation of Dok7 causes congenital myasthenic syndromes, presumably due to impairment of the MuSK-Dok7 interaction [93, 94]. There are at least two explanations for the requirement for Dok7 in agrin-induced MuSK activation. First, Dok7 may be required for agrin function, either as an effector through an unknown intermediate or in concert with agrin to activate MuSK. Alternatively, Dok7 may serve as an adaptor or co-factor of MuSK and have no direct relationship with agrin. For instance, Dok7 binding to MuSK may affect its conformation in a manner that facilitates autophosphorylation.
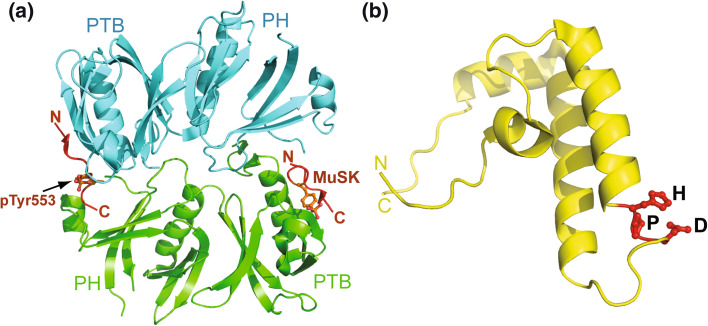
The PH domain of Dok7 binds to membrane phosphoinositides and localizes the protein to the plasma membrane [95], whereas the PTB domain binds to the phosphorylated Tyr553 residue in the NPXY motif of the MuSK juxtamembrane region [92]. The N-terminal PH-PTB region of Dok7 (residue 1–220) has been co-crystallized with a 13-residue MuSK peptide containing pTyr553 [95]. Both the PH and PTB domains of Dok7 adopt a core 7-stranded β sandwich structure with a C-terminal α helix (Fig. 4a). The two domains are connected with an extensive buried surface, which contrasts with the small interface shared by the tandem PH–PTB domains of IRS1 [96]. The MuSK-pTry553 peptide was located in the phosphopeptide-binding groove of the PTB domain, where the phosphate group of pTry553 was stabilized by multiple PTB domain residues (Fig. 4a). This finding is consistent with a previous predication that the pTry553 residue of MuSK might serve as a protein binding site [52].
Fig. 4.
Structures of MuSK adaptor proteins. a The N-terminal region of Dok7, containing a PH and a PTB domain, was co-crystallized with a 13-amino acid peptide of the MuSK juxtamembrane region (in orange), which contained a phosphotyrosine Y553 (pTyr553). The orientation of the MuSK peptide is indicated by its N- and C-termini. The pTyr553 peptide is bound in the phosphopeptide-binding groove of the Dok7 PTB domain. The Dok7-peptide complex forms a dimer in the crystal asymmetric unit. The dimerized Dok7 fragments are colored in green and cyan, respectively. b The NMR structure of human Tid1 shows a 4-helix fold, highly similar to other DnaJ domains in the HSP40 family. The conserved HPD tripeptide motif (in red) between the second and third helices may be critical for the AChR clustering function of Tid1
The most striking feature of the Dok7–MuSK complex is that it forms a dimer in the crystal asymmetric unit, with face-to-face alignment of the two phosphopeptides [95] (Fig. 4a). The major protein–protein interaction between the two protomers occurs within the PH domain of Dok7, suggesting that, in addition to membrane association, the PH domain may mediate Dok7 dimerization [95]. Based on the structure, Dok7–MuSK is predicated to interact as a 2:2 complex, which is consistent with the behavior of the proteins in solution. This arrangement allows the two PH domains of Dok7 to be oriented for phosphoinositide binding while the PTB domains interact with the juxtamembrane region of MuSK. Mutagenesis of key residues on the dimer interface and the Dok7–MuSK interface have also confirmed that dimerization of the two proteins is critical for MuSK autophosphorylation and activation.
The dimerization of the Dok7–MuSK complex raises the possibility that Dok7 alone may be able to dimerize and activate MuSK intracellularly. However, it is unclear whether such a mechanism would have physiological relevance. It is possible that Dok7-triggered MuSK activation, which would be independent of agrin binding, could facilitate the pre-patterning of MuSK and AChRs [58]. Alternatively, Dok7 may simply be required to overcome the strong autoinhibition of MuSK once agrin is bound. These scenarios assume that the juxtamembrane Tyr553 in MuSK is already phosphorylated to allow Dok7 association; however, the exact mechanism by which Tyr553 is phosphorylated is not yet clear.
Tid1 is another MuSK adaptor protein
A rat homolog of Drosophila tumorous imaginal disc 1 (Tid1) was identified recently as an effector of the agrin–MuSK pathway [97]. Tid1 is constitutively associated with the juxtamembrane region of MuSK and is required for MuSK–Dok7 signaling. Although Tid1 is not directly involved in agrin-dependent activation of MuSK, it could play a role downstream of MuSK activation. For example, Tid1 regulates the activity of GTPases Rac and Rho, and induces tyrosine phosphorylation of the AChR β subunits necessary for interactions with the adaptor protein rapsyn, which directly anchors AChR clusters to the synaptic sites [97]. Therefore, Tid1 seems to bridge the gap between MuSK and rapsyn.
Tid1 is a member of the heat shock protein 40 (HSP40) family; these proteins contain a DnaJ domain that interacts with and activates HSP70 [98, 99]. The DnaJ domain of Tid1 thus brings heat shock proteins into the agrin–LRP4–Dok7–Tid1 signaling complex. An NMR structure of the human Tid1 DnaJ domain is available in the Protein Data Bank (PDB code 2DN9, unpublished), which shows a 4-helix architecture that is highly conserved among the DnaJ-containing chaperones (Fig. 4b) [100, 101]. A turn connecting the second and third helices contains a histidine-proline-aspartate (HPD) tripeptide motif, which is conserved in the HSP40 family and is essential for interaction with HSP70 [102]. A mutation within the HPD motif of the human Tid1 DnaJ domain, H121Q, prevents activation of HSP70 [103]. Interestingly, the same mutation in rat Tid1 failed to rescue Tid1 shRNA inhibition of AChR clustering, suggesting the Tid1 DnaJ domain may regulate AChR clustering by mediating the activation of HSP70 [97]. It should be noted that the discovery of the Tid1’s interaction with MuSK and Dok7 was based on the cultured C2C12 myotubes. In vivo evidence is required to confirm the functional role of the MuSK–Dok7–Tid1 complex in NMJ differentiation.
Future perspectives
Agrin, LRP4, and MuSK are all large, complex proteins with multiple modular domains. Although the structures of several individual domains or relatively short fragments are now available, structures of larger fragments encompassing multiple domains are still needed to extrapolate in vitro structural findings to an in vivo system. Moreover, future investigations should focus on protein–protein interactions not only among agrin, LRP4, and MuSK, but also with their various binding partners.
Solving the structure of the agrin–LRP4–MuSK complex will be a major challenge that will reveal how MuSK is activated by the ligand–co-receptor complex, and create a new paradigm for RTK activation. Similarly, structural studies of the interaction of Wnt proteins with LRP4 and MuSK will contribute to our understanding of the role of Wnt signaling in NMJ development. Inside the muscle cells, we do not yet know how Tid1 is constitutively associated with the juxtamembrane region of MuSK, how this interaction is coordinated with the Dok7–MuSK complex, and whether other signaling proteins may be involved.
Rapsyn directly anchors AChR clusters to the synaptic sites, and thus is an important component of the agrin-induced signaling cascade [104–106]. The structure of rapsyn, which contains various domains for membrane targeting, AChR interaction, self-association, and MuSK binding, may reveal how it functions at the NMJ. Across the postsynaptic membrane, the activated multiprotein complex, agrin–LRP4–MuSK–Dok7–Tid1, is presumably responsible for localizing rapsyn to the synaptic sites. Rapsyn may interact with the intracellular region of MuSK directly or with the ectodomain of MuSK through a putative transmembrane component, RATL (rapsyn-associated transmembrane linker). Structural and biochemical studies of the MuSK–rapsyn association will contribute to our understanding of the events occurring between agrin-induced MuSK activation and AChR clustering.
Acknowledgments
We apologize to our colleagues whose work could not be individually cited due to space limitations. R.J. acknowledges support from the Alfred P. Sloan Foundation.
Abbreviations
- AChR
Acetylcholine receptor
- Fz-CRD
Frizzled cysteine-rich domain
- Ig-like
Immunoglobulin-like
- LG
Laminin globular
- LRP4
Low-density lipoprotein receptor-related protein 4
- MuSK
Muscle-specific kinase
- NMJ
Neuromuscular junction
- NtA
N-terminal agrin
- PH
Pleckstrin homology
- PTB
Phosphotyrosine binding
- RTK
Receptor tyrosine kinase
- Tid1
Tumorous imaginal disc 1
References
- 1.Slater CR. Reliability of neuromuscular transmission and how it is maintained. Handb Clin Neurol. 2008;91:27–101. doi: 10.1016/S0072-9752(07)01502-3. [DOI] [PubMed] [Google Scholar]
- 2.Madhavan R, Peng HB. Molecular regulation of postsynaptic differentiation at the neuromuscular junction. IUBMB Life. 2005;57:719–730. doi: 10.1080/15216540500338739. [DOI] [PubMed] [Google Scholar]
- 3.Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci. 2001;2:791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
- 4.Fertuck HC, Salpeter MM. Quantitation of junctional and extrajunctional acetylcholine receptors by electron microscope autoradiography after 125I-alpha-bungarotoxin binding at mouse neuromuscular junctions. J Cell Biol. 1976;69:144–158. doi: 10.1083/jcb.69.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burden SJ. Building the vertebrate neuromuscular synapse. J Neurobiol. 2002;53:501–511. doi: 10.1002/neu.10137. [DOI] [PubMed] [Google Scholar]
- 6.Ferraro E, Molinari F, Berghella L. Molecular control of neuromuscular junction development. J Cachexia Sarcopenia Muscle. 2012;3:13–23. doi: 10.1007/s13539-011-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, Xiong WC, Mei L. To build a synapse: signaling pathways in neuromuscular junction assembly. Development. 2010;137:1017–1033. doi: 10.1242/dev.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witzemann V. Development of the neuromuscular junction. Cell Tissue Res. 2006;326:263–271. doi: 10.1007/s00441-006-0237-x. [DOI] [PubMed] [Google Scholar]
- 9.Ngo ST, Noakes PG, Phillips WD. Neural agrin: a synaptic stabiliser. Int J Biochem Cell Biol. 2007;39:863–867. doi: 10.1016/j.biocel.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Sigoillot SM, Bourgeois F, Lambergeon M, Strochlic L, Legay C. ColQ controls postsynaptic differentiation at the neuromuscular junction. J Neurosci. 2010;30:13–23. doi: 10.1523/JNEUROSCI.4374-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amenta AR, et al. Biglycan is an extracellular MuSK binding protein important for synapse stability. J Neurosci. 2012;32:2324–2334. doi: 10.1523/JNEUROSCI.4610-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singhal N, Martin PT. Role of extracellular matrix proteins and their receptors in the development of the vertebrate neuromuscular junction. Dev Neurobiol. 2011;71:982–1005. doi: 10.1002/dneu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghazanfari N, Fernandez KJ, Murata Y, Morsch M, Ngo ST, Reddel SW, Noakes PG, Phillips WD. Muscle specific kinase: organiser of synaptic membrane domains. Int J Biochem Cell Biol. 2011;43:295–298. doi: 10.1016/j.biocel.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim N, et al. Lrp4 is a receptor for agrin and forms a complex with MuSK. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weatherbee SD, Anderson KV, Niswander LA. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development. 2006;133:4993–5000. doi: 10.1242/dev.02696. [DOI] [PubMed] [Google Scholar]
- 17.Song Y, Balice-Gordon R. New dogs in the dogma: lrp4 and Tid1 in neuromuscular synapse formation. Neuron. 2008;60:526–528. doi: 10.1016/j.neuron.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Antolik C, Catino DH, Resneck WG, Bloch RJ. The tetratricopeptide repeat domains of rapsyn bind directly to cytoplasmic sequences of the muscle-specific kinase. Neuroscience. 2006;141:87–100. doi: 10.1016/j.neuroscience.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 19.McMahan UJ. The agrin hypothesis. Cold Spring Harb Symp Quant Biol. 1990;55:407–418. doi: 10.1101/SQB.1990.055.01.041. [DOI] [PubMed] [Google Scholar]
- 20.Nitkin RM, Smith MA, Magill C, Fallon JR, Yao YM, Wallace BG, McMahan UJ. Identification of agrin, a synaptic organizing protein from Torpedo electric organ. J Cell Biol. 1987;105:2471–2478. doi: 10.1083/jcb.105.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bezakova G, Ruegg MA. New insights into the roles of agrin. Nat Rev Mol Cell Biol. 2003;4:295–308. doi: 10.1038/nrm1074. [DOI] [PubMed] [Google Scholar]
- 22.Ferns MJ, Campanelli JT, Hoch W, Scheller RH, Hall Z. The ability of agrin to cluster AChRs depends on alternative splicing and on cell surface proteoglycans. Neuron. 1993;11:491–502. doi: 10.1016/0896-6273(93)90153-I. [DOI] [PubMed] [Google Scholar]
- 23.Hoch W, Campanelli JT, Scheller RH. Structural domains of agrin required for clustering of nicotinic acetylcholine receptors. EMBO J. 1994;13:2814–2821. doi: 10.1002/j.1460-2075.1994.tb06575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahan UJ, Horton SE, Werle MJ, Honig LS, Kroger S, Ruegg MA, Escher G. Agrin isoforms and their role in synaptogenesis. Curr Opin Cell Biol. 1992;4:869–874. doi: 10.1016/0955-0674(92)90113-Q. [DOI] [PubMed] [Google Scholar]
- 25.Cohen NA, Kaufmann WE, Worley PF, Rupp F. Expression of agrin in the developing and adult rat brain. Neuroscience. 1997;76:581–596. doi: 10.1016/S0306-4522(96)00345-4. [DOI] [PubMed] [Google Scholar]
- 26.Kroger S, Mann S. Biochemical and functional characterization of basal lamina-bound agrin in the chick central nervous system. Eur J Neurosci. 1996;8:500–509. doi: 10.1111/j.1460-9568.1996.tb01234.x. [DOI] [PubMed] [Google Scholar]
- 27.Ksiazek I, et al. Synapse loss in cortex of agrin-deficient mice after genetic rescue of perinatal death. J Neurosci. 2007;27:7183–7195. doi: 10.1523/JNEUROSCI.1609-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McFarlane AA, Stetefeld J. An interdomain disulfide bridge links the NtA and first FS domain in agrin. Protein Sci. 2009;18:2421–2428. doi: 10.1002/pro.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascarenhas JB, Ruegg MA, Sasaki T, Eble JA, Engel J, Stetefeld J. Structure and laminin-binding specificity of the NtA domain expressed in eukaryotic cells. Matrix Biol. 2005;23:507–513. doi: 10.1016/j.matbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Stetefeld J, et al. The laminin-binding domain of agrin is structurally related to N-TIMP-1. Nat Struct Biol. 2001;8:705–709. doi: 10.1038/90422. [DOI] [PubMed] [Google Scholar]
- 31.Caterina NC, Windsor LJ, Yermovsky AE, Bodden MK, Taylor KB, Birkedal-Hansen H, Engler JA. Replacement of conserved cysteines in human tissue inhibitor of metalloproteinases-1. J Biol Chem. 1997;272:32141–32149. doi: 10.1074/jbc.272.51.32141. [DOI] [PubMed] [Google Scholar]
- 32.Huang W, Meng Q, Suzuki K, Nagase H, Brew K. Mutational study of the amino-terminal domain of human tissue inhibitor of metalloproteinases 1 (TIMP-1) locates an inhibitory region for matrix metalloproteinases. J Biol Chem. 1997;272:22086–22091. doi: 10.1074/jbc.272.35.22086. [DOI] [PubMed] [Google Scholar]
- 33.Mascarenhas JB, Ruegg MA, Winzen U, Halfter W, Engel J, Stetefeld J. Mapping of the laminin-binding site of the N-terminal agrin domain (NtA) EMBO J. 2003;22:529–536. doi: 10.1093/emboj/cdg041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornish T, Chi J, Johnson S, Lu Y, Campanelli JT. Globular domains of agrin are functional units that collaborate to induce acetylcholine receptor clustering. J Cell Sci. 1999;112(Pt 8):1213–1223. doi: 10.1242/jcs.112.8.1213. [DOI] [PubMed] [Google Scholar]
- 35.Gesemann M, Denzer AJ, Ruegg MA. Acetylcholine receptor-aggregating activity of agrin isoforms and mapping of the active site. J Cell Biol. 1995;128:625–636. doi: 10.1083/jcb.128.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng CN, Zhang L, Wu SL, Wang WF, Wang ZZ, Cascio M. Asparagine of z8 insert is critical for the affinity, conformation, and acetylcholine receptor-clustering activity of neural agrin. J Biol Chem. 2010;285:27641–27651. doi: 10.1074/jbc.M110.130625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng CN, Zhang L, Cascio M, Wang ZZ. Calcium plays a critical role in determining the acetylcholine receptor-clustering activities of alternatively spliced isoforms of Agrin. J Biol Chem. 2003;278:17236–17245. doi: 10.1074/jbc.M300282200. [DOI] [PubMed] [Google Scholar]
- 38.Tidow H, Mattle D, Nissen P. Structural and biophysical characterisation of agrin laminin G3 domain constructs. Protein Eng Des Sel. 2011;24:219–224. doi: 10.1093/protein/gzq082. [DOI] [PubMed] [Google Scholar]
- 39.Stetefeld J, et al. Modulation of agrin function by alternative splicing and Ca2+ binding. Structure. 2004;12:503–515. doi: 10.1016/j.str.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Rudenko G, Hohenester E, Muller YA. LG/LNS domains: multiple functions—one business end? Trends Biochem Sci. 2001;26:363–368. doi: 10.1016/S0968-0004(01)01832-1. [DOI] [PubMed] [Google Scholar]
- 41.Godfrey EW, Roe J, Heathcote RD. Agrin fragments differentially induce ectopic aggregation of acetylcholine receptors in myotomal muscles of Xenopus embryos. J Neurobiol. 2000;44:436–445. doi: 10.1002/1097-4695(20000915)44:4<436::AID-NEU6>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 42.Ruegg MA, Tsim KW, Horton SE, Kroger S, Escher G, Gensch EM, McMahan UJ. The agrin gene codes for a family of basal lamina proteins that differ in function and distribution. Neuron. 1992;8:691–699. doi: 10.1016/0896-6273(92)90090-Z. [DOI] [PubMed] [Google Scholar]
- 43.Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E. Structure and function of laminin LG modules. Matrix Biol. 2000;19:309–317. doi: 10.1016/S0945-053X(00)00072-X. [DOI] [PubMed] [Google Scholar]
- 44.O’Toole JJ, Deyst KA, Bowe MA, Nastuk MA, McKechnie BA, Fallon JR. Alternative splicing of agrin regulates its binding to heparin alpha-dystroglycan, and the cell surface. Proc Natl Acad Sci USA. 1996;93:7369–7374. doi: 10.1073/pnas.93.14.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campanelli JT, Gayer GG, Scheller RH. Alternative RNA splicing that determines agrin activity regulates binding to heparin and alpha-dystroglycan. Development. 1996;122:1663–1672. doi: 10.1242/dev.122.5.1663. [DOI] [PubMed] [Google Scholar]
- 46.Maselli RA, et al. LG2 agrin mutation causing severe congenital myasthenic syndrome mimics functional characteristics of non-neural (z-) agrin. Hum Genet. 2011;131(7):1123–1135. doi: 10.1007/s00439-011-1132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopf C, Hoch W. Dimerization of the muscle-specific kinase induces tyrosine phosphorylation of acetylcholine receptors and their aggregation on the surface of myotubes. J Biol Chem. 1998;273:6467–6473. doi: 10.1074/jbc.273.11.6467. [DOI] [PubMed] [Google Scholar]
- 49.Xie MH, Yuan J, Adams C, Gurney A. Direct demonstration of MuSK involvement in acetylcholine receptor clustering through identification of agonist ScFv. Nat Biotechnol. 1997;15:768–771. doi: 10.1038/nbt0897-768. [DOI] [PubMed] [Google Scholar]
- 50.Glass DJ, et al. Agrin acts via a MuSK receptor complex. Cell. 1996;85:513–523. doi: 10.1016/S0092-8674(00)81252-0. [DOI] [PubMed] [Google Scholar]
- 51.DeChiara TM, et al. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/S0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- 52.Till JH, Becerra M, Watty A, Lu Y, Ma Y, Neubert TA, Burden SJ, Hubbard SR. Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation. Structure. 2002;10:1187–1196. doi: 10.1016/S0969-2126(02)00814-6. [DOI] [PubMed] [Google Scholar]
- 53.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/S0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 54.Songyang Z, Margolis B, Chaudhuri M, Shoelson SE, Cantley LC. The phosphotyrosine interaction domain of SHC recognizes tyrosine-phosphorylated NPXY motif. J Biol Chem. 1995;270:14863–14866. doi: 10.1074/jbc.270.25.14863. [DOI] [PubMed] [Google Scholar]
- 55.Uhlik MT, Temple B, Bencharit S, Kimple AJ, Siderovski DP, Johnson GL. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J Mol Biol. 2005;345:1–20. doi: 10.1016/j.jmb.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 56.Jones G, Moore C, Hashemolhosseini S, Brenner HR. Constitutively active MuSK is clustered in the absence of agrin and induces ectopic postsynaptic-like membranes in skeletal muscle fibers. J Neurosci. 1999;19:3376–3383. doi: 10.1523/JNEUROSCI.19-09-03376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X, Arber S, William C, Li L, Tanabe Y, Jessell TM, Birchmeier C, Burden SJ. Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron. 2001;30:399–410. doi: 10.1016/S0896-6273(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 58.Lin W, Burgess RW, Dominguez B, Pfaff SL, Sanes JR, Lee KF. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature. 2001;410:1057–1064. doi: 10.1038/35074025. [DOI] [PubMed] [Google Scholar]
- 59.Grassot J, Gouy M, Perriere G, Mouchiroud G. Origin and molecular evolution of receptor tyrosine kinases with immunoglobulin-like domains. Mol Biol Evol. 2006;23:1232–1241. doi: 10.1093/molbev/msk007. [DOI] [PubMed] [Google Scholar]
- 60.Zhou H, Glass DJ, Yancopoulos GD, Sanes JR. Distinct domains of MuSK mediate its abilities to induce and to associate with postsynaptic specializations. J Cell Biol. 1999;146:1133–1146. doi: 10.1083/jcb.146.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Q, Zhang B, Wang YE, Xiong WC, Mei L. The Ig1/2 domain of MuSK binds to muscle surface and is involved in acetylcholine receptor clustering. Neurosignals. 2008;16:246–253. doi: 10.1159/000111567. [DOI] [PubMed] [Google Scholar]
- 62.Stiegler AL, Burden SJ, Hubbard SR. Crystal structure of the agrin-responsive immunoglobulin-like domains 1 and 2 of the receptor tyrosine kinase MuSK. J Mol Biol. 2006;364:424–433. doi: 10.1016/j.jmb.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ultsch MH, Wiesmann C, Simmons LC, Henrich J, Yang M, Reilly D, Bass SH, de Vos AM. Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC. J Mol Biol. 1999;290:149–159. doi: 10.1006/jmbi.1999.2816. [DOI] [PubMed] [Google Scholar]
- 64.Jennings CG, Dyer SM, Burden SJ. Muscle-specific trk-related receptor with a kringle domain defines a distinct class of receptor tyrosine kinases. Proc Natl Acad Sci USA. 1993;90:2895–2899. doi: 10.1073/pnas.90.7.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masiakowski P, Yancopoulos GD. The Wnt receptor CRD domain is also found in MuSK and related orphan receptor tyrosine kinases. Curr Biol. 1998;8:R407. doi: 10.1016/S0960-9822(98)70263-5. [DOI] [PubMed] [Google Scholar]
- 66.Xu YK, Nusse R. The Frizzled CRD domain is conserved in diverse proteins including several receptor tyrosine kinases. Curr Biol. 1998;8:R405–R406. doi: 10.1016/S0960-9822(98)70262-3. [DOI] [PubMed] [Google Scholar]
- 67.Jing L, Lefebvre JL, Gordon LR, Granato M. Wnt signals organize synaptic prepattern and axon guidance through the zebrafish unplugged/MuSK receptor. Neuron. 2009;61:721–733. doi: 10.1016/j.neuron.2008.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banerjee S, Gordon L, Donn TM, Berti C, Moens CB, Burden SJ, Granato M. A novel role for MuSK and non-canonical Wnt signaling during segmental neural crest cell migration. Development. 2011;138:3287–3296. doi: 10.1242/dev.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strochlic L, et al. Wnt4 participates in the formation of vertebrate neuromuscular junction. PLoS One. 2012;7:e29976. doi: 10.1371/journal.pone.0029976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang B, Liang C, Bates R, Yin Y, Xiong WC, Mei L. Wnt proteins regulate acetylcholine receptor clustering in muscle cells. Mol Brain. 2012;5:7. doi: 10.1186/1756-6606-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stiegler AL, Burden SJ, Hubbard SR. Crystal structure of the frizzled-like cysteine-rich domain of the receptor tyrosine kinase MuSK. J Mol Biol. 2009;393:1–9. doi: 10.1016/j.jmb.2009.07.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature. 2001;412:86–90. doi: 10.1038/35083601. [DOI] [PubMed] [Google Scholar]
- 73.Carron C, Pascal A, Djiane A, Boucaut JC, Shi DL, Umbhauer M. Frizzled receptor dimerization is sufficient to activate the Wnt/beta-catenin pathway. J Cell Sci. 2003;116:2541–2550. doi: 10.1242/jcs.00451. [DOI] [PubMed] [Google Scholar]
- 74.Oishi I, et al. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8:645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- 75.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.May P, Woldt E, Matz RL, Boucher P. The LDL receptor-related protein (LRP) family: an old family of proteins with new physiological functions. Ann Med. 2007;39:219–228. doi: 10.1080/07853890701214881. [DOI] [PubMed] [Google Scholar]
- 77.Hussain MM. Structural, biochemical and signaling properties of the low-density lipoprotein receptor gene family. Front Biosci. 2001;6:D417–D428. doi: 10.2741/Hussain1. [DOI] [PubMed] [Google Scholar]
- 78.Go GW, Mani A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J Biol Med. 2012;85:19–28. [PMC free article] [PubMed] [Google Scholar]
- 79.Willnow TE, Nykjaer A, Herz J. Lipoprotein receptors: new roles for ancient proteins. Nat Cell Biol. 1999;1:E157–E162. doi: 10.1038/14109. [DOI] [PubMed] [Google Scholar]
- 80.Simon-Chazottes D, Tutois S, Kuehn M, Evans M, Bourgade F, Cook S, Davisson MT, Guenet JL. Mutations in the gene encoding the low-density lipoprotein receptor LRP4 cause abnormal limb development in the mouse. Genomics. 2006;87:673–677. doi: 10.1016/j.ygeno.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 81.Johnson EB, Steffen DJ, Lynch KW, Herz J. Defective splicing of Megf7/Lrp4, a regulator of distal limb development, in autosomal recessive mulefoot disease. Genomics. 2006;88:600–609. doi: 10.1016/j.ygeno.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 82.Zong Y, et al. Structural basis of agrin-LRP4-MuSK signaling. Genes Dev. 2012;26:247–258. doi: 10.1101/gad.180885.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bourhis E, et al. Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6. Structure. 2011;19:1433–1442. doi: 10.1016/j.str.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 84.Takagi J, Yang Y, Liu JH, Wang JH, Springer TA. Complex between nidogen and laminin fragments reveals a paradigmatic beta-propeller interface. Nature. 2003;424:969–974. doi: 10.1038/nature01873. [DOI] [PubMed] [Google Scholar]
- 85.Choi HY, Dieckmann M, Herz J, Niemeier A. Lrp4, a novel receptor for Dickkopf 1 and sclerostin, is expressed by osteoblasts and regulates bone growth and turnover in vivo. PLoS One. 2009;4:e7930. doi: 10.1371/journal.pone.0007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leupin O, et al. Bone overgrowth-associated mutations in the LRP4 gene impair sclerostin facilitator function. J Biol Chem. 2011;286:19489–19500. doi: 10.1074/jbc.M110.190330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang W, Coldefy AS, Hubbard SR, Burden SJ. Agrin binds to the N-terminal region of Lrp4 protein and stimulates association between Lrp4 and the first immunoglobulin-like domain in muscle-specific kinase (MuSK) J Biol Chem. 2011;286:40624–40630. doi: 10.1074/jbc.M111.279307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gomez AM, Burden SJ. The extracellular region of Lrp4 is sufficient to mediate neuromuscular synapse formation. Dev Dyn. 2011;240:2626–2633. doi: 10.1002/dvdy.22772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cai D, Dhe-Paganon S, Melendez PA, Lee J, Shoelson SE. Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J Biol Chem. 2003;278:25323–25330. doi: 10.1074/jbc.M212430200. [DOI] [PubMed] [Google Scholar]
- 90.Mashima R, Hishida Y, Tezuka T, Yamanashi Y. The roles of Dok family adapters in immunoreceptor signaling. Immunol Rev. 2009;232:273–285. doi: 10.1111/j.1600-065X.2009.00844.x. [DOI] [PubMed] [Google Scholar]
- 91.Inoue A, Setoguchi K, Matsubara Y, Okada K, Sato N, Iwakura Y, Higuchi O, Yamanashi Y. Dok-7 activates the muscle receptor kinase MuSK and shapes synapse formation. Sci Signal. 2009;2(59):ra7. doi: 10.1126/scisignal.2000113. [DOI] [PubMed] [Google Scholar]
- 92.Okada K, et al. The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science. 2006;312:1802–1805. doi: 10.1126/science.1127142. [DOI] [PubMed] [Google Scholar]
- 93.Hamuro J, et al. Mutations causing DOK7 congenital myasthenia ablate functional motifs in Dok-7. J Biol Chem. 2008;283:5518–5524. doi: 10.1074/jbc.M708607200. [DOI] [PubMed] [Google Scholar]
- 94.Beeson D, et al. Dok-7 mutations underlie a neuromuscular junction synaptopathy. Science. 2006;313:1975–1978. doi: 10.1126/science.1130837. [DOI] [PubMed] [Google Scholar]
- 95.Bergamin E, Hallock PT, Burden SJ, Hubbard SR. The cytoplasmic adaptor protein Dok7 activates the receptor tyrosine kinase MuSK via dimerization. Mol Cell. 2010;39:100–109. doi: 10.1016/j.molcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dhe-Paganon S, Ottinger EA, Nolte RT, Eck MJ, Shoelson SE. Crystal structure of the pleckstrin homology-phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1. Proc Natl Acad Sci USA. 1999;96:8378–8383. doi: 10.1073/pnas.96.15.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Linnoila J, Wang Y, Yao Y, Wang ZZ. A mammalian homolog of Drosophila tumorous imaginal discs, Tid1, mediates agrin signaling at the neuromuscular junction. Neuron. 2008;60:625–641. doi: 10.1016/j.neuron.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gaestel M. Molecular chaperones in signal transduction. Handb Exp Pharmacol. 2006;172:93–109. doi: 10.1007/3-540-29717-0_4. [DOI] [PubMed] [Google Scholar]
- 99.Lu B, Garrido N, Spelbrink JN, Suzuki CK. Tid1 isoforms are mitochondrial DnaJ-like chaperones with unique carboxyl termini that determine cytosolic fate. J Biol Chem. 2006;281:13150–13158. doi: 10.1074/jbc.M509179200. [DOI] [PubMed] [Google Scholar]
- 100.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hennessy F, Boshoff A, Blatch GL. Rational mutagenesis of a 40 kDa heat shock protein from Agrobacterium tumefaciens identifies amino acid residues critical to its in vivo function. Int J Biochem Cell Biol. 2005;37:177–191. doi: 10.1016/j.biocel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 102.Tsai J, Douglas MG. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 103.Syken J, De-Medina T, Munger K. TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc Natl Acad Sci USA. 1999;96:8499–8504. doi: 10.1073/pnas.96.15.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee Y, Rudell J, Ferns M. Rapsyn interacts with the muscle acetylcholine receptor via alpha-helical domains in the alpha, beta, and epsilon subunit intracellular loops. Neuroscience. 2009;163:222–232. doi: 10.1016/j.neuroscience.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moransard M, Borges LS, Willmann R, Marangi PA, Brenner HR, Ferns MJ, Fuhrer C. Agrin regulates rapsyn interaction with surface acetylcholine receptors, and this underlies cytoskeletal anchoring and clustering. J Biol Chem. 2003;278:7350–7359. doi: 10.1074/jbc.M210865200. [DOI] [PubMed] [Google Scholar]
- 106.Ramarao MK, Bianchetta MJ, Lanken J, Cohen JB. Role of rapsyn tetratricopeptide repeat and coiled-coil domains in self-association and nicotinic acetylcholine receptor clustering. J Biol Chem. 2001;276:7475–7483. doi: 10.1074/jbc.M009888200. [DOI] [PubMed] [Google Scholar]