Abstract
The blood-brain barrier (BBB) is a critical structure that serves as the gatekeeper between the central nervous system and the rest of the body. It is the responsibility of the BBB to facilitate the entry of required nutrients into the brain and to exclude potentially harmful compounds; however, this complex structure has remained difficult to model faithfully in vitro. Accurate in vitro models are necessary for understanding how the BBB forms and functions, as well as for evaluating drug and toxin penetration across the barrier. Many previous models have failed to support all the cell types involved in the BBB formation and/or lacked the flow-created shear forces needed for mature tight junction formation. To address these issues and to help establish a more faithful in vitro model of the BBB, we have designed and fabricated a microfluidic device that is comprised of both a vascular chamber and a brain chamber separated by a porous membrane. This design allows for cell-to-cell communication between endothelial cells, astrocytes, and pericytes and independent perfusion of both compartments separated by the membrane. This NeuroVascular Unit (NVU) represents approximately one-millionth of the human brain, and hence, has sufficient cell mass to support a breadth of analytical measurements. The NVU has been validated with both fluorescein isothiocyanate (FITC)-dextran diffusion and transendothelial electrical resistance. The NVU has enabled in vitro modeling of the BBB using all human cell types and sampling effluent from both sides of the barrier.
I. INTRODUCTION
The blood-brain barrier (BBB) is a critical structure acting as the gatekeeper between the central nervous system (CNS) and the rest of the body. Not only does this unique barrier regulate traffic of small molecules, hormones, and nutrients between the body and the brain but it is also the principal means of regulating drug and toxin access into the brain. Because of these features, the ability to model the BBB is essential to testing the delivery and safety of drugs designed to act in the CNS.
The BBB is also a complex structure comprised of multiple cell types, of which endothelial cells have received the most attention. Tight junctions between endothelial cells form a physical link and prevent the passage of molecules from the blood directly into the brain. It has been shown that both cell-to-cell interactions and diffusible cues from the CNS, primarily originating from pericytes and astrocytes, are necessary for cell polarity and the proper formation of tight junctions.1,2 In addition to multiple cell types, true formation of a BBB requires shear forces experienced by endothelial cells from blood circulation, as well as the differential serum concentration across the BBB.3,4 Thus, recapitulation of this complicated and dynamic barrier of the BBB requires innovation for high fidelity modeling.
Current animal models, while informative, are expensive and have been shown to lack consistent predictive value for humans.5–7 Recent research has shown that microvascular cells have species-specific properties that may account for these differences between the human BBB and those of other mammalian species. Current computer models of the BBB are excellent for high-throughput studies and limit cost, but they are only useful for predicting basic permeability based on drug structure—they cannot provide information as to the efficacy of the drug in question or whether the drug itself may influence BBB permeability.8,9 Traditional transwell cell culture models have the advantage of being able to use actual human cells with relatively high throughput, but it has been demonstrated that many of the cell types used in these experiments do not maintain their terminally differentiated state under these culture conditions, causing perturbations and abnormalities within the BBB.6,10–13 Three-dimensional dynamic models overcome the differentiation issues seen within transwells, but do not faithfully recapitulate the lumen and sub-lumen spaces which allow for endothelial cell specialization14–18 and polarization.5,19,20 In addition, these static models lack continually refreshed media.14,21–23 All of the current models add to our understanding of the BBB formation and function; however, each still leaves room for improvement to better represent this complex, multi-dimensional, multi-cellular brain structure.
The organ-on-chip community is expending significant effort on the development of advanced, tissue-engineered microphysiological systems, typically using microfluidics.24–27 Planar microfluidic BBB models are excellent for examining the BBB in cross-section, particularly to observe leukocyte extravasation, but lack the cell number to support in-depth secretome characterization.28–31 To address the limitations of the classic BBB models and extend the capabilities of planar microfluidic ones, we have developed a self-contained microfluidic bioreactor that recreates the in vivo-like conditions of the BBB in vitro. Our BBB organ-on-a-chip system, termed the NeuroVascular Unit (NVU), supports growth and development of primary cell-derived human neurons, astrocytes, pericytes, and microvascular endothelial cells, all of which are involved in the proper formation of the BBB. The NVU not only allows these cells to differentiate and survive but also recreates such important microenvironmental conditions as cell-to-fluid volume ratios, spatial gradients, and proper fluid flows with appropriate shear stresses. We believe that our organ-on-a-chip NVU system will enable bridging the gap between animal and human models of the BBB and help in understanding how various assaults and manipulations can perturb the function of the BBB.
II. MATERIALS AND METHODS
A. NVU design
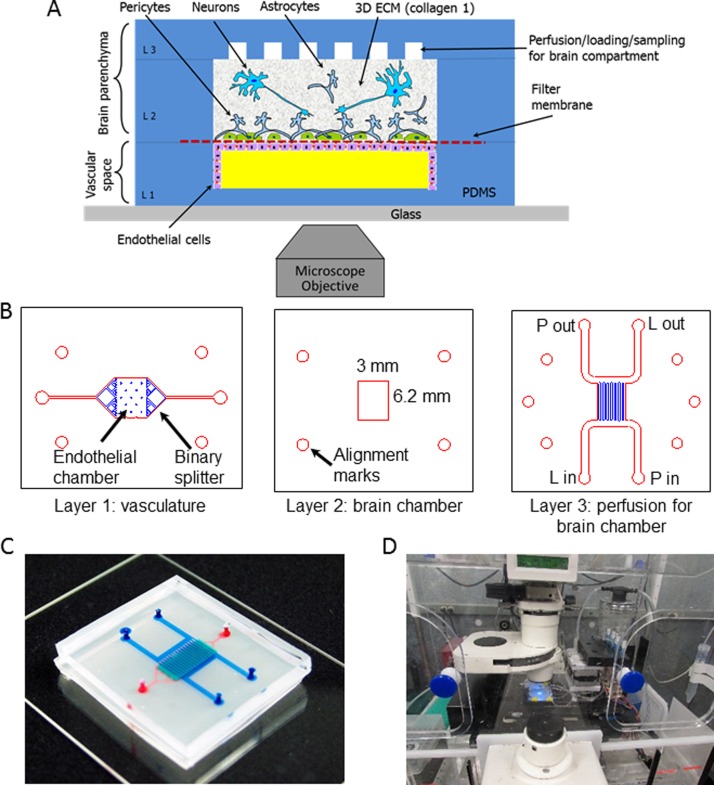
Our NVU (Figure 1(a)) is designed to be approximately one-millionth the size of an adult human brain.32 The NVU is made up of three polydimethylsiloxane (PDMS) layers (Figure 1(b)) plus the 0.2 μm pore polycarbonate membrane that divides the two chambers. At present, we use track-etched polycarbonate membranes, similar to those found in commercially available transwells, to provide the scaffold for vascular cells and nervous system cells to grow in a two-compartment arrangement. The membrane is bonded between layers of PDMS to create the microfluidic vasculature and brain compartments (Figures 1(a) and 1(c)). The NVU has two perfusion streams, one for the vascular media supply to the endothelial cells in layer 1 (Figure 1(c), on bottom in red), with an inlet and outlet port, and another for the brain media supply to layers 2 and 3 (in blue, Figure 1(c)), with four ports, so that one diagonal pair can be used for collagen and cell loading and the other for subsequent perfusion of the brain compartment. While not in use, these ports are closed with knotted tubing. The design of layer 3 was chosen to facilitate even and uniform loading of collagen gel containing cells. The volumes of the vascular and brain chambers are 2.91 μl and 17.5 μl, respectively. The two chambers share a common outline, eliminating problems that might arise in crossed-channel bioreactors wherein only a portion of the two chambers overlap, creating larger populations of cells that do not share important signals.28,30 Because we need to grow cells on each side of the membrane, it is necessary to perfuse the NVU device first with the vascular side of the membrane facing up to seed the endothelial cells. Once the cells adhere to the membrane, the device can be turned over so that the vascular side faces down, allowing the seeding of astrocytes and pericytes to the other side of the membrane. It is always a challenge to invert transwell inserts for this purpose, but for the NVU we designed a flippable backpack that maintains the feed and collection vials in an upright position regardless of the orientation of the NVU device (Supplementary Figures 1(a) and 1(b)).69 This backpack enables culture of the NVU in either of its orientations while still maintaining a standard 6-well footprint for ease of imaging (Figure 1(d)).
FIG. 1.
Layout and design of the NeuroVascular Unit (NVU). (a) Schematic view of the NVU indicating major components, cell types, and their spatial arrangement: endothelial cells lining the lower, vascular chamber; astrocytes and pericytes lining the other side of the filter membrane, with neurons in the collagen gel in the upper brain chamber. (b) Photolithographic masks used to fabricate the three-layered NVU. L in and L out—ports used for cell and extracellular matrix (ECM) loading; P in and P out—ports used for perfusion. (c) A photograph of the assembled NVU loaded with colored dyes indicating different compartments: red = vasculature; semi-transparent white = filter membrane; turquoise = brain compartment; blue = brain perfusion. (d) NVU device with this perfusion system on an incubated microscope stage.
B. NVU fabrication and assembly
We used standard soft-lithographic replica molding to create the three layers of PDMS that comprise the NVU.33 The vascular layer and brain perfusion (or “H” layer, given its overall shape) (Figure 1(b), layers 1 and 3, respectively) were produced by pouring PDMS onto a mold created photolithographically with SU-8 2100 photoresist (MicroChem, Newton, MA, USA) on Si wafers. The channel height was 100 μm. The final thicknesses of the cured PDMS for the vascular layer and “H” layer were 1 mm and 3 mm, respectively. The brain compartment (Figure 1(b), layer 2) was fabricated by curing PDMS sandwiched between two large polycarbonate plates separated by 500 μm spacers with a block that defined the layer 2 brain chamber. This resulted in a PDMS membrane of uniform thickness and a well-defined brain culture chamber that is 4.75 mm long × 6.2 mm wide × 0.5 mm deep. For all steps, we used a standard 10:1 ratio mixture of PDMS (Dow Corning).
The following procedure was used to assemble the NVU. After the PDMS was poured onto the Si/SU-8 molds, it was allowed to cure overnight at 65 °C. All layers were then removed from their molds, trimmed to size, washed with isopropanol, and dried with nitrogen gas. We used a Harrick model PDC-001 plasma oven to activate the PDMS surface and irreversibly bond layers to each other and the glass slide. First, the vascular layer 1 was plasma-treated and bonded to a 50 mm × 75 mm microscope glass slide with the chamber facing upward. Next, four access ports in layer 3 were punched for 1.5 mm OD Tygon tubing, and both layers 2 and 3 were bonded to each other. Then the polycarbonate filter membrane (Sigma CAT #111106) with 0.2 μm pores was bonded to layers 2 and 3 with 5% solution of 3-aminopropyltriethoxysilane (APTES, 99% Cat #440140, Sigma-Aldrich, St. Louis, MO, USA) using the protocol described in Ref. 34. Once the filter membrane was bonded, two additional ports were punched through layers 2 and 3 to create access to load and perfuse the endothelial cells in layer 1. Finally, both layer 1, attached to a glass slide, and combined layers 2 and 3 with the APTES-treated filter membrane were exposed to oxygen plasma again and bonded together, creating a sealed NVU device. Immediately following assembly, while the surfaces were still hydrophilic, both compartments of the NVU were loaded with deionized (DI) water. Care was taken not to trap any air bubbles within the rectangular brain compartment. If any did get trapped, the devices were placed under house vacuum until the bubbles were dissipated through the PDMS walls. All NVU bioreactors were gamma sterilized and stored under water until use.
C. Cell loading and culture in microfluidic devices
Primary human brain-derived microvascular endothelial cells (hBMVEC) from Applied Cell Biology (Kirkland, WA, USA) were maintained in endothelial basal medium (EBM)-2 (Lonza, Basel, Switzerland) containing 5% fetal bovine serum (FBS) and their growth bullet kit; however, penicillin streptomycin 1× was substituted for the gentamicin to prevent neuronal toxicity. Pericytes and astrocytes from ScienCell (Carlsbad, CA, USA) and ATCC (Manassas, VA, USA), respectively, both primary cells, were maintained in Dulbecco's modified Eagle's medium (DMEM) F-12% + 10% FBS. All cells, except human neurons (described next), were maintained in T-25 flasks under standard culture procedure35 until collected for seeding into the NVU.36–38
Human induced pluripotent stem cells (hiPSCs) from de-identified normal subjects were generated and maintained as previously described.39,40 From these, human cortical glutamatergic neurons were differentiated in multi-well plates coated with Matrigel for 80–90 days by an 11-day dual-SMAD inhibition protocol to generate early PAX6-positive forebrain neuroprogenitors, followed by a cortical glutamatergic neuron induction protocol, as previously described. Immediately prior to seeding into the NVU, the resulting human glutamatergic neurons were harvested using Accutase (Innovative Cell Technologies, Inc., San Diego, CA, USA).
Prior to cell seeding, NVU devices were first coated with laminin at 9.6 μg/ml for 24 h at 37 °C. On Day 0, the devices were washed three times with warm EBM-2 media to remove unbound laminin, and hBMVECs were loaded into the vascular compartment at 1 × 106 cells/ml. The NVU was then inverted and incubated at 37 °C overnight to allow attachment of the endothelial cells to the porous membrane. On Day 1, still inverted, the NVU was connected to a constant 2 μl/min flow of EBM-2 media delivered by either a syringe or peristaltic pump and maintained this way until Day 12. On Day 12, the device was placed right-side up, and astrocytes at 2 × 106 cells/ml and pericytes at 1 × 106 cells/ml were mixed and loaded into the brain chamber and maintained under flow for two days. On Day 14, hiPSC-derived neurons were suspended within a collagen I matrix (Invitrogen) at 1 × 106 cells/ml, loaded into the brain chamber, and allowed to solidify for 2 h before flow was restarted at 2 μl/min. This complete device was cultured for 3 days in the presence of Rho-associated coiled-coil kinase (ROCK) inhibitor (10 μM, Tocris) Y-27632 (Sigma-Aldrich, St. Louis, MO, USA) on the brain side to help the neurons survive. Y-27632 (inhibitor for p160-ROCK) markedly diminishes dissociation-induced anoikis (detachment-induced apoptosis) of dissociated single hiPSCs.41 It has been shown that ROCK inhibitor also increases the survival rate during thawing and re-plating of dissociated hiPSC-derived neural precursors and mature neurons and supports regrowth of neurites after injury.42–45 The process of loading the neurons into the NVU requires their removal from the original culture dish in which they were differentiated and their dissociation into single cells. Thus, ROCK-inhibitor Y-27632 was added to the medium for the first 3 days after seeding the neurons into the NVU to boost their survival. Once the ROCK inhibitor has been removed, the blood-brain barrier within the NVU is considered mature because (1) tight junctions have formed, (2) all barrier cells are at confluence, and (3) the neurons no longer need ROCK inhibition for survival. Finally, all cells were perfused with EBM-2 media, and the device was ready for testing.
D. Establishing a cellular blood-brain barrier
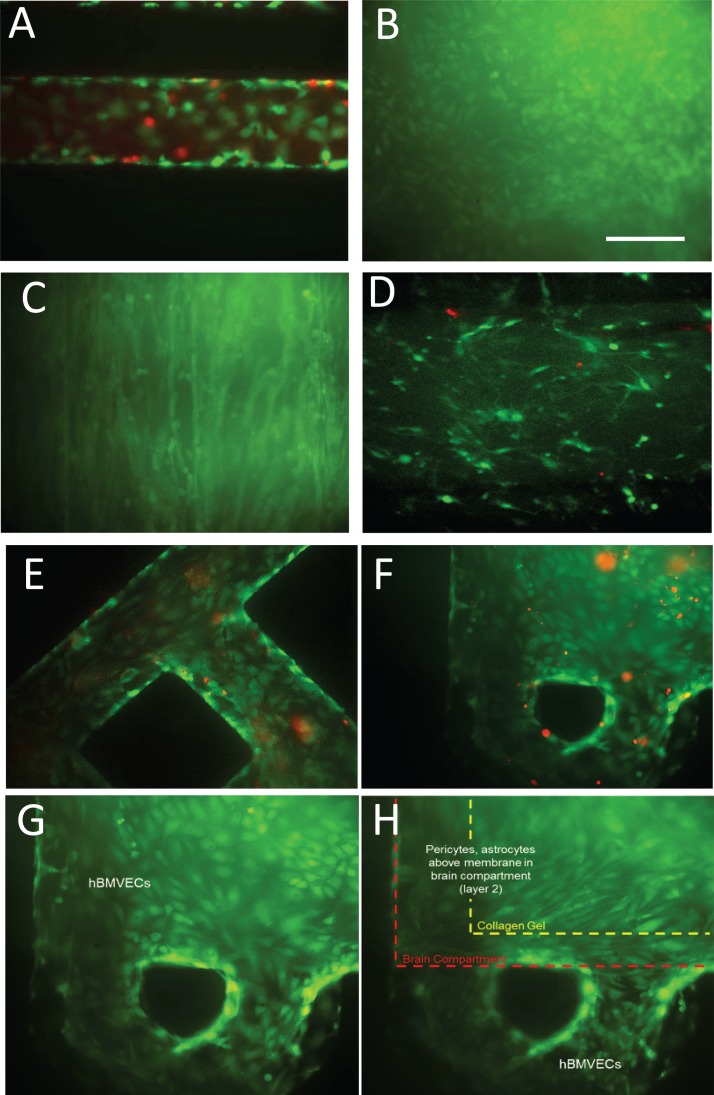
To truly recapitulate the complexity of the BBB, we felt it imperative that its three major cell types be present (endothelial cells, astrocytes, and pericytes), as well as neurons to evaluate neuronal response to changes in BBB permeability. To this end we lined the vascular chamber with HBMVECs (Figure 2(a)). The HBMVECs were given 12–14 days to reach confluence (Figure 2(b)) and establish tight junctions. After the HBMVECs were established, the NVU was then placed right-side up and a 2:1 mix of primary human astrocytes and pericytes (Figure 2(c)) was loaded into the brain chamber and given 1–2 days to reach confluence before seeding with collagen gel containing 4 × 106 human neurons/ml differentiated from hiPSCs (Figure 2(d)). Once the cell seeding was completed and the four different cell cultures were established, cell viability remained stable for all cell types between week 2 and 3, as determined by live/dead analysis (>80% cell viability, Figures 2(a)–2(h)). Taken together, these findings demonstrate that we can achieve a three-dimensional microenvironment capable of supporting multiple cell types for long periods in culture without the loss of cell viability.
FIG. 2.
Live/dead stain of cell culture within the NVU. (a) Human brain-derived microvascular endothelial cells in the perfusion channel of the NVU. Live = green; dead = red (DIV 14; DIV = days in vivo). (b) Endothelial cells coating the vascular chamber (DIV 14). (c) Pericytes and astrocytes on the membrane in the brain chamber. (d) Neurons in the brain chamber suspended in collagen gel (DIV 14). (e) Endothelial cells in the perfusion channels of the NVU (DIV 21). (f) Live/dead and (g) live-only staining of endothelial cells coating the vascular chamber side of the membrane (DIV 21). Over 80% viability is maintained after 3 weeks in culture (DIV 21, live only). (h) Pericytes and astrocytes on the brain side of the same membrane region as in (g) and (h), with the collagen shrinkage shown.
E. FITC-dextran diffusion across the BBB
Solutions of 10 KDa or 70 KDa fluorescein isothiocyanate (FITC)-dextran (Sigma-Aldrich, St. Louis, MO, USA) were prepared at 1 μm/ml (100 nM for 10 KDa and 700 nM for 70 KDa) in cell culture media. Then the vascular compartment of the NVU was perfused with either 70 or 10 KDa solution for 23 h. At the 23 h mark, the flows through both the vascular and brain compartments were stopped for 1 h, allowing the dextran to diffuse across the BBB and accumulate in the brain compartment. After a 1 h pause, perfusion of both chambers was restarted and individual effluents were collected for fluorescent intensity analysis using a plate reader (TECAN M1000). By measuring FITC-dextran diffusion across the membrane, we are able to evaluate the effectiveness of our BBB.46,47
F. Ascorbate transport across the BBB
Twenty-four hours before testing, the NVUs were switched into media containing no vitamin C. At time 0, 100 μM ascorbate was added to the vascular media, and samples were collected for both FITC-dextran analysis and ascorbate transport every 15 min for 1 h. Perfusate samples for ascorbate (20 μl) were treated with 20 μl of 25% metaphosphoric acid, mixed by vortexing, and treated with 80 μl of 0.1 M Na2HPO4 containing 0.05 mM EDTA, pH 8.0. After vortexing briefly, the samples were centrifuged at 4 °C for 1 min at 13 000× g. The supernatant was taken for assay of ascorbate by high performance liquid chromatography as previously described.48
G. Cold shock and glutamate exposure
We used cold shock and exposure to glutamate to evaluate the quality of the BBB established within the NVU and its physiological responses to chemical and environmental perturbations. For glutamate exposure, NVUs were first maintained under normal culture conditions for 14 days to develop a mature BBB, then the media perfusing the vascular compartment was switched to one containing 1 mM of glutamate (Sigma-Aldrich, St. Louis, MO, USA) and flowed through the reactor for 1 h. At the end of a 1 h exposure to glutamate, the state of the BBB was evaluated using FITC-dextran as described above. For cold shock exposure, the NVUs were cultured normally for 18 days and then were placed for 12 h at 33 °C. The disruption of the BBB was evaluated as changes in transendothelial electrical resistance (TEER) measured between the vascular compartment and the “H” layer.
H. TEER measurements in the NVU
TEER measurements were performed using our custom-built impedance analyzer based on an AD5933 chip (Analog Devices, Nashua, NH, USA) and utilizing a four-probe approach.49 Electrical connections to the NVU chambers were created by incorporating 5 mm long sections of 23 ga stainless steel tubing into the media-supply Tygon tubing 2.5 cm away from the NVU inlets (Supplementary Figure 2).69 While it might still suffer from the cable properties outlined in Ref. 50 these measures did change as the biology of the cells changed, indicating its biological relevance. The current source probes were connected between the inlet of the “H” layer and the vascular chamber outlet, ensuring that the excitation current flowed across the brain compartment and through the endothelial layer. The sensing voltage probes were connected between the vascular chamber inlet and the “H” layer outlet. Unlike commercially available TEER instruments, such as the WPI Evom2, where impedance measurements are performed at a single frequency of 12.5 Hz, we have the ability to monitor impedance as a function of probe frequency between 3 and 100 KHz, allowing us to determine the range of frequencies with the highest sensitivity to cell-to-cell junction formation. Impedance measurements were taken once a week with the full range frequency sweep.51 Changes of impedance at 15 kHz showed the largest change as a function of BBB maturation.
I. Imaging and data analysis
Fluorescent images of the cells stained within the NVU were collected using a Zeiss Axiovert 200 automated microscope equipped with a CoolSnap CCD camera. Collected images were analyzed with ImageJ.
Tight junction evaluation was conducted as detailed in Ref. 12 using ZO-1 (Invitrogen) directly conjugated to Alexa 488. In brief, greyscale measurements of the border between cells were taken for 30 different cell-to-cell junctions and an average intensity was derived for each culture condition.
Actin orientation assessment was conducted using phalloidin staining of actin fibers.52 Actin filaments were defined as a line longer than half the diameter of the cell in which they were being measured. They were scored as being parallel with flow if their orientation had fallen within 60° of the direction of flow (±30°), or perpendicular if they were within 60° (±30°) normal to the flow. The remaining actin fibers were classified as having orientation independent of the flow. Bundles were assigned a straight line vector for analysis.53,54 For a given cell, 10 filaments were chosen at random for each cell counted.
For both tight junction and actin orientation, four fields of view for each bioreactor were used for statistical analysis. Statistical analysis was performed using unpaired Student's t test for p values N > 8. For sample sizes of less than 8, the Mann-Whitney U test was performed to generate p values.55
III. RESULTS AND DISCUSSION
A. Development of a multi-chambered perfusable microfluidic device
Current in vitro BBB models, such as the popular transwell-based cell culture method,56 are predominantly static, i.e., they lack flow and therefore also the shear forces needed for mature barrier formation.56 These models also suffer from high media-to-cell volume ratios that dilute out important co-culture signals. By restricting flow to the microfluidic chamber, BBB devices can provide the requisite flow velocities and shear forces to polarize endothelial cells.29 In developing our NVU device, we attempted to incorporate the best elements of the transwell culture, e.g., ease of cell culture of multiple cell types, while also reducing the media volume-to-cell ratio and introducing the shear force needed for the formation of mature tight junctions. The reduction in media volume is critical to any studies that will couple different organ chips to avoid dilution of metabolites and paracrine, autocrine, or endocrine signals, both to ensure that they are present at physiological levels and to aid in their detection.57 An additional concern for cell biologists (and a challenge in building a BBB) is the ability to easily and in a clean and sterile manner invert their culture devices. As our NVU chip is self-contained, closed, and can operate in any orientation, we designed a flippable backpack for the feed and waste media vials (Supplementary Figures 1(a) and 1(b)).69 Taken together, the current NVU has four crucial design features that help it to model the BBB: (1) adjustable flow on both sides of the barrier, (2) low media-to-cell volume, (3) scaffolding to orient and support multiple cell types, and (4) easy adjustment of orientation of the device.
B. Characterizing the blood brain barrier with the NVU
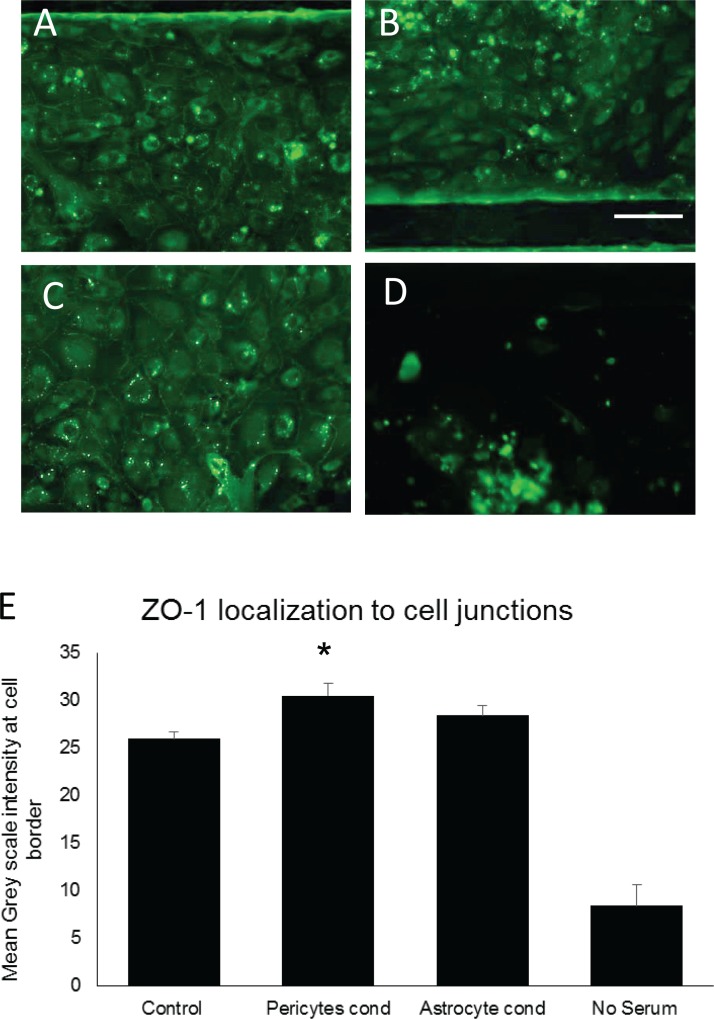
Once we had achieved stable cell viability within our NVU device over a 3-week time period, the next challenge was to evaluate how effectively it generated the mature, tight junctions critical to the BBB's selective permeability. Previous culture methods for generating in vitro BBBs often lacked either the shear forces necessary to maintain a polarized and differentiated state within the endothelial cells,3,10,19 or the appropriate paracrine factors from astrocytes and pericytes for stable and selective function of tight junctions. Experiments using gravity-fed microfluidic chambers show that with the addition of flow, and exposure to pericyte- or astrocyte-conditioned media, we can establish high levels of tight junction staining that are stable over the course of two weeks after the tight junctions have formed (Figures 3(a)–3(d)). When ZO-1 staining was quantified using mean greyscale intensity at the cell border of thirty different cell-to-cell interactions, we found that pericyte-conditioned media significantly increased ZO-1 intensity (p = 0.002, N = 30) (Figure 3(e)). Astrocyte-conditioned media trended toward an increase but was not statically significant due to variability (Supplementary Figure 3).69 It should be noted, however, that flow alone was sufficient for tight junction staining to be visible. Further, all cultures required the presence of at least 5% serum to maintain cell viability. These experiments suggest that our NVU device, which combines both the shear force of flow and access to paracrine factors from pericytes and astrocytes, is ideal for generating and maintaining tight junctions58–60 in brain-derived endothelial cells.
FIG. 3.
ZO-1 staining of HBMVECs in microfluidic channels under flow and under different culture conditions. (a) Control culture using EBM-2 media, showing higher intensity ZO-1 staining around the cell border that indicates tight junction staining. (b) Astrocyte-conditioned EBM-2 media, showing a trend towards increased tight junctions. (c) Pericyte-conditioned EBM-2 media, showing significant increases in staining around the cell border (p = 0.002, N = 10). (d) EBM-2 media with no serum, showing that the absence of serum reduces cell survival. (e) Mean intensity of ZO-1 staining at cell borders. 10 cells for each condition were evaluated and could not share a border with a cell that had already been counted.
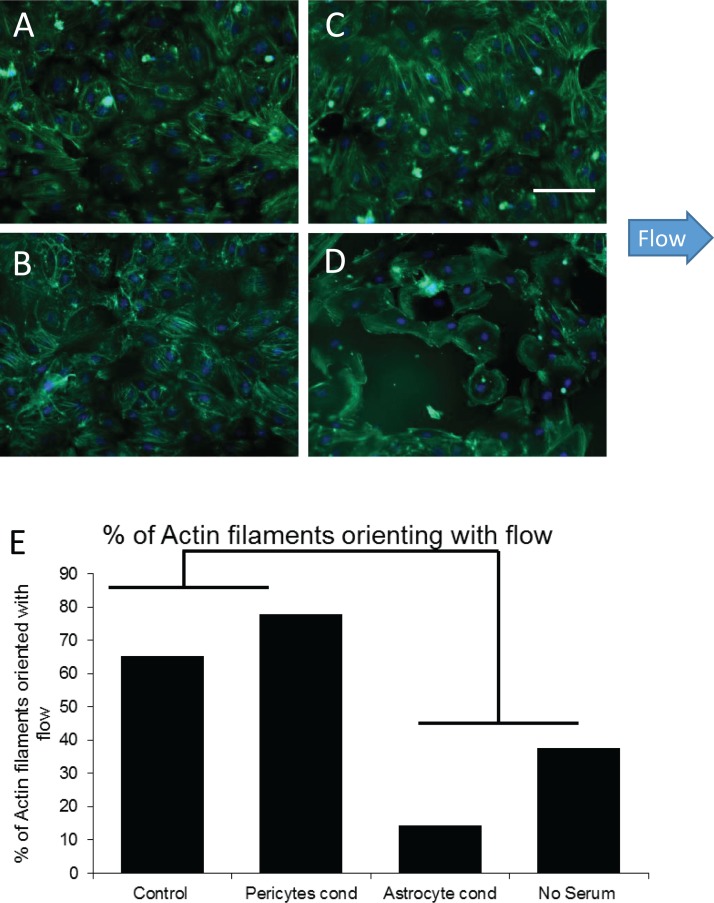
In addition to markers for tight junction formation, another vital component for BBB formation is polarization of the endothelial cells in the direction of flow.4,19,61 To evaluate cell polarity, actin filaments in endothelial cells under our various culture conditions were stained with phalloidin. We then assessed the percent of actin filaments whose orientation was parallel to the direction of flow. Filaments were considered to have an orientation parallel to flow if they were within 32.5° (60° arc) of the flow vector (Figures 4(a)–4(d)). Control cultures under flow show 65% of actin filaments orienting in the direction of flow. This number increased to 78% with the addition of pericyte-conditioned media. Interestingly, astrocyte-conditioned media encouraged polarization of actin filaments perpendicular to the direction of flow. Cells maintained in no-serum media showed little, if any, actin bundle formation at all (Figure 4(e)). These findings demonstrate the need for flow, serum, and pericyte-generated factors for the formation and orientation of actin filaments in the direction of flow in the development of a mature BBB.
FIG. 4.
Actin staining of HBMVECs under flow and under different culture conditions. (a) Control culture with EBM-2 media, showing actin polarization with 65% of the actin filaments oriented in the direction of flow. (b) Astrocyte-conditioned EBM-2 media, demonstrating actin polarization filaments oriented perpendicular to the direction of flow. (c) Pericyte-conditioned EBM-2 media, showing actin polarization with 78% of the actin filaments oriented in the direction of flow. (d) EBM-2 media with no serum, showing reduced cell survival and neither actin polarization nor actin bundle formation. (e) Percent of actin filaments that orient in the direction of flow. For each condition, 30 cells were evaluated for actin bundle orientation.
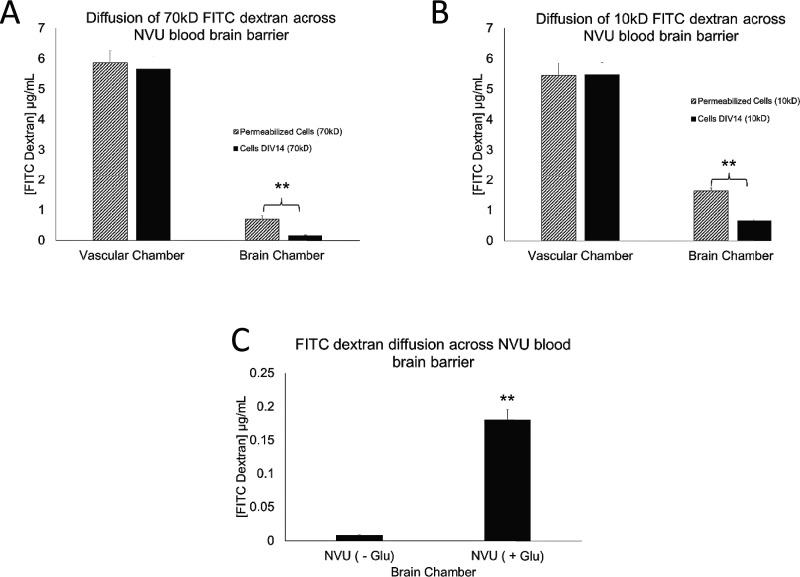
Having established that the necessary components for BBB formation are indeed present and maintained in our NVU device, we then tested the function of this in vitro BBB. One of the classical ways to evaluate the BBB function is the diffusion of sucrose molecules across the barrier. The smaller the molecule, the more easily it should diffuse through the gaps between cells; however, a functional BBB significantly reduces the diffusion of even very small molecules.2,7,61–63 To evaluate the effectiveness of our BBB, we tested the permeability of two different molecular weight fluorescent dextrans. The fluorescent dextran was diluted in the vascular side media feed, and effluent from both the vascular and the brain chambers was then evaluated for the presence of fluorescence after 1 h diffusion and collection. FITC-dextran diffusion tests showed that our NVU BBB significantly blocked both 70 kD and 10 kD FITC-dextran from diffusing from the vascular chamber into the brain chamber (p = 0.01). We also showed that disruption of the BBB by exposing it to 1 mM glutamate, which is known to considerably disrupt tight junctions, does indeed significantly increase diffusion of FITC-dextran across the BBB (Figures 5(a)–5(c)). While both are significant effects, the first compares the cellular barrier only to the mechanical properties of the device, while the second is a biologically relevant disruption (but does not destroy the BBB such that the graphs in Figure 2 have differing scales). In combination, these experiments demonstrate the generation of a functional cellular barrier within our NVU.
FIG. 5.
FITC-dextran diffusion across the complete NVU BBB with all four cell types. (a) The concentration of 70 kD FITC-dextran and (b) 10 kD dextran in the vascular chamber is significantly less than across the membrane in the brain chamber (both p = 0.01, N = 6), as compared to the device whose cells were permeabilized with Triton. (c) Glutamate exposure for 1 h significantly increases the concentration of 10 kD FITC-dextran across the membrane (p = 0.01, N = 6).
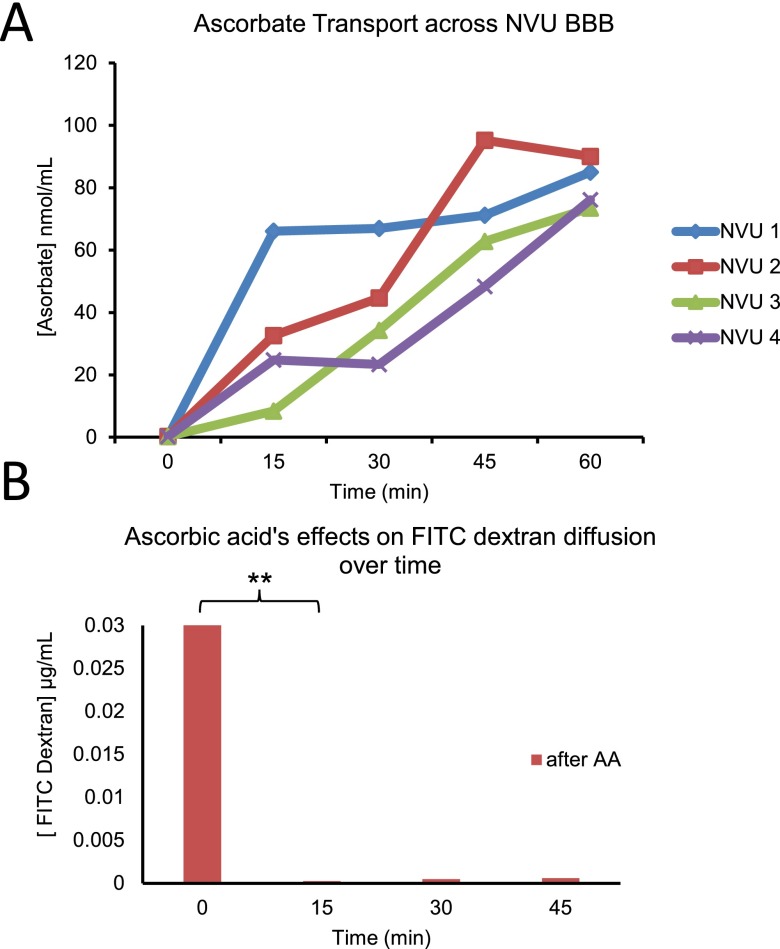
Next, we next undertook to show that active transport—another critical component of the BBB often overlooked in cellular BBB models46—could be shown in our system. To this end, we used ascorbate, which is known both to tighten the BBB64 and be actively transported across the barrier,65,66 as an indication of active transport. In looking at ascorbate concentration over time, we saw a significant increase in all four NVUs tested (p = 0.01) (Figure 6(a)). In contrast, FITC-dextran diffusion across the barrier was significantly reduced (p = 0.0012) by the addition of ascorbate, and then remained steady over time (Figure 6(b)). Since diffusion cannot account for the increase in ascorbate on the brain side, this is an indication of active transport.
FIG. 6.
Ascorbate transport across NVU. (a) The ascorbate concentration across the BBB for 4 separate NVU devices, showing increases over time (p = 0.01). (b) 10 kD FITC-dextran diffusion across the BBB, showing no change (15–60 min). After the addition of ascorbate to the vascular side at time 0, time 0–15 min shows a significant reduction in diffusion (p = 0.0012, N = 4).
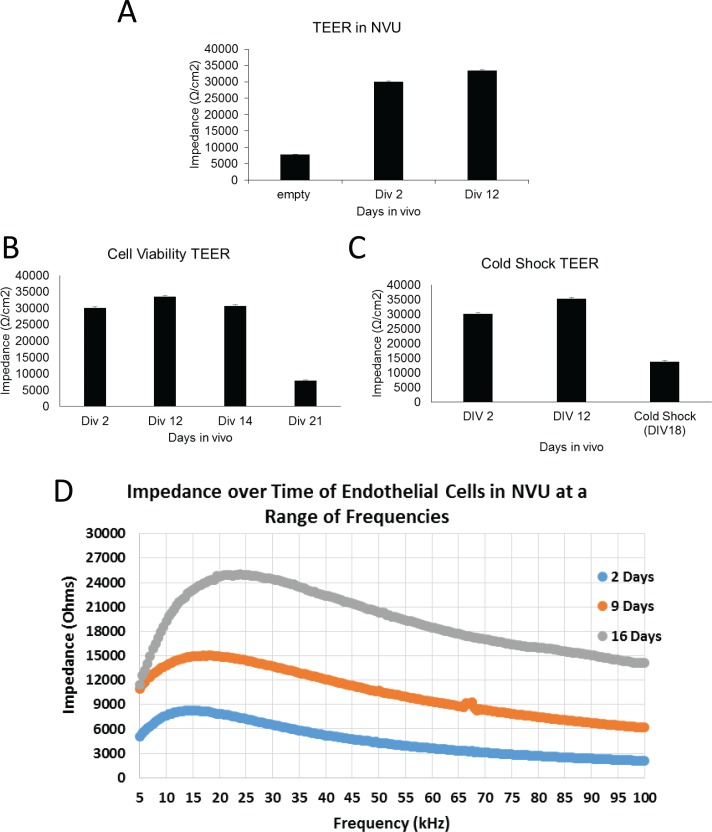
In addition to testing fluorescent molecule diffusion to evaluate the effectiveness of our in vitro BBB, we have developed a custom TEER measuring device specifically for the NVU to evaluate resistance across the endothelial cell membrane (Supplementary Figure 2)69 at a range of frequencies over the time course in which NVUs were in cell culture. As was seen with staining for tight junction markers, there is a significant increase in TEER around Day 12 of approximately 30% (p = 0.05) (Figure 7(a)). The percent increase is typical of reports for TEER and tight junctions,51 although our values are higher, as the NVU itself has a high natural impedance. This correlates well with the histology verifying tight junction formation (Figure 3). In addition to detecting tight junction formation, TEER was also useful for evaluating cell viability. In devices in which perfusion was occluded by 50% at Day 14 in culture, TEER showed a large drop at Day 21, which was later shown to correlate with cell survival (Figure 7(b)). When tight junctions (but not cell survival) were impaired via cold shock of 33 °C for 12 h, TEER decreased significantly (p = 0.001) (Figure 7(c)). Furthermore, these measures of impedance show that the NVU device acts as a capacitor, with its peak impedance between 15–20 kHz, and that we can use the shape of this capacitance to monitor the health of our devices over time (Figure 7(d)). These findings demonstrate not only the reliability of our custom TEER device in our NVU but also show that our BBB is functioning as expected with regard to its endothelial cell resistance.
FIG. 7.
TransEndothelial Electrical Resistance (TEER) measurement of barrier integrity in the NVU as an indication of tight junction formation. (a) TEER shows a significant increase (30%, p = 0.05, N = 6) compared to before tight junction formation at Day 2 and after Day 12. (b) TEER provides cell viability information within the device, as demonstrated by the drop in both TEER (60%) and cell viability (p = 0.001, N = 6) when perfusion was occluded by 50% after Day 14. (c) Cold shock of 33 °C for 12 h reduces tight junctions significantly, as evidenced by TEER (p = 0.001, N = 6). (d) The output of a prototype swept-frequency TEER instrument, showing significant increases in TEER for the NVU device over time for a range of frequencies.
IV. LIMITATIONS
One of the limitations of our approach was the use of PDMS to create the NVU, given the propensity of hydrophobic molecules to adsorb to the PDMS.67,68 We are proceeding to develop an NVU that is fabricated out of harder thermoplastics to avoid this problem.
The measurement of TEER in a transwell device benefits from the large volume of culture media on both sides of the membrane, in that the conductivity of the media ensures that each side of the membrane is an electrical isopotential, and hence, a four-probe TEER measurement records the potential drop across the membrane. With microfluidic devices, the access impedance of the media in the narrow channels can be significant, and the measurement of TEER then corresponds to the measurement of the properties of a distributed electrical cable with a short length constant. The TEER measurement is then most sensitive to the membrane properties closest to the current injection electrodes.50 While our TEER measurement demonstrates the expected decrease in barrier permeability as the cells grow to confluence and form tight junctions, the exact extent of the region of the barrier being quantified is as yet unknown. For this reason, we believe that the FITC-dextran measurements may be more representative of the barrier function over the entire membrane in this type of NVU.
V. CONCLUSION
Our NeuroVascular Unit is a novel organ-on-a-chip bioreactor that combines microfluidics and three-dimensional cell culture to successfully recreate structure and behavior of the BBB and is a substantial improvement for modeling the human BBB. This unique microfluidic device provides an environment for all cell types involved in BBB formation, the shear forces necessary for barrier formation, and the paracrine factors needed for long-term stable differentiation of the BBB. This device also provides the structures necessary for growing human neurons, so that the effects of drugs on neuronal function can be evaluated in the context of a BBB to account for drug permeability and effects on the BBB itself. The dual perfusion nature of this device allows for manipulation of either side of the BBB, as well as differential delivery of drugs and nutrients to either the vascular or the brain chamber. Taken together, these innovations provide a novel platform for modeling of BBB function and testing of drug toxicity and permeability with regard to the CNS.
ACKNOWLEDGMENTS
Funding for this work was provided in part by NIH/NIEHS R1 ES016931 (A.B.B.), NIH DK50435 (J.M.M.), and, through the NIH Common Fund, NCATS Grant No. 5UH3TR000491-04 (J.P.W.). We thank Dr. Hak-Joon Sung for his willingness to explore electrospun barrier membranes and the use of his lab's microplate fluorimeter, Daniel A. Balikov for the technical assistance with the fluorimeter, and Allison Price for her editorial assistance. We appreciate detailed discussions with Dr. Damir Janigro, particularly with regard to the cable properties of small channel BBB models.
References
- 1. Al Ahmad A., Taboada C. B., Gassmann M., and Ogunshola O. O., J. Cereb. Blood Flow Metab. 31(2), 693 (2011). 10.1038/jcbfm.2010.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vandenhaute E., Dehouck L., Boucau M. C., Sevin E., Uzbekov R., Tardivel M., Gosselet F., Fenart L., Cecchelli R., and Dehouck M. P., Curr. Neurovasc. Res. 8(4), 258 (2011). 10.2174/156720211798121016 [DOI] [PubMed] [Google Scholar]
- 3. Cucullo L., Hossain M., Puvenna V., Marchi N., and Janigro D., BMC Neurosci. 12(1), 40 (2011). 10.1186/1471-2202-12-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Germann B., Neuhaus W., Hofer-Warbinek R., and Noe C. R., Pharmazie 63(4), 303 (2008). [PubMed] [Google Scholar]
- 5. Molino Y., Jabes F., Lacassagne E., Gaudin N., and Khrestchatisky M., J. Visualized Exp. 2014, e51278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shawahna R., Decleves X., and Scherrmann J. M., Curr. Drug Metab. 14(1), 120 (2013). 10.2174/138920013804545232 [DOI] [PubMed] [Google Scholar]
- 7. Vandenhaute E., Sevin E., Hallier-Vanuxeem D., Dehouck M. P., and Cecchelli R., Drug Discovery Today 17(7–8), 285 (2012). 10.1016/j.drudis.2011.10.006 [DOI] [PubMed] [Google Scholar]
- 8. Filipovic N., Ghimire K., Saveljic I., Milosevic Z., and Ruegg C., “ Computational modeling of shear forces and experimental validation of endothelial cell responses in an orbital well shaker system,” Comput. Methods Biomech. Biomed. Eng. (published online 2015). 10.1080/10255842.2015.1051973 [DOI] [PubMed]
- 9. Naik P. and Cucullo L., J. Pharm. Sci. 101(4), 1337 (2012). 10.1002/jps.23022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown R. C., Morris A. P., and O'Neil R. G., Brain Res. 1130(1), 17 (2007). 10.1016/j.brainres.2006.10.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cucullo L., Aumayr B., Rapp E., and Janigro D., Curr. Opin. Drug Discovery Dev. 8(1), 89 (2005). [PubMed] [Google Scholar]
- 12. Cao X. F., Lin H. X., Muskhelishvili L., Latendresse J., Richter P., and Heflich R. H., Respir. Res. 16, 30 (2015). 10.1186/s12931-015-0191-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vernon H., Clark K., and Bressler J. P., Methods Mol. Biol. 758, 153 (2011). 10.1007/978-1-61779-170-3_10 [DOI] [PubMed] [Google Scholar]
- 14. Bowman P. D., Betz A. L., Ar D., Wolinsky J. S., Penney J. B., Shivers R. R., and Goldstein G. W., In Vitro 17(4), 353 (1981). 10.1007/BF02618147 [DOI] [PubMed] [Google Scholar]
- 15. Gaillard P. J., Voorwinden L. H., Nielsen J. L., Ivanov A., Atsumi R., Engman H., Ringbom C., de Boer A. G., and Breimer D. D., Eur. J. Pharm. Sci. 12(3), 215 (2001). 10.1016/S0928-0987(00)00123-8 [DOI] [PubMed] [Google Scholar]
- 16. Sun Z. Z., Worden M., Wroczynskyj Y., Yathindranath V., van Lierop J., Hegmann T., and Miller D. W., Int. J. Nanomed. 9, 3013 (2014). 10.2147/IJN.S62260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeshita Y., Obermeier B., Cotleur A., Sano Y., Kanda T., and Ransohoff R. M., J. Neurosci. Methods 232, 165 (2014). 10.1016/j.jneumeth.2014.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Z. Q., McGoron A. J., Crumpler E. T., and Li C. Z., Appl. Biochem. Biotechnol. 163(2), 278 (2011). 10.1007/s12010-010-9037-6 [DOI] [PubMed] [Google Scholar]
- 19. Cucullo L., Hossain M., Tierney W., and Janigro D., BMC Neurosci. 14, 18 (2013). 10.1186/1471-2202-14-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakagawa S., Deli M. A., Kawaguchi H., Shimizudani T., Shimono T., Kittel A., Tanaka K., and Niwa M., Neurochem. Int. 54(3–4), 253 (2009). 10.1016/j.neuint.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 21. Easton A. S. and Abbott N. J., Brain Res. 953(1–2), 157 (2002). 10.1016/S0006-8993(02)03281-X [DOI] [PubMed] [Google Scholar]
- 22. Hurst R. D. and Fritz I. B., J. Cell Physiol. 167(1), 89 (1996). [DOI] [PubMed] [Google Scholar]
- 23. Kusch-Poddar M., Drewe J., Fux I., and Gutmann H., Brain Res. 1064(1–2), 21 (2005). 10.1016/j.brainres.2005.10.014 [DOI] [PubMed] [Google Scholar]
- 24. Moraes C., Mehta G., Lesher-Perez S. C., and Takayama S., Annu. Biomed. Eng. 40(6), 1211 (2012). 10.1007/s10439-011-0455-6 [DOI] [PubMed] [Google Scholar]
- 25. van der Meer A. D. and van den Berg A., Integr. Biol. 4(5), 461 (2012). 10.1039/c2ib00176d [DOI] [PubMed] [Google Scholar]
- 26. Wikswo J. P., Exp. Biol. Med. 239, 1061 (2014). 10.1177/1535370214542068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wikswo J. P. and Porter A. P., Exp. Biol. Med. 240, 3 (2015). 10.1177/1535370214564534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Griep L. M., Wolbers F., de Wagenaar B., ter Braak P. M., Weksler B. B., Romero I. A., Couraud P. O., Vermes I., van der Meer A. D., and van den Berg A., Biomed. Microdevices 15(1), 145 (2013). 10.1007/s10544-012-9699-7 [DOI] [PubMed] [Google Scholar]
- 29. Prabhakarpandian B., Shen M. C., Nichols J. B., Mills I. R., Sidoryk-Wegrzynowicz M., Aschner M., and Pant K., Lab Chip 13(6), 1093 (2013). 10.1039/c2lc41208j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Booth R. and Kim H., Annu. Biomed. Eng. 42(12), 2379 (2014). 10.1007/s10439-014-1086-5 [DOI] [PubMed] [Google Scholar]
- 31. Kim J. A., Kim H. N., Im S. K., Chung S., Kang J. Y., and Choi N., Biomicrofluidics 9(2), 024115 (2015). 10.1063/1.4917508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wikswo J., Curtis E. L., Eagleton Z. E., Evans B. C., Kole A., Hofmeister L. H., and Matloff W. J., Lab Chip 13(18), 3496 (2013). 10.1039/c3lc50243k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duffy D. C., McDonald J. C., Schueller O. J. A., and Whitesides G. M., Anal. Chem. 70(23), 4974 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- 34. Aran K., Sasso L. A., Kamdar N., and Zahn J. D., Lab Chip 10(5), 548 (2010). 10.1039/b924816a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y., Markov D., Wikswo J., and McCawley L., Biomed. Microdevices 13(5), 837 (2011). 10.1007/s10544-011-9554-2 [DOI] [PubMed] [Google Scholar]
- 36. Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., and Studer L., Nat. Biotechnol. 27(3), 275 (2009). 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi Y. C., Kirwan P., and Livesey F. J., Nat. Protoc. 7(10), 1836 (2012). 10.1038/nprot.2012.116 [DOI] [PubMed] [Google Scholar]
- 38. Shi Y. C., Kirwan P., Smith J., Robinson H. P. C., and Livesey F. J., Nat. Neurosci. 15(3), 477 (2012). 10.1038/nn.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aboud A. A., Tidball A. M., Kumar K. K., Neely M. D., Han B. Y., Ess K. C., Hong C. C., Erikson K. M., Hedera P., and Bowman A. B., Neurobiol. Dis. 73, 204 (2015). 10.1016/j.nbd.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tidball A. M., Bryan M. R., Uhouse M. A., Kumar K. K., Aboud A. A., Feist J. E., Ess K. C., Neely M. D., Aschner M., and Bowman A. B., Hum. Mol. Genet. 24(7), 1929 (2015). 10.1093/hmg/ddu609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J. B., Nishikawa S., Nishikawa S., Muguruma K., and Sasai Y., Nat. Biotechnol. 25(6), 681 (2007). 10.1038/nbt1310 [DOI] [PubMed] [Google Scholar]
- 42. Boissart C., Poulet A., Georges P., Darville H., Julita E., Delorme R., Bourgeron T., Peschanski M., and Benchoua A., Transl. Psychiatry 3, e294 (2013). 10.1038/tp.2013.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lamas N. J., Johnson-Kerner B., Roybon L., Kim Y. A., Garcia-Diaz A., Wichterle H., and Henderson C. E., PLoS One 9(10), e110324 (2014). 10.1371/journal.pone.0110324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roloff F., Scheiblich H., Dewitz C., Dempewolf S., Stern M., and Bicker G., PLoS One 10(2), e0118536 (2015). 10.1371/journal.pone.0118536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tang X., Zhou L., Wagner A. M., Marchetto M. C., Muotri A. R., Gage F. H., and Chen G., Stem Cell Res. 11(2), 743 (2013). 10.1016/j.scr.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bickel U., NeuroRX 2(1), 15 (2005). 10.1602/neurorx.2.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hoffmann A., Bredno J., Wendland M., Derugin N., Ohara P., and Wintermark M., Transl. Stroke Res. 2(1), 106 (2011). 10.1007/s12975-010-0049-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. May J. M., Qu Z. C., and Mendiratta S., Arch. Biochem. Biophys. 349(2), 281 (1998). 10.1006/abbi.1997.0473 [DOI] [PubMed] [Google Scholar]
- 49. Sarro E., Lecina M., Fontova A., Sola C., Godia F., Cairo J. J., and Bragos R., Biosens. Bioelectron. 31(1), 257 (2012). 10.1016/j.bios.2011.10.028 [DOI] [PubMed] [Google Scholar]
- 50. Odijk M., van der Meer A. D., Levner D., Kim H. J., van der Helm M. W., Segerink L. I., Frimat J. P., Hamilton G. A., Ingber D. E., and van den Berg A., Lab Chip 15(3), 745 (2015). 10.1039/C4LC01219D [DOI] [PubMed] [Google Scholar]
- 51. Benson K., Cramer S., and Galla H. J., Fluids Barriers CNS 10(1), 5 (2013). 10.1186/2045-8118-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Newell-Litwa K. A., Badoual M., Asmussen H., Patel H., Whitmore L., and Horwitz A. R., J. Cell Biol. 210(2), 225 (2015). 10.1083/jcb.201504046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McCue S., Dajnowiec D., Xu F., Zhang M., Jackson M. R., and Langille B. L., Circ. Res. 98(7), 939 (2006). 10.1161/01.RES.0000216595.15868.55 [DOI] [PubMed] [Google Scholar]
- 54. Wojciak-Stothard B. and Ridley A. J., J. Cell Biol. 161(2), 429 (2003). 10.1083/jcb.200210135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zar J. H., Biostatistical Analysis, 2nd ed. ( Prentice-Hall, Englewood Cliffs, NJ, 1984). [Google Scholar]
- 56. Hatherell K., Couraud P. O., Romero I. A., Weksler B., and Pilkington G. J., J. Neurosci. Methods 199(2), 223 (2011). 10.1016/j.jneumeth.2011.05.012 [DOI] [PubMed] [Google Scholar]
- 57. Wikswo J. P., Block F. E. III, Cliffel D. E., Goodwin C. R., Marasco C. C., Markov D. A., McLean D. L., McLean J. A., McKenzie J. R., Reiserer R. S., Samson P. C., Schaffer D. K., Seale K. T., and Sherrod S. D., IEEE Trans. Biomed. Eng. 60(3), 682 (2013). 10.1109/TBME.2013.2244891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamagata K., Tagami M., Nara Y., Mitani M., Kubota A., Fujino H., Numano F., Kato T., and Yamori Y., Clin. Exp. Pharmacol. Physiol. 24(9–10), 710 (1997). 10.1111/j.1440-1681.1997.tb02117.x [DOI] [PubMed] [Google Scholar]
- 59. Pan W., Stone K. P., Hsuchou H., Manda V. K., Zhang Y., and Kastin A. J., Curr. Pharm. Des. 17(33), 3729 (2011). 10.2174/138161211798220918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shimizu F., Sano Y., Saito K., Abe M. A., Maeda T., Haruki H., and Kanda T., Neurochem. Res. 37(2), 401 (2012). 10.1007/s11064-011-0626-8 [DOI] [PubMed] [Google Scholar]
- 61. Gonzalez-Burgos E., Carretero M. E., and Gomez-Serranillos M. P., Planta Med. 79(16), 1545 (2013). 10.1055/s-0033-1350797 [DOI] [PubMed] [Google Scholar]
- 62. Stanimirovic D. B., Bani-Yaghoub M., Perkins M., and Haqqani A. S., Expert Opin. Drug Discovery 10(2), 141 (2015). 10.1517/17460441.2015.974545 [DOI] [PubMed] [Google Scholar]
- 63. Walsh T. G., Murphy R. P., Fitzpatrick P., Rochfort K. D., Guinan A. F., Murphy A., and Cummins P. M., J. Cell Physiol. 226(11), 3053 (2011). 10.1002/jcp.22655 [DOI] [PubMed] [Google Scholar]
- 64. May J. M. and Qu Z. C., Free Radical Res. 44(11), 1359 (2010). 10.3109/10715762.2010.508496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Agus D. B., Gambhir S. S., Pardridge W. M., Spielholz C., Baselga J., Vera J. C., and Golde D. W., J. Clin. Invest. 100(11), 2842 (1997). 10.1172/JCI119832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. May J. M., Qu Z. C., and Qiao H., Am. J. Physiol. Cell 297(1), C169–C178 (2009). 10.1152/ajpcell.00674.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Toepke M. W. and Beebe D. J., Lab Chip 6(12), 1484 (2006). 10.1039/b612140c [DOI] [PubMed] [Google Scholar]
- 68. Wang J. D., Douville N. J., Takayama S., and ElSayed M., Annu. Biomed. Eng. 40(9), 1862 (2012). 10.1007/s10439-012-0562-z [DOI] [PubMed] [Google Scholar]
- 69.See supplementary material at http://dx.doi.org/10.1063/1.4934713E-BIOMGB-9-024505 for additional information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4934713E-BIOMGB-9-024505 for additional information.