Abstract
The main goal of our study was to evaluate the effect of the individual administration of five lyophilized lactic acid bacteria strains (Lactobacillus fermentum 428ST, Lactobacillus rhamnosus E4.2, Lactobacillus plantarum FCA3, Lactobacillus sp. 34.1, Weissella paramesenteroides FT1a) against the in vitro simulated microbiota of the human colon using the GIS1 system. The influence on the metabolic activity was also assessed by quantitative determination of proteins and polysaccharides at each segment of human colon. The obtained results indicated that the lactic acid bacteria L. rhamnosus E4.2 and W. paramesenteroides FTa1 had better efficiency in synthesising exopolysaccharides and also a better probiotic potential and therefore could be recommended for use in probiotics products or food industry.
Keywords: Lactic acid bacteria, Probiotics, Prebiotics, Fermentation biotechnology
Introduction
The human gastrointestinal tract (GIT) represents the densest, complex and diverse community of microorganisms. The complex found inside the human gut is an ecosystem where the microbiota, the nutrients and the host cells interact intensively. Many researchers have detailed the positive interactions between the commensal microbiota and the human body, considering the microbiota as a powerful partner for a good health [1].
The very complex microbiota is known as an active element of the colon physiology having numerous functions, the most important being: trophic functions, metabolic functions and “barrier effect”. The gut microbiota is critical to human health, a microbial imbalance called dysbiosis leading to the apparition of diseases; however, it is not always easy to determine whether the changes in the gut microbiota are a cause or a consequence of a disease. The relationship between the microbiota and the health condition is of great importance for the positive modulation of the gut flora by administering live bacteria (probiotics) or non-digestible compounds (prebiotics) to prevent and sometimes to cure several diseases associated to dysbiosis [2].
The Food and Agriculture Organization of the United Nations (FAO) defined probiotics as “live microorganisms, that, when administered in adequate amounts, confer a health benefit on the host” [3, 4]. Probiotics exert several beneficial effects on human health by stimulating the immune system, producing antimicrobial substances, improving the barrier function of the intestinal mucosa and competing with pathogenic bacteria to receptors on epithelial cells [5].
Probiotics formed from microorganisms belonging to the genera Lactobacillus (L.) and Bifidobacterium (B.) are a group of functional products that are intended to be consumed by larger groups of population. Viability, the main characteristic of probiotics, is determined by their ability to survive in a large number after the transit through the stomach (acid pH) and the small intestine (enzymes and bile acids), respectively. Moreover, the capability to release substances having antimicrobial effect and to form biofilms is extremely important as they can increase the persistence in the colon segments [6].
Lactic acid bacteria (LAB) producing exopolysaccharides have gained considerable attention in the dairy industry due to their thickening properties that contribute to the improvement of the texture and appearance of the dairy products obtained by fermentation (yogurt, cheese). Moreover, these exopolysaccharides have beneficial effects on human health by lowering cholesterol assimilation, by their immunostimulatory effects and prebiotic effects on the intestinal microflora [7, 8]. Ruas-Madiedo and De Los Reyes-Gavilán [9] have correctly noted that the ability to produce exopolysaccharides is widespread among LAB. Therefore there is need to better understand their physiological role and to search new strains able to produce exopolysaccharides.
The viability of these strains during the industrial manufacturing process and also during the shelf life of food must be carefully examined, at the end the total number of viable bacteria should be 105 CFU/g. Functional food has to meet a series of sensory characteristics when compared to a lyophilized product. Accordingly, it is expected that a probiotic product composed by one or more bacterial strains meet all these requirements, naturally increasing the degree of consumer acceptability. Therefore viability is also influenced by the interactions between strains that in some cases may cause additional losses [10, 11].
The aim of the present work was to study the effect of administering several lyophilized lactic acid bacteria strains on the GIS1 system used for in vitro simulation of the colon. The analysed strains were previously selected for their probiotic potential by in vitro assays involving antimicrobial activity, adherence to eukaryotic cell lines, immunomodulatory activity and resistance to different pH values and bile salts concentrations (unpublished data). The comparative study aimed to determine the strain(s) that have a significant positive influence on the number of beneficial strains (especially the number of LAB) correlated to a decrease in potentially pathogenic strains. The influence on the metabolic activity was also monitored at each segment of the human colon. The studies were carried out in parallel with the administration of the two lyophilized control strains—Kluyveromyces marxianus and L. casei 431.
Materials and Methods
Microbial Strains
Five lactic acid bacteria strains were used for the tests: L. fermentum 428ST, L. rhamnosus E4.2, L. plantarum FCA3, Lactobacillus sp. 34.1, Weissella (W.) paramesenteroides FT1a and the control yeast K. marxianus from the Faculty of Biology collection, University of Bucharest, Romania. For comparison, the strain L. casei 431 was isolated from diary product—Probiotic Yogurt Drink Vivacto, Tesco-Hungary [12]. The bacterial strains were maintained in the de Man Rogosa Sharpe (MRS) broth (Oxoid, Basingstoke, Hampshire, England) at 37 °C. The yeast strain was maintained in YPG broth at 37 °C. The cells were collected by centrifugation at 5000×g for 10 min and washed three times. The isolated biomass was freeze-dried in a Christ-Alpha 1-2 LD plus freeze dryer (Martin Christ Gefriertrocknungsanlagen GmbH). After lyophilization all the strains were introduced into hard gelatine capsules that were gastro-resistant in order to simulate as accurate as possible the effect when in vivo administered. Each capsule contained approximately 0.25 g of lyophilized biomass having a minimum viability of 107 CFU/g.
In Vitro Human Colon Simulation
The conditions in the colon were replicated in a single-chamber system, GIS1, inoculated with 10 % (wt:v) fecal homogenate from a child (7 years old) in peptonate water (control). After inoculation, the GIS1 was left for approximately 24 h to allow stabilisation. The system was operated for 20 h. The GIS1 system was described in previous studies [12, 13]. Each bacterial strain was individually tested for better characterization.
Metabolic Activity Analysis
Exopolysaccharides separation was performed by precipitation with ethanol, according to the method presented by Dimitrijević et al. [14], while the quantification was performed according to a recent work of authors [15]. Protein concentration in the extracellular extracts of LAB was determined using the Bradford method [16]. The cell free solution was obtained by centrifuging the samples at 5000 rpm for 20 min followed by the filtration of the supernatant through a 0.2 µm pore size PTFE syringe driven filter unit (Macherey–Nagel). The chemical reaction with specific reactive and the absorbance of each sample were quantified at 595 nm. The standard curve of protein concentration was obtained using a series of dilutions of the Bovine Serum Albumin (BSA) protein standard stock solution.
Microbiological Analysis
The analyses were made by serial dilution of the culture sample in physiological saline solution (pH 7.0). The two highest dilutions were then plated on specific media and evaluated by an automated colony counter, Colony Quant, with the corresponding software [12, 13].
The total number of anaerobes was determined by using Brain-Heart Infusion (BHI) agar, Mc. Conkey agar for coliforms; Azide blood agar base for enterococci; Mannitol salt phenol red agar for staphylococci; Tryptose sulfite cycloserine agar for clostridia; Bifidus Selective Medium (BSM) agar for bifidobacteria; Rogosa agar for lactobacilli. The media used were purchased from Sigma-Aldrich, Germany.
Statistical Analysis
All the microbiological and biochemical experiments were performed in triplicate, and the results were expressed as mean ± SD values.
Results and Discussion
The probiotic effect of the newly isolated lactic acid bacteria strains depends on their ability to preserve viability after the gastrointestinal transit and also on their influence on the colonic microbiota. The persistence along the colon segments depends on the dose and the way of administration that further will directly affect the homeostasis of the bacteria population at each level [17].
The five newly isolated lactic acid bacteria strains were selected based on: antimicrobial activity against bacterial strains belonging to Escherichia, Salmonella, Bacillus, Staphylococcus, Klebsiella and Campylobacter genera; adherence to eukaryotic cell lines (HCT8 and H29) and competition with pathogenic microbial strains; immunomodulatory activity; lack of cytotoxicity; resistance to different pH values and resistance to bile salts concentrations (unpublished data).
Two groups and four bacteria genera were influenced by the administration of probiotics and/or prebiotics (Table 1). The main bacterial groups have shown good stability for all the tested biomass. The yeast K. marxianus was an exception as its administration resulted in an average decrease of 0.5 log CFU/mL in the transverse and descending colon segments.
Table 1.
The effect of administration of lyophilised probiotic strains on the simulated colon microbiota
| Sample | Number of microorganisms (log CFU/mL) | |||||
|---|---|---|---|---|---|---|
| Total anaerobes | Total aerobes | Enterococci | Coliforms | Clostridia | Bifidobacteria | |
| Control (without any strain administration) | ||||||
| Ascending colon | 5.26 ± 0.01 | 4.74 ± 0.65 | 4.27 ± 0.40 | 5.07 ± 0.10 | 5.60 ± 0.70 | 4.96 ± 0.40 |
| Transverse colon | 5.64 ± 0.35 | 4.93 ± 0.70 | 4.57 ± 0.01 | 5.87 ± 0.60 | 5.83 ± 0.80 | 5.29 ± 0.60 |
| Descending colon | 5.74 ± 1.05 | 4.68 ± 0.95 | 4.73 ± 2.30 | 5.97 ± 1.20 | 5.81 ± 1.35 | 5.05 ± 1.25 |
| Control—K. marxianus | ||||||
| Ascending colon | 5.20 ± 1.00 | 4.98 ± 1.60 | 4.50 ± 2.40 | 5.03 ± 0.01 | 5.27 ± 1.60 | 4.71 ± 0.30 |
| Transverse colon | 4.99 ± 2.60 | 4.76 ± 1.00 | 4.76 ± 1.20 | 4.88 ± 0.10 | 5.01 ± 0.55 | 4.79 ± 1.00 |
| Descending colon | 4.56 ± 0.30 | 4.72 ± 0.50 | 4.72 ± 2.20 | 5.82 ± 1.40 | 5.69 ± 0.70 | 4.56 ± 1.00 |
| Control—L. casei 431 | ||||||
| Ascending colon | 5.62 ± 0.70 | 5.32 ± 1.00 | 5.05 ± 0.70 | 5.09 ± 0.20 | 5.60 ± 0.05 | 5.26 ± 0.15 |
| Transverse colon | 5.05 ± 0.50 | 4.96 ± 0.01 | 4.74 ± 0.01 | 4.94 ± 0.00 | 4.89 ± 0.10 | 5.02 ± 0.30 |
| Descending colon | 4.92 ± 0.75 | 4.81 ± 0.20 | 4.60 ± 1.00 | 4.70 ± 0.04 | 4.75 ± 0.01 | 4.86 ± 0.55 |
| L. plantarum FCA3 | ||||||
| Ascending colon | 5.01 ± 0.20 | 5.34 ± 0.10 | 4.67 ± 0.01 | 5.87 ± 0.30 | 5.37 ± 0.40 | 4.81 ± 0.02 |
| Transverse colon | 5.52 ± 0.00 | 5.65 ± 0.55 | 4.77 ± 0.50 | 5.85 ± 0.60 | 5.88 ± 0.02 | 5.16 ± 0.40 |
| Descending colon | 5.90 ± 1.10 | 5.19 ± 0.80 | 4.59 ± 0.00 | 5.90 ± 0.00 | 5.35 ± 1.00 | 5.20 ± 1.10 |
| Lactobacillus sp. 34.1 | ||||||
| Ascending colon | 5.23 ± 0.65 | 4.90 ± 0.70 | 5.44 ± 0.05 | 5.50 ± 1.00 | 5.29 ± 0.44 | 4.98 ± 0.35 |
| Transverse colon | 5.63 ± 0.44 | 4.93 ± 0.06 | 5.54 ± 0.40 | 5.69 ± 1.25 | 4.90 ± 0.25 | 4.88 ± 0.50 |
| Descending colon | 5.90 ± 0.01 | 5.57 ± 0.68 | 5.23 ± 0.35 | 5.58 ± 1.05 | 5.47 ± 0.45 | 5.23 ± 0.00 |
| L. fermentum 428ST | ||||||
| Ascending colon | 4.95 ± 1.02 | 5.00 ± 1.40 | 4.63 ± 0.70 | 4.68 ± 0.48 | 5.55 ± 0.35 | 4.71 ± 0.01 |
| Transverse colon | 5.81 ± 1.10 | 4.94 ± 0.60 | 4.78 ± 0.40 | 4.38 ± 0.35 | 5.54 ± 0.28 | 5.24 ± 0.44 |
| Descending colon | 5.84 ± 1.40 | 4.98 ± 0.45 | 4.98 ± 0.60 | 4.67 ± 0.00 | 4.92 ± 0.25 | 5.41 ± 0.36 |
| L. rhamnosus E4.2 | ||||||
| Ascending colon | 4.89 ± 0.00 | 4.85 ± 0.30 | 4.82 ± 0.45 | 4.54 ± 0.68 | 4.93 ± 0.60 | 4.95 ± 0.04 |
| Transverse colon | 4.69 ± 0.05 | 4.68 ± 0.00 | 4.47 ± 0.03 | 4.36 ± 0.48 | 4.47 ± 0.00 | 4.62 ± 0.05 |
| Descending colon | 4.64 ± 0.01 | 4.77 ± 0.36 | 4.83 ± 0.00 | 4.36 ± 0.56 | 4.88 ± 0.00 | 4.30 ± 0.00 |
| W. paramesenteroides FT1a | ||||||
| Ascending colon | 4.88 ± 0.40 | 4.66 ± 0.10 | 4.25 ± 0.70 | 4.27 ± 1.60 | 4.60 ± 0.30 | 4.49 ± 0.86 |
| Transverse colon | 4.92 ± 0.08 | 4.79 ± 0.37 | 4.20 ± 1.30 | 4.55 ± 0.76 | 4.34 ± 0.00 | 4.70 ± 1.25 |
| Descending colon | 4.89 ± 0.00 | 4.65 ± 0.45 | 4.44 ± 1.46 | 4.20 ± 1.40 | 4.81 ± 0.70 | 4.59 ± 0.35 |
A major change in the structure of the simulated colon microbiota was observed when the strains L. fermentum 428ST, L. rhamnosus E4.2 and L. plantarum FT1a were administered. A decrease in the number of potentially pathogenic strains and Bifidobacterium species was noticed together with the stabilisation of the total number of anaerobic microorganisms.
The L. fermentum 428ST strain distinguishes as it stabilizes the bifidobacteria in the descending colon with an increase of 0.36 log CFU/mL and it decreases the coliform bacteria colonies in all the colon segments. The L. rhamnosus E4.2 strain lowers the number of coliforms and enterococci in the entire colon, while L. plantarum FT1a strain acts against all pathogenic bacteria. To conclude, a stabilisation was noticed between the determined bacteria.
The stability of the microbiota was determined for all tested strains. The L. fermentum 428ST and L. plantarum FCA3 strains cause a significant persistence of Bifidobacterium strains in the descending segment of the colon when compared to the reference strain L. casei 431. In the case of lyophilized strains, the decrease in bifidobacteria number was explained as a balance of the number of beneficial anaerobic strains, directly correlated to the number of lactobacilli. As the aim is to increase and stabilise the number of favourable strains, the effective lyophilic compound (e.g. L. fermentum 428ST) must have an antagonistic activity. This effect was also observed by the decrease in the coliforms number, decrease that was not achieved for the strains Lactobacillus sp. 34.1 and W. paramesenteroides FT1a. Moreover, the L. casei 431 strain has no antagonistic activity on strains from Clostridium genus. These minor variations are comparable to those determined in untreated microbiota (the first control).
The presence of the strains belonging to Clostridium genus showed higher levels (over 5 log CFU/mL) when lyophilized L. plantarum FCA3 was administered and partially for L. fermentum 428ST (without the terminal segment). The obtained results were comparable to the lyophilized yeast strain that was not able to reduce the microbial species. When L. rhamnosus E4.2 and W. paramesenteroides FT1a were administered a significant reduction (over 0.5 log CFU/mL) in the transverse section of the colon was observed. For several strains it was noticed a loss in the antagonist effect on this microbial group in the descending segment. A maximum increase of 0.5 log CFU/mL resulted for this section of the colon.
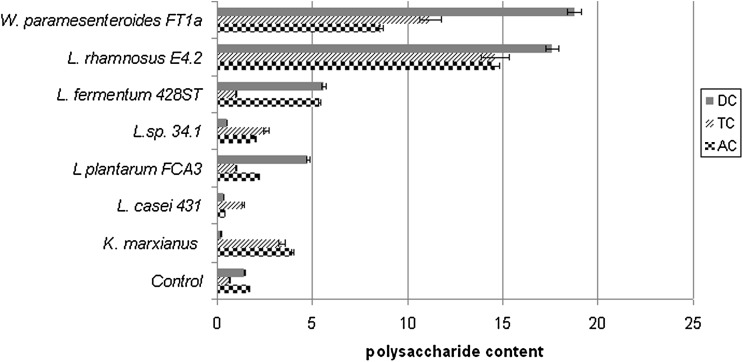
The amount of quantified exopolysaccharides for the seven tested bacterial strains had variations between the strains and randomly on the three sections of the colon where their production was simulated (Fig. 1). In the ascending colon, the strains L. rhamnosus E4.2 and W. paramesenteroides FTa1 had significantly synthesised higher amount of exopolysaccharides compared to control (without the inoculated strain) and reference strains, L casei 431 and K. marxianus, respectively. The same trend was observed even for the transverse and descending colon. Consequently, it could be concluded that L. rhamnosus E4.2 and W. paramesenteroides FTa1 have better capacity in the synthesis of exopolysaccharides together with a good probiotic effect such reduction of Clostridium cells numbers. Lactic acid bacteria exopolysaccharides are important for probiotic effect due to their involvement in the adherence to human or animal eukaryotic cells and therefore exclusion of pathogenic microbial cells, immunomodulatory and anticarcinogenic activity. During the last decades exopolysaccharides were used in order to obtain prebiotic and symbiotic products [18, 19]. The capacity of the two lactic acid strains to produce exopolysaccharides could be important from medical point of view if the strains are used as probiotics or for food industry.
Fig. 1.
The amount of polysaccharides from simulated media (mg/L). DC descending colon, TC transverse colon, AC ascending colon
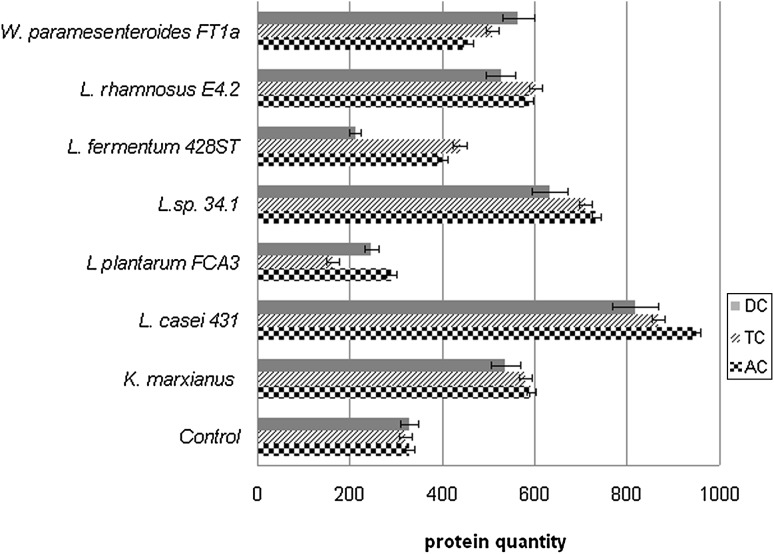
The experimental results indicated higher quantities of released extracellular proteins for the bacterial strains Lactobacillus sp. 34.1, L. rhamnosus E4.2 and W. paramesenteroides FT1a than for control for all the three colon segments, but lower than the reference strain L. casei 431 (Fig. 2). The differences that appeared in the exopolysaccharides amount obtained from Lactobacillus sp. 34.1, L. rhamnosus E4.2 and W. paramesenteroides FT1a compared to the amount obtained from L. casei 431 (strain isolated from Probiotic Yogurt Drink Vivacto) could be the result of the higher capacity of the three strains to biosynthesise exopolysaccharides under stress conditions (unpublished data). A possible explanation of the high amount of extracellular proteins obtained for L. casei 431 could be explained by the higher number of the total viable microbial cells as shown in Table 1.
Fig. 2.
The amount of proteins from simulated media (mg/L). DC descending colon, TC transverse colon, AC ascending colon
The extracellular proteins secreted by the lactic acid bacteria (in vitro in the culture medium corresponding to in vivo release to the intestinal lumen) can diffuse through the mucous layer covering the surface of the colon and interact with the epithelial and immune cells of the host organism. The role of LAB secreted proteins is not fully understood-can increase mucus barrier, have immunomodulating action, increase the absorption of electrolytes through the gastric mucosa. If the role of LAB secreted proteins will be understood than the researchers will better understand how probiotics act on the health of the host organism and then treat various inflammatory bowel diseases, allergies and autoimmune diseases [5].
Using the in vitro system GIS1, the interactions between human microbiota and epithelial cells or components of the immune system could not been simulated [20]. These technological limitations make necessary, at the end, an in vivo study to validate the effects of administering a dietary supplement that will contain lyophilized biomass. The effect of the individual administration of a LAB strain on the composition of the microbiota that is in formation had emphasised that the formulation of a competitive probiotic product cannot be made unless associating microbiological and biochemical effects of several LAB strains.
Due to the fact that the amount of lactobacilli in the microbiota is considered to be approximately 1 % [21], the increase in anaerobic microorganism number can be considered as an exceeding of this percentage coming from lyophilized compounds administration (Table 1). In terms of persistence in the colon sections, the exopolysaccharides synthesis adds to other physiological phenomena that will favour certain selected strains. These can cause an increase in the intake of nutrients, but also provides a decrease in cellular density (Fig. 1). The strains W. paramesenteroides FT1a and L. rhamnosus E4.2, which are in this situation, have caused the isolation of significant amounts of exopolysaccharides in the colon segments, this being explained as an improved persistence at the level of descending colon. This metabolic behaviour was noticed in the case of gastrointestinal transit for the same strains (unpublished data) and is in opposition to previous studies that have reported a higher number of favourable microorganisms in the ascending colon [22]. Moreover, the increase in the level of lactobacilli is correlated to the decrease in the level of bifidobacteria, generating equilibrium between the strains that formed the tested microbiota.
In this scientific research, the reduction of Bifidobacterium number, after the administration of W. paramesenteroides FT1a and L. rhamnosus E4.2 was inversed with the amount of exopolysaccharides. Other study demonstrated the similar reduction of the viable cells related to the inflammatory effect and proliferation of pathogenic strains [23]. An increase in the metabolic activity could determine a secondary synthesis of other compounds with antimicrobial effects.
Stability of Bifidobacterium species is very important in the microbiota composition as they could stabilize the microbiota of the host [24] by several mechanisms (production of inhibitory substances, competition with pathogenic strains, metabolic activity, toxins inhibitory etc.). These mechanisms could interact with the positive strains from the normal microbiota and it is possible to determine the disruption of equilibrium from the different strains. Interaction can appear between probiotic strains and therefore it is important to determine the incompatibility after the ingestion of the end product.
The human colon has a direct influence on health due to the significant fermentative activities that occur inside. Therefore, it can be assumed that a negative change in the fermentative balance can cause nutritional dysfunction that further can transform into obesity and/or diabetes [25]. The viability and colonisation ability at colon level are influenced by a series of physico-chemical factors, the microbiota fermentative capacity playing a significant role. The action mechanism is not completely understood, but it primarily depends on the number of viable strains after the passage through the stomach and small intestine. It is possible that the low pH, the enzymes and the action of biliary acids to influence the probiotic activity [26].
An easy way to improve the health status at gastrointestinal tract level is to administer different lyophilized complexes (consisting of compounds having prebiotic effect and bacterial strains that have modulatory capacity when applied to microbiota) to target groups of people [27].
Conclusions
The high level of beneficial microorganisms and the synthesis of bioactive compounds have transformed the tested strains into an important element in developing consortia having probiotic properties for normalising the unbalanced colon microbiota.
In vitro studies have addressed the role that every lyophilized strain has on the human microbiota. Moreover, these preliminary studies have to be continued with in vivo studies that involve biochemical and molecular biology analysis.
Acknowledgments
This research was supported by the National Centre for Program Management PN-II-PT-PCCA-2011-3.1-0969/2012 coordinated by Romanian Agency for Scientific Research.
References
- 1.Ljung A, Wadstrom T. Lactic acid bacteria as probiotics. Curr Issues Intest Microbiol. 2006;7:73–90. [PubMed] [Google Scholar]
- 2.Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Ther Adv Gastroenterol. 2013;6:295–308. doi: 10.1177/1756283X13482996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FAO/WHO (2001) Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Cordoba, Argentina, October 1–4
- 4.Araya MML, Reid G, Sanders ME, Stanton C (2002) Guidelines for the evaluation of probiotics in food. Jt. FAO/WHO Work. Group: London, UK; Ontario, Canada
- 5.Sanchez B, Urdaci M, Margolles A. Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa–bacteria interactions. Microbiology. 2010;156:3232–3242. doi: 10.1099/mic.0.044057-0. [DOI] [PubMed] [Google Scholar]
- 6.Boesten RJ, De Vos WM. Interactomics in the human intestine: Lactobacilli and Bifidobacteria make a difference. J Clin Gastroenterol. 2008;42:S163–S167. doi: 10.1097/MCG.0b013e31817dbd62. [DOI] [PubMed] [Google Scholar]
- 7.Makelainen H, Forssten S, Olli K, Granlund L, Rautonen N, Ouwehand AC. Probiotic lactobacilli in a semi-soft cheese survive in the simulated human gastrointestinal tract. Int Dairy J. 2009;19:675–683. doi: 10.1016/j.idairyj.2009.06.005. [DOI] [Google Scholar]
- 8.Xu X, Xu P, Ma C, Tang J, Zhang X. Gut microbiota, host health, and polysaccharides. Biotechnol Adv. 2013;31:318–337. doi: 10.1016/j.biotechadv.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Ruas-Madiedo P, De Los Reyes-Gavilán CG. Methods for the screening, isolation and characterization of exopolysaccharides produced by lactic acid bacteria. J Dairy Sci. 2005;88:843–856. doi: 10.3168/jds.S0022-0302(05)72750-8. [DOI] [PubMed] [Google Scholar]
- 10.Doleyres Y, Schaub L, Lacroix C. Comparison of functionality of exopolysaccharides produced in situ or added as bio-ingredients on yoghurt properties. J Dairy Sci. 2005;88:4146–4156. doi: 10.3168/jds.S0022-0302(05)73100-3. [DOI] [PubMed] [Google Scholar]
- 11.Silva PDLD, Bezerra MDF, Santos KMOD, Correia RTP. Potentially probiotic ice cream from goat’s milk: characterization and cell viability during processing, storage and simulated gastrointestinal conditions. LWT Food Sci Technol. 2015;62:452–457. doi: 10.1016/j.lwt.2014.02.055. [DOI] [Google Scholar]
- 12.Vamanu E, Pelinescu D, Marin I, Vamanu A. Study of probiotic strains viability from PROBAC product in a single chamber gastrointestinal tract simulator. Food Sci Biotechnol. 2012;21:979–985. doi: 10.1007/s10068-012-0128-8. [DOI] [Google Scholar]
- 13.Vamanu E, Pelinescu D, Avram I, Nita S, Vamanu A. Study of PROBAC product influence on infant microbiota in a single-chamber colonic fermentation model GIS1. Ann Microbiol. 2013;63:1029–1038. doi: 10.1007/s13213-012-0558-9. [DOI] [Google Scholar]
- 14.Dimitrijević A, Velickovkc D, Rikalovic M, Avramovic N, Milosavic N, Jankov R, Karadzic I. Simultaneous production of exopolysaccharide and lipase from extremophylic Pseudomonas aeruginosa san-ai strain: a novel approach for lipase immobilization and purification. Carbohydr Polym. 2011;83:1397–1401. doi: 10.1016/j.carbpol.2010.10.005. [DOI] [Google Scholar]
- 15.Vamanu E, Nita S (2013) Antioxidant capacity and the correlation with major phenolic compounds, anthocyanin, and tocopherol content in various extracts from the wild edible Boletus edulis mushroom. Biomed Res Int, Article ID 313905. doi:10.1155/2013/313905 [DOI] [PMC free article] [PubMed]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;7:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Delzenne NM, Cani PD. Interaction between obesity and the gut microbiota: relevance in nutrition. Ann Rev Nutr. 2011;31:15–31. doi: 10.1146/annurev-nutr-072610-145146. [DOI] [PubMed] [Google Scholar]
- 18.Nwodo UU, Green E, Okoh AI. Bacterial exopolysaccharides: functionality and prospects. Int J Mol Sci. 2012;13:14002–14015. doi: 10.3390/ijms131114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel S, Majumder A, Goyal A. Potentials of exopolysaccharides from lactic acid bacteria. Indian J Microbiol. 2012;52:3–12. doi: 10.1007/s12088-011-0148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shamekhi F, Shuhaimi M, Ariff A, Manap YA. Cell viability of microencapsulated Bifidobacteriumanimalis subsp. lactis under freeze-drying, storage and gastrointestinal tract simulation conditions. Folia Microbiol. 2013;58:91–101. doi: 10.1007/s12223-012-0183-9. [DOI] [PubMed] [Google Scholar]
- 21.Nelson-Dooley C, Olmstead SF (2015) The microbiome and overall health part 5: the oropharyngeal microbiota’s far-reaching role in immunity, gut health, and cardiovascular disease. Complementary Prescriptions. http://www.cpmedical.net/newsletter/the-microbiome-and-overall-health-part-5-the-oropharyngeal. Accessed 12 June 2015
- 22.Probert HM, Gibson GR. Development of a fermentation system to model sessile bacterial populations in the human colon. Biofilms. 2004;1:13–19. doi: 10.1017/S1479050503001029. [DOI] [Google Scholar]
- 23.Al-Okbi SY, Mohamed DA, Donya SM, Abd El Khalek AB. Role of Bifidobacterium bifidum and plant food extracts in improving microflora and biochemical and cytogenetic parameters in adjuvant arthritis. Grasas Aceites. 2011;62:308–320. doi: 10.3989/gya.089810. [DOI] [Google Scholar]
- 24.Prakash S, Tomaro-Duchesneau C, Saha S, Cantor A (2011) The gut microbiota and human health with an emphasis on the use of microencapsulated bacterial cells. J Biomed Biotechnol, Article ID 981214. doi:10.1155/2011/981214 [DOI] [PMC free article] [PubMed]
- 25.Desai AR, Powell IB, Shah NP (2004) Survival and activity of probiotic Lactobacilli in skim milk containing prebiotics. J Food Sci 69:FMS57–FMS60. doi:10.1111/j.1365-2621.2004.tb13371.x
- 26.Soccol CR, Vandenberghe LPS, Spier MR, Medeiros ABP, Yamaguishi CT, De Dea Lindner J, Pandey A, Thomaz-Soccol V. The potential of probiotics: a review. Food Technol Biotechnol. 2010;48:413–434. [Google Scholar]
- 27.Butel MJ. Probiotics, gut microbiota and health. Med Mal Infect. 2014;44:1–8. doi: 10.1016/j.medmal.2013.10.002. [DOI] [PubMed] [Google Scholar]