Abstract
The rhizospheric bacteria play key role in plant nutrition and growth promotion. The effects of increased nitrogen inputs on plant rhizospheric soils also have impacted on whole soil microbial communities. In this study, we analyzed the effects of applied nitrogen (urea) on rhizospheric bacterial composition and diversity in a greenhouse assay using the high-throughput sequencing technique. To explore the environmental factors driving the abundance, diversity and composition of soil bacterial communities, the relationship between soil variables and the bacterial communities were also analyzed using the mantel test as well as the redundancy analysis. The results revealed significant bacterial diversity changes at different amounts of applied urea, especially between the control treatment and the N fertilized treatments. Mantel tests showed that the bacterial communities were significantly correlated with the soil nitrate nitrogen, available nitrogen, soil pH, ammonium nitrogen and total organic carbon. The present study deepened the understanding about the rhizospheric soil microbial communities under different amounts of applied urea in greenhouse conditions, and our work revealed the environmental factors affecting the abundance, diversity and composition of rhizospheric bacterial communities.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-015-0551-7) contains supplementary material, which is available to authorized users.
Keywords: Greenhouse assay, Nitrogen fertilizer, Rhizospheric soil, Bacterial communities, High throughput sequencing, Environmental factors
Introduction
Nitrogen is regarded as the important macroelements to maintain the growth and development of plants [1]. Currently, anthropogenic nitrogen input is estimated to be from 30 to 50 %, which is bigger than that from natural sources [2]. Nitrogen fertilizer plays crucial roles in the production of grains, vegetables and fruits. In China, the polytunnel greenhouse-based vegetable production is a very important pillar industry and a greatly intensive cropping pattern [3]. The inputs of chemical fertilizers, pesticides and other agricultural chemicals are maintained at a high level for a long time [4]. Unreasonable fertilization, in particular, is an excess application of nitrogen fertilizer which caused serious problems in the greenhouse-based vegetable field, such as the phenomenon of acidic soil and the secondary salinization of soil [5]. These problems caused by unreasonable use of nitrogen fertilizer may influence the growth of vegetables and even raise the risk of NO3− contamination of the groundwater [6].
Some soil microorganisms colonize in the vicinity of plant roots, and some of which are obviously beneficial to the plant growth and development [7]. These bacteria include nitrogen fixing bacteria, phosphate-solubilizing bacteria or potassium-solubilizing bacteria, and antagonistic soil bacteria, etc. [8–11]. On the one hand, the rhizospheric bacteria can promote plant growth and productivity directly by biological nitrogen fixation, phosphate or potassium solubilization, phytohormone production and so on. On the other hand, they can promote plant growth indirectly through inducing resistance to pathogens. The healthy growth of plants partially depends on its interaction with these microorganisms, including both beneficial microorganisms and pathogens [12]. The composition and activity of the soil microbial community are influenced by farming practices in agroecosystem [2]. However, the effects of nitrogen fertilizer dosages on the soil microbial communities in greenhouse-based arable soil are still unclear.
Microorganisms in nature are mostly in a viable but non-culturable state. They cannot grow on the traditional microbiological media, but they are still alive [13]. Therefore, we cannot detect these microorganisms by the conventional method of plate dilution. In order to study the microbial distribution and organization in the root zones, a series of modern molecular techniques are employed, such as the polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) technique, the phospholipid fatty acid (PLFA) technique, clone library technique, and the high-throughput sequencing technique, etc. [14–17]. Among them, the high-throughput sequencing technique has been regarded as the most appropriate technique to evaluate the bacterial diversity and community composition in nature. It supplies a fast and large-scale sequencing approach. Thus, the high throughput sequencing will enhance our comprehensively understanding of the microbial communities of environmental samples.
In the present study, the Miseq high throughput sequencing method was used to study the bacterial communities in the soil applied with different dosages of nitrogen fertilizer to the polytunnel greenhouse vegetable land. We compared the microbial community structure, diversity, and bacterial phylogeny in different treatments, and evaluated the influence of different nitrogen application rates on the rhizospheric bacterial communities in the greenhouse. In addition, the key environmental factors controlling the distribution of bacterial communities were also determined.
Materials and Methods
Description of the Study Site and Soil Sampling
The experimental fields located in the experimental base of Shenyang Agricultural University (N41°50′, E123°34′), Liaoning Province, China. The soil samples were collected from the polytunnel greenhouse vegetable land in the experimental field, where supplied rotate crops of tomatoes, beans and celeries. N fertilizers at 4 levels in terms of the traditional amounts: (1) 0 kg N ha−1 year−1 (CK treatment); (2) 840 kg N ha−1 year−1 (N1PK treatment); (3) 630 kg N ha−1 year−1 (N2PK treatment); and (4) 420 kg N ha−1 year−1 (N3PK treatment) were supplied to the crops. The nitrogen amendment was urea. Other conditions of the designed treatments were kept the same.
The rhizospheric soil samples were collected at a tomato harvest. Three soil cores collected from one plot were randomly mixed to form an independent sample. Soil samples collected from three plots of one treatment were regarded as three independent replicates. Samples were collected and placed in the plastic bags and then quickly carried to the lab. Then the samples were sieved to 2 mm and subdivided into two sub-samples. One sub-sample for the determination of soil properties was stored at 4 °C, while the other for DNA extraction was stored at −20 °C.
Soil Nutrient Analysis
Determination soil pH was performed using the glass electrode (shaking the soil (1: 5 w v−1) mixed solution for 30 min). And the soil moisture was determined using the gravimetric method. The total organic carbon (TOC) was determined with a TOC analyzer (Analytikjena HT1300, Germany) [18]. The available nitrogen was determined as described previously [19]. The nitrate nitrogen and ammonium nitrogen of the soils were measured as described by Bremner and Keeney [20]. The available phosphorus was determined using the previously described methods [21].
Soil DNA Extraction
The total DNA extraction from rhizospheric soil samples was performed employing a MO-BIO PowerSoil® DNA Isolation Kit (MOBIO, USA) according to the instruction. Extracted DNA solution was examined through the agarose gel electrophoresis and stained with Goldview (LaBEST, China). Concentrations and purity of the total DNA were measured by Nanodrop 2000 (Quawell, USA).
PCR Amplification and High Throughput Sequencing
Polymerase chain reaction (PCR) amplification of 16S rDNA fragments and subsequent high throughput sequencing were carried out at the Novogene Bioinformatics Technology Co. Ltd (Beijing, China). The PCR amplifications were conducted using the primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) [22]. Reactions were run using the following cycling parameters: 30 cycles consisting of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; with the final step of 10 min at 72 °C. The primers were selected as they exhibited few biases and should generate precise information about bacterial phylogeny and taxonomy. DNA fragments were amplified as described by Caporaso et al. Sequencing was performed using the Illumina MiSeq platform.
Processing of High Throughput Sequencing Data
Read pairs were merged from the original DNA fragments using the software FLASH. The merged reads were assigned into each sample in terms of the barcode peculiar to each sample [23]. Sequences were first filtered by QIIME quality filters [24]. Then we used UPARSE pipeline to pick OTUs (operational taxonomic units) by making an OTU table [25]. The sequences were defined as OTUs at 97 % sequence similarity. Then we picked typical sequences for every OTU and assigned taxonomic data to each typical sequence through the RDP classifier [26]. In addition, we customized Perl scripts to study α-diversity and β-diversity. To compute α-diversity, we rarified the OTU table and used it to calculate three metrics: the Chao1 metric, the Observed Species metric and the Rarefaction curves.
Statistical Analysis
Mantel test was employed to explore the soil variables that significantly correlated to the bacterial abundance of OTUs. These parameters were used to establish a soil variables matrix used for redundancy analysis (RDA) using the Canoco for Windows 4.5 software [27].
Results
Rarefaction Analysis
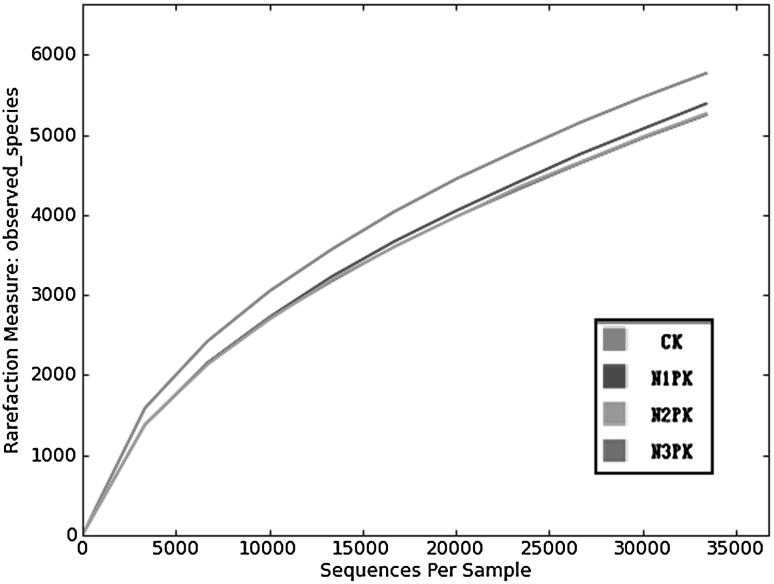
Of all the soil samples, 232,857 readings and 21,677 OTUs in total were obtained from the 4 treated samples by use of Miseq analysis. Each library included 52,184–70,637 reads, and the phylogenetic OTUs in every library ranged from 5248 to 5770. All the 4 rarefaction curves were inclined to be close to the saturation plateau. The curve of CK treatment located upper than the other curves, which indicated a larger variation within the total number of OTUs in the N-amended soil samples compared with CK (decreases in species richness were detected), but there were no obvious variation in those among different nitrogen fertilization rates (Fig. 1).
Fig. 1.
Rarefaction analysis of the different samples. Rarefaction curves of OTUs clustered at 97 % phylotype similarity level. Soil samples were treated with different nitrogen fertilization rate, CK (without nitrogen fertilizer), N1PK (840 kg N ha−1 year−1), N2PK (630 kg N ha−1 year−1), N3PK (420 kg N ha−1 year−1)
Bacterial Community Composition
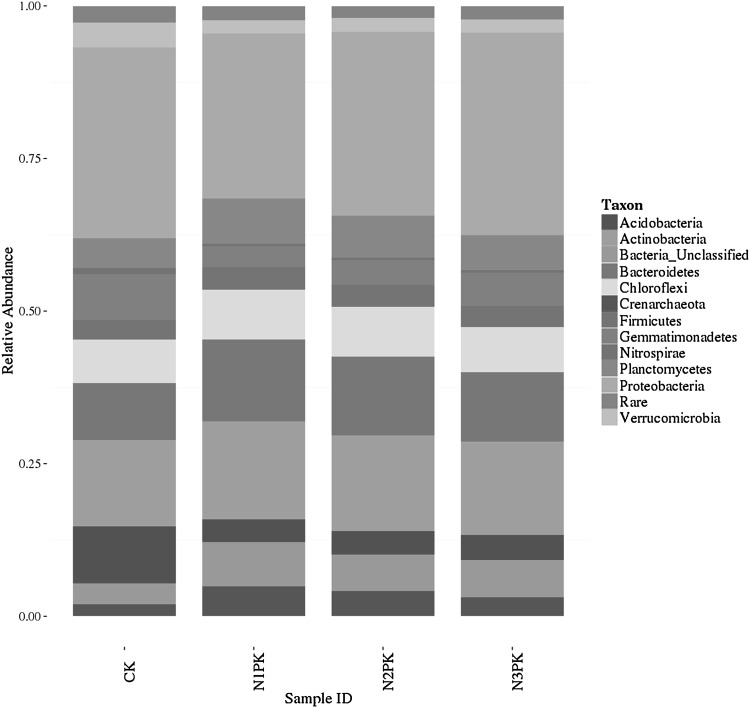
All samples consisted of different out numbers and OTU abundances. The whole bacterial OTUs could be classified into 44 known phyla, 251 known families or 283 known genera. Sequences which could not belong to known groups were defined as Bacteria_unclassified and sequences with too low abundances were assigned as Rare. Thirty-two of 44 phyla were common to the whole 4 libraries, occupying 96.58, 92.67, 93.94 and 93.68 % of the total OTUs in the libraries of CK, N1PK, N2PK and N3PK, respectively. Only 6, 5, 5, 6 phyla maintained OTUs abundance more than 5 % in the CK samples, N1PK samples, N2PK samples and N3PK samples, respectively. However, they jointly hold 75.80 % of the total OTUs. Proteobacteria was the most abundant division (Fig. 2; Table 1), comprising approximately 30.38 % (6585) OTUs of all samples, whereas the members from Actinobacteria (15.29 %, 3315 OTUs), Bacteroidetes (11.76 %, 2549 OTUs), Acidobacteria (5.33 %, 1155 OTUs), Gemmatimonadetes (5.22 %, 1132 OTUs), Chloroflexi (7.66 %, 1660 OTUs), Planctomycetes (6.22 %, 1348 OTUs) were also considerable in the whole libraries. The average OTUs of Bacteria_unclassified group accounted for 5.68 %, and fluctuated slightly in different samples. There was also a certain amount of Verrucomicrobia (2.70 %, 585 OTUs), Firmicutes (3.49 %, 757 OTUs), Crenarchaeota (3.50 %, 760 OTUs) and Nitrospirae (0.51 %, 110 OTUs). The other lineages represented a small proportion of (ca. 2.25 %) of the bacterial communities.
Fig. 2.
Mean relative abundances of dominant bacterial phyla in all soils from each nitrogen treatment. Sequences that could not be classified into any known group were assigned as Bacteria_unclassified and sequences with too low abundances were assigned as Rare. CK: without nitrogen fertilizer, N1PK: 840 kg N ha−1 year−1, N2PK: 630 kg N ha−1 year−1, N3PK: 420 kg N ha−1 year−1
Table 1.
The mean relative proportions of different phyla (%)
| Proteobacteria | Actinobacteria | Bacteroidetes | Acidobacteria | Gemmatimonadetes | Chloroflexi | Planctomycetes | |
|---|---|---|---|---|---|---|---|
| CK | 31.24a | 14.19a | 9.40a | 9.31a | 7.64a | 7.13a | 4.91a |
| N3PK | 33.16a | 15.32b | 11.39b | 4.10b | 5.55b | 7.37a | 5.76ab |
| N2PK | 30.09ab | 15.66b | 12.97c | 3.85b | 4.09c | 8.07b | 6.86bc |
| N1PK | 27.03b | 16.09b | 13.44c | 3.71b | 3.42c | 8.10b | 7.44c |
Superscript letters a, b and c mean significant difference at 0.05 level
Generally, each sample contained approximately similar phyla, and the samples without nitrogen fertilizer treatment showed higher phylotypic richness comparing with the treated samples. For example, the CK (without nitrogen fertilizer, mean 36 phyla) samples showed relatively lower phylotypic richness than the N3PK (420 kg N ha−1 year−1, 40 phyla) samples. Moreover, the relative proportions of different phyla varied as the application of nitrogen fertilizer amount changed. Strikingly, the portion of the dominant Proteobacteria generally decreased with the elevated application of nitrogen fertilizer (Table 1). The second-predominant phylum, Actinobacteria showed an obviously increasing trend along with the application of N fertilizer (Table 1). In addition, Bacteroidetes, Chloroflexi and Planctomycetes showed readily increasing trends along with the application of N fertilizer. However, Acidobacteria and Gemmatimonadetes showed obviously declining trends as the dosage of N fertilizer increased. Relative abundances of main phyla showed significant differences among communities with various amounts of applied urea, especially between the control and the N fertilized treatments (Table 1).
Further phylogenetic analysis revealed that the members of Gammaproteobacteria dominated the Proteobacteria, occupying 11.48 % (2489 OTUs) of total OTUs in the four libraries. The subdivision of Alphaproteo bacteria accounted for 8.05 % (1745 OTUs) of the total OTUs, while Betaproteobacteria and Deltaproteobacteria accounted for 6.45 % (1398 OTUs) and 4.08 % (883 OTUs) of the total phylotypes, respectively. The Epsilonproteobacteria only accounted for 0.002 % OTUs of the total OTUs. Statistically, Gammaproteobacteria contributed 38.73 % to the total OTUs of Proteobacteria, and the variation of Proteobacteria was strongly influenced by the Gammaproteobacteria in all samples, especially in the N3PK and N2PK samples. The members of Actinobacteria was the most abundant, making up 10.69 % (2318 OTUs) of the total OTUs, and Thermoleophilia and Acidimicrobiia accounted for 2.15 % (466 OTUs) and 1.82 % (395 OTUs) of total phylotypes, respectively. Bacteroidetes was dominated by the members of Sphingobacteriia (10.07 %, 2182 OTUs). For Acidobacteria, the most common groups were Acidobacteria-6 and Chloracidobacteria, with 1.85 and 1.56 % OTUs, respectively. For Gemmatimonadetes (phylum), the most common group was Gemmatimonadetes (Class) which contributed 2.78 % OTUs to the total libraries.
Bacterial Community Variation with Nitrogen Fertilizer Dosage
Hierarchal clustering double dendogram was founded based on relative percentages of the top 35 bacteria at genus level (Y-axis) under the treatments of different dosages of nitrogen fertilizer (X-axis) (Fig S1). The hierarchical heat map generally indicated that the treatment with low nitrogen fertilizer was similar to the CK treatment (without N fertilizer). With the increase of nitrogen application, the difference between the treatment and the CK treatment became greater. Comparison of the relative abundance revealed significant differences among the treatments at the genus level. We could divide all the bacterial communities into three groups: nitrogen-like group, nitrogen-tolerant group, and nitrogen-sensitive group (Table 2). In detail, the relative abundances of Candidatus_Solibacter Catellatospora, DA101, Gemmata, Halomonas, Hydrogenophaga, Methylibium, Nitrospira, and Ramlibacter decreased with the elevation of nitrogen application, therefore they could be defined as the nitrogen-sensitive group. However, the relative abundances of A4, A17, Aeromicrobium, Arenibacter, Candidatus_Nitrososphaera, Hyhomicrobium, Kribbella, Micromonospora, Nocardioides, and Thermomonas, showed opposite trends, which were regarded as the nitrogen-like group. In addition, the relative abundances of some genera (e.g. Aquicella, Dokdonella, Kaistobacter, Luteimonas, Rhodanobacter, and Steroidobacter) increased under low dosage of nitrogen fertilizer, whereas declined again as the nitrogen application increased further, so they were defined as the nitrogen-tolerant ones.
Table 2.
Classification of the bacterial communities in the genus level based on their preference for nitrogen
| Nitrogen-sensitive | Nitrogen-like | Nitrogen-tolerant |
|---|---|---|
|
Candidatus_Solibacter Catellatospora
DA101 Gemmata Halomonas Hydrogenophaga Methylibium Nitrospira |
A4
A17 Aeromicrobium Arenibacter Candidatus_Nitrososphaera Hyhomicrobium Kribbella Micromonospora |
Aquicella
Dokdonella Kaistobacter Luteimonas Rhodanobacter Steroidobacter |
| Ramlibacter | Nocardioides | |
| Thermomonas |
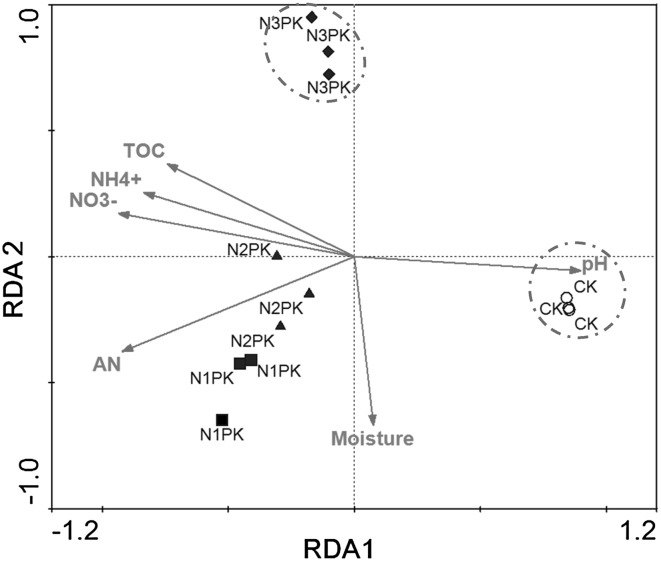
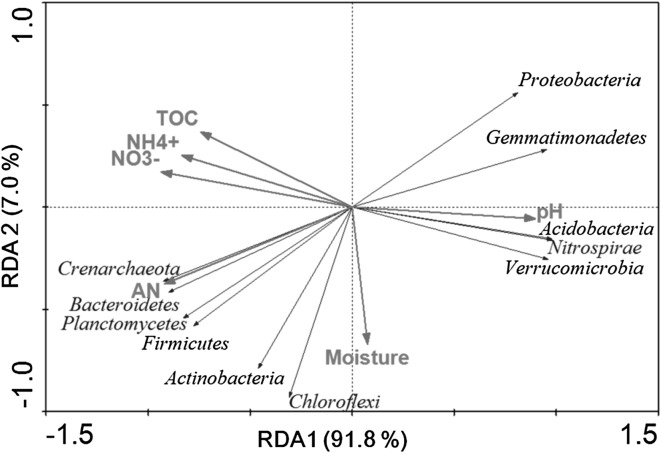
Bacterial communities were obviously different among various nitrogen gradient, and mantel tests showed that bacterial community composition was significantly correlated to nitrate nitrogen, available nitrogen, soil pH, ammonium nitrogen and total organic carbon (Table 3). Among the soil variables examined, nitrate nitrogen showed the highest correlation with the bacterial communities (r = 0.807, P = 0.002). The soil properties correlated with the bacterial communities were picked to perform a redundancy analysis (RDA), and the results showed that soil nitrate nitrogen and pH have stronger effects on the bacterial communities (Fig. 3). Here, soil pH and the available nitrogen (especially the nitrate nitrogen) can be regarded as the fundamental factors affecting the bacterial community composition (Fig. 3). Meanwhile, relative abundance of main phyla revealed difference among communities at nitrogen gradients, and the bacterial communities of two fertilized treatments were well separated from the other two treatments (Fig. 3). Furthermore, we also performed a canonical correspondence analysis to study the correlation between the main phyla and the soil properties. The pH indicated a positive correlation with the relative abundance of the phyla Acidobacteria, Verrucomicrobia, and Nitrospirae (Fig. 4). While three other soil characteristics (nitrate nitrogen, ammonium nitrogen, and total organic carbon) had negative correlation to the relative abundance of Acidobacteria, Verrucomicrobia, and Nitrospirae. The pH exhibited an opposite correlative pattern to the bacterial community composition, compared to the nitrate nitrogen, ammonium nitrogen, and total organic carbon. The available nitrogen showed a positive correlation to Bacteroidetes, Planctomycetes, Firmicutes, and Crenarchaeota, while it had the negative correlation to Proteobacteria and Gemmatimonadetes.
Table 3.
The correlation between the soil properties and the bacterial communities
| Variables | r | P value |
|---|---|---|
| Nitrate nitrogen | 0.807 | 0.002** |
| Available nitrogen | 0.788 | 0.002** |
| pH | 0.739 | 0.002** |
| Ammonium nitrogen | 0.645 | 0.006** |
| Total organic carbon | 0.516 | 0.01** |
| Moisture | 0.037 | 0.608 |
The correlation (r) and significance (P value) were calculated between the environmental variables and community structure using the method of mantel test. P value is from the Montel Carlo Permutation Tests
** Significant difference at 0.01 level
Fig. 3.
The redundancy analysis (RDA) of the bacterial communities with symbols coded by the nitrogen gradient. NO3 −, nitrate nitrogen; AN, available nitrogen; NH4 +, ammonium nitrogen; TOC, total organic carbon
Fig. 4.
Canonical correspondence analysis bi-plot of the dominant phyla and soil properties. NO3 −, Nitrate nitrogen; AN, available nitrogen; NH4 +, ammonium nitrogen; TOC, total organic carbon
Discussion
High-throughput sequencing technology has been employed extensively to research the bacterial communities in various habitats and environmental samples [22, 28]. Based on our results, the predominant phyla in the polytunnel greenhouse vegetable land were Proteobacteria (30.38 %, 6585 OTUs) Actinobacteria (15.29 %, 3315 OTUs), Bacteroidetes (11.76 %, 2549 OTUs), Acidobacteria (5.33 %, 1155 OTUs), Gemmatimonadetes (5.22 %, 1132 OTUs), Chloroflexi (7.66 %, 1660 OTUs), Planctomycetes (6.22 %, 1348 OTUs). The most dominant bacteria in the greenhouse-based vegetable land were Proteobacteria, which was in agreement with a summary report for several soils [29]. The members of Proteobacteria cover enormous morphological and metabolic diversity, playing a crucial role in the global carbon, nitrogen and sulphur cycles [30]. The relative abundances of Proteobacteria in the whole libraries decreased with the increase of the nitrogen fertilizer application in this study, which would have an impact on vegetable production. Among Proteobacteria, the relative abundance of Gamma-, Alpha-, Beta-, and Delta- subdivisions averaged to 38.73 % (2489 OTUs), 26.51 % (1745 OTUs), 21.23 % (1398 OTUs), and 13.42 % (883 OTUs), respectively. However, the Epsilonproteobacteria only accounted for 0.002 % OTUs of the total OTUs, which was a very small proportion compared with other reports [29]. The second predominant phylum of Actinobacteria showed an obviously increasing trend with the elevated application of N fertilizer. The important role of Actinobacteria in soil function is well-known because of their function of decomposition of organic materials, such as cellulose. Therefore, Actinobacteria plays key roles in process of organic matter turnover and carbon cycling. Another interesting point in this study was the increase of Bacteroidetes and Chloroflexi with the increased application of N fertilizer. This point particularly emphasized the ability of these bacteria to survive in the environment with very low pH or high concentration of nitrogen.
In the present study, the results showed that bacterial diversity was significantly reduced in nitrogen fertilizer-managed soils (N3PK, N2PK, N1PK) compared to the CK sample. Among the whole profiles, bacterial communities living in the low concentration of nitrogen were more diverse, therefore these bacteria shared more taxa than those in the higher pH conditions. Despite of a large variation within the total OTU numbers in the N-amended soil samples compared with the CK samples (decreases in species richness were detected), there was no large variation in total of OTU numbers among different nitrogen fertilization rates. The ordination analysis indicated that the treatment of low nitrogen fertilizer was similar to the CK treatment (without nitrogen fertilizer), while the difference between the nitrogen-treated samples and the CK one was greater when the nitrogen application increased. In general, the bacterial diversity (OTUs numbers) decreased with the increase of nitrogen application.
Nitrogen gradient is regarded as one of the most fundamental and important patterns in plant and crops study. It was observed that soil bacterial diversity was apparently affected by the nitrogen gradient in the greenhouse-based agriculture. These results suggested that the pattern bacterial distribution might also follow the basic patterns of plants along with the nitrogen gradient. Both the bacterial diversity and community structure of were greatly different among the nitrogen gradient in our work. According to the response of different groups of bacteria to nitrogen, we divided the bacterial communities into three groups: nitrogen-like group, nitrogen-tolerant group, and nitrogen-sensitive group (Table 2). Factors including nitrate nitrogen, available nitrogen, soil pH, ammonium nitrogen and total organic carbon played important roles in determining the bacterial communities.
When the organic nitrogen (urea) was applied to the soil, it was transferred into inorganic ammonium nitrogen through ammonification. Then the ammonium nitrogen was changed into nitrate nitrogen through nitrification [1]. In this study, nitrate nitrogen revealed the highest positive correlation with bacterial communities (r = 0.807, P = 0.002), indicating that nitrification increased significantly over that of the control soil. Meanwhile, the relative abundance of nitrobacteria among the bacterial community should be increased.
Besides nitrate nitrogen, other soil properties like soil pH, TOC and ammonium nitrogen were also correlated to the bacterial communities (Table 2). The results showed that bacterial communities were significantly correlated to the soil pH. The importance of soil pH in controlling soil communities has currently been verified across all kinds of spatial scales, continental scales included [31]. The nitrogen can result in the accumulation of net proton in soils, which contributes to the pH decrease of soils [32]. The lower soil pH is unfavorable for the growth of microorganisms sensitive to acidic environment. This situation causes a large number of microorganisms losing competitive advantages in the nitrogen-amended soils and eventually leads to the decrease of microbial diversity. At the level of genus, comparison of relative abundance revealed significant variations among different treatments. The relative abundances of Catellatospora, Candidatus_Solibacter, DA101, Gemmata, Halomonas, Hydrogenophaga, Methylibium, Nitrospira, and Ramlibacter decreased with the increase of nitrogen application, which indicated that these bacteria were highly susceptible to the ecological changes caused by nitrogen fertilizer. Among these genera, Nitrospira is one of the most important members of ammonia-oxidizing bacteria (AOB). Previous studies reported that Nitrosospira was the most common bacteria in arable soils and steppe soils [33, 34]. Results indicated that high dosage of nitrogen in the polytunnel greenhouse vegetable land might result in non-preferable growth of AOB. The relative abundances of A4, A17, Candidatus_Nitrososphaera, Hyhomicrobium, Kribbella, Micromonospora, Nocardioides, Thermomonas, Aeromicrobium, and Arenibacter increased along with the nitrogen application, which showed that the nitrogen element and the lower pH caused by the application of nitrogen fertilizer were useful for the growth of these bacteria. Moreover, the abundance of some bacteria (e.g. Aquicella, Dokdonella, Kaistobacter, Luteimonas, Rhodanobacter, and Steroidobacter) increased under the conditions of low dosage of nitrogen fertilizer, but decreased when the nitrogen application increased further. This situation indicated that a low concentration of nitrogen could promote the growth of these bacteria, but a high concentration of nitrogen would inhibit their growth. The healthy growth of the plant is closely related to the beneficial rhizospheric microorganisms and the rhizospheric pathogens, so it is critical to understand the change of the microorganisms with nitrogen gradient [35]. Therefore, further studies need to focus on the changes of beneficial and harmful rhizospheric microorganisms with the nitrogen fertilizer concentration.
The survey aimed at indigenous microbial communities (including the diversity and community composition) of the greenhouse-based vegetable land and the possible influences of nitrogen on them were extremely important for the scientific fertilization in the greenhouse soil in China and for improving the efficiency of nitrogen fertilizer. We studied the effects of the nitrogen application on the rhizospheric soil bacterial communities in a greenhouse assay and determined the key environmental factors controlling the distribution of bacterial communities. It is conducive to deepen the current understanding about influence of nitrogen application on the the bacterial communities in greenhouse conditions, and to reveal the environmental factors driving the diversity and abundance changes of the rhizospheric soil bacterial communities.
The present research studied the soil properties, bacterial community abundance and diversity at various dosages of nitrogen fertilizer in a greenhouse assay. The correlation between soil properties and bacterial communities was also studied to determine the key environmental factors controlling the distribution of bacterial communities. The results showed that excess application of nitrogen fertilizer regimes reduced the biodiversity and abundance of bacteria. It was also found that bacterial communities were significantly correlated with the soil nitrate nitrogen, available nitrogen, soil pH, ammonium nitrogen and total organic carbon. The present work enhances our understanding of the effects of applied nitrogen amount on the rhizospheric soil bacterial communities under greenhouse conditions. These results underline the importance of reasonable nitrogen fertilization to soil microbial community in greenhouse conditions.
Electronic supplementary material
Acknowledgments
This work was supported by the Program of the National Science Foundation of China (41171192).
Contributor Information
Shuanghua Shang, Email: shuanghuashang@sina.com.
Yanli Yi, Email: yiyanli@126.com.
References
- 1.Vitousek PM, Hättenschwiler S, Olander L, Allison S. Nitrogen and nature. Ambio. 2002;31:97–101. doi: 10.1579/0044-7447-31.2.97. [DOI] [PubMed] [Google Scholar]
- 2.Ramirez KS, Craine JM, Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Change Biol. 2012;18:1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x. [DOI] [Google Scholar]
- 3.Shen W, Lin X, Shi W, Min J, Gao N, Zhang H, Yin R, He X. Higher rates of nitrogen fertilization decrease soil enzyme activities, microbial functional diversity and nitrification capacity in a Chinese polytunnel greenhouse vegetable land. Plant Soil. 2010;337:137–150. doi: 10.1007/s11104-010-0511-2. [DOI] [Google Scholar]
- 4.Shen W, Lin X, Gao N, Zhang H, Yin R, Shi W, Duan Z. Land use intensification affects soil microbial populations, functional diversity and related suppressiveness of cucumber Fusarium wilt in China’s Yangtze River Delta. Plant Soil. 2008;306:117–127. doi: 10.1007/s11104-007-9472-5. [DOI] [Google Scholar]
- 5.Jorquera MA, Martínez OA, Marileo LG, Acuña JJ, Saggar S, Mora ML. Effect of nitrogen and phosphorus fertilization on the composition of rhizobacterial communities of two Chilean Andisol pastures. World J Microbiol Biotechnol. 2013;30:99–107. doi: 10.1007/s11274-013-1427-9. [DOI] [PubMed] [Google Scholar]
- 6.Spalding RF, Exner ME. Occurrence of nitrate in groundwater—a review. J Environ Qual. 1993;22:392–402. doi: 10.2134/jeq1993.00472425002200030002x. [DOI] [Google Scholar]
- 7.Caballero-Mellado J, Onofre-Lemus J, Estrada-de los Santos P, Martínez-Aguilar L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl Environ Microbiol. 2007;73:5308–5319. doi: 10.1128/AEM.00324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocking EC. Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil. 2003;252:169–175. doi: 10.1023/A:1024106605806. [DOI] [Google Scholar]
- 9.Rodríguez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv. 1999;17:319–339. doi: 10.1016/S0734-9750(99)00014-2. [DOI] [PubMed] [Google Scholar]
- 10.Basak B, Biswas D. Influence of potassium solubilizing microorganism (Bacillus mucilaginosus) and waste mica on potassium uptake dynamics by sudan grass (Sorghum vulgare Pers.) grown under two Alfisols. Plant Soil. 2009;317:235–255. doi: 10.1007/s11104-008-9805-z. [DOI] [Google Scholar]
- 11.De Boer W, Wagenaar AM, Klein Gunnewiek PJ, Van Veen JA. In vitro suppression of fungi caused by combinations of apparently non-antagonistic soil bacteria. FEMS Microbiol Ecol. 2007;59:177–185. doi: 10.1111/j.1574-6941.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- 12.Brahim B. Bacteria for plant growth promotion and disease management. In: Dinesh K, editor. Bacteria in agrobiology: disease management, Chap. 2. Berlin: Springer; 2013. pp. 15–47. [Google Scholar]
- 13.Oh YM, Kim M, Lee-Cruz L, Lai-Hoe A, Go R, Ainuddin N, Rahim RA, Shukor N, Adams JM. Distinctive bacterial communities in the rhizoplane of four tropical tree species. Microb Ecol. 2012;64:1018–1027. doi: 10.1007/s00248-012-0082-2. [DOI] [PubMed] [Google Scholar]
- 14.Osés SM, Diez AM, Melero B, Luning PA, Jaime I, Rovira J. Characterization by culture-dependent and culture-independent methods of the bacterial population of suckling-lamb packaged in different atmospheres. Food Microbiol. 2013;36:216–222. doi: 10.1016/j.fm.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Kakumanu ML, Cantrell CL, Williams MA. Microbial community response to varying magnitudes of desiccation in soil: a test of the osmolyte accumulation hypothesis. Soil Biol Biochem. 2013;57:644–653. doi: 10.1016/j.soilbio.2012.08.014. [DOI] [Google Scholar]
- 16.Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, Brostoff J, Parkhill J, Dougan G, Petrovska L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Mirobiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamaki H, Wright CL, Li X, Lin Q, Hwang C, Wang S, Thimmapuram J, Kamagata Y, Liu W. Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform. PLoS ONE. 2011;6:e25263. doi: 10.1371/journal.pone.0025263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Ye D, Wang X, Settles ML, Wang J, Hao Z, Zhou L, Dong P, Jiang Y, Ma ZS. Soil bacterial communities of different natural forest types in Northeast China. Plant Soil. 2014;383:203–216. doi: 10.1007/s11104-014-2165-y. [DOI] [Google Scholar]
- 19.Aviva H, Larissa K, Mustafa G, Emine EK. Rates of decomposition of plant residues and available nitrogen in soil, related to residue composition through simulation of carbon and nitrogen turnover. Soil Biol Biochem. 1984;36:255–266. [Google Scholar]
- 20.Bremner J, Keeney D. Determination and isotope-ratio analysis of different forms of nitrogen in soils: 3. Exchangeable ammonium, nitrate, and nitrite by extraction-distillation methods. Soil Sci Soc Am J. 1966;30:577–582. doi: 10.2136/sssaj1966.03615995003000050015x. [DOI] [Google Scholar]
- 21.Fernanda C, Hernán EE, Luis AA. Soil available phosphorus status determines indigenous mycorrhizal colonization of field and glasshouse-grown spring wheat from Argentina. Appl Soil Ecol. 2007;35:1–9. doi: 10.1016/j.apsoil.2006.06.001. [DOI] [Google Scholar]
- 22.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA. 2013;110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ter Braak C, Smilauer P. CANOCO reference manual and User’s guide to Canoco for windows: software for canonical community ordination (version 4.5)) Cajo JF ter Braak and Petr Smilauer. Netherlands: Centre for Biometry, Wageningen University; 1998. [Google Scholar]
- 28.Qiu M, Zhang R, Xue C, Zhang S, Li S, Zhang N, Shen Q. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol Fertil Soils. 2012;48:807–816. doi: 10.1007/s00374-012-0675-4. [DOI] [Google Scholar]
- 29.Yang S, Wen X, Jin H, Wu Q. Pyrosequencing investigation into the bacterial community in permafrost soils along the China-Russia Crude Oil Pipeline (CRCOP) PLoS ONE. 2012;7:e52730. doi: 10.1371/journal.pone.0052730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E. Introduction to the Proteobacteria. In: Martin D, editor. The prokaryotes. New York: Springer; 2006. pp. 3–37. [Google Scholar]
- 31.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernes-Debuyser A, Tessier D. Soil physical properties affected by long-term fertilization. Eur J Soil Sci. 2004;55:505–512. doi: 10.1111/j.1365-2389.2004.00614.x. [DOI] [Google Scholar]
- 33.He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di H. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol. 2007;9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 34.Shen X-Y, Zhang L-M, Shen J-P, Li L-H, Yuan C-L, He J-Z. Nitrogen loading levels affect abundance and composition of soil ammonia oxidizing prokaryotes in semiarid temperate grassland. J Soil Sediment. 2011;11:1243–1252. doi: 10.1007/s11368-011-0375-y. [DOI] [Google Scholar]
- 35.Rodrigo M, Paolina G, Jos MR. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.