Abstract
In recent years, microalgae have attracted considerable interest as a biofuel resource owing to their rapid growth, tolerance to harsh conditions, and ability to accumulate a large amount of triacylglycerols (TAGs). However, the economic effectiveness of algal biofuel is still low. In this study, we attempted to increase oil production of the microalga Scenedesmus quadricauda by elevating intracellular malonyl-CoA and glycerol-3-phosphate (G3P) pools. To increase intracellular oil content, yeast-derived genes encoding acetyl-CoA carboxylase (ACC1), glycerol kinase (GPD1), and glycerol-3-phosphate dehydrogenase (GUT1) were overexpressed under the control of CaMV 35S and NOS promoters with SV40 large T antigen components. Fatty acid profiling, G3P content, and the number of cells with high oil content were analyzed by gas chromatography-mass spectrometry, G3P assay kit, and flow cytometry, respectively. Overexpression of ACC1 increased the total fatty acid content by 1.6-fold. Overexpression of GPD1 and GUT1 increased intracellular G3P content by 1.6- and 1.9-fold, respectively. Multi-gene expression of ACC1, GPD1, and GUT1 increased the number of cells with high oil content by 1.45-fold compared with that observed with the wild-type. This study is the first to report increased oil production by overexpression of the key genes (ACC1, GPD1, and GUT1) for TAG biosynthesis in microalgae.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-015-0546-4) contains supplementary material, which is available to authorized users.
Keywords: Oil production, Microalgae, Gene overexpression, Total fatty acid content, Glycerol-3-phosphate content
Introduction
In recent years, interest in liquid biofuels has grown rapidly due to depletion of excess crude oil savings and global warming caused by greenhouse gases [1, 2]. Currently, first-generation biofuels are produced from corn starch, sugar cane, sugar beet, palm, and oilseed rape. However, the use of biofuels is still limited, because large areas of agricultural land are required to grow these crops and they compete with food crops for arable land; moreover, the energy efficiency of these biofuels is still very low [2–4].
To overcome these problems with first-generation biofuels, many potential new sources of biofuels have been evaluated. Among these, microalgae have attracted considerable interest as a biofuel resource, because some species of microalgae have higher productivities than terrestrial crops, can be grown on marginal land using waste or salt water, and can accumulate a large amount of triacylglycerols (TAGs), which can be used for the production of biofuel [1, 5–7]. The theoretical oil productivity of microalgae is estimated to be from 47,000 to 308,000 L ha−1 annum−1, whereas the productivity of oil palm is 5950 L ha−1 annum−1 of biodiesel [8].
In order to increase the economic viability of algal biofuel by improving algal oil productivity, biochemical and genetic engineering approaches have been attempted [6]. Biosynthesis of TAGs and other lipids, which are the sources of biofuels, can be physically divided into three steps: de novo fatty acid synthesis in the plastid, glycerol assembly in the endoplasmic reticulum (ER), and final packaging into the oil bodies [9].
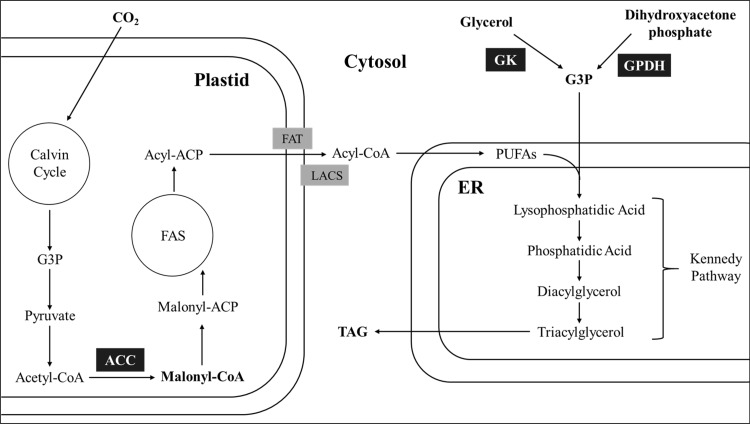
In the plastids, fatty acids, which are the building blocks of TAGs and other lipids, are synthesized from acetyl-CoA (Fig. 1). The first reaction in fatty acid biosynthesis is the formation of malonyl-CoA by the carboxylation of acetyl-CoA, which is catalyzed by acetyl-CoA carboxylase (ACC) [6]. Malonyl-CoA, which is derived from the first reaction, is converted to malonyl-ACP by a malonyl-CoA:acyl carrier protein malonyltransferase and then introduced into the fatty acid synthesis cycle [9–12]. Fatty acid synthase drives two-carbon chain-elongating reactions until a 16- or 18-carbon fatty acid is formed. The chain-elongation reaction is terminated by an acyl-ACP thioesterase. The synthesized free fatty acids are converted to acyl-CoA by long chain acyl-CoA synthetase and transferred into the cytosol [13, 14]. Acyl-CoA chains in the cytosol are esterified with structural phospholipids of the ER to be converted into polyunsaturated fatty acids (PUFAs), which are used as building blocks for the formation of TAGs via the Kennedy pathway. In the first step of the Kennedy pathway, PUFAs are esterified with glycerol-3-phosphate (G3P) by diacylglycerol acyltransferase (DGAT) for the formation of lysophosphatidic acid (LPA). LPA undergoes acylation to yield phosphatidic acid and diacylglycerol (DAG), catalyzed by lysophosphatidic acid acyltransferase and lysophosphatidylcholine acyltransferase, respectively. DAG is converted into triacylglycerol (TAG) via acylation catalyzed by DGAT. TAG molecules form oil droplets and eventually bud off from the ER [14].
Fig. 1.
A simplified scheme showing lipid synthesis in microalgae. Abbreviations: ACC acetyl-CoA carboxylase, GK glycerol kinase, FAS fatty acid synthase, GPDH glycerol-3-phosphate dehydrogenase, FAT fatty acyl-ACP thioesterase, LACS long-chain acyl-CoA synthetase, G3P glycerol-3-phosphate, ER endoplasmic reticulum, PUFAs polyunsaturated fatty acids, TAG triacylglycerol
Improving the fuel properties of lipid-derived biofuels depends on increasing the level of TAG accumulation. Previous studies revealed that the exposure of microalgae to stress condition accumulates the intracellular lipid pool, and eventually increases the level of TAG [15, 16]. Recently, Anand and Arumugam reported that nitrogen starvation stress increased the lipid yield by 2.27-fold in Scenedesmus quadricauda [17]. Moreover, the increasing of the TAG proportion the total lipid content was observed under nitrogen depletion condition. In addition, genetic engineering could also be used to increase TAG production. To enhance the TAG level, increasing the levels of the main precursors for TAG (malonyl-CoA and G3P) has been considered as a potentially effective approach. There have been several trials to increase the pools of those precursors, which have shown positive results [18].
To increase the level of the malonyl-CoA pool, ACC was overexpressed in the yeast Yarrowialipolytica and lipid production was successfully increased [19]. G3P can be synthesized through two different pathways, either from glycerol via glycerol kinase (GK)-mediated phosphorylation or from dihydroxyacetone phosphate via glycerol-3-phosphate dehydrogenase (GPDH)-mediated reduction [20, 21]. Overexpression of GK in the yeast Saccharomycescerevisiae improved lipid productivity [22]. The yeast GPDH was overexpressed in the seeds of Brassica napus and it increased lipid content by 40 % [23].
Among many microalgae, members of the Scenedesmus genus have been identified as potential biofuel feedstock owing to favorable characteristics such as rapid growth, high lipid content, and tolerance to high alkalinity and salinity [24, 25]. However, to our knowledge, no trials have yet attempted to increase oil content through overexpression of the key enzymes (ACC, G3P dehydrogenase, and glycerol kinase) for malonyl-CoA and G3P synthesis in Scenedesmus spp.
In the present study, three genes ACC1, GPD1, and GUT1 were amplified from S. cerevisiae and separately overexpressed in the microalga S. quadricauda. The intracellular content of glycerol-3-phosphate, total oil productivity, and transcribed levels of these genes in transgenic S. quadricauda were determined. Finally, the three genes, ACC1, GPD1, and GUT1, were overexpressed together in S. quadricauda, and then the total oil content was determined via flow cytometry analysis.
Materials and Methods
Algal Culture
An algal strain was isolated from the fresh water sample collected from a small pool next to Deokgok-Je reservoir in Jeollanam-do Province, South Korea. The isolated algal strain was cultivated in Beijerinck medium [26]. Based on morphological characteristics and 18S rDNA sequencing analysis, the algal strain was identified as S. quadricauda. The purified S. quadricauda was grown in 30 ml Jaworski/Euglena gracilis 1:1 medium [27] at 24 °C under continuous illumination (light intensity: 8,000 Lm).
Construction of Gene Expression Cassettes
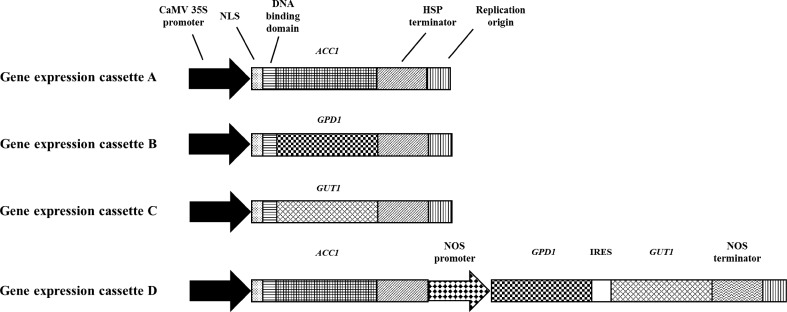
The PCR primers used for gene expression cassette construction are listed in Table S1. The genes GDP1 (encoding glycerol-3-phosphate dehydrogenase), ACC1 (encoding acetyl-CoA carboxylase), and GUT1 (encoding glycerol kinase) were amplified from yeast chromosomal DNA of the strain S. cerevisiae S288c. The CaMV 35S promoter and HSP-terminators were amplified from the pRI 201 vector (TaKaRa Bio, Japan), and the IRES fragment was amplified from pIRES2-EGFP (Clontech, USA). The NOS-promoter and NOS-terminator were amplified from the pRI 909 vector (TaKaRa Bio, Japan). SV40 genomic DNA (ATCC accession: 45019D) was used as a template source for the NLS, DNA binding domain, and replication origin (ori). The structures of each gene expression cassette are shown in Fig. 2. All DNA fragments for gene expression cassettes A, B, C, and D were connected by overlap extension PCR and the detailed procedure is shown in Figs. S1 and S2. The overlapping PCR products were examined by electrophoresis on 1.5 % agarose gels and DNA bands of the correct size were extracted using the QIAquick Gel Extraction Kit (Qiagen, USA). All PCR reactions were carried out using high fidelity DNA polymerase (TLA polymerase) (Bioneer, Republic of Korea).
Fig. 2.
Schematic diagrams of the gene cassettes used in current study
Chromosomal Integration of Gene Expression Cassettes by Electroporation
The gene expression cassettes described above were used to transform S. quadricauda by electroporation. Scenedesmus cells in the exponential phase were collected by centrifugation at 1300×g for 10 min. Scenedesmus cells were washed out with 20 ml of HEPES-glucose buffer (7 mM HEPES and 252 mM glucose, pH 7.0) two times. The equivalent of 1 × 105Scenedesmus cells was resuspended in 2 ml of sterilized distilled water and kept on ice for 2 h. Suspension aliquots of 80 μl were mixed with 40 ng of each gene expression cassette then transferred into an electroporation cuvette. Electroporation was carried out under the following conditions: electric field 1,800 V/cm and shunt 200 Ω. After electroporation, cells were kept on ice for 5 min and resuspended in 5 ml Jaworski/Euglena gracilis 1:1 medium containing 1 M mannitol and 1 M sorbitol, and then incubated in the dark for 24 h. After incubation, 1 ml of suspension was transferred into 30 ml Jaworski/Euglena gracilis 1:1 medium and cultivated for 24 h under continuous illumination. Afterwards, cells were collected by centrifugation at 1300×g for 5 min and resuspended in 1 ml Jaworski/Euglena gracilis 1:1 medium. Finally, the cells were spread on Petri plates in Jaworski/Euglena gracilis 1:1 medium.
PCR Screening for Gene Expression Cassette Integration
To determine the integration of the gene expression cassettes in transgenic S. quadricauda, the transformants were cultivated in 30 ml Jaworski/Euglena gracilis 1:1 medium. Genomic DNA was isolated from transgenic S. quadricauda according to previously described methods [28]. PCR analysis was conducted using Ex Taq™ (TaKaRa Bio, China) with a specific primer pair targeting the CaMV 35S promoter (35Sf/35Sr producing 843-bp).
Transcription Analysis of Transgenic S. quadricauda
In order to confirm the transcription of gene expression cassette in transgenic S. quadricauda by reverse-transcriptase PCR (RT-PCR), transgenic S. quadricauda were cultivated in 30 ml of Jaworski/Euglena gracilis 1:1 medium and Scenedesmus cells were collected from 4 ml culture. Total RNA was extracted using Isol-RNA Lysis reagent (TaKaRa Bio, Japan) and then cDNA was synthesized using Maxima First Strand cDNA Synthesis Kit (Thermo Scientific Fermentas, USA) with 1 μg of total RNA as a template. PCR was carried out using AccuTaq polymerase (Invitrogen, USA) with specific primer pairs for ACC1, GPD1, and GUT1: ACCRTf/ACCRTr, GPDRTf/GPDRTr, and GUTRTf/GUTRTr, respectively. All primer pairs were designed to produce 200-bp DNA fragments.
Analysis of Cell Growth
To determine cell growth of transgenic S. quadricauda, each transgenic line was cultivated in Jaworski/Euglena gracilis 1:1 medium for 8 days and the optical density was measured at 680 nm on days 1, 2, 3, 4, 6, and 8. All measurements were repeated three times and their mean values were used to determine the growth rate.
Analysis of Fatty Acids
To determine the profile of fatty acids, lipid samples were transesterified using a method from a previous study [29]. The resulting fatty acid methyl esters (FAME) were analyzed using a gas chromatography-mass spectrometer (890A GC system, Agilent, USA) equipped with a BPX70 fused-silica capillary column (50 × 0.32 mm, 0.25 μm film thickness, SGE, Austin, USA). The temperatures were as follows: injector, 250 °C; detector, 250 °C; oven, 205 °C (programmed to start at 35 °C, held at this temperature for 5 min, and then heated at a rate of 2.5 °C/min to 205 °C). The reference FAME standard FAMQ005 was obtained from AccuStandard (New Haven, USA) and diluted in dichloromethane (1:10). The resulting data were analyzed using ChemStation software (Agilent). Identification of fatty acids was based upon the retention time and the mass to char ratio of standard FAME mixtures. The area of each peak was calculated and related to 1 g cell dry weight. Each sample was measured three times.
Analysis of Intracellular G3P Content
Intracellular G3P content was determined using a glycerol-3-phosphate assay kit (Assay Biotechnology, USA) according to the manufacturer’s manual. For the determination of intracellular G3P content, transgenic S. quadricauda were grown in Jaworski/Euglena gracilis 1:1 medium for 10 days and 5 ml alga samples were collected. Algal cells were suspended in 300 μl distilled water and boiled for 1 h, and then centrifuged at 1300×g for 10 min. Fifty-microliter aliquots were taken from the supernatant for the determination of intracellular G3P content. The intracellular content of G3P as nM (nanomolar).
Determination of Lipid Content by Flow Cytometry Analysis
Nile red fluorescence (NRFL) was determined to measure the oil content of S. quadricauda [30]. Before flow cytometry (FC) analysis, samples were incubated in the dark for 5 h. A stock solution of nile red (NR) (Sigma-Aldrich, St. Louis, USA) was prepared by adding 2 mg of NR into 2 ml of dimethyl sulfoxide (DMSO). A working solution was prepared by adding 0.4 ml of stock solution to 1.6 ml DMSO. The concentration of NR in the working solution was 0.1 μg/μl. The solutions were stored in the dark at 4 °C.
A 999 μl aliquot from each sample was transferred to a micro centrifuge tube containing 13 μl of NR working solution and vortexed for 3 s, then incubated at 36 °C in the dark for 10 min.
All samples were analyzed using a FACSCalibur Flow Cytometer (BD Biosciences, USA) equipped with a 488 nm argon laser. The conditions for cytometric analysis were: acquisition setting was 10,000 events, AMPs/Detector-FSC E00-SSC 288, all detectors in log mode, threshold FSC 80, compensation FL3 12.7 % FL2. All settings were first optimized using a pure bacterial culture in liquid medium and a control algal sample. Neutral lipid analysis was carried out using an FL3 versus FL2 dot plot and a counts versus FL2 histogram. All flow cytometry data were analyzed using Flowing Software version 2 (Turku Centre for Biotechnology, Finland).
Analysis of Stability of the Gene Expression Cassette in Transgenic S. quadricauda
To determine the stability of gene expression cassette in transgenic S. quadricauda, the ACC1/GPD1/GUT1-overexpressing S. quadricauda was cultivated at subculture, low-, and normal-temperature conditions. For the determination of gene expression cassette stability under subculture condition, the ACC1/GPD1/GUT1-overexpressing S. quadricauda was serially subcultured 4 times with 6 days intervals at 24 °C in 30 ml Jaworski/Euglena gracilis 1:1 medium, and the presence of 35S CaMV promoter was determined at each subculture cycle by PCR. For the determination of gene expression cassette stability under different temperature conditions, the ACC1/GPD1/GUT1-overexpressing S. quadricauda was cultivated for 24 days without subculture at 4 and 24 °C under continuous illumination. The presence of 35S CaMV promoter under different temperature conditions was determined at 6 days intervals after inoculation by PCR.
Results
Molecular Analysis of Transgenic S. quadricauda
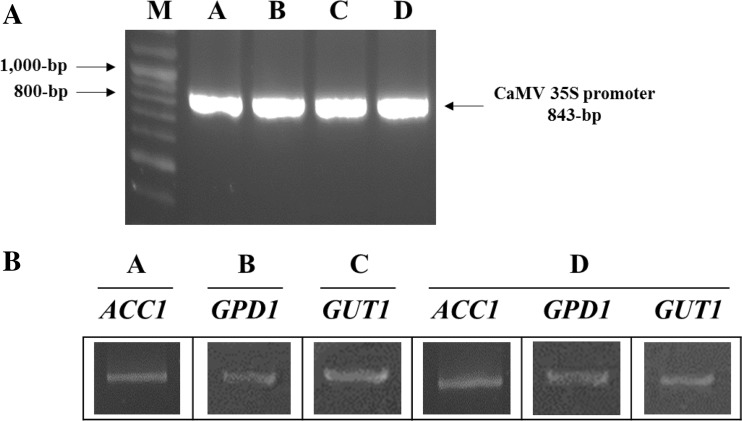
The gene expression cassettes were introduced into cells of S. quadricauda by electroporation. S. quadricauda transformants were regenerated on Jaworski/Euglena gracilis 1:1 agar medium, and then each transgenic line of S. quadricauda was cultivated in liquid medium. To confirm the integration of the gene expression cassette into the chromosome of S. quadricauda, CaMV 35S promoter was amplified with a specific primer pair, which produced an 843-bp PCR product, using the genomic DNA from transgenic S. quadricauda as a template. The 843-bp PCR products were amplified from all transgenic S. quadricauda containing gene expression cassettes A, B, C, or D (Fig. 3a).
Fig. 3.
PCR analysis for the detection of CaMV 35S promoter in the chromosomal DNA of transgenic S. quadricauda (a) and transcription analysis for ACC1, GPD1, and GUT1 in transgenic S. quadricauda (b). A transgenic S. quadricauda transfected with gene expression cassette A; B transgenic S. quadricauda transfected with gene expression cassette B; C transgenic S. quadricauda transfected with gene expression cassette C; D, transgenic S. quadricauda transfected with gene expression cassette D
In order to verify the expression of each gene expression cassette in transgenic S. quadricauda, the mRNA levels of the genes encoding AAC1, GPD1, and GUT1 were analyzed by RT-PCR. For the analysis of gene transcription, each transgenic line of S. quadricauda was grown in Jaworski/Euglena gracilis 1:1 medium and cells were collected. Total RNA was isolated from each sample and subjected to RT-PCR. Figure 3b shows that ACC1, GPD1, and GUT1 were successfully expressed in transgenic S. quadricauda containing gene expression cassette A, B, or C. In the transgenic S. quadricauda containing the gene expression cassette D, three genes, ACC1, GPD1, and GUT1, were also successfully expressed.
Cell Growth of Transgenic S. quadricauda
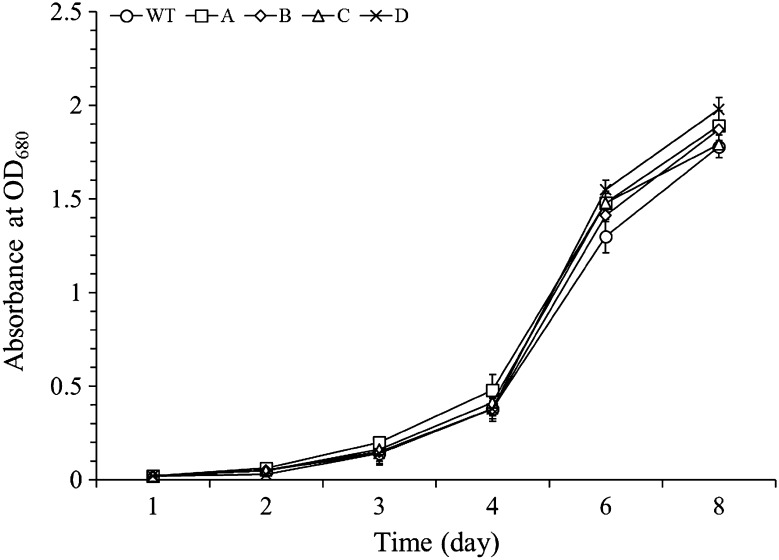
Transgenic S. quadricauda were cultivated in Jaworski/Euglena gracilis 1:1 medium for 8 days under continuous illumination and their growth was determined by measuring optical density at 680 nm (Fig. 4). A lag phase was observed in the growth of all transgenic lines during the first 2 days. Exponential growth started on day 3 and was maintained until day 8. There were no significant differences in growth observed among the different transgenic S. quadricauda lines.
Fig. 4.
Cell growth analysis of transgenic S. quadricauda. WT the wild-type S. quadricauda (circle); A transgenic S. quadricauda transfected with gene expression cassette A (square); B transgenic S. quadricauda transfected with gene expression cassette B (diamond); C transgenic S. quadricauda transfected with gene expression cassette C (triangle); D transgenic S. quadricauda transfected with gene expression cassette D (times)
Fatty Acid Profiling of ACC1-Overexpressing S. quadricauda
To determine the effect of ACC1-overexpression, total fatty acid profiling was performed using GC–MS in the wild-type and ACC1-overexpressing S. quadricauda (Table 1). The amount of total fatty acids was increased in the ACC1-overexpressing S. quadricauda by 1.61-fold compared with the wild-type. Except tridecylic acid, pentadecylic acid, elaidic acid methyl ester, and tricosylic acid, most fatty acids were increased by 1.5–7.3-fold by overexpression of ACC1.
Table 1.
Comparison of the fatty acid contents of ACC1-overexpressing and wild-type S. quadricauda
| Fatty acid | ACC1 | Wild-type |
|---|---|---|
| C10:0 (capric acid) | 0.16 ± 0.02 | 0.02 ± 0.00 |
| C11:0 (undecylic acid) | 0.37 ± 0.03 | 0.14 ± 0.01 |
| C13:0 (tridecylic acid) | 0 | 0.12 ± 0.02 |
| C15:0 (pentadecylic acid) | 0 | 0.62 ± 0.03 |
| C16:0 (palmitic acid) | 7.24 ± 0.18 | 4.26 ± 0.04 |
| C16:1n7 (palmitoleic acid) | 0.35 ± 0.04 | 0.18 ± 0.02 |
| C18:1n9t (elaidic acid methyl ester) | 0 | 0.13 ± 0.09 |
| C18:1n9c (oleic acid methyl ester) | 7.10 ± 0.08 | 4.25 ± 0.03 |
| C18:2n6t (linolelaidic acid methyl ester) | 3.91 ± 0.25 | 1.85 ± 0.14 |
| C18:2n6c (linoleic acid methyl ester) | 10.55 ± 0.18 | 5.66 ± 0.07 |
| C18:3n6 (gamma-linolenic acid methyl ester) | 1.76 ± 0.13 | 0.95 ± 0.10 |
| C18:3n3 (linolenic acid methyl ester) | 15.73 ± 0.11 | 10.67 ± 0.15 |
| C22:0 (behenic acid) | 0.41 ± 0.05 | 0.27 ± 0.02 |
| C23:0 (tricosylic acid) | 0 | 0.42 ± 0.09 |
| Total fatty acids | 47.58 ± 1.07 | 29.57 ± 0.81 |
The GC–MS measurements used for comparisons were the specific peak areas (peak area/sample dry weight in g)
Intracellular G3P Contents of GPD1- and GUT1-Overexpressing S. quadricauda
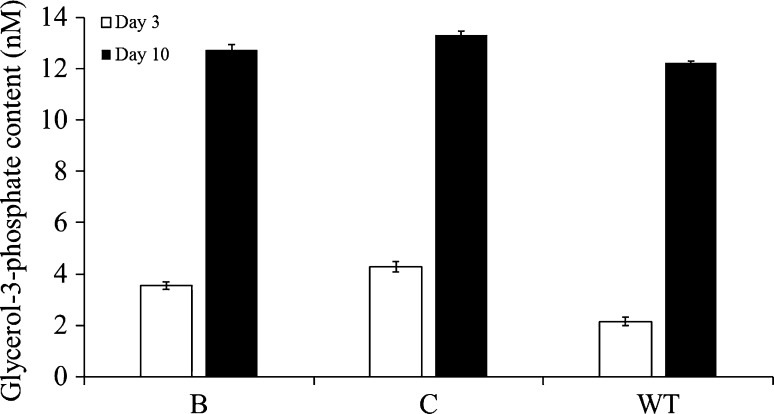
Intracellular G3P contents of the GPD1- or GUT1-overexpressing S. quadricauda were determined using G3P enzymatic assay kit at day 3 and day 10 (Fig. 5). On day 3, the intracellular G3P contents of the transgenic S. quadricauda with gene expression cassettes B and C were increased 1.6-fold and 1.9-fold (3.5 ± 0.15 nM and 4.2 ± 0.21 nM) compared with the wild-type (2.2 ± 0.17 nM). On day 10, the intracellular G3P contents of the transgenic S. quadricauda with gene expression cassettes B and C were increased 1.0-fold and 1.1-fold (12.74 ± 0.2 nM and 13.33 ± 0.13 nM) compared with the wild-type (10.24 ± 0.06 nM).
Fig. 5.
Intracellular glycerol-3-phosphate contents of transgenic S. quadricauda at day 3 (square) and day 10 (filled square). WT the wild-type S. quadricauda; B transgenic S. quadricauda transfected with gene expression cassette B; C transgenic S. quadricauda transfected with gene expression cassette C
Oil Content of the Multi-gene Overexpressing S. quadricauda
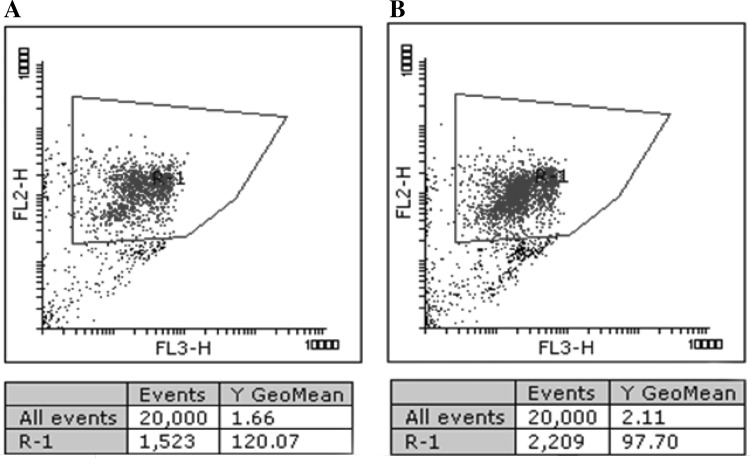
To increase total lipid content, ACC1, GPD1, and GUT1 were expressed together in S. quadricauda, and then the average level of lipid content and number of cells with high lipid content were determined by flow cytometry. Upon excitation by a 488 nm argon laser, NR emits fluorescence at 580 and 610 nm when dissolved in neutral and polar lipids, respectively [31], which are detected by the FL2 (590 ± 25 nm) and FL3 (675 ± 20 nm) channels. In this analysis, the average level of oil content and number of cells with high oil content were determined. Results from flow cytometry analysis showed that the number of cells with a high oil content was increased from 1523 to 2209 by the combined overexpression of ACC1, GPD1, and GUT1 (Fig. 6).
Fig. 6.
Oil content of the wild-type sample (a) and the ACC1/GPD1/GUT1-overexpressing S. quadricauda sample (b)
Stability of the Gene Expression Cassette in the Multi-gene Overexpressing S. quadricauda
To analyze stability of the gene expression cassette in transgenic S. quadricauda, the ACC1/GPD1/GUT1-overexpressing S. quadricauda was cultivated under different conditions and presence of 35S CaMV promoter was verified by PCR (Fig. S3). For PCR analysis of the gene expression cassette under subculture condition, the multi-gene overexpressing S. quadricauda was subcultured 4 times. The PCR results demonstrated that the gene expression cassette was maintained at all subculture cycles (Fig. S3A). The multi-gene overexpressing S. quadricauda was also cultivated under low- (4 °C) and normal-temperature (24 °C) conditions for 24 days and presence of the gene expression cassette was analyzed by PCR. PCR analysis demonstrated that the cassette was stably maintained during cultivation (Fig. S3B), however, it was not possible to analyze presence of the cassette in the transgenic S. quadricauda at 4 °C condition because S. quadricauda showed no growth at that temperature.
Discussion
In order to improve the economic efficiency of biofuels, many kinds of biomass resources have been studied as potential new sources of biofuel. At present, microalgae are considered the most energy effective material for biofuel production, because of their rapid growth rate and strong tolerance to harsh conditions. However, the normal oil content of microalgae is not high enough to achieve cost-effectiveness of algal biofuel. To overcome this problem, biochemical and genetic engineering approaches for increasing cellular oil content have been attempted in various microalgae [6].
TAGs are feedstocks for biofuel production and malonyl-CoA and G3P are major precursors for TAG biosynthesis. Malonyl-CoA is synthesized by carboxylation of acetyl-CoA, which is catalyzed by ACC [6], and it is used for fatty acid synthesis. The previous studies reported that the elevated malonyl-CoA level led to an increase in intracellular fatty acid content [32, 33]. G3P is derived from two precursors, glycerol and dihydroxyacetone phosphate, and their conversion into G3P is catalyzed by GK and GPDH, respectively [20, 21]. Previous reports have shown that experimentally increasing intracellular G3P enhanced the production of triacylglycerol [22, 34]. Following their successful results, there were several trials of approaches to improve cellular oil production via overexpression of ACC, GK, or GPDH in plants, microalgae, and yeast [19–23]. However, to our knowledge, no previous reports have attempted to overexpress these three genes together.
In the current study, to improve the cellular oil content, we selected the Scenedesmus genus due to their rapid growth, high lipid content, and tolerance to harsh conditions. Then, we tried to increase intracellular malonyl-CoA and G3P pools via overexpression of the key genes for malonyl-CoA and G3P biosynthesis.
To identify the effects of overexpression of ACC1, GPD1, and GUT1 on fatty acid content and G3P content in S. quadricauda, three gene overexpression cassettes (gene expression cassettes A, B, and C) were constructed and were transferred into S. quadricauda by electroporation. PCR and RT-PCR analysis results confirmed that the gene expression cassettes were successfully integrated into the chromosomal DNA of S. quadricauda and expressed.
GC–MS analysis of fatty acids demonstrated that the wild-type S. quadricauda contains 14 fatty acids and their isomers and the overexpression of ACC1 increased the total fatty acid content by 1.6-fold compared with that of the wild-type. A previous study reported that overexpression of ACC1 significantly reduced the growth rate of yeast [35]. However, suppression of cell growth was not observed in the ACC1-overexpressing S. quadricauda. The significant increasing of the intracellular G3P content by overexpression of GPD1 and GUT1 was observed at day 3. The intracellular G3P content of GPD1- and GUT1-overexpressing S. quadricauda was increased by 1.6- and 1.9-fold, respectively, compared with the wild-type. However, increasing rate of G3P in GPD1- and GUT1-overexpressing S. quadricauda was decreased to 1.0- and 1.1-fold, respectively, compared with the wild-type at day 10. These results demonstrated that the overexpression of GPD1 and GUT1 worked effectively in early-growth stage. Moreover, the results from the total fatty acid and the intracellular G3P content analysis showed the possibility of engineering a combined increase in the malonyl-CoA and G3P pools to enhance TAG production.
Based on the results from the overexpression of ACC1, GPD1, and GUT1 separately, these three genes were subcloned into a single gene expression cassette and transferred into the wild-type S. quadricauda. Transcription analysis showed that the three genes were successfully expressed in transgenic S. quadricauda. The number of cells with high oil content was analyzed by flow cytometry and it was found that the ACC1/GPD1/GUT1-overexpressing S. quadricauda had more cells with high oil content than did the wild-type.
Additionally, stability of the gene expression cassette under subculture and continuous culture conditions was analyzed in the ACC1/GPD1/GUT1-overexpressing S. quadricauda by PCR and analysis results demonstrated that the gene expression cassette was stably maintained in the transgenic S. quadricauda under both long-term culture conditions.
In conclusion, this study is the first to report an experimental increase in oil production through elevating intracellular fatty acids and G3P together in microalgae. Moreover, no negative effects were observed to be caused by the gene overexpression cassettes. We believe that the novel gene expression strategy for increasing oil content in the microalga S. quadricauda that we have reported here may contribute to improving the cost-effectiveness of algal biofuel.
Electronic supplementary material
Acknowledgments
This research was supported by a grant from the National Research Foundation (NRF) of Korea Grant funded by the Korean Government (MOE) (MEST) [NRF-2010-0024596].
Footnotes
Ahmed E. Gomma and Sung-Kwon Lee equally contributed to this work.
Contributor Information
Sung-Kwon Lee, Email: saintk-lee@hanmail.net.
Seung Hwan Yang, Email: ymichigan@mju.ac.kr.
Gyuhwa Chung, Email: chung@chonnam.ac.kr.
References
- 1.Brennan L, Owendea P. Biofuels from microalgae—a review of technologies for production, processing, and extractions of biofuels and co-products. Renew Sust Energy Rev. 2010;14:557–577. doi: 10.1016/j.rser.2009.10.009. [DOI] [Google Scholar]
- 2.Scott SA, Davey MP, Dennis JS, Horst I, Howe CJ, Lea-Smith DJ, Smith AG. Biodiesel from algae: challenges and prospects. Curr Opin Biotechnol. 2010;21:277–286. doi: 10.1016/j.copbio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental economic and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA. 2006;103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephenson AL, Dennis JS, Scott SA. Improving the sustainability of the production of biodiesel from oilseed rape in the UK. Chem Eng Res Des. 2008;86:427–440. doi: 10.1016/j.cherd.2007.12.006. [DOI] [Google Scholar]
- 5.Sheehan J, Dunahay T, Benemann J, Roessler P (1998) A look back at the US Department of Energy’s Aquatic Species Program-biodiesel from algae. NREL/TP‐580‐24190, National Renewable Energy Laboratory, Golden, CO, USA
- 6.Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. Genetic engineering of algae for enhanced biofuel production. Eukaryot Cell. 2010;9:486–501. doi: 10.1128/EC.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah GC, Yadav M, Tiwari A. Assessment for the higher production of biodiesel from Scenedesmusdimorphus algal species. Erud J Biotechnol. 2012;1:1–9. [Google Scholar]
- 8.Ahmad AL, Yasin NHM, Derek CJC, Lim JK. Microalgae as a sustainable energy source for biodiesel production: a review. Sustain Energy Rev. 2011;15:584–593. doi: 10.1016/j.rser.2010.09.018. [DOI] [Google Scholar]
- 9.Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- 10.Blatti JL, Beld J, Behnke CA, Mendez M, Mayfield SP, Burkart MD. Manipulating fatty acidbiosynthesis in microalgae for biofuel through protein–protein interactions. PLoS ONE. 2012;7:e42949. doi: 10.1371/journal.pone.0042949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenwell HC, Laurens LML, Shields RJ, Lovitt RW, Flynn KJ. Placing microalgae on the biofuels priority list: a review of the technological challenges. J R Soc Interface. 2010;7:703–726. doi: 10.1098/rsif.2009.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subrahmanyam S, Cronan JE. Overproduction of a functional fatty acid biosynthetic enzyme blocks fatty acid synthesis in Escherichia coli. J Bacteriol. 1998;180:4596–4602. doi: 10.1128/jb.180.17.4596-4602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellou S, Baeshen MN, Elazzazy AM, Aggeli D, Sayegh F, Aggelis G. Microalgal lipids biochemistry and biotechnological perspectives. Biotechnol Adv. 2014;32:1476–1493. doi: 10.1016/j.biotechadv.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Li-Beisson Y, Shorrosh B, Beisson B, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, Franke RB, Graham IA, Katayama K, Kelly AA, Larson T, Markham JE, Miquel M, Molina I, Nishida I, Rowland O, Samuels L, Schmid KM, Wada H, Welti R, Xu C, Zallot R, Ohlrogge J. Acyl-lipid metabolism. Arabidopsis Book. 2010;8:e0133. doi: 10.1199/tab.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths MJ, Harrison STL. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J Appl Phycol. 2009;21:493–507. doi: 10.1007/s10811-008-9392-7. [DOI] [Google Scholar]
- 16.Rismani-Yazdi H, Haznedaroglu BZ, Hsin C, Peccia J. Transcriptomic analysis of the oleaginous microalga Neochlorisoleoabundans reveals metabolic insights into triacylglyceride accumulation. Biotechnol Biofuels. 2012;5:74. doi: 10.1186/1754-6834-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anand J, Arumugam M. Enhanced lipid accumulation and biomass yield of Scenedesmus quadricauda under nitrogen starved condition. Bioresour Technol. 2015;188:190–194. doi: 10.1016/j.biortech.2014.12.097. [DOI] [PubMed] [Google Scholar]
- 18.Thevenieau F, Nicaud JM. Microorganisms as sources of oils. OCL. 2013;20:D603. doi: 10.1051/ocl/2013034. [DOI] [Google Scholar]
- 19.Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowialipolytica for biofuel production. Metab Eng. 2013;15:1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Kang L, Li J, Zhao T, Xiao F, Tang X, Thilmony R, He S, Zhou JM. Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc Natl Acad Sci USA. 2003;100:3919–3924. doi: 10.1073/pnas.0630495100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding JW, Pyeritz EA, Copeland ES, White HB. Role of glycerol 3-phosphate dehydrogenase in glyceride metabolism. Effect of diet on enzyme activities in chicken liver. Biochem J. 1975;146:223–229. doi: 10.1042/bj1460223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu KO, Jung J, Ramzi AB, Choe SH, Kim SW, Park C, Han SO. Development of a Saccharomycescerevisiae strain for increasing the accumulation of triacylglycerol as a microbial oil feedstock for biodiesel production using glycerol as a substrate. Biotechnol Bioeng. 2013;110:343–347. doi: 10.1002/bit.24623. [DOI] [PubMed] [Google Scholar]
- 23.Vigeolas H, Waldeck P, Zank T, Geigenberger P. Increasing seed oil content in oil-seed rape (Brassicanapus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol J. 2007;5:431–441. doi: 10.1111/j.1467-7652.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- 24.Yang F, Long L, Sun X, Wu H, Li T, Xiang W. Optimization of medium using response surface methodology for lipid production by Scenedesmus sp. Mar Drugs. 2014;12:1245–1257. doi: 10.3390/md12031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannon M, Gimpel J, Tran M, Rasala B, Mayfield S. Biofuels from algae: challenges and potential. Biofuels. 2010;1:763–784. doi: 10.4155/bfs.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein JR. Growth and mating of Goniumpectoral (Volvocales) in defined media. J Phycol. 1996;2:23–28. doi: 10.1111/j.1529-8817.1966.tb04587.x. [DOI] [PubMed] [Google Scholar]
- 27.CCAP . Culture collection of algae and protozoa: catalogue of strains. Kendal: Titus Wilson & Son Ltd; 1998. [Google Scholar]
- 28.Dawson HN, Burlingame R, Cannons AC. Stable transformation of Chlorella: rescue of nitrate reductase-deficient mutants with the nitrate reductase gene. Curr Microbiol. 1997;35:356–362. doi: 10.1007/s002849900268. [DOI] [PubMed] [Google Scholar]
- 29.Furuhashi T, Weckwerth W (2013) Introduction to lipid (FAME) analysis in algae using gas chromatography-mass spectrometry. The Handbook of Plant Metabolomics 215–225
- 30.Da Silva TL, Santos CA, Reis A. Multi-parameter flow cytometry as a tool to monitor heterotrophic microalgal batch fermentations for oil production towards biodiesel. Biotechnol Bioprocess Eng. 2009;14:330–337. doi: 10.1007/s12257-008-0228-8. [DOI] [Google Scholar]
- 31.Andrade R, Leal R, Roseiro J, Reis A, Silva TL. Monitoring Rhodosporidiumtoruloides NCYC 921 batch fermentations growing under carbon and nitrogen limitation by flow cytometry. W J Microbiol Biotechnol. 2012;28:1175–1184. doi: 10.1007/s11274-011-0920-2. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Guo D, Cheng Y, Zhu F, Deng Z, Liu T. Overproduction of fatty acids in engineered Saccharomycescerevisiae. Biotechnol Bioeng. 2014;111:1841–1852. doi: 10.1002/bit.25239. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Chen H, Yu O. A plant malonyl-CoA synthetase enhances lipid content and polyketide yield in yeast cells. Appl Microbiol Biotechnol. 2014;98:5435–5447. doi: 10.1007/s00253-014-5612-z. [DOI] [PubMed] [Google Scholar]
- 34.Yu WL, Ansari W, Schoepp NG, Hannon MJ, Mayfield SP, Burkart MD. Modifications of the metabolic pathways of lipid and triacylglycerol production in microalgae. Microb Cell Fact. 2011;10:91. doi: 10.1186/1475-2859-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi S, Chen Y, Siewers V, Nielsen J. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. MBio. 2014;5:e01130-14. doi: 10.1128/mBio.01130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.