Abstract
Metal homeostasis is a crucial cellular function for nearly all organisms. Some heavy metals (e.g., Fe, Zn, Co, Mo) are essential because they serve as cofactors for enzymes or metalloproteins, and chlorophototrophs such as cyanobacteria have an especially high demand for iron. At excessive levels, however, metals become toxic to cyanobacteria. Therefore, a tight control mechanism is essential for metal homeostasis. Metal homeostasis in microorganisms comprises two elements: metal acquisition from the environment and detoxification or excretion of excess metal ions. Different families of metal-sensing regulators exist in cyanobacteria and each addresses a more or less specific set of target genes. In this study the regulons of three Fur-type and two ArsR-SmtB-type regulators were investigated in a comparative approach in the cyanobacterium Synechococcus sp. PCC 7002. One Fur-type regulator controls genes for iron acquisition (Fur); one controls genes for zinc acquisition (Zur); and the third controls two genes involved in oxidative stress (Per). Compared to other well-investigated cyanobacterial strains, however, the set of target genes for each regulator is relatively small. Target genes for the two ArsR-SmtB transcriptional repressors (SmtB (SYNPCC7002_A2564) and SYNPCC7002_A0590) are involved in zinc homeostasis in addition to Zur. Their target genes, however, are less specific for zinc and point to roles in a broader heavy metal detoxification response.
Keywords: iron homeostasis, zinc homeostasis, Zur, Per, SmtB, ArsR, transcriptome profiling, photosynthesis
Introduction
Metal uptake and homeostasis are important functions for most bacteria and are very important for chlorophototrophs because of the extensive use of metals in photosynthetic complexes. Iron availability is a key issue for marine microorganisms because iron availability limits microbial growth and productivity in marine environments (Coale et al., 1996; Morel, 2008; Behrenfeld and Milligan, 2013). Fur-type transcriptional regulators are widespread among prokaryotes and are involved in regulating the uptake and homeostasis of several metal ions and related functions (e.g., oxidative stress response; Lee and Helmann, 2007; Fillat, 2014; O’Brian, 2015; Porcheron and Dozois, 2015). Control of iron uptake was the first function to be described for such a regulator, and subsequently the protein was named ferric uptake regulator (Fur) (Hantke, 1981; Bagg and Neilands, 1987a,b). Fur-type regulators are homodimers with both DNA and metal-binding domains (Bagg and Neilands, 1987a). These regulators have commonly been described as transcriptional repressors that bind to the DNA operator sites when loaded with co-repressor (Lee and Helmann, 2007; Osman and Cavet, 2010), but there are also examples in which Fur-type regulators act as transcriptional activators (Delany et al., 2001). Indirect activation of gene expression can also occur via small RNAs: the Fur-type regulator acts as a transcriptional repressor for a sRNA, and as long as the sRNA level is low, the target genes are expressed, whereas de-repression of the sRNA results in a degradation of target RNAs (Massé and Gottesman, 2002). Small RNAs have been identified to be involved in regulation of iron homeostasis in the cyanobacterium Synechocystis sp. PCC 6803 (Hernández-Prieto et al., 2012). Besides iron homeostasis, Fur-type regulators have been described for zinc, manganese and nickel homeostasis; have been described as regulators for oxidative stress responses; and play direct or indirect roles in several other cellular functions (Lee and Helmann, 2007; Fillat, 2014). The amino acid sequences of these regulators are highly similar, and subtle changes can alter the specificity of metal binding. For example, it has been demonstrated that an exchange of a single amino acid transformed an iron-responsive Fur-type regulator into a protein as sensitive to H2O2 as a natural, peroxide-sensing Fur-type regulator (Parent et al., 2013). Besides the Fur-type transcriptional regulators there are transcriptional repressors of the ArsR-SmtB family that are also involved in regulating metal homeostasis. ArsR-SmtB family repressors respond to high levels of metal ions that are stress-inducing or toxic for the cell; metal ions bind to the regulator as a de-repressor for this repressor family (Busenlehner et al., 2003; Osman and Cavet, 2010).
Synechococcus sp. strain PCC 7002 (hereafter Synechococcus 7002) is a euryhaline, unicellular cyanobacterium that is capable of growth over a wide range of NaCl concentrations and is extremely tolerant of high light intensities (Batterton and Van Baalen, 1971; Nomura et al., 2006; Bernstein et al., 2014). It is a very fast growing and well-characterized model organism that is naturally transformable, and convenient methods for the heterologous expression of genes have been developed (Stevens and Porter, 1980; Xu et al., 2011). Cyanobacteria have been shown to possess different Fur-type transcriptional regulators, and studies in various cyanobacterial strains have assigned functions and regulation patterns to single Fur-type regulators (Ghassemian and Straus, 1996; Kobayashi et al., 2004; Hernández et al., 2006; González et al., 2010; Napolitano et al., 2012). The genome of Synechococcus 7002 encodes three genes that are annotated as Fur-type regulators. In this study we performed deletion mutagenesis and, on the basis of transcriptome profiling, we assigned functions to these three transcriptional regulators of this cyanobacterium. In addition to the fur genes of Synechococcus 7002, we investigated the regulons of two transcriptional repressors of the ArsR-SmtB family that are involved in responses to heavy metal stress and toxicity.
Materials and Methods
Bacterial Strains and Culture Conditions
Wild-type and mutant strains of Synechococcus sp. PCC 7002 were grown in liquid culture and on 1.5% (w/v) agar plates in medium A supplemented with 1 mg NaNO3 ml-1 (designated as medium A+) as previously described (Stevens and Porter, 1980; Ludwig and Bryant, 2011). Liquid cultures were grown in tubes containing medium (25 mL) at 38°C with continuous irradiation with 250 μmol photons m-2 s-1, and the cultures were sparged with 1% (v/v) CO2 in air. The following antibiotic concentrations were added to the medium whenever appropriate: spectinomycin, 50 μg mL-1; erythromycin, 20 μg mL-1. Unless otherwise stated, cultures for growth rate experiments were grown without antibiotics, whereas cultures for RNA analyses were grown in the presence of the respective antibiotics. Cell growth was monitored by measuring the optical density at 730 nm (OD730 nm; OD730 nm = 1.0 is equivalent to 1.0 ± 0.2 × 108 cells mL-1) with a Genesys 10 spectrophotometer (ThermoSpectronic, Rochester, NY, USA). Cultures for RNA analyses were inoculated at an OD730 nm between 0.05 and 0.1 from pre-cultures that had been grown under the same conditions. When these cultures reached an OD730 nm = 0.7, three independently grown, replicate cultures were pooled. Cells derived from 25 mL aliquots of the cultures were rapidly centrifuged (5 min, 5000 × g, 4°C), and the cell pellets were rapidly frozen in liquid nitrogen and stored at -80°C until required.
Fe-limitation was induced by adding the Fe-binding chelator, deferoxamine mesylate B, to the culture as previously described (Ludwig and Bryant, 2012a). At OD730 nm = 0.35 deferoxamine mesylate B (720 μM final concentration) was added to a culture in A+ medium (containing 14.4 μM iron). The resulting culture was grown under standard conditions until OD730 nm = 0.7.
Medium A+ contains 2.3 μM Zn2+, and to induce zinc stress, aliquots of a ZnCl2 stock solution (100 mM ZnCl2, buffered in 1 M Tris at pH 8.0) were added to produce final Zn2+ concentrations of 12.3 μM, 22.3 μM, 52.3 μM, 102 μM, and 202 μM. Cultures subjected to Zn-limitation were grown in modified A+ medium without added ZnCl2 (ZnCl2 was omitted from the trace element solution). After several transfers on “Zn-free” A+ medium (to dilute any residual Zn2+), the heavy metal chelator TPEN was added at a final concentration of 50 μM to lower the availability of divalent cations (Anderegg et al., 1977). TPEN was prepared as a 50 mM stock solution in dimethylsulfoxide.
Construction of Knock-out And Knock-down Strains
For inactivation of SynPCC7002_A1649 (fur), SynPCC7002_A1836 (per), SynPCC7002_A2498 (zur), SynPCC7002_A0590, and SynPCC7002_A2564 (smtB), ∼1000-bp regions upstream and downstream of the respective genes were amplified by PCR and ligated to drug marker cassettes. Primers #1 and #2 were used to amplify the upstream region and #3 and #4 for amplification of the downstream region of SynPCC7002_A1649 (see Table 1 for oligonucleotide sequences). Primers #5 and #6 (upstream region) and primers #7 and #8 (downstream region) were used for amplification of SYNPCC7002_A1836 flanking regions; and primers #9 and #10 (upstream region) and primers #11 and #12 (downstream region) were used to amplify the flanking regions of SYNPCC7002_A2498, respectively. Oligonucleotides #13 and #14 and oligonucleotides #15 and #16 were used for amplification of the SYNPCC7002_A0590 upstream and downstream flanking regions; and for amplification of SYNPCC7002_A2564 flanking regions primers #17 and #18 (upstream fragment) and #19 and #20 (downstream fragment) were used. The PCR fragments were digested using the enzymes listed in Table 1. The aadA gene, conferring streptomycin and spectinomycin resistance, was excised as a 1040-bp Eco53kI/HindIII fragment from plasmid pSRA81 (Frigaard et al., 2004b) or as a 1091-bp Eco53kI fragment from plasmid pSRA2 (Frigaard et al., 2004a). The ermC gene, conferring erythromycin resistance, was excised from pRL409 (Elhai and Wolk, 1988) as a 1503-bp EcoRV-HindIII fragment. The respective fragments were mixed at a 3:1:3 ratio of the upstream flank to the antibiotic resistance cassette to the downstream flanking region and ligated with T4 DNA ligase. The ligation reactions were directly used to transform Synechococcus 7002 as described previously (Frigaard et al., 2004b).
Table 1.
Oligonucleotide primers used in this study.
| Number | Sequence (relevant restriction sites underlined, 5′ to 3′) |
|---|---|
| #1 | AAATCTAGAGCAATGGTGCTAATCTCCTC (XbaI site underlined) |
| #2 | TTTGATATCATATTTTTCAACAAGTTCAG (EcoRV site underlined) |
| #3 | AAAAAGCTTAATCGAGGCGATCGCATTTGACGGTAAC (HindIII site underlined) |
| #4 | TTTCTCGAGGCACTCCGCAGGGCTAAATC (XhoI site underlined) |
| #5 | AAATCTAGACCGCAATCTGGGTGCCGAGG (XbaI site underlined) |
| #6 | TTTGATATCAAGGCTTGTTCGTTGCAAAATC (EcoRV site underlined) |
| #7 | AAAGATATCTAACTTGTTTAAATAATCTG (EcoRV site underlined) |
| #8 | TTTCTCGAGGGGTATCGCTTTGGGCACAG (XhoI site underlined) |
| #9 | AAATCTAGATGATGGCGGGAATCCTCGTC (XbaI site underlined) |
| #10 | TTTGATATCATTAGGCTTCTGGTTGGGTG (EcoRV site underlined) |
| #11 | ACCCAAGCTTCAGGCCGATAATGCTGAATG (HindIII site underlined) |
| #12 | TTTCTCGAGATATAGGAGTGGCCATCCTC (XhoI site underlined) |
| #13 | AAATCTAGAACTGCTCGCCGCCGTGGAACAG (XbaI site underlined) |
| #14 | CCCCCCGGGCATTAAATCAAAAATAATTGATGC (SmaI site underlined) |
| #15 | AAAAAGCTTAAAACCCCAACCCAGAATATAAATC (HindIII site underlined) |
| #16 | CCCCTCGAGAGCTGCATCTTCATAGGTAAATCCC (XhoI site underlined) |
| #17 | TGCTCTAGACAAGGGCAATATGAACTGGC (XbaI site underlined) |
| #18 | TCCGATATCTTGGCATCCCCCAAGACACC (EcoRV site underlined) |
| #19 | CCCAAGCTTACACACCCACTAATCAACGC (HindIII site underlined) |
| #20 | CCGCTCGAGAAGGTTACGAAGTTCATGGC (XhoI site underlined) |
RNA Preparation, RNA Sequencing, and Data Analysis
RNA samples for cDNA library construction were prepared from frozen cell pellets derived from 25-ml aliquots of liquid culture (see above). The RNA preparation and quantitation was performed as described previously (Ludwig and Bryant, 2011), and construction of cDNA libraries and SOLiDTM sequencing was performed in the Genomics Core Facility at The Pennsylvania State University (University Park, PA). The cDNA libraries were constructed using SOLiDTM Whole Transcriptome Analysis Kit (Applied Biosystems) and were barcoded by using the SOLiDTM Transcriptome Multiplexing Kit (Applied Biosystems). SOLiDTM ePCR Kit and SOLiDTM Bead Enrichment Kit (both Applied Biosystems) were used for processing the samples for sequencing, and the SOLiDTM 4 or SOLiDTM 5500 protocol (Applied Biosystems) was used for sequencing. The sequence data have been submitted to the NCBI Sequence Read Archive (SRA) under accession numbers SRR833548, SRR1057998–SRR1058000, SRR2179721–SRR2179730.
The cDNA sequences were mapped using the Burrows–Wheeler algorithm (Li and Durbin, 2009). The Synechococcus 7002 genome and the sequences for the antibiotic resistance cassettes served as the reference set; four mismatches within the 50-bp reads were allowed (>90% sequence identity), and sequences mapping to non-unique locations were excluded. Further processing, analysis and comparisons of data sets were performed as described previously (Ludwig and Bryant, 2011). Statistical analysis was performed using the chi-square test if one data set was available for each condition or the z-test for comparisons to the WT standard sample, for which three independent data sets were available. The data for all protein-coding open reading frames (ORFs) derived from these analyses are listed in the Supplementary Tables S1 and S2.
Measurement of Reactive Oxygen (ROS) and Nitrogen Species (RNS)
The cytosolic ROS/RNS contents in cells was probed using the membrane-permeant indicator CM-H2DCFDA from Molecular Probes, Invitrogen (Eugene, OR), which reacts with ROS and RNS and is detected by fluorescence emission. The protocol recommended by the manufacturer was used. Cultures of WT and mutant strains were grown to OD730 nm = 0.7 under standard conditions, then CM-H2DCFDA was added at a final concentration of 5.0 μM, and the cultures were incubated under high irradiance (830 μmol photons m-2 s-1 (at 38°C and standard sparging conditions). For the subsequent fluorescence measurements the OD730 nm was adjusted to 0.5 for all samples and the fluorescence was measured using an SLM8000-based spectrofluorometer modernized for computerized, solid-state operation by On-Line Instrument Systems Inc., (Bogart, GA). The excitation wavelength was 493 nm, and the emission spectrum was recorded from 510 to 610 nm; the amplitude at 523 nm was used for calculations and comparisons (Zhu et al., 2010).
Results
Deletion of Genes Encoding Fur-type Regulators
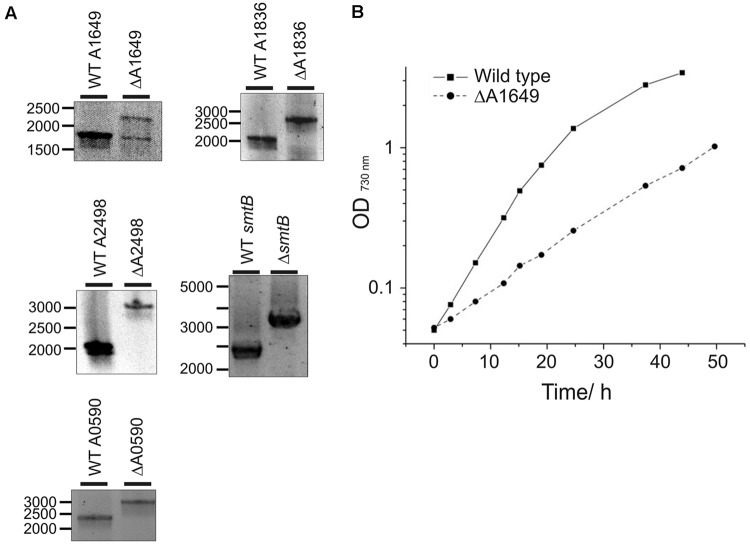
The Synechococcus 7002 genome encodes three members of the helix-turn-helix family of Fur-type metalloregulators. One gene was originally annotated as the fur gene based on sequence homology (locus tag number SYNPCC7002_A1649) (pfam 01475), whereas the other two paralogs (locus tag numbers SYNPCC7002_A1836 and SYNPCC7002_A2498) were not specifically assigned functions during annotation. To explore the function of these three transcriptional regulators, deletion mutants were constructed for all three fur homologs as described in the Section “Materials and Methods”. Wild-type and mutant alleles in transformants for the deletion of the SYNPCC7002_A1836 and SYNPCC7002_A2498 genes segregated fully, which demonstrates that the products of these genes are not essential for growth under standard conditions. However, PCR analysis of the WT and mutant alleles in transformants for the deletion of SYNPCC7002_A1649 showed a ∼1:1 ratio of WT and mutant amplicons that was stable over numerous transfers and varying light conditions; the agar plates used for selection were incubated under low irradiance levels (Figure 1A). This result indicates that SYNPCC7002_A1649 is essential for growth at least under the conditions that were tested in this study.
FIGURE 1.
Segregation analysis of deletion mutants and growth of the merodiploid SYNPCC7002_A1649 strain. (A) Electrophoretic analysis of PCR amplicons produced using DNA templates derived from the wild-type (WT) strain and SYNPCC7002_A1649 (ΔA1649), SYNPCC7002_A1836 (ΔA1836), SYNPCC7002_A2498 (ΔA2498), SYNPCC7002_A2564 (ΔsmtB), and SYNPCC7002_A0590 (ΔA0590) mutant strains of Synechococcus 7002. For the SYNPCC7002_A1649 (fur) mutant, the expected amplicon size was 1.82 kb for WT and 2.35 kb for the mutant, respectively. Note that the mutant is a stable merodiploid with approximately equal copies of each allele (fur and fur::aadA). For SYNPCC7002_A1836 (perR), the expected amplicon size was 2.12 kb for WT and 2.72 kb for the mutant. For SYNPCC7002_A2498 (zur), the expected amplicon size was 1.99 kb for WT and 3.11 kb for the deletion mutant, respectively. The amplicon sizes for smtB were 2.33 kb (WT) and 3.14 kb (mutant), and for SYNPCC7002_A0590 amplicons had 2.29 kb (WT) and 3.02 kb (mutant), respectively. (B) Growth of Synechococcus 7002 WT and the merodiploid SYNPCC7002_A1649 (fur)::aadA strain in A+ medium. The growth medium for the latter strain contained 50 μg/mL spectinomycin for continuous selection. The cultures were grown under standard growth conditions, and representative growth curves for these two strains are displayed. Growth of the other mutant strains investigated in this study was wild type-like.
Strains in which SYNPCC7002_A1836 or the SYNPCC7002_A2498 were deleted grew identically to the WT under standard temperature and light conditions in A+ growth medium (data not shown). However, the growth of the merodiploid SYNPCC7002_A1649 strain was severely impaired compared to the WT and all other mutant strains (Figure 1B). Although antibiotics were not necessary for strains deleted for SYNPCC7002_A1836 and SYNPCC7002_A2498, antibiotics had to be added to the culture medium for the SYNPCC7002_A1649 merodiploid strain to maintain selection on the drug marker that was introduced to delete the SYNPCC7002_A1649 gene. Because streptomycin was present during growth of this strain, and because the target gene was only partially deleted, calculation of a valid growth rate is not possible for this particular strain. However, in general we have found that the aadA gene and streptomycin have at most a very modest effect on the growth of mutants in Synechococcus 7002.
Transcriptome Profiling of Mutant Strains for the Fur Family
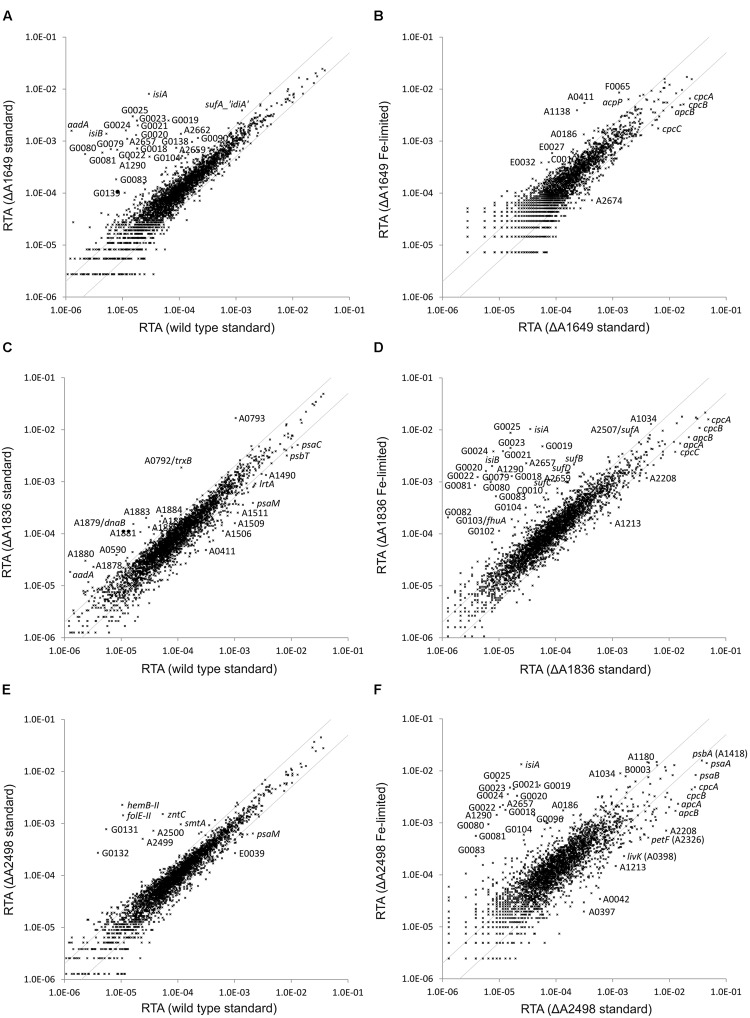
Global transcriptome profiling of the regulator mutants was performed in order to identify target genes and their functions in the regulons for each of the three Fur-type regulators. The transcriptome of the merodiploid SYNPCC7002_A1649 strain was compared to that obtained for the WT when both strains were cultivated under standard growth conditions (Figure 2A). Many genes showed much higher transcript levels in the mutant strain compared to the WT (see Supplementary Tables S2 and S3). Those genes encode proteins and enzymes involved in iron acquisition (siderophore biosynthesis, components of different types of Fe-transporters, etc.) or encode alternative proteins that replace proteins containing Fe-S clusters [e.g., isiB (flavodoxin)] (Leonhardt and Straus, 1992). Transcript levels for all of these genes have been reported to increase under iron limitation in Synechococcus 7002 (Ludwig and Bryant, 2012a), and in fact the comparison of mutant to WT under standard conditions closely resembles the comparison of an iron-limited WT profile to the WT profile under standard growth conditions (Ludwig and Bryant, 2012a). Transcript levels resulting from the SYNPCC7002_A1649 gene were about 2.5-fold lower in the merodiploid strain compared to the WT, which approximately reflects the 1:1 ratio of mutant and WT gene that was detected in the PCR analysis (Figure 1A). Fe-limitation of the merodiploid SYNPCC7002_A1649 strain had almost no effect on the global transcriptome of that strain when compared to standard growth conditions (Figure 2B). Only very few genes encoding enzymes/proteins related to iron uptake or metabolism showed further induction under Fe-limitation of a culture of the merodiploid SYNPCC7002_A1649 strain. Genes that have been previously reported to have decreased mRNA levels upon limitation of several nutrients (e.g., nitrogen, sulfur, phosphorous, and carbon) (Ludwig and Bryant, 2012a) displayed lower transcript levels in the SYNPCC7002_A1649 merodiploid strain upon Fe-limitation (e.g., cpcA, cpcB, cpcC, apcB, etc.; see Supplementary Table S3).
FIGURE 2.
Relative transcript abundances (RTAs) for the SYNPCC7002_A1649 (fur), SYNPCC7002_A1836 (perR) and SYNPCC7002_A2498 (zur) mutant strains compared to WT Synechococcus 7002 and of the mutant strains subjected to iron limitation compared to standard growth conditions. The scatter plots show (A) the RTAs of transcripts from the merodiploid SYNPCC7002_A1649 (fur) strain compared to the wild type when both strains were grown under standard conditions; (B) RTAs of the merodiploid SYNPCC7002_A1649 strain subjected to iron limitation compared to standard growth conditions; (C) the RTAs of the SYNPCC7002_A1836 (perR) deletion mutant compared to the wild type when both were grown under standard conditions; (D) the RTAs of SYNPCC7002_A1836 deletion mutant subjected to iron limitation compared to those for standard growth conditions; (E) the RTAs of the SYNPCC7002_A2498 (zur) deletion mutant strain compared to a wild type culture, both grown under standard conditions; and (F) the RTAs of an iron-limited culture of the SYNPCC7002_A2498 deletion mutant compared to a culture grown under standard conditions. The values for the wild type under standard conditions were calculated as the mean of three independent biological replicates. The gray lines show twofold changes in either direction. Selected genes are identified by gene locus or abbreviated locus tag number (e.g., G0025 = SynPCC7002_G0025). For further details, see text.
The global transcriptome of the SYNPCC7002_A1836 deletion strain did not show many differences compared to the WT when the two strains were grown under standard conditions (Figure 2C). In fact, excluding some prophage genes (see below), strongly increased transcript levels were only observed for two genes: SYNPCC7002_A0792 and SYNPCC7002_A0793. BLAST searches and searches for conserved domains revealed that SYNPCC7002_A0792 encodes thioredoxin reductase (trxB) and that SYNPCC7002_A0793 encodes peroxiredoxin. The products of these two genes form an electron transfer chain in which peroxiredoxin inactivates H2O2 by reduction (Lu and Holmgren, 2014). Oxidized peroxiredoxin receives electrons from reduced thioredoxin, which in turn receives electrons from the cellular NADPH pool via thioredoxin reductase. Interestingly, the transcript levels of thioredoxin genes, however, were not more abundant in the mutant strain compared to the WT. A gene cluster comprising several prophage genes also showed increased transcript abundance in the SYNPCC7002_A1836 mutant strain. However, we did not observe cell lysis that would indicate activation of a lytic phage in our experiments with Synechococcus 7002. In WT cells, the mRNA levels of those genes (SYNPCC7002_A1876 to SYNPCC7002_A1888) were very low, and the transcript levels were still rather low even in the mutant strain. The significance of these latter changes is not clear at this time.
Fe-limitation of a culture of the SYNPCC7002_A1836 deletion strain had a very large effect on the relative transcript abundance (RTA) (Figure 2D). Many genes had strongly increased mRNA levels compared to a culture that was grown under standard conditions. Those genes (e.g., isiA, isiB, sufA, sufB, sufD, SYNPCC7002_G0018-G0025, SYNPCC7002_G0079-G0082) have been previously reported to be induced upon Fe-limitation in the WT (Ludwig and Bryant, 2012a), and these genes are not further induced upon Fe-limitation in the SYNPCC7002_A1649 merodiploid strain because they are already de-repressed to the maximum in the knock-down strain. Another set of genes showed slightly lower transcript levels in Fe-limited cells of the SYNPCC7002_A1836 mutant (e.g., cpcA, cpcB, cpcC, apcA, apcB). These genes generally have lower transcript levels under any nutrient limitation (Ludwig and Bryant, 2012a), and their transcript levels were also lower in the SYNPCC7002_A1649 merodiploid strain. To summarize, the transcriptome profile of iron-limited SYNPCC7002_A1836 mutant was essentially the same as iron-starved WT.
The transcriptome analysis of the third deletion mutant (SYNPCC7002_A2498) also showed increased mRNA levels of some genes compared to the WT when both strains were cultivated under standard conditions (Figure 2E). Those genes are basically located in two clusters. The first cluster is located on the chromosome adjacent to the SYNPCC7002_A2498 regulator (SYNPCC7002_A2499-SYNPCC7002_A2500-SYNPCC7002_A2501 (zntC)), and these genes are predicted to encode the components of an ABC-transporter for zinc. The second cluster, composed of hemB-II, folE-II, SYNPCC7002_G0131 and SYNPCC7002_G0132 is located on plasmid pAQ7. These genes encode a delta-aminolevulinic acid dehydratase paralog, a GTP cyclohydrolase I, a conserved hypothetical protein (BLAST and conserved domain searches showed that it harbors a methylmalonyl-coenzyme A mutase domain and has similarities to GTPases), and an FAD-dependent oxidoreductase, respectively. Interestingly, two genes located within this cluster, SYNPCC7002_G0129 and SYNPCC7002_G0130, did not show higher transcript levels in the SYNPCC7002_A2498 mutant strain. SYNPCC7002_G0129 is annotated as phosphoesterase, and it has homologs in other cyanobacterial strains, whereas SYNPCC7002_G0130 does not show conserved domains and no homologs were found in other organisms. In addition to these two clusters there is a single gene, smtA, which is more highly expressed in the SYNPCC7002_A2498 deletion strain compared to the WT. The smtA gene encodes a metallothionein, which is a cysteine-rich, heavy metal-binding protein (Ruttkay-Nedecky et al., 2013).
The transcriptome of the SYNPCC7002_A2498 deletion mutant was also probed under iron limitation and compared to that of a culture grown under standard conditions (Figure 2F). This comparison closely resembles that obtained for the SYNPCC7002_A1836 deletion strain and that of the WT when comparing Fe-limitation to standard growth (see above).
Accumulation of Reactive Oxygen Species in fur Mutant Strains
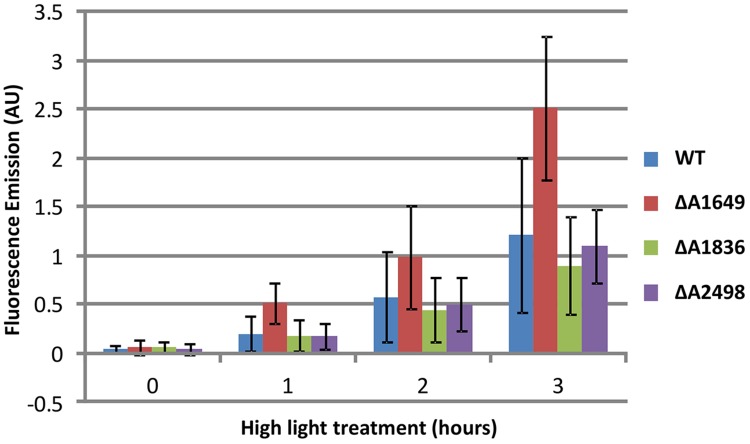
To investigate further the function of the single Fur-type regulators, the relative ability of each strain to prevent formation and accumulation of ROS/RNS was compared to the WT. Cultures of Synechococcus 7002, which are known to be tolerant to very high light intensities (Sakamoto and Bryant, 2002; Nomura et al., 2006; Bernstein et al., 2014), were exposed to a high irradiance (830 μmol photons m-2 s-1) after growth under standard conditions. This treatment results in the enhanced production of ROS/RNS species, which can be detected by a fluorescent dye as described in the Section “Materials and Methods” (also see Eruslanov and Kusmartsev, 2010; Zhu et al., 2010). The dye reacts with ROS/RNS to form a fluorescent compound, and over the course of 3 h of high irradiance treatment an increase of ROS/RNS was observed for the WT (Figure 3). The SYNPCC7002_A1836 and SYNPCC7002_A2498 deletion mutants accumulated ROS/RNS at about the same levels as the WT, whereas the merodiploid SYNPCC7002_A1649 strain displayed a strong increase of ROS/RNS accumulation compared to the WT and the other Fur-type regulator mutants. The SYNPCC7002_A1836 mutant appeared to have slightly lower ROS/RNS levels than the WT.
FIGURE 3.
Accumulation of ROS/RNS as detected by CM-H2DCFDA in cultures of Synechococcus 7002 wild type and SYNPCC7002_A1649 (fur), SYNPCC7002_A1836 and SYNPCC7002_A2498 mutant strains exposed to high light. Cultures of wild type Synechococcus 7002 and SYNPCC7002_A1649 (fur), SYNPCC7002_A1836, and SYNPCC7002_A2498 mutant strains were grown to OD730 nm = 0.7 under standard conditions, and then CM-H2DCFDA was added and the cultures were exposed to high light (830 μmol photons m-2 s-1). Samples were taken from two independent cultures and measured in duplicate after 0 h, 1 h, 2 h, and 3 h of high light incubation. The cultures were adjusted to OD730 nm = 0.5 for subsequent fluorescence emission measurements.
Zinc Tolerance and Effect of Zinc Limitation in Synechococcus 7002
To investigate the effect of zinc on Synechococcus 7002 cells, cultures were subjected to increased zinc levels and attempts were also made to induce zinc limitation. The standard A+ growth medium used for this study contains 2.3 μM Zn2+. Additional ZnCl2 (10 μM, 20 μM, 50 μM, 100 μM, and 200 μM), was added to cultures of the WT and the SYNPCC7002_A2498 mutant, and cultures were grown under otherwise standard conditions. Both the WT and the SYNPCC7002_A2498 mutant failed to grow at a Zn2+ concentration of 200 μM. At Zn2+ concentrations of 102 μM and below, both WT and mutant grew equally well, and even at 102 μM Zn2+ the WT Synechococcus 7002 showed the same growth behavior compared to growth in normal A+ medium (data not shown). Attempts to subject Synechococcus 7002 cultures to zinc limitation were performed in zinc-free A+ medium and by adding TPEN as a chelator for divalent cations. TPEN as a chelator, however, has an even higher affinity to the divalent forms of nickel, copper, cobalt and cadmium, and Fe2+ is also complexed by TPEN, though with lower affinity (Anderegg et al., 1977; Sigdel et al., 2006). However, no difference in growth of the WT and the SYNPCC7002_A2498 mutant was observed, even after several transfers.
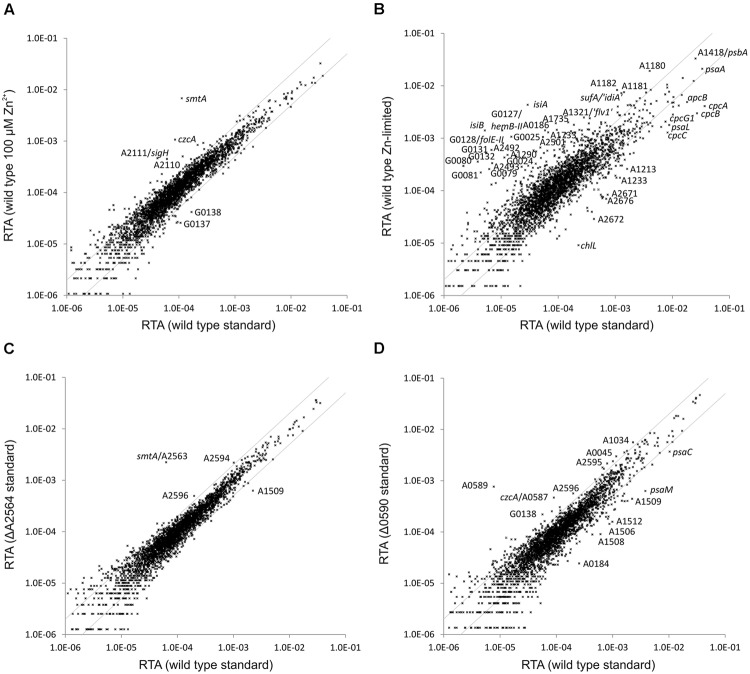
Synechococcus 7002 cultures grown at high Zn2+ concentration (100 μM) and a culture grown in the presence of 50 μM TPEN were subjected to transcriptome profiling. Very few genes showed a variation of the transcript level in the culture grown at a high zinc level compared to a culture grown in A+ medium under standard conditions (Figure 4A): smtA (encoding a metallothionein, see above), czcA (annotated as cation efflux system protein CzcA) and, interestingly, sigH (sigma factor H; SYNPCC7002_A2111) and the adjacent ORF SYNPCC7002_A2110 (hypothetical protein; not well conserved according to BLAST searches) had higher transcript levels. The sigH gene and ORF SYNPCC7002_A2110 have been reported to be co-transcribed, but the function of the latter remained obscure (Inoue-Sakamoto et al., 2007). SYNPCC7002_G0137 (annotated as MotA/TolQ/ExbB proton channel family protein, a biopolymer transport protein) and SYNPCC7002_G0138 (annotated as TonB-dependent siderophore receptor) showed lower transcript levels.
FIGURE 4.
Relative transcript abundances for Synechococcus 7002 wild type cultures grown at altered zinc levels compared to standard growth conditions and of the SYNPCC7002_A2564 (smtB) and SYNPCC7002_A0590 deletion mutants compared to the Synechococcus 7002 wild type. The scatter plots show (A) the RTAs of a wild type Synechococcus 7002 culture grown in presence of high zinc (102 μM) compared to a culture grown under standard conditions; (B) the RTAs of a culture of wild type Synechococcus 7002 culture grown under zinc limitation compared to a culture grown under standard conditions; (C) RTAs of the SYNPCC7002_A2564 (smtB) deletion mutant compared to the wild type, both grown under standard conditions; (D) RTAs of the SYNPCC7002_A0590 deletion mutant compared to a wild type culture, both grown under standard conditions. The values for the wild type under standard conditions were calculated as the mean of three independent biological replicates. The gray lines show twofold changes in either direction. Selected genes are identified by gene locus or abbreviated locus tag number. For details, see text.
The RTA pattern of a culture grown in the presence of 50 μM TPEN (to induce zinc limitation) showed more differences relative to a culture grown in A+ medium under standard conditions (Figure 4B). The pattern resembles the one obtained for iron-limited cultures: several genes related to iron uptake and low iron adaptation (e.g., isiA, isiB, SYNPCC7002_G0079–SYNPCC7002_G0081) were induced and genes encoding components of the light harvesting complex (apcB, cpcA, cpcB, cpcC) displayed a lower mRNA level. In addition there are two sets of genes hemB-II (delta-aminolevulinic acid dehydratase), folE-II (GTP cyclohydrolase), SYNPCC7002_G0131–SYNPCC7002_G0132 (a conserved hypothetical protein and a FAD-dependent oxidoreductase, respectively) located on plasmid pAQ7 and SYNPCC7002_A2499–SYNPCC7002_A2501 (encoding components of a zinc-ABC transporter) located on the chromosome having a sharply induced mRNA level upon TPEN addition. These two clusters also showed a higher expression level in the SYNPCC7002_A2498 deletion mutant strain compared to the WT (see above).
Effect of smtB Deletion on the Global Transcriptome
In presence of a high Zn2+ level and in the SYNPCC7002_A2498 deletion mutant strain (in A+ medium under standard conditions), transcript levels for smtA (encoding metallothionein; SYNPCC7002_A2563) increased dramatically. A gene annotated as metallothionein repressor (smtB; SYNPCC7002_A2564) occurs adjacent to smtA, an organization that exists for an orthologous regulator in other cyanobacterial strains, either in combination with the gene encoding metallothionein or with a zinc exporter (Morby et al., 1993; Thelwell et al., 1998). In order to investigate the regulon of this transcriptional repressor in Synechococcus 7002, the smtB gene was deleted. The WT and mutant alleles for the smtB deletion mutant segregated fully in Synechococcus 7002, and the resulting mutant strain grew similarly to the WT (data not shown). The smtB deletion mutant strain was subjected to transcriptome profiling under standard growth conditions, and the resulting transcriptome was compared to that of the WT (Figure 4C). Only a single gene (smtA) had a much higher mRNA level in the smtB deletion strain compared to the WT. Transcript levels of essentially all other genes with relatively high expression levels changed less than twofold compared to the WT. Transcript levels for SYNPCC7002_A2594 and SYNPCC7002_A2596 (both hypothetical proteins) were slightly above the twofold higher level, and transcript levels for SYNPCC7002_A1509 (annotated as putative membrane protein) were about 3.5-fold lower compared to the WT.
Effect of SYNPCC7002_A0590 Deletion on the Global Transcriptome
The czcA gene (annotated as cation efflux system protein CzcA) showed the second highest increase of the mRNA level in cells of WT Synechococcus 7002 in the presence of high Zn2+. The czcA gene is located within a gene cluster (SYNPCC7002_A0585 –SYNPCC7002_A0591) that also harbors a gene annotated as arsenical resistance operon repressor, ArsR family (SYNPCC7002_A0590). To characterize the regulon of SYNPCC7002_A0590, a deletion mutant was constructed for this gene. Mutant and WT alleles fully segregated in the transformants, showing that the product of SYNPCC7002_A0590 is not required for viability under standard growth conditions. Consistent with this, the mutant strain grew similarly to the WT in A+ medium under standard conditions (data not shown). The transcriptome of the SYNPCC7002_A0590 mutant strain was determined for a culture grown under standard conditions and compared to that of the WT (Figure 4D). Two genes within the cluster (SYNPCC7002_A0589, annotated as arsenite efflux pump ACR3, and SYNPCC7002_A0588 annotated as low molecular weight phosphotyrosine protein phosphatase), which are located just downstream of the regulator SYNPCC7002_A0590, showed a much higher expression level in the SYNPCC7002_A0590 deletion mutant compared to the WT (about 150- and 100-fold higher, respectively). The next gene within the cluster, czcA, had an about fivefold higher mRNA level in the mutant strain. Other genes located in the same gene cluster displayed only minor changes in the transcript levels in the deletion mutant relative to the WT: these are SYNPCC7002_A0591 (annotated as efflux transporter, RND family, MFP subunit), which is located upstream of SYNPCC7002_A0590, SYNPCC7002_A0586 (annotated as probable dioxygenase of extradiol dioxygenase family) and SYNPCC7002_A0585 (annotated as outer membrane efflux protein), both located downstream from czcA. A few other genes with relatively high transcript levels showed slightly higher transcript levels in the SYNPCC7002_A0590 deletion mutant compared to the WT; among those were SYNPCC7002_A2595 and SYNPCC7002_A2596, both annotated as conserved hypothetical proteins, which exhibited a ∼3-fold increase in transcript levels. Interestingly, those genes displayed slightly higher transcript levels in the smtB mutant. An elevated zinc level (100 μM zinc in the culture or SYNPCC7002_A2498 deletion), however, left the transcript level of those genes unaffected, whereas zinc limitation induced by TPEN caused ∼1.5-fold and ∼4-fold lower expression levels for SYNPCC7002_A2595 and SYNPCC7002_A2596, respectively. Among those genes that had a relatively high transcript abundance and that showed somewhat lower transcript levels in the SYNPCC7002_A0590 mutant strain relative to the WT, were SYNPCC7002_A1506 to SYNPCC7002_A1512, which showed up to ∼7-fold lower expression levels; these genes included glycosyl transferases, a pyruvyltransferase, and a sulfotransferase.
Discussion
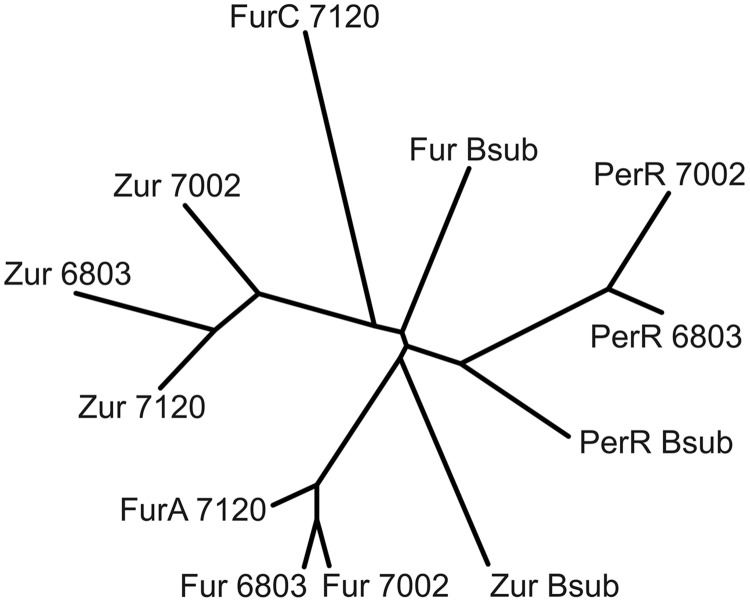
Fur-type regulators are present in many cyanobacterial strains, and target genes and functions have been assigned to specific regulators in a few cyanobacterial strains (Ghassemian and Straus, 1996; Kobayashi et al., 2004; Li et al., 2004; López-Gomollón et al., 2009; González et al., 2010, 2011, 2014; Napolitano et al., 2012). However, no previous study has investigated all of the Fur-type regulators in a single cyanobacterium using the same experimental conditions. A phylogenetic comparison of the three Fur-type regulators of Synechococcus 7002 with those of Synechocystis sp. PCC 6803, Anabaena sp. PCC 7120 and Bacillus subtilis shows that SYNPCC7002_A1649 clusters with Anabaena 7120 FurA and Fur from Synechocystis 6803 (Figure 5). SYNPCC7002_A2498 clusters with Zur from Anabaena 7120 and from Synechocystis 6803; however, the similarity is not that high compared to the first cluster comprising of Fur/FurA. The third Fur-type regulator, SYNPCC7002_A1836, is similar to PerR from Synechocystis 7002. Interestingly, the third Fur-type regulator (FurC) from Anabaena 7120 does not fall into this cluster and is distinct from the cyanobacterial clusters. As expected, the Fur-type regulators from B. subtilis, which were used as outgroups, are somewhat different from the respective cyanobacterial clusters.
FIGURE 5.
Phylogenetic tree of Fur-type regulators from three cyanobacterial strains. Amino acid sequences of the three Fur-type regulators from Anabaena 7120, Synechocystis 6803, and Synechococcus 7002 were aligned using ClustalW and a phylogenetic tree was generated using (Protein alignment, BLOSUM matrix, default settings; unrooted neighbor-joining tree with branch lengths). The three respective Fur-type regulators (Fur, Zur, and PerR) from Bacillus subtilis were used as an outgroup.
In this study the targets for these three Fur-type regulators were investigated in Synechococcus 7002, and to our knowledge this is the first transcriptome profiling study on a merodiploid knock-down strain of an iron-responsive Fur-type regulator gene. The transcriptome profiling results clearly show that all three Fur-type regulators are transcriptional repressors and that one (SYNPCC7002_A1649, annotated as fur) is an important regulator of iron homeostasis. Transcript levels of genes, which are specifically responsive to iron limitation (Ludwig and Bryant, 2012a), were maximally de-repressed in the merodiploid SYNPCC7002_A1649 strain, and expression levels of genes that respond to any other type of nutrient limitation were unchanged as long as no iron limitation was induced. Similar to the results obtained in this study, previous studies of the fur gene in other cyanobacteria reported that it was not possible to delete this gene, indicating that Fur is required for cell viability (Ghassemian and Straus, 1996; González et al., 2011). Studies in Anabaena 7120 based on in silico predictions, promoter binding experiments and proteomic studies on over-expression of FurA in its natural host revealed a number of target genes in that cyanobacterial strain (González et al., 2011, 2014). Interestingly, besides iron homeostasis those target genes in Anabaena 7120 belonged to additional functional categories, e.g., photosynthesis, respiration, heterocyst differentiation, oxidative stress response and light-dependent signal transduction mechanisms. These findings are clearly different from the results obtained here for a fur knock-down strain of Synechococcus 7002, for which the Fur regulon seems to be restricted to the principal function, iron homeostasis. The fact that we could achieve a maximal de-repression by having just a 2.5-fold lower transcript level of the regulator was surprising. Assuming that the mRNA quantity approximately translates to the protein level, this would mean that the cellular level or Fur protein is below the threshold that is required for a full transcriptional repression and that even excess iron as co-repressor could not compensate for that. A low level of the co-repressor iron – as induced in the iron limitation experiment – did not result in significantly higher de-repression, suggesting that only low levels of the functional repressor can be present in the merodiploid SYNPCC7002_A1649 strain.
Discounting the viral genes of unknown function, the second Fur-type regulator, SYNPCC7002_A1836, has only two target genes in Synechococcus 7002, which were de-repressed in the deletion mutant. Those two genes encode peroxiredoxin and thioredoxin reductase, both of which involved in detoxification of ROS. A study of a homologous gene, named perR in Synechocystis 6803, produced similar results; however, microarray analysis indicated that the number of genes in the PerR regulon of Synechocystis 6803 was much higher, and they were functionally more diverse (Li et al., 2004). On the other hand, a global transcriptome study of oxidative stress induced by treatment with methyl viologen in Synechococcus 7002 (Ludwig and Bryant, 2012b), also showed a very limited response to oxidative stress.
Deletion of the third Fur-type regulator gene (SYNPCC7002_A2498) resulted in de-repression of genes encoding zinc-transporters and enzymes that are paralogs of zinc-containing enzymes encoded elsewhere in the genome. The regulon of the zinc-uptake regulator (Zur) has been well studied in Anabaena 7120 (Napolitano et al., 2012). In Anabaena 7120 a total of 23 genes, organized in six operons and six single transcriptional units, are targets of Zur, whereas in Synechococcus 7002 the Zur regulon comprises only two clusters with a total of seven target genes. Similar to the target genes of the other two Fur-type regulators, the functional diversity of Zur targets seems to be more limited in Synechococcus 7002. Functional assignments of the target genes in Synechococcus 7002 included only a subset of those reported for Anabaena 7120 (Napolitano et al., 2012).
Based on the similarities of the three Fur-type regulators from Synechococcus 7002 to those of Anabaena 7120 and Synechocystis 6803 on the amino acid sequence level and on the functional level (target genes), we propose the following names for the three genes encoding Fur-type regulators in Synechococcus 7002: SYNPCC7002_A1649 is fur, SYNPCC7002_A1836 perR, and SYNPCC7002_A2498 is named zur.
Metal homeostasis and oxidative stress are intimately related, and thus it was not surprising that oxidative stress was exacerbated in some of the mutants produced in this study. In particular, inappropriate expression of the Fur regulon strongly exacerbated oxidative stress induced by high light. ROS/RNS levels in cells accumulated much higher in the fur merodiploid strain than in the WT after a 3-h exposure to high irradiance. This is probably due to increased iron uptake, resulting in a higher cellular Fe level, which may lead to increased ROS formation via the Fenton reaction (Kozlowski et al., 2014). This observation provides a likely explanation for why the fur gene cannot be fully deleted: the resulting intracellular iron levels, and thus the resulting ROS/RNS levels, might exceed the cellular capacity to inactivate ROS/RNS—even for an extremely stress tolerant strain like Synechococcus 7002. Inappropriate expression of the Zur regulon did not enhance ROS/RNS production in response to high irradiance. Derepression of peroxiredoxin and thioredoxin reductase in the mutant lacking PerR had at most a very small effect in reducing ROS/RNS.
Synechococcus 7002 cultures can grow in the presence of Zn2+ levels as high as 100 μM, and growth was almost unaffected up to this level. However, absolutely no growth was observed at 200 μM Zn2+. Interestingly, the zur mutant strain grew equally well compared to the WT, although transcriptome data indicated an increased expression of the zinc regulon. This probably means that detoxification mechanisms for intracellular zinc/heavy metals, metallothionein (SmtA) and the cation efflux system protein CzcA, are still able to balance the increased zinc import to maintain homeostasis. Zinc limitation could be induced in Synechococcus 7002 cultures by adding the chelator TPEN, resulting in an increase of transcript levels of genes under control of Zur. This is consistent with data obtained for zinc limitation in Anabaena 7120 (Napolitano et al., 2012). However, using TPEN as a chelator for divalent cations for Synechococcus 7002 also clearly resulted in iron limitation as indicated by increased transcript levels of genes related to iron uptake and the Fur regulon. This result is very similar to observations made in a transcriptome profiling study on metal-limitation in Escherichia coli (Sigdel et al., 2006). The lack of specificity of TPEN makes it difficult to prepare and study cells that are solely zinc-limited.
In addition to studying the regulons of the three Fur-type regulators, we investigated the targets of two other transcriptional repressors of the ArsR-SmtB family that are related to a heavy metal stress response. The SmtB transcriptional repressor of Synechococcus 7002 has only one target gene, smtA, encoding metallothionein. This regulation pattern is the same as for Synechococcus sp. PCC 7942 (Morby et al., 1993); however, in that study additional targets cannot be excluded since no global approach was performed. Synechocystis 6803 also encodes smtB, but in this cyanobacterium a zinc exporter, ziaA, is the target gene rather than smtA (Thelwell et al., 1998), also this study did not include a global screen for other potential targets.
The transcriptional regulator SYNPCC7002_A0590, annotated as an arsenical resistance repressor, ArsR, is a repressor for four genes within a cluster adjacent to SYNPCC7002_A0590 in Synechococcus 7002. The highest de-repression was observed for the two genes just downstream of the regulator, one of which encodes an arsenite efflux pump ACR3. One of the next two genes further downstream, which showed a lower level of de-repression, encodes the cation efflux system protein CzcA. This is an interesting observation, because SYNPCC7002_A0590 seems to regulate multiple functions, at least according to its target genes. The ArsR-SmtB family of transcriptional repressors displays a broad variety both with respect to the metal ions, which de-repress these regulators, and to the target genes (Busenlehner et al., 2003; Osman and Cavet, 2010). The main function for SYNPCC7002_A0590 seems to be the regulation of arsenite efflux as indicated by the high level of de-repression of the gene encoding an arsenite efflux pump. However, in Synechococcus 7002 this ArsR-type transcriptional repressor has additional target genes, although they are de-repressed to a lesser extent. Finally, the target genes of both SmtB and SYNPCC7002_A0590, smtA and czcA, respectively, are derepressed in response to elevated zinc level identifying zinc at least as one de-repressor of both transcriptional regulators. Therefore, together with Zur, these two ArsR-SmtB family regulators form a regulatory network that controls zinc homeostasis in Synechococcus 7002.
Conclusion
In this study we demonstrated the target genes of the three Fur-type regulators Fur, PerR and Zur of Synechococcus 7002, which were fewer in number compared to the respective regulons that have been reported for other cyanobacterial strains through microarray studies. Furthermore, we presented the target genes for two additional transcriptional repressors that are involved in zinc homeostasis in addition to Zur. We suggest that the Fur transcription factor is essential for cyanobacterial viability because de-repression of the Fur regulon can exacerbate oxidative stress in response to high irradiance.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Dr. Craig Praul and Candace Price for advice and oversight of cDNA sequencing in the Genomics Core Facility (Huck Institutes for the Life Sciences, Penn State University) and Dr. Zhenfeng Liu helpful discussions concerning the data processing pipeline.
Abbreviations
- CM-H2DCFDA
5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester
- PCR
polymerase chain reaction
- ROS/RNS
reactive oxygen/nitrogen species
- sRNA
small regulatory RNA
- TPEN
N,N,N’,N’-tetrakis-(2-pyridylmethyl)ethane-1,2-diamine
- WT
wild type
Footnotes
Funding. This study was supported by the Air Force Office of Scientific Research (MURI Grant FA9550-05-1-0365) and by the National Science Foundation (MCB-1021725).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01217
References
- Anderegg G., Hubmann E., Podder N. G., Wenk F. (1977). Pyridinderivate als Komplexbildner. XI. Die thermodynamik der metallkomplexbildung mit bis-, tris- und tetrakis [(2-pyridyl) methyl]-aminen. Helv. Chem. Acta 60 123–140. 10.1002/hlca.19770600115 [DOI] [Google Scholar]
- Bagg A., Neilands J. (1987a). Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26 5471–5477. 10.1021/bi00391a039 [DOI] [PubMed] [Google Scholar]
- Bagg A., Neilands J. (1987b). Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol. Rev. 51 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batterton J. C., Jr., Van Baalen C. (1971). Growth responses of blue-green algae to sodium chloride concentration. Arch. Mikrobiol. 76 151–165. 10.1007/BF00411789 [DOI] [PubMed] [Google Scholar]
- Behrenfeld M. J., Milligan A. J. (2013). Photophysiological expressions of iron stress in phytoplankton. Ann. Rev. Mar. Sci. 5 217–246. 10.1146/annurev-marine-121211-172356 [DOI] [PubMed] [Google Scholar]
- Bernstein H. C., Konopka A., Melnicki M. R., Hill E. A., Kucek L. A., Zhang S., et al. (2014). Effect of mono-and dichromatic light quality on growth rates and photosynthetic performance of Synechococcus sp. PCC 7002. Front. Microbiol. 5:488 10.3389/fmicb.2014.00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busenlehner L. S., Pennella M. A., Giedroc D. P. (2003). The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27 131–143. 10.1016/S0168-6445(03)00054-8 [DOI] [PubMed] [Google Scholar]
- Coale K. H., Johnson K. S., Fitzwater S. E., Gordon R. M., Tanner S., Chavez F. P., et al. (1996). A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean. Nature 383 495–501. 10.1038/383495a0 [DOI] [PubMed] [Google Scholar]
- Delany I., Spohn G., Rappuoli R., Scarlato V. (2001). The Fur repressor controls transcription of iron-activated and-repressed genes in Helicobacter pylori. Mol. Microbiol. 42 1297–1309. 10.1046/j.1365-2958.2001.02696.x [DOI] [PubMed] [Google Scholar]
- Elhai J., Wolk C. P. (1988). A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68 119–138. 10.1016/0378-1119(88)90605-1 [DOI] [PubMed] [Google Scholar]
- Eruslanov E., Kusmartsev S. (2010). Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 594 57–72. 10.1007/978-1-60761-411-1_4 [DOI] [PubMed] [Google Scholar]
- Fillat M. F. (2014). The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546 41–52. 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- Frigaard N.-U., Li H., Milks K. J., Bryant D. A. (2004a). Nine mutants of Chlorobium tepidum each unable to synthesize a different chlorosome protein still assemble functional chlorosomes. J. Bacteriol. 186 646–653. 10.1128/JB.186.3.646-653.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N.-U., Sakuragi Y., Bryant D. A. (2004b). Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol. Biol. 274 325–340. [DOI] [PubMed] [Google Scholar]
- Ghassemian M., Straus N. A. (1996). Fur regulates the expression of iron-stress genes in the cyanobacterium Synechococcus sp. strain PCC 7942. Microbiology 142 1469–1476. 10.1099/13500872-142-6-1469 [DOI] [PubMed] [Google Scholar]
- González A., Angarica V. E., Sancho J., Fillat M. F. (2014). The FurA regulon in Anabaena sp. PCC 7120: in silico prediction and experimental validation of novel target genes. Nucl. Acids Res. 42 4833–4846. 10.1093/nar/gku123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Bes M. T., Barja F., Peleato M. L., Fillat M. F. (2010). Overexpression of FurA in Anabaena sp. PCC 7120 reveals new targets for this regulator involved in photosynthesis, iron uptake and cellular morphology. Plant Cell Physiol. 51 1900–1914. 10.1093/pcp/pcq148 [DOI] [PubMed] [Google Scholar]
- González A., Bes M. T., Peleato M. L., Fillat M. F. (2011). Unravelling the regulatory function of FurA in Anabaena sp. PCC 7120 through 2-D DIGE proteomic analysis. J. Proteomics 74 660–671. 10.1016/j.jprot.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Hantke K. (1981). Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182 288–292. 10.1007/BF00269672 [DOI] [PubMed] [Google Scholar]
- Hernández J., López-Gomollón S., Muro-Pastor A., Valladares A., Bes M., Peleato M., et al. (2006). Interaction of FurA from Anabaena sp. PCC 7120 with DNA: a reducing environment and the presence of Mn2+ are positive effectors in the binding to isiB and furA promoters. BioMetals 19 259–268. 10.1007/s10534-005-7750-3 [DOI] [PubMed] [Google Scholar]
- Hernández-Prieto M. A., Schön V., Georg J., Barreira L., Varela J., Hess W. R., et al. (2012). Iron deprivation in Synechocystis: inference of pathways, non-coding RNAs, and regulatory elements from comprehensive expression profiling. G3 (Bethesda) 2 1475–1495. 10.1534/g3.112.003863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue-Sakamoto K., Gruber T. M., Christensen S. K., Arima H., Sakamoto T., Bryant D. A. (2007). Group 3 sigma factors in the marine cyanobacterium Synechococcus sp. strain PCC 7002 are required for growth at low temperature. J. Gen. Appl. Microbiol. 53 89–104. 10.2323/jgam.53.89 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Ishizuka T., Katayama M., Kanehisa M., Bhattacharyya-Pakrasi M., Pakrasi H. B., et al. (2004). Response to oxidative stress involves a novel peroxiredoxin gene in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 45 290–299. 10.1093/pcp/pch034 [DOI] [PubMed] [Google Scholar]
- Kozlowski H., Kolkowska P., Watly J., Krzywoszynska K., Potocki S. (2014). General aspects of metal toxicity. Curr. Med. Chem. 21 3721–3740. 10.2174/0929867321666140716093838 [DOI] [PubMed] [Google Scholar]
- Lee J.-W., Helmann J. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20 485–499. 10.1007/s10534-006-9070-7 [DOI] [PubMed] [Google Scholar]
- Leonhardt K., Straus N. A. (1992). An iron stress operon involved in photosynthetic electron transport in the marine cyanobacterium Synechococcus sp. PCC 7002. J. Gen. Microbiol. 138 1613–1621. 10.1099/00221287-138-8-1613 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Singh A. K., Mcintyre L. M., Sherman L. A. (2004). Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 186 3331–3345. 10.1128/JB.186.11.3331-3345.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Gomollón S., Sevilla E., Bes M., Peleato M., Fillat M. (2009). New insights into the role of Fur proteins: FurB (All2473) from Anabaena protects DNA and increases cell survival under oxidative stress. Biochem. J. 418 201–207. 10.1042/BJ20081066 [DOI] [PubMed] [Google Scholar]
- Lu J., Holmgren A. (2014). The thioredoxin antioxidant system. Free Rad. Biol. Med. 66 75–87. 10.1016/j.freeradbiomed.2013.07.036 [DOI] [PubMed] [Google Scholar]
- Ludwig M., Bryant D. A. (2011). Transcription profiling of the model cyanobacterium Synechococcus sp. strain PCC 7002 by NextGen (SOLiDTM) sequencing of cDNA. Front. Microbiol. 2:41 10.3389/fmicb.2011.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M., Bryant D. A. (2012a). Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front. Microbiol. 3:145 10.3389/fmicb.2012.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M., Bryant D. A. (2012b). Synechococcus sp. strain PCC 7002 transcriptome: acclimation to temperature, salinity, oxidative stress, and mixotrophic growth conditions. Front. Microbiol. 3:354 10.3389/fmicb.2012.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M., Pandelia M. E., Chew C. Y., Zhang B., Golbeck J. H., Krebs C., et al. (2014). ChlR protein of Synechococcus sp. PCC 7002 is a transcription activator that uses an oxygen-sensitive [4Fe-4S] cluster to control genes involved in pigment biosynthesis. J. Biol. Chem. 289 16624–16639. 10.1074/jbc.M114.561233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Gottesman S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99 4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morby A. P., Turner J. S., Huckle J. W., Robinson N. J. (1993). SmtB is a metal-dependent repressor of the cyanobacterial metallothionein gene smtA: identification of a Zn inhibited DNA-protein complex. Nucl. Acids Res. 21 921–925. 10.1093/nar/21.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel F. M. (2008). The co-evolution of phytoplankton and trace element cycles in the oceans. Geobiology 6 318–324. 10.1111/j.1472-4669.2008.00144.x [DOI] [PubMed] [Google Scholar]
- Napolitano M., Rubio M. Á, Santamar, ía-Gómez J., Olmedo-Verd E., Robinson N. J., Luque I. (2012). Characterization of the response to zinc deficiency in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 194 2426–2436. 10.1128/JB.00090-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura C. T., Sakamoto T., Bryant D. A. (2006). Roles for heme-copper oxidases in extreme high-light and oxidative stress response in the cyanobacterium Synechococcus sp. PCC 7002. Arch. Microbiol. 185 471–479. 10.1007/s00203-006-0107-7 [DOI] [PubMed] [Google Scholar]
- O’Brian M. (2015). Perception and homeostatic control of iron in the Rhizobia and related bacteria. Ann. Rev. Microbiol. 10.1146/annurev-micro-091014-104432 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Osman D., Cavet J. S. (2010). Bacterial metal-sensing proteins exemplified by ArsR–SmtB family repressors. Nat. Prod. Rep. 27 668–680. 10.1039/b906682a [DOI] [PubMed] [Google Scholar]
- Parent A., Caux-Thang C., Signor L., Clémancey M., Sethu R., Blondin G., et al. (2013). Single glutamate to aspartate mutation makes ferric uptake regulator (Fur) as sensitive to H2O2 as peroxide resistance regulator (PerR). Angew Chem. Int Ed. Engl. 125 10529–10533. 10.1002/ange.201304021 [DOI] [PubMed] [Google Scholar]
- Porcheron G., Dozois C. M. (2015). Interplay between iron homeostasis and virulence: Fur and RyhB as major regulators of bacterial pathogenicity. Vet. Microbiol. 179 2–14. 10.1016/j.vetmic.2015.03.024 [DOI] [PubMed] [Google Scholar]
- Ruttkay-Nedecky B., Nejdl L., Gumulec J., Zitka O., Masarik M., Eckschlager T., et al. (2013). The role of metallothionein in oxidative stress. Int. J. Mol. Sci. 14 6044–6066. 10.3390/ijms14036044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Bryant D. A. (2002). Synergistic effect of high-light and low temperature on cell growth of the delta12 fatty acid desaturase mutant in Synechococcus sp. PCC 7002. Photosynth. Res. 72 231–242. 10.1023/A:1019820813257 [DOI] [PubMed] [Google Scholar]
- Sigdel T. K., Easton J. A., Crowder M. W. (2006). Transcriptional response of Escherichia coli to TPEN. J. Bacteriol. 188 6709–6713. 10.1128/JB.00680-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S. E., Jr., Porter R. D. (1980). Transformation in Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. U.S.A. 77 6052–6056. 10.1073/pnas.77.10.6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelwell C., Robinson N. J., Turner-Cavet J. S. (1998). An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc. Natl. Acad. Sci. U.S.A. 95 10728–10733. 10.1073/pnas.95.18.10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Alvey R. M., Byrne P. O., Graham J. E., Shen G., Bryant D. A. (2011). Expression of genes in cyanobacteria: adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. Methods Mol. Biol. 684 273–293. 10.1007/978-1-60761-925-3_21 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Graham J. E., Ludwig M., Xiong W., Alvey R. M., Shen G., et al. (2010). Roles of xanthophyll carotenoids in protection against photoinhibition and oxidative stress in the cyanobacterium Synechococcus sp. strain PCC 7002. Arch. Biochem. Biophys. 504 86–99. 10.1016/j.abb.2010.07.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.