Abstract
Background:
Plasmid mediated quinolone resistance (PMQR) has been shown to play an important role in resistance not only to quinolones, but also β-lactams and aminoglycosides. In fact, qnr genes are frequently carried along with β-lactamase determinants on the same plasmids. We studied the prevalence of qnrA, qnrB, qnrS and aac(6’)-Ib-cr genes among quinolone and cephalosporin resistant clinical isolates of Klebsiella pneumoniae (K. pneumoniae), as well as the association between PMQR genes with resistance to quinolones, cephalosporins and aminoglycosides.
Methods:
The study was conducted on 79 K. pneumoniae clinical isolates collected from Imam Hussein hospital in Tehran between July 2010 and January 2011, based on their resistance to quinilones and cephalosporins. Antibacterial susceptibility was determined to 15 antibiotics by disc diffusion. Presence of qnrA, qnrB, qnrS and aac(6’)-Ib-cr genes were investigated using specific primers and PCR.
Results:
Of the 79 K. pneumoniae isolates, 47 (59.5%) carried the PMQR determinants. Among these, 42 (89.4%) carried aac(6’)-Ib-cr of which, 21 (50%) also harbored qnrB. Three isolates carried qnrB alone, two (4.2%) harbored qnrS and none had qnrA. Resistance to aminoglycosides and cephalosporins was significantly higher in the isolates carrying both qnrB and aac(6’)-Ib-cr genes compared to aac(6’)-Ib-cr alone.
Conclusion:
This study showed a high prevalence of aac(6’)-Ib-cr and qnrB genes among the Iranian K. pneumoniae clinical isolates as well as co-carriage of the two genes. There was a significant association between qnrB gene carriage and resistance to quinolones, cephalosporins, and aminoglycosides.
Keywords: Klebsiella pneumoniae, Quinolone resistance, Qnr protein, Iran
Introduction
Klebsiella pneumoniae is an opportunistic pathogen responsible for up to 10% of nosocomial infections.1 Extensive use of extended-spectrum cephalosporins, fluoroquinolones and carbapenems for treatment of these infections, has led to a significant increase of resistance in these bacteria. Resistance mechanisms such as production of β-lactamases, plasmid-mediated quinolone resistance (PMQR) and carbapanemases have caused serious therapeutic problems.2,3
Quinolones were introduced into clinical use in 1962 in the form of nalidixic acid, a fully synthetic agent with bactericidal effects on most members of Enterobacteriaceae. These broad-spectrum antibacterial agents are commonly used for treatment of infections, both in humans and in veterinary medicine. As a result, enhanced levels of quinolone resistance has occurred in recent years.4 For example, ciprofloxacin, previously shown to have excellent activity against clinical isolates of Klebsiella, has become less effective due to its extensive use.5,6 Early studies showed that quinolone resistance arises by mutations occurring in topoisomerase subunits, changes in the expression of efflux pumps and porins that provide entry for these agents into the bacterial cell.7 PMQR-mediated resistance was discovered in the late 1990s in clinical isolates of Enterobacteriaceae, including K. pneumoniae. It has been shown to play not only an important role in quinolone resistance, but also resistance to other antibiotics, particularly β-lactams and aminoglycosides.7,8 The first PMQR gene in K. pneumoniae was reported in 1998 in USA. Since then, plasmids harboring qnr genes have been found in clinical isolates of Enterobacteriaceae worldwide.8-11 PMQR determinants include: Qnr proteins (QnrA, QnrB, QnrS, QnrC, and QnrD), which protect DNA gyrase and topoisomerase IV from inhibition by quinolones, the aminoglycoside acetylteransferase, Aac(6’)-Ib-cr, and more recently, the fluoroquinolone specific efflux pump protein, QepA.12 The aac(6′)-Ib-cr (cr for ciprofloxacin resistance) is a variant of aac(6’)-Ib (responsible for resistance to kanamycin, tobramycin and amikacin) with two amino acid substitutions compared to the wild-type allowing it to acetylate and subsequently reduce the activity of norfloxacin and ciprofloxacin.13-15 Therefore, in addition to quinolone resistance, PMQR determinants can play an important role in resistance to other antibiotics, particularly β-lactams and aminoglycosides. In fact, a number of studies have shown plasmids which carry qnr genes along with various β-lactamase determinants.15,16 We studied the prevalence of qnrA, qnrB, qnrS and aac(6’)-Ib-cr genes among quinolone and cephalosporin resistant clinical isolates of Klebsiela pneumoniae, as well as the association between PMQR gene carriage with resistance to quinolones, cephalosporins and aminoglycosides.
Materials and Methods
Bacterial Isolates
Seventy-nine isolates of K. pneumoniae, which were resistant to quinolones and/or cephalosporins, were chosen from a collection of 104 clinical isolates obtained from Imam Hussein hospital in Tehran between July 2010 and January 2011. Majority of the specimens were from urine (44.3%), followed by sputum (25.3%), catheters (11.4%), wound and blood (7.6% each), and other miscellaneous sources (3.8%). All isolates were identified by conventional biochemical tests and were maintained at -20°C in brain heart infusion broth (Oxoid, UK) containing 10% (v/v) dimethyl sulfoxide (Merck, Germany) until use.
Antimicrobial Susceptibility
Susceptibility to 15 antibiotics was determined by disc diffusion using the CLSI recommendations.17 The antibiotics (Himedia, India) were: amoxicillin/clavulanic acid (AMC, 20+10 µg), cefepime (CPM, 30 µg), cefotaxime (CTX, 30 µg), ceftazidime (CAZ, 30 µg), aztreonam (ATM, 30 µg), imipenem (IPM, 10 µg), ciprofloxacin (CP, 30 µg), levofloxacin (LOM, 5 µg), norfloxacin (NOR, 10 µg), ofloxacin (OFX, 5 µg), nalidixic acid (NA, 30 µg), amikacin (AK, 30 µg), gentamicin (GM, 10 µg), kanamycin (KM, 30 µg) and nitrofurantoin (NF, 300 µg). K. pneumoniae ATCC 10031 (obtained from Persian type culture collection, Iran) was used as control for antimicrobial susceptibility.
ESBL Production
The isolates were initially screened for ESBL production by the double disc synergy test (DDST) using cefotaxime (30 µg), ceftazidime (30 µg) and cefepime (30 µg), placed 20 mm (center to center) from an amoxicillin/clavulanic acid disc (20+10 µg). ESBL production was detected when synergy was observed between the inhibition zones of cephalosporins and amoxicillin/clavulanic acid and was further confirmed by the phenotypic confirmatory test (PCT) using ceftazidime and cefotaxime alone or in combination with clavulanic acid.18,19
DNA Extraction and PCR Amplifications
Extraction of plasmid DNA was performed using an improved phenol/chloroform method where the cells were directly lysed by phenol.20 Presence of qnrA, qnrB, qnrS and aac(6’)-Ib-cr genes were detected by PCR using the primers shown in table 1.21,22
Table 1.
Primers used for detection of qnr and aac (6’)-Ib-cr genes
| Gene | Primer | Primer sequence | Product size (bps) | Reference |
|---|---|---|---|---|
| qnrA | Forward | TTCTCACGCCAGGATTTGAG | 571 | (21) |
| Reverse | TGCCAGGCACAGATCTTGAC | |||
| qnrB | Forward | TGGCGAAAAAATTGAACAGAA | 594 | (21) |
| Reverse | GAGCAACGATCGCCTGGTAG | |||
| qnrS | Forward | GACGTGCTAACTTGCGTGAT | 388 | (21) |
| Reverse | AACACCTCGACTTAAGTCTGA | |||
| aac (6’)-Ib-cr | Forward | TTGCGATGCTCTATGAGTGGCTA | 482 | (22) |
| Reverse | CTCGAATGCCTGGCGTGTTT |
PCR reaction mixtures (25 µl) contained 1.5 µl DNA template, 1.5 mM MgCl2, 0.25 mM of dNTP mix, 1 unit of DFS-Taq DNA polymerase (Bioron, Germany) and 20 pmol of each primer. PCR amplifications were performed in a thermal cycler (Bioer TC25/H, Bioer Technology, China) using the following program. Initial denaturation at 94ºC for 5 min followed by 30 cycles of 1 min at 94ºC, 1 min at annealing temperature (57ºC for qnrA, qnrB and qnrS, 54ºC for aac(6’)-Ib-cr genes), 1 min at 72ºC and a final extension period of 10 min at 72ºC. The amplified PCR products were resolved by electrophoresis on 1.5% agarose gel and visualized after staining with ethidium bromide. Antibiotic resistance rates in PMQR harboring isolates were analyzed by the two-way ANOVA test.
Results
Antibiotic resistance rates (including intermediate resistance) of the isolates were: amoxicillin/clavulanic acid (100%), ceftazidime (81%), cefotaxime (74.7%) cefepime (72.1%), aztreonam (73.4%), nitrofurantoin (59.5%), kanamycin (78.5%), gentamicin (53.2%), amikacin (20.2%), norfloxacin (74.7%), ciprofloxacin (58.2%), nalidixic acid (45.6%), ofloxacin (43%) and levofloxacin (38%). All isolates were susceptible to imipenem.
Overall, PMQR determinants were found in 47 isolates (59.5%). figure 1 shows the amplification product of the qnr genes. Among these, 24 isolates (30.4%) carried qnrB of which, the majority (n=21, 87.5%) also co-harbored the aac(6’)-Ib-cr gene. Two isolates (2.5%) harbored qnrS and none carried the qnrA gene. figure 2 presents the PCR product of the aac(6’)-Ib-cr gene. Forty two isolates (53.2%) carried aac(6’)-Ib-cr of which, half also had the qnrB as mentioned above. There was no association between the specimen source and gene carriage (data not shown).
Figure 1.
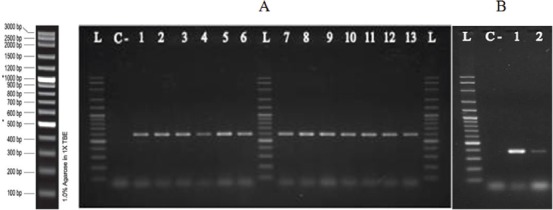
Shows gel electrophoresis of qnrB (A; lanes 1-6, and 7-13) and qnrS (B: lanes 1 and 2) genes in some isolates of K. pneumoniae by PCR. L: 1Kb Ladder, C-: negative control.
Figure 2.
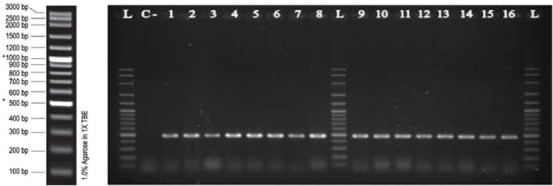
Shows gel electrophoresis of aac(6’)-Ib-cr gene in some isolates of K. pneumoniae by PCR (lanes 1-8 and 9-16). L: 1Kb Ladder, C-: negative control.
Four K. pneumoniae isolates were detected as ESBL producers, half of which harbored qnrS and the other two carried aac(6’)-Ib-cr.
figure 3 compares the antibiotic resistance profiles of the K. pneumoniae isolates carrying qnrB alone, with strains harboring qnrB+aac(6’)-Ib-cr genes. As observed, resistance to nalidixic acid, ciprofloxacin, levofloxacin, norfloxacin, cefotaxime, ceftazidime, cefepime, gentamycin, amikacin and kanamycin were almost twice as high in the isolates carrying both genes compared to the isolates, which harbored qnrB alone.
Figure 3.
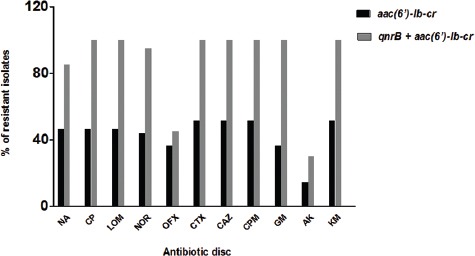
Shows comparison of antibiotic resistance profiles in K. pneumoniae isolates carrying only qnrB (black bars) with the isolates which harbored both qnrB+aac(6’)-Ib-cr (grey bars). NA: Nalidixic acid, CP: Ciprofloxacin, LOM: Levofloxacin, NOR: Norfloxacin, OFX: Ofloxacin, CTX: Cefotaxime, CAZ: Ceftazidime, CPM: Cefepime, GM: Gentamycin, AK: Amikacin, KM: Kanamycin.
Discussion
Presence of PMQR determinants in quinolone resistant E. coli and K. pneumoniae is increasingly reported worldwide. Most of these studies report a higher prevalence of PMQR genes in K. pneumoniae compared to E. coli.3,16,23-27 However, there are variations found in PMQR carriage rates from different parts of the world, mostly because the selection of the test bacteria is based on resistance to different antibiotics (e.g., nalidixic acid, ciprofloxacin). Despite that, most studies have shown a higher rate of aac(6’)-Ib-cr carriage compared to qnr genes.3,16,21-26 We found a high rate of qnrB and aac(6’)-Ib-cr co-carriage among our isolates. These results are supported by the results of previous studies.3,23,25,27 Co-carriage of qnr and aac(6’)-Ib-cr genes have been reported.3,23,25,27 Presence of qnrB and aac(6’)-Ib-cr has also been shown in K. pneumoniae, but not at the high frequency which was found in our study.27 Interestingly, the isolates which carried both qnrB and aac(6’)-Ib-cr were significantly more resistant to aminoglycosides, cephalosporins and quinolones (P<0.0001) in comparison to the isolates that harbored the aac(6’)-Ib-cr alone, suggesting the important role of qnrB. Additional resistance mechanisms such as mutations in chromosomal gyrA and gyrC genes or presence of efflux pumps such as QepA, can also be responsible for the observed high resistance levels.12,28
The association of PMQR determinants with ESBL production has been shown by a number of investigators. However, the rates of PMQR carriage vary considerably in ESBL producing Enterobacteriaceae (mostly E. coli and K. pneumoniae). For example, PMQR genes were observed in 48% of the isolates in Thailand, 30.5% in Canada, 10% in Australia, 4.9% in Spain and 1.6% in France.9,16,27,29,30 Our results showed 5.1% PMQR carriage in ESBL producing K. pneumoniae similar to the results of the Spanish study. Co-carriage of PMQR determinants along with ESBL-encoding genes on conjugative plasmids have also been reported by a number of investigators.16,21,23,29,30
We did not find any reports on the presence of PMQR genes in K. pneumoniae isolates from Iran. Other Iranian studies have shown the presence of qnr genes in E. coli and Salmonella.31,32 To our knowledge, the only other report on qnr gene carriage in Iranian clinical isolates of K. pneumoniae was a poster presentation which showed that qnrB was the predominant gene (13.4%) followed by qnrA and qnrS (1.2% each). Presence of aac(6′)-Ib-cr was not determined in that study, but co-presence of qnrB and qnrS was observed in one isolate (1.2%).33 Our results are important in presenting the prevalence of PMQR determinants (specifically aac(6′)-Ib-cr and qnrB) in clinical isolates of K. pneumoniae and their association with multiple drug resistance. However, due to the lack of research in this area, there is no basis for comparison of these results with other local or national studies. Hence, further research should be carried out in different parts of Iran to obtain a realistic rate of PMQR gene carriage on the national level. Considering the fact that these genes are often carried on mobile genetic elements and could easily be spread among the members of Enterobacteriaceae, such information is needed for choosing a proper antibiotic therapy and may prevent the dissemination of these resistance determinants among important Gram-negative pathogens.
Conclusion
The results of this study showed a high prevalence of aac(6’)-Ib-cr and qnrB, as well as a high rate of co-carriage of the two genes among the K. pneumoniae isolates in Tehran. There was a significant association between aac(6’)-Ib-cr and qnrB co-carriage with resistance to quinolones, cephalosporins and aminoglycosides.
Acknowledgment
The authors wish to thank the Shahid Beheshti University Research Council for providing the financial support of this research.
Conflicts of Interest: None declared.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11:589–603. doi: 10.1128/cmr.11.4.589. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–86. doi: 10.1055/s-2003-37919. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karah N, Poirel L, Bengtsson S, Sundqvist M, Kahlmeter G, Nordmann P, et al. Plasmid-mediated quinolone resistance determinants qnr and aac(6’)-Ib-cr in Escherichia coli and Klebsiella spp. from Norway and Sweden. Diagn Microbiol Infect Dis. 2010;66:425–31. doi: 10.1016/j.diagmicrobio.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–40. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 5.Fuursted K, Schumacher H. Significance of low-level resistance to ciprofloxacin in Klebsiella pneumoniae and the effect of increased dosage of ciprofloxacin in vivo using the rat granuloma pouch model. J Antimicrob Chemother. 2002;50:421–4. doi: 10.1093/jac/dkf148. [DOI] [PubMed] [Google Scholar]
- 6.Cheung TK, Chu YW, Chu MY, Ma CH, Yung RW, Kam KM. Plasmid-mediated resistance to ciprofloxacin and cefotaxime in clinical isolates of Salmonella enterica serotype Enteritidis in Hong Kong. J Antimicrob Chemother. 2005;56:586–9. doi: 10.1093/jac/dki250. [DOI] [PubMed] [Google Scholar]
- 7.Hooper DC. Mechanisms of quinolone resistance. In: Hooper DC, Rubinstein E, editors. Quinolone antimicrobial agents. 3rd ed. Washington DC: Am Soc Microbiol Press; 2003. pp. 41–67. [Google Scholar]
- 8.Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–9. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 9.Poirel L, Cattoir V, Nordmann P. Is plasmid-mediated quinolone resistance a clinically significant problem? Clin Microbiol Infect. 2008;14:295–7. doi: 10.1111/j.1469-0691.2007.01930.x. [DOI] [PubMed] [Google Scholar]
- 10.Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci U S A. 2002;99:5638–42. doi: 10.1073/pnas.082092899. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Martinez L, Eliecer Cano M, Manuel Rodriguez-Martinez J, Calvo J, Pascual A. Plasmid-mediated quinolone resistance. Expert Rev Anti Infect Ther. 2008;6:685–711. doi: 10.1586/14787210.6.5.685. [DOI] [PubMed] [Google Scholar]
- 12.Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2007;51:3354–60. doi: 10.1128/AAC.00339-07. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vetting MW, Park CH, Hegde SS, Jacoby GA, Hooper DC, Blanchard JS. Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6’)-Ib and its bifunctional, fluoroquinolone-active AAC(6’)-Ib-cr variant. Biochemistry. 2008;47:9825–35. doi: 10.1021/bi800664x. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006;12:83–8. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 15.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6’)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006;50:3953–5. doi: 10.1128/AAC.00915-06. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavilla S, Gonzalez-Lopez JJ, Sabate M, Garcia-Fernandez A, Larrosa MN, Bartolome RM, et al. Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J Antimicrob Chemother. 2008;61:291–5. doi: 10.1093/jac/dkm448. [DOI] [PubMed] [Google Scholar]
- 17.Franklin R, Cockerill Performance Standards for Antimicrobial Susceptibility Testing, Twenty-First Informational Supplement M100-S21. Clinical and Laboratory Standard Institute. 2011;31:68–80. [Google Scholar]
- 18.Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008;14:90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- 19.Health Protection Agency. Laboratory detection and reporting of bacteria with extended spectrum β-lactamases. National standard method QSOP 51. London, United Kingdom: Health Protection Agency; 2008. pp. 1–13. [Google Scholar]
- 20.Cheng HR, Jiang N. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett. 2006;28:55, 9. doi: 10.1007/s10529-005-4688-z. [DOI] [PubMed] [Google Scholar]
- 21.Bouchakour M, Zerouali K, Gros Claude JD, Amarouch H, El Mdaghri N, Courvalin P, et al. Plasmid-mediated quinolone resistance in expanded spectrum beta lactamase producing Enterobacteriaceae in Morocco. J Infect Dev Ctries. 2010;4:779–803. doi: 10.3855/jidc.796. [DOI] [PubMed] [Google Scholar]
- 22.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006;50:2872–4. doi: 10.1128/AAC.01647-05. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Zhou Z, Qian Y, Wei Z, Yu Y, Hu S, et al. Plasmid-mediated quinolone resistance determinants qnr and aac(6’)-Ib-cr in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J Antimicrob Chemother. 2008;61:1003–6. doi: 10.1093/jac/dkn063. [DOI] [PubMed] [Google Scholar]
- 24.Briales A, Rodriguez-Martinez JM, Velasco C, de Alba PD, Rodriguez-Bano J, Martinez-Martinez L, et al. Prevalence of plasmid-mediated quinolone resistance determinants qnr and aac(6’)-Ib-cr in Escherichia coli and Klebsiella pneumoniae producing extended-spectrum beta-lactamases in Spain. Int J Antimicrob Agents. 2012;39:431–4. doi: 10.1016/j.ijantimicag.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Deepak RN, Koh TH, Chan KS. Plasmid-mediated quinolone resistance determinants in urinary isolates of Escherichia coli and Klebsiella pneumoniae in a large Singapore hospital. Ann Acad Med Singapore. 2009;38:1070–3. [PubMed] [Google Scholar]
- 26.Magesh H, Kamatchi C, Vaidyanathan R, Sumathi G. Identification of plasmid-mediated quinolone resistance genes qnrA1, qnrB1 and aac(6’)-1b-cr in a multiple drug-resistant isolate of Klebsiella pneumoniae from Chennai. Indian J Med Microbiol. 2011;29:262–8. doi: 10.4103/0255-0857.83910. [DOI] [PubMed] [Google Scholar]
- 27.Pitout JD, Wei Y, Church DL, Gregson DB. Surveillance for plasmid-mediated quinolone resistance determinants in Enterobacteriaceae within the Calgary Health Region, Canada: the emergence of aac(6’)-Ib-cr . J Antimicrob Chemother. 2008;61:999–1002. doi: 10.1093/jac/dkn068. [DOI] [PubMed] [Google Scholar]
- 28.Fendukly F, Karlsson I, Hanson HS, Kronvall G, Dornbusch K. Patterns of mutations in target genes in septicemia isolates of Escherichia coli and Klebsiella pneumoniae with resistance or reduced susceptibility to ciprofloxacin. APMIS. 2003;111:857–66. doi: 10.1034/j.1600-0463.2003.1110904.x. [DOI] [PubMed] [Google Scholar]
- 29.Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob Agents Chemother. 2005;49:3523–5. doi: 10.1128/AAC.49.8.3523-3525.2005. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Martinez JM, Poirel L, Pascual A, Nordmann P. Plasmid-mediated quinolone resistance in Australia. Microb Drug Resist. 2006;12:99–102. doi: 10.1089/mdr.2006.12.99. [DOI] [PubMed] [Google Scholar]
- 31.Pakzad I, Ghafourian S, Taherikalani M, Sadeghifard N, Abtahi H, Rahbar M, et al. qnr Prevalence in Extended Spectrum Beta-lactamases (ESBLs) and None-ESBLs Producing Escherichia coli Isolated from Urinary Tract Infections in Central of Iran. Iran J Basic Med Sci. 2011;14:458–64. [ PMC Free Article] [PMC free article] [PubMed] [Google Scholar]
- 32.Saboohi R, Siadat SD, Aghasadeghi MR, Razavi MR, Rajaei B, Sepehri Rad N, et al. Molecular Detection of qnrA, qnrB and qnrS Resistance Genes among Salmonella spp. in Iran. Current Research in Bacteriology. 2012;5:24–30. [Google Scholar]
- 33.Azadpour M, Solymani Y. Prevalence of qnr genes in Klebsiella pneumoniae in Khorramabad, Iran. Poster presentation in the 14th International Congress of Microbiology. 2013 Aug 28-30; Tehran, Iran. [Google Scholar]


