Abstract
Radiation and microgravity exposure have been proven to induce abnormal apoptosis in microRNA (miRNA) and mRNA expression, but whether space conditions, including radiation and microgravity, activate miRNAs to regulate the apoptosis is undetermined. For that purpose, we investigated miRNome and mRNA expression in the ced-1 Caenorhabditis elegans mutant vs the wild-type, both of which underwent spaceflight, spaceflight 1g-centrifuge control and ground control conditions during the Shenzhou-8 mission. Results showed that no morphological changes in the worms were detected, but differential miRNA expression increased from 43 (ground control condition) to 57 and 91 in spaceflight and spaceflight control conditions, respectively. Microgravity altered miRNA expression profiling by decreasing the number and significance of differentially expressed miRNA compared with 1 g incubation during spaceflight. Alterations in the miRNAs were involved in alterations in apoptosis, neurogenesis larval development, ATP metabolism and GTPase-mediated signal transduction. Among these, 17 altered miRNAs potentially involved in apoptosis were screened and showed obviously different expression signatures between space conditions. By integrated analysis of miRNA and mRNA, miR-797 and miR-81 may be involved in apoptosis by targeting the genes ced-10 and both drp-1 and hsp-1, respectively. Compared with ground condition, space conditions regulated apoptosis though a different manner on transcription, by altering expression of seven core apoptotic genes in spaceflight condition, and eight in spaceflight control condition. Results indicate that, miRNA of Caenorhabditis elegans probably regulates apoptotic gene expression in response to space environmental stress, and shows different behavior under microgravity condition compared with 1 g condition in the presence of space radiation.
Keywords: apoptosis, spaceflight, microRNA, core apoptotic gene
INTRODUCTION
The space environment, which is characterized by intense radiation, microgravity and other factors, profoundly differs from the environment on Earth [1, 2]. Space radiation and microgravity are recognized as the primary and inevitable risk factors for humans traveling in space [3–6]. Previous studies report that the damage to living organisms occurring during spaceflight is often concomitant with abnormal apoptosis at cellular and/or molecular levels [7–10]. Apoptosis is vital for development, tissue formation and the survival of multicellular organisms, because it exerts protective effects against inflammatory responses or autoimmune diseases in tissues by eliminating abnormal or unnecessary cells [11] and maintaining genomic stability during DNA damage response (DDR) [12, 13]. Although ground-based studies report a variety of modality induced by ionizing radiation (IR) or modeled microgravity (MMG) on apoptosis [14–18], the modality by which microgravity incubation affects IR-induced apoptosis remains controversial [19–22].
MicroRNAs (miRNAs), a class of small non-coding RNA, mediate post-transcriptional regulation of specific target mRNAs in various cellular processes [23]. Several studies have reported that IR induces changes in miRNA expression, in vitro and in vivo, according to cell type, radiation dose and linear energy transfer (LET) [24–26], and suggested that miRNA expression is regulated in DDR, including apoptosis [27]. Changes in miRNAs, e.g. miR-9, 150, also represent the post-transcriptional responses to microgravity using human lymphoblastoid cells [28, 29]. Girardi et al. found that eight miRNAs were deregulated by the combined action of radiation and MMG [24], suggesting that miRNA expression represents environmental specificity. While the response of miRNAs involved in apoptosis during spaceflight has not been studied yet, it should give important information about the risks of the exposure to space environment.
It is noteworthy that the core apoptotic pathway includes decision, execution, engulfment and DNA degradation [12]. Therefore, there are some limitations to studying integrated apoptosis alterations in an individual metazoan by observing the response of cellular material. Caenorhabditis elegans (C. elegans) is a well-characterized metazoan in the field of apoptosis study, and abundant research with C. elegans has established the conserved core apoptotic machinery under genetic control [12, 30]. C. elegans also has been used for several space biological studies and shown good tolerance during spaceflights [31, 32]. Research has shown that the transcriptional profiling of C. elegans could alter in response to spaceflight conditions [33]; mutant strains responded to space conditions in a different manner compared with wild-type C. elegans [34–36]. However, the checkpoints and physiological apoptosis in germ cells proceeded normally, both in the ced-1 mutant and wild-type C. elegans [31]. We have also investigated the mRNA and miRNA expression profiling by using diapause dauer larvae of C. elegans. Our previous results have shown changes in miRNA expression profiling in response to different space conditions [37], as well as in the expression of non-core apoptotic genes [38]. Given the apoptotic gene expression dynamics regulated by miRNAs [39, 40], miRNAs of C. elegans were speculated to regulate the cellular tolerance/resistance to apoptosis during spaceflight.
In the present study, we focus on the changes in apoptosis induced by space conditions at the post-transcriptional level, and the difference in post-transcriptional regulation between wild-type and an abnormal apoptotic mutant during spaceflight. For these purposes, we investigated the differences in miRNA expression profiling and in core apoptotic mRNA expression between ced-1 mutant and wild-type C. elegans at the dauer stage. By using diapause dauer larvae, which present better resistance to reactive oxygen species (ROS) damage and genomic instability [41–43], we can avoid the influence of metagenesis and the different development stages during spaceflight. In C. elegans, CED-1 triggers a signaling pathway in phagocytic cells that promotes cell corpse engulfment, phagosome maturation and apoptotic cell degradation [44, 45]. Dysfunctional mutation of the ced-1 gene produces weak defects but does not totally block cell engulfing [46], because ced-1 functioning with ced-6-ced-7 forms a partially redundant pathway with ced-2-ced-5-ced-12 for controlling engulfment [12].
MATERIALS AND METHODS
Sample preparation and spaceflight experiments
The wild-type strain and ced-1 (n1995) of C. elegans were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN, USA). As described in our previous study [38], worms were maintained on solid nematode growth medium (NGM) [47] and approximately 105 synchronized dauer larvae were separately loaded into static and 1g-centrifuge experimental containers (Table 1). During the 16.5-day Shenzhou-8 mission (1–17 November 2011), the environment was maintained at a temperature of 23 ± 0.5°C, and a relative humidity of 20.79–56.35%. Space radiation doses were measured at 1.92 mGy (static slot) and 2.27 mGy (centrifuge slot). The corresponding ground control conditions were performed in parallel at the Payload Integration Test Centre in Beijing two days later. Seven hours after landing, several worms were transfer to new NGM with E. coli OP50 for morphology observation (L4 stage) after ∼10 h [47], and others were collected and kept in liquid nitrogen for further studies.
Table 1.
Experimental groups setting and indicated meaning
| No. | Group | Treatment | Radiationa | Microgravity |
|---|---|---|---|---|
| 1 | Spaceflight | Spaceflight static slot | 1.92 mGy | μg |
| 2 | Spaceflight control | Spaceflight 1g-centrifuge slot | 2.27 mGy | 1g |
| 3 | Ground control | Ground static slot | – | 1g |
aSpace radiations dose was measured by the thermoluminescent detector during spaceflight.
Total RNA isolation
As described in our previous study, about 2000 worms from each group were collected, and total RNA was isolated using InvitrogenTM TRIzol (Invitrogen, Carlsbad, CA, USA) and the miRNeasy mini kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions [38]. Quality and purity of the RNA preparations were assessed with the OD260/OD280 ratio and quantification of the ratios of 28S:18S ribosomal RNA using the NanoDrop 2000 (Thermo Fisher, Wilmington, DE, USA) and the GelDoc-ItTM 310 Imaging System (UVP, Cambridge, CA, EUA), respectively.
miRNA expression analysis and target prediction
The NimbleGen Gene Expression Profiling service and miRCURY™ LNA Array microRNA Expression Profiling service were performed by KangChen Bio-tech Inc. (Shanghai, China) as previously described [38]. Differentially expressed miRNAs (>1.5-fold change and <0.67) and mRNAs (>2-fold change and <0.5) were identified through fold-change filtering. To predict the putative target genes of miRNA, computational analysis was performed with at least two combinations of MicroRNA (www.microrna.org/microrna/home.do), Microcosm Targets (Version 5, www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), PicTar (pictar.mdc-berlin.de/cgi-bin/new_PicTar_nematode.cgi?species = nematode) and Targetscan Worm 6.2 (www.targetscan.org/worm_52/). We performed a gene ontology (GO) analysis using the DAVID tool (david.abcc.ncifcrf.gov/) to identify which biological processes were significantly enriched among the target genes (P < 0.05).
Apoptotic miRNA identification and anti-correlated analysis of miRNA and genes
To predict which C. elegans miRNAs were involved in the apoptosis process, we used the miRò tool (ferrolab.dmi.unict.it/miro/index.php) to screen human miRNAs involved in apoptosis by GO analysis, then compared the similarity of mature miRNA sequences between human and C. elegans according to Ibanez's study in order to obtain potential apoptotic miRNAs in C. elegans [48]. To identify the most likely targets, we integrated gene and miRNA expression data obtained for the same biological sample to screen the anti-correlated putative pairs of miRNA and mRNA.
Verification of microarray data by qRT-PCR
The data of the mRNA and of the miRNA microarray were validated by quantitative real-time polymerase chain reaction (qRT-PCR). Total RNA independent from that used in microarrays was extracted from ∼2000 worms with InvitrogenTM TRIzol (Invitrogen, Carlsbad, CA, USA). qRT-PCR was performed with the SuperScript® III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and SYBR® Green Master Mix (Applied Biosystems, Foster City, CA, USA) as described in previous studies [38]. The miRNA levels were adjusted to that of internal standard U6, and the results were statistically analyzed by the 2−△△Ct method.
RESULTS
Morphology observation
As shown in Fig. 1, after a 16.5-day spaceflight and ∼10 h development on ground, dauer larvae could molt to the L4 stage. There were no obvious or microscopic morphological abnormalities observed in the wild-type or ced-1 mutant worms.
Fig. 1.
Image of L4 stage larva of C. elegans after spaceflight experiment. Scale bars represent 100 μm. N2, wild-type; ced-1, ced-1 mutant strain. SF, spaceflight; SC, spaceflight control; GC, ground control. Due to malfunctions CCD device in the spaceflight landing scene, the images showed darker background brightness with white noise when checking “N2 SC” and “ced-1 SF” worm.
Analysis of miRNA expression profiling and target genes
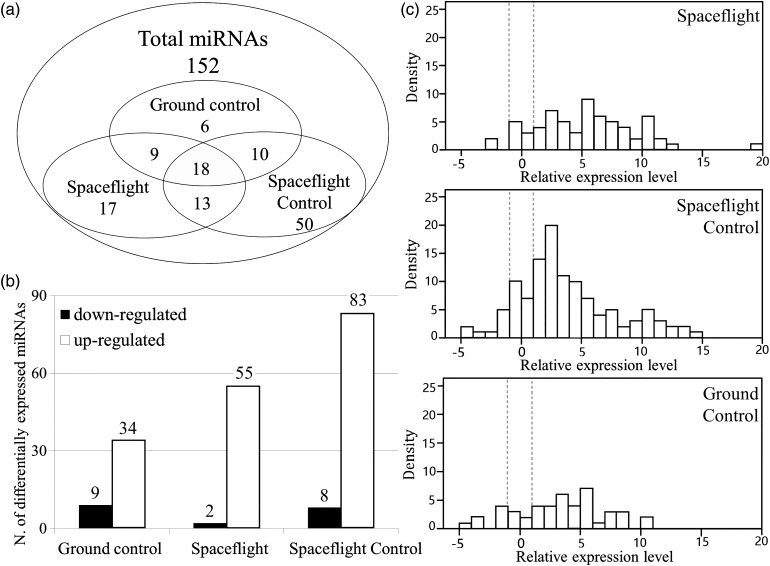
miRNA expression profiling was carried out in the ced-1 mutant strain vs wild-type that underwent spaceflight (SF) condition, spaceflight control (SC) condition and ground control (GC) condition. The results showed that 43 out of 152 miRNAs in the microarray were differentially expressed in the GC condition, and the number of differentially expressed miRNAs increased to 57 and 91 in SF and SC conditions, respectively (Fig. 2a). Greater than 80% of the altered miRNAs were upregulated in each condition (Fig. 2b and c). The distribution frequency of the miRNA expression profiling indicated that space conditions differentially enhanced the number and the significance of the expression level affected by the ced-1 mutation in ground samples, while microgravity combined with space radiation decreased them compared with spaceflight 1g incubation (Fig. 2c).
Fig. 2.
Analysis of miRNA profiling of ced-1 mutant C. elegans under different conditions. (a) Venn diagram depicts the numbers of miRNAs being significantly affected by spaceflight (SF), spaceflight control (SC) and ground control (GC), with overlaps apparent among these groups. (b) The number of upregulated and downregulated miRNAs in each condition. (c) Distribution frequency of the 152 miRNAs affected by ced-1 mutation in different conditions.
To analyze the biological processes regulated by miRNA, we predicted the target genes of differentially expressed miRNAs and determined the most enriched biological processes by GO analysis in relation to the different conditions. The results showed that nematode larval development was the most enriched biological process in each condition, and other processes differed depending on the conditions. In GC conditions, with the exception of nematode larval development and purine nucleoside triphosphate metabolic processes, genes involved in cation transport and ATP synthesis coupled with proton transport were regulated by downregulated miRNAs, and positive regulation of growth rate and cell part morphogenesis was regulated by upregulated miRNAs (Fig. 3a). There were four biological processes, including nematode larval development common to SF and SC conditions (Fig. 3b), small GTPase-mediated signal transduction, cation transcription and intracellular signaling cascades, which were enriched only in SF conditions. In SC conditions, enrichment occurred in categories of neuronal generation, cell migration, cell part morphogenesis and endocytosis.
Fig. 3.
GO analysis of putative target genes regulated by altered miRNAs. (a) The biological processes affected by upregulated miRNA (white) and downregulated miRNA (black) in ground group. (b) The biological processes regulated by altered miRNAs in spaceflight group (blue) and spaceflight control group (red).
Integrated analysis of miRNA and genes involved in apoptosis
In order to investigate the effects of spaceflight on apoptosis transcriptionally, we predicted the miRNAs involved in apoptosis and investigated known apoptotic gene expression profiling. By sequence similarity comparison with human miRNAs, 17 differentially expressed miRNAs related to apoptosis were identified (Table 2). By integrated analysis of these 17 miRNA and putative target genes, we found that miR-81 and 797 were anti-correlated to apoptotic genes drp-1, hsp-1 and ced-10 (Fig. 4).
Table 2.
Analysis of sequence similarity of miRNA involved in apoptosis between C.elegans and Homo sapiens
| cel-miR | Change fold (log 1.5) |
hsa- miR | Seq. blast (5′-3′, top-cel, bottom-hsa) | Pairing% | ||
|---|---|---|---|---|---|---|
| SF | SC | GC | ||||
| miR-1 | 7.69 | / | / | miR-1 | UGGAAUGUaaagaaguaugua UGGAAUGUaaagaaguauguau |
100 |
| miR-256 | 5.12 | 2.23 | −4.76 | miR-1 | UGGAAUGCauagaagacugua UGGAAUGUaaagaaguauguau |
81.0 |
| miR-796 | 2.30 | −0.52 | −1.98 | miR-1 | UGGAAUGUaguugagguuaguaa UGGAAUGUaaagaaguauguau |
69.6 |
| miR-48 | / | 13.83 | / | let-7c | UGAGGUAGgcucaguagaugcga UGAGGUAGuagguuguaugguu |
47.8 |
| miR-84 | 3.11 | 7.00 | 3.21 | let-7c | UGAGGUAGuauguaauauuguaga UGAGGUAGuagguuguaugguu |
75.0 |
| miR-795 | −2.19 | 5.13 | 1.95 | let-7c | UGAGGUAGauugaucagcgagcuu UGAGGUAGuagguuguaugguu |
41.6 |
| miR-81 | 10.31 | 6.87 | 10.04 | miR-143 | UGAGAUCAucgugaaagcuagu UGAGAUGAagcacuguagcuc |
63.6 |
| miR-82 | 11.95 | 11.62 | 7.96 | miR-143 | UGAGAUCAucgugaaagccagu UGAGAUGAagcacuguagcuc |
54.5 |
| miR-787 | 0.22 | −3.84 | −1.42 | miR-99a | UAAGCUCGuuuuaguaucuuucg CAAGCUCGcuucuaugggucug |
60.9 |
| miR-73 | / | 7.06 | 7.65 | miR-31 | UGGCAAGAuguaggcaguucagu AGGCAAGAugcuggcauagcu |
65.2 |
| miR-235 | / | −1.30 | 8.63 | miR-92a | UAUUGCACucuccccggccuga UAUUGCACuugucccggccugu |
81.8 |
| miR-34 | / | 7.42 | / | miR-34 | AGGCAGUGugguuagcugguug UGGCAGUGucuuagcugguugu |
86.4 |
| miR-791 | 6.39 | / | / | miR-96 | UUUGGCACuccgcagauaaggcaa UUUGGCACuagcacauuuuugcu |
58.3 |
| miR-83 | 7.40 | 6.72 | 5.53 | miR-29a | UAGCACCAuauaaauucaguaa UAGCACCAucugaaaucgguua |
68.1 |
| miR-90 | 4.99 | 6.54 | 5.14 | miR-190 | UGAUAUGUuguuugaaugccccu UGAUAUGUuugauauauuaggu |
60.9 |
| miR-124 | 1.40 | 0.17 | −1.15 | miR-124 | GCAUGCACccuagugacuuuagu UAAGGCACgcggugaaugcc |
47.8 |
| miR-797 | / | 1.63 | / | miR-499 | / | a |
cel = C. elegans, hsa = Homo sapiens, Seq. = mature miRNA sequence, SF = spaceflight, SC = spaceflight control, GC = ground control, Pairing = the number of matched base in mature cel-miRNA/the total number of base in mature cel-miRNA, ‘/’ = The low intensity differentially expressed miRNAs are filtered though median normalization method. The seed sequence are capitalized. a cel-miR-797 and hsa-miR-499 were identified with 5′ end sequence homology [48].
Fig. 4.
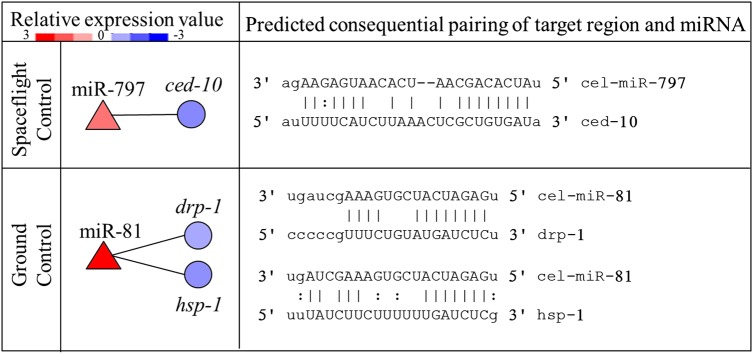
Integrated analysis of differentially expressed miRNA and target genes involved in apoptosis. The left column shows relative expression level of altered miRNA (red triangle) and the anti-correlated target genes (blue circle), and the right shows the predicted consequential pairing of target gene region and miRNAs.
Analysis of apoptotic gene expression
Gene expression profiling of core apoptotic machinery showed that 10 of 26 genes functioning at different stages in the apoptotic pathway were differentially expressed, including ced-4, ced-9, ced-1, ced-10, cdc-42, tra-1 and unc-73 in GC samples, eor-2 in the SF samples, and tra-1 and ced-10 in the SC samples (Table 3). Importantly, space conditions altered the expression value of genes in ground condition. According to the difference in the relative gene expression between space conditions and ground condition (difference value ≥1), seven genes were altered in SF condition, and eight genes in SC condition. Except for ced-4, most of the altered genes were upregulated by SF or SC conditions, indicating that space conditions could alter apoptosis on transcription.
Table 3.
The genes expression of core apoptotic pathway in different conditions
| Gene | Description | Ensembl ID | Change fold (log 2) |
||||
|---|---|---|---|---|---|---|---|
| SF | SC | GC | |||||
| Decision | |||||||
| eor-1 | egl-1 suppressor/ DiO uptake defective/ Raf enhancer, BTB /zinc-finger transcription factor | R11E3.6 | −0.39 | 0.18 | −0.16 | ||
| eor-2 | egl-1 suppressor/ DiO uptake defective/ Raf enhancer | C44H4.7 | 1.09 | ↑ | 0.00 | −0.35 | |
| tra-1 | Transformer, repress egl-1 to inhibit neurons apoptosis | Y47D3A.6 | 0.08 | ↑ | 1.55 | ↑ | −1.09 |
| cep-1 | C. elegans P53-like protein, an ortholog of p53 | F52B5.5 | 0.14 | ↑ | 0.95 | ↑ | −0.82 |
| Execution | |||||||
| ced-3 | Cell death abnormality, a cysteine-aspartate caspase, | C48D1.2 | 0.46 | −0.31 | 0.39 | ||
| ced-4 | Cell death abnormality, adaptor | C35D10.9 | −0.21 | ↓ | −0.33 | ↓ | 1.47 |
| ced-9 | Cell death abnormality, cell-death inhibitor | T07C4.8 | −0.56 | 0.80 | ↑ | −1.08 | |
| Engulfment | |||||||
| ced-1 | Cell death abnormality, transmembrane protein for phagocytosis | Y47H9C.4 | −0.37 | 0.38 | ↑ | −1.18 | |
| ced-2 | Cell death abnormality, adaptor for phagocytosis | Y41D4B.13 | −0.03 | −0.20 | −0.87 | ||
| ced-5 | Cell death abnormality, scaffolding protein | C02F4.1 | 0.32 | −0.26 | −0.34 | ||
| ced-6 | Cell death abnormality, transmembrane protein for phagocytosis | F56D2.7 | 0.18 | −0.22 | 0.43 | ||
| ced-7 | Cell death abnormality, transporter protein | C48B4.4 | −0.74 | 0.18 | ↑ | −0.84 | |
| ced-10 | Cell death abnormality, GTPase for phagocytosis | C09G12.8 | −0.14 | ↑ | −1.23 | −1.40 | |
| ced-12 | Cell death abnormality, PH-domian protein | Y106G6E.5 | −0.30 | 0.26 | 0.00 | ||
| pat-2 | Paralysed arrest at two- fold, alpha integrin subunit | F54F2.1 | −0.27 | −0.13 | 0.65 | ||
| pat-3 | Paralysed arrest at two- fold, beta-integrin subunit | ZK1058.2 | 0.07 | 0.18 | 0.03 | ||
| cdc-42 | Cell division cycle related, Rho GTPase | R07G3.1 | −0.17 | ↑ | 0.14 | ↑ | −2.02 |
| mig-2 | Abnormal cell migration, Rho family of GTP-binding proteins | C35C5.4 | −0.05 | −0.28 | −0.01 | ||
| unc-73 | Uncoordinated, guanine nucleotide exchange factor | F55C7.7 | 0.12 | ↑ | 0.18 | ↑ | −1.15 |
| nex-1 | An nexin family, phospholipid binding proteins | ZC155.1 | −0.13 | 0.25 | −0.46 | ||
| psr-1 | Phosphatidylserine Receptor family | F29B9.4 | 0.06 | −0.07 | 0.54 | ||
| dyn-1 | Dynamin related | C02C6.1 | −0.37 | 0.13 | −0.06 | ||
| DNA degradation | |||||||
| nuc-1 | Abnormal Nuclease, DNase II | C07B5.5 | −0.21 | −0.38 | −0.18 | ||
| cps-6 | CED-3 protease suppressor, mitochondrial endonuclease | C41D11.8 | −0.25 | 0.06 | −0.93 | ||
| wah-1 | Worm AIF homolog, oxidoreductase | Y56A3A.32 | −0.01 | 0.02 | 0.21 | ||
| crn-1 | Cell-death-related nuclease, 5′-3' exonuclease and endonuclease activity | Y47G6A.8 | 0.12 | 0.19 | −0.57 | ||
DiO= diet-induced obesity, Raf = Rapidly Accelerated Fibrosarcoma, BTB= Broad complex, Tramtrack and Bric-a-Brac, GLI= glioblastoma, AIF = apoptosis inducing factor, SF= spaceflight, SC= spaceflight control, GC= ground control.
Gene was upregulated (↑) induced by SF/ SC condition when ( change fold SF/SC- change fold GC) >1; gene was downregulated(↓) when (change fold SF/SC- change fold GC) < 1.
Verification of microarray expression levels by qRT-PCR
The microarray data from miRNA expression profiling was validated by qRT-PCR for five miRNAs, including miR-55 (GC), miR-56 (SC), miR-73 (SC), miR-84 (SF) and miR-124 (SF) (Table 4). From the independent worm samples used in the microarray, the qRT-PCR experiment showed that miR-84 and 73 were upregulated (>1.5-fold change), respectively; miR-55, 56 and 124 were unchanged (1.5 >fold change >0.67). The gene expression profiling was also validated by qRT-PCR, as carried out in our previous study [38].
Table 4.
Validation of microarray results by using qRT-PCR
| miRNA | Primer (5′→3′) | Group | Relative expression (mean ± sd) |
|
|---|---|---|---|---|
| microarray | qRT-PCR | |||
| U6 | F:GGAACAATACAGAGAAGATTAGCA | SF/SC/GC | 1 | 1 |
| cel-miR-55 | GSP:GCCCATACCCGTATAAGTTTCT | GC | 1.55 | 0.91 ± 0.12 |
| cel-miR-56 | GSP:GCCACTACCCGTAATGTTTCC | SC | 2.45 | 1.02 ± 0.09 |
| cel-miR-73 | GSP:GGTAAGGCACGCGGTGA | SC | 17.53 | 2.43 ± 0.64 |
| cel-miR-84 | GSP:GGGGGGTGAGGTAGTATGTAAT | SF | 3.53 | 2.53 ± 1.17 |
| ced-miR-124 | GSP:GGATGGCAAGATGTAGGCAG | SF | 1.77 | 0.75 ± 0.32 |
GSP = gene specific primer, SF = spaceflight, SC = spaceflight control, GC = ground control.
DISCUSSION
Whether space radiation and microgravity activate miRNAs to regulate apoptosis in vivo during spaceflight is undetermined. In the present study, we analyzed the differences in miRNA and core apoptotic gene expression profiles between wild-type and ced-1 mutant C. elegans in response to space radiation and/or microgravity, by investigating dauer C. elegans under spaceflight, spaceflight 1g-control and ground control conditions during a 16.5-day spaceflight.
Previous studies have shown that worms can mate, reproduce, develop and undergo radiation-induced mutations during spaceflight experiments [32, 49]. In the present study, the morphology of wild-type and ced-1 mutant space-flown worms was not changed (Fig. 1). This is likely due to the resistance of somatic cells to apoptosis, both under normal developmental conditions and during radiation [12, 50], as well as to the high resistance of dauer larvae to ROS damage and genomic instability [41–43]. However, the miRNA expression profiling showed differences between the ced-1 mutant strain and the wild-type in GC condition, and space conditions increased the number and significance of differentially expressed miRNAs (Fig. 2). The altered miRNA regulated nematode development, growth and energy metabolic process in each condition (Fig. 3). These results are consistent with results from previous gene array studies on spaceflight using human lymphoblastoid cells [17], mice [51] and C. elegans [33], indicating metabolic and developmental perturbations may be conserved to assess the risk induced by spaceflight [33].
Similar to the results reported for irradiated human lymphocytes [24] and our previous study using wild-type C. elegans [38] (Supplementary Table 2), we found that microgravity decreased the number of miRNAs when compared with 1 g condition in the presence of radiation, and it is speculated to induce different categories of biological processes between SF and SC conditions. In the SF samples, most of the target genes enriched in intracellular signaling cascades were involved in small GTPase-mediated signal transduction. While in the SC samples, miRNA regulated endocytosis, cell migration and neurons compared with the response in GC samples (Supplementary Table 1). Small GTPases, including Ras, Rho, Rab, ARF and Ran, are a family of hydrolase enzymes that can bind and hydrolyze GTP and can determine the temporal aspects of a broad variety of signaling events involved in numerous cellular processes and responses [52]. Studies have shown that both microgravity and radiation modulated the cytoskeleton by regulating the activation of Rho [53–55]; Ras-related small GTPases regulated apoptosis and DNA repair in an irradiated cell line and in irradiated C. elegans [56, 57]. CED-10 within an engulfing cell is also a small GTPase [58]. Therefore, small GTPase signaling transduction may be activated by miRNA in response to SF condition, including space radiation and microgravity stress. The results in the SC samples mainly relate to the apoptotic process, suggesting that apoptosis plays a vital role in neurogenesis during development [59], and that being exposed to space radiation may enhance the impact resulting from engulfment defects in the ced-1 mutant C. elegans [46, 60]. In general, we speculate that microgravity could alter the miRNA expression signature [24, 29] and the regulation of small GTPase-mediated signal transduction when compared with 1 g incubation in the presence of space radiation.
Focusing on apoptosis, we first screened 17 altered miRNAs related to apoptosis by comparison to the sequence similarity of human apoptotic miRNAs, since core apoptotic pathways and most miRNAs are well conserved from C. elegans to human beings [61, 62]. As shown in Table 2, the obvious differences in the miRNA expression signature between the three groups indicates that spaceflight might regulate apoptosis at the post-transcriptional level, and that microgravity may be involved in the apoptotic process, combined with space radiation. Worth noting is that cel-miR-1/256/796, cel-miR-48/84/795 and cel-miR-81/82 have high homology to hsa-miR-1, hsa-let-7 and hsa-miR-143, which have been demonstrated to function in apoptosis in response to IR or microgravity [63–68]. Other human miRNAs having sequence similarity to C. elegans were also proven to be involved either directly or indirectly in apoptosis [39, 69–72]. In order to refine the functional miRNA–mRNA relationship, we performed integrated analysis of the miRNA and mRNA expression profiles and found two pairs of significantly anti-correlated miRNA and apoptotic genes, including miR-797 and ced-10 in SC conditions and miR-81 and drp-1, hsp-1 in GC conditions (Fig. 4). cel-miR-797 has a high sequence similarity to hsa-miR-499 [48], which could protect cardiomyocytes from H2O2-induced apoptosis and regulate the expression of the tumor suppressors Forkhead box O 4 (FOXO4) and programmed cell death 4 (PDCD4) [69, 73]; the target gene ced-10 functions with ced-2 and -5 to promote formation of the polarized cell extensions associated with cell corpse engulfment during apoptosis [58]. cel-miR-81 may regulate apoptosis according to the sequence-similar hsa-miR-143, which has been shown to regulate the apoptotic process by affecting MAPK/Extracellular signal-regulated kinase (ERK) signaling transduction and the expression of Bcl-2, pro-caspase-3 and -9 [56, 57]; the target gene drp-1, encoding dynamin-related protein 1 (DRP-1), regulates distinct cell-death execution pathways downstream of ced-3 and independent of ced-9 [74]; another target gene hsp-1 encodes Hsp70A, which is involved in receptor-mediated endocytosis and apoptosis by GO identification. In addition, a previous study reported that the heat shock protein HSP72 accumulated in response to the space environment in goldfish [75]. Hence, we speculated that the apoptotic response to space radiation might be attributable to ced-10-mediated engulfment regulated by miR-797, while miR-81 might be involved in apoptosis by regulating drp-1 and hsp-1.
It has been reported that checkpoints and physiological apoptosis in germ cells proceed normally in space-flown C. elegans by evaluating the expression of apoptotic execution and checkpoint genes [31]. Ground-based studies have reported that MMG inhibited apoptosis in irradiated lymphocyte cells by delaying double-strand break re-joining or amplifying ROS impacts [20, 21]. However, the transcriptional response of the integrated core apoptotic pathway to space conditions in vivo has not been documented. In our study, we found that space conditions could regulate various stages of apoptosis. Possibly due to the resistance to ROS and genomic instability in dauer larvae, there was little difference between the ced-1 mutant and the wild-type larvae in space conditions. When compared with GC samples, space conditions both altered decision and execution of apoptosis and promoted engulfment transcriptionally (Table 3). Upregulated genes acting in the engulfment stage indicated that apoptotic engulfment were promoted in both space conditions, but genes in other stages showed different change manners. In SC condition, apoptosis was more likely to decrease according to the expression of execution genes ced-4, and ced-9 and decision gene tra-1, although the pro-apoptotic gene cep-1 was upregualted. Alteration tendency of apoptosis in SF condition was not determined, because the opposing regulation by cep-1, eor-2, and tra-1. It is speculated that these changes on apoptosis result from interaction of environmental factors and genetic factors. Ground-based study has shown that apoptosis is reduced by protons but induced by γ-rays at the gene expression level during long-term spaceflight [76], indicating that the composition of complex types of space radiation may contribute to the perturbation of apoptosis. Environmentally, although the dose of space radiation is relative low in our study, the effect of space radiation can't be denied, because space radiation has been proven to induce obvious effects, even though the dose is three orders of magnitude lower than that of ground-based simulation [36]. Microgravity also may contribute to the complex regulation of apoptosis in the presence of space radiation [21, 22]. Genetically, combined with the changes in miRNA, increased engulfment, affected by miR-797, may help protect against the abnormal apoptosis induced by space conditions. The results suggest that there may be a feedback loop for the apoptotic process to promote cell survival, supplied by miRNAs and apoptotic genes [39, 40], e.g. miR-81, or by the different stages of apoptosis, e.g. decision, execution and engulfment. Further study on the function of miRNAs and target genes during apoptosis will be performed. In generally, apoptosis might undergo an adaptive modification at the transcription level in response to space conditions [77], and miRNA could be involved in different stages of the process.
In conclusion, miRNAs are probably involved in apoptosis during short-term spaceflight. Space conditions increase changes in the expression signature of miRNAs that could affect development, energy metabolism, apoptosis and signaling transduction in C. elegans. In comparable 1g incubation in the presence of space radiation, microgravity decreases the number and significance of differentially expressed miRNAs, and regulates the expression signature of potential apoptotic miRNAs and genes acting in the decision, execution and engulfment stages in the core apoptotic pathway.
SUPPLEMENTARY DATA
Supplementary data are available at the Journal of Radiation Research online.
FUNDING
This work was supported by the National Natural Science Foundation of China (No. 31270903) and Fundamental Research Funds for the Central Universities (No. 3132014306). Funding to pay the Open Access publication charges for this article was provided by the National Natural Science Foundation of China (No.31270903).
Supplementary Material
REFERENCES
- 1.Ohnishi T, Takahashi A, Ohnishi K. Studies about space radiation promote new fields in radiation biology. J Radiat Res 2002;43 Suppl:S7–12. [DOI] [PubMed] [Google Scholar]
- 2.Horneck G. Impact of microgravity on radiobiological processes and efficiency of DNA repair. Mutat Res 1999;430:221–8. [DOI] [PubMed] [Google Scholar]
- 3.Fumio Yatagai NI. Are biological effects of space radiation really altered under the microgravity environment. Life Sci Space Res 2014;3:76–89. [Google Scholar]
- 4.Honda Y, Honda S, Narici M, et al. Spaceflight and ageing: reflecting on Caenorhabditis elegans in space. Gerontology 2014;60:138–42. [DOI] [PubMed] [Google Scholar]
- 5.Maalouf M, Durante M, Foray N. Biological effects of space radiation on human cells: history, advances and outcomes. J Radiat Res 2011;52:126–46. [DOI] [PubMed] [Google Scholar]
- 6.Nichols HL, Zhang N, Wen X. Proteomics and genomics of microgravity. Physiol Genomics 2006;26:163–71. [DOI] [PubMed] [Google Scholar]
- 7.Novoselova EG, Lunin SM, Khrenov MO, et al. Changes in immune cell signalling, apoptosis and stress response functions in mice returned from the BION-M1 mission in space. Immunobiology 2015;220:500–9. [DOI] [PubMed] [Google Scholar]
- 8.Naumenko VS, Kulikov AV, Kondaurova EM, et al. Effect of actual long-term spaceflight on BDNF, TrkB, p75, BAX and BCL-XL genes expression in mouse brain regions. Neuroscience 2015;284:730–6. [DOI] [PubMed] [Google Scholar]
- 9.Reschke MF, Cohen HS, Cerisano JM, et al. Effects of sex and gender on adaptation to space: neurosensory systems. J Womens Health (Larchmt) 2014;23:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi A, Suzuki H, Omori K, et al. Expression of p53-regulated proteins in human cultured lymphoblastoid TSCE5 and WTK1 cell lines during spaceflight. J Radiat Res 2012;53:168–75. [DOI] [PubMed] [Google Scholar]
- 11.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell 2010;140:619–30. [DOI] [PubMed] [Google Scholar]
- 12.Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol 2006;7:97–108. [DOI] [PubMed] [Google Scholar]
- 13.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature 2000;408:433–9. [DOI] [PubMed] [Google Scholar]
- 14.Cucinotta FA, Durante M. Cancer risk from exposure to galactic cosmic rays: implications for space exploration by human beings. Lancet Oncol 2006;7:431–5. [DOI] [PubMed] [Google Scholar]
- 15.Mao XW, Pecaut MJ, Stodieck LS, et al. Spaceflight environment induces mitochondrial oxidative damage in ocular tissue. Radiat Res 2013;180:340–50. [DOI] [PubMed] [Google Scholar]
- 16.Pietrofesa RA, Turowski JB, Arguiri E, et al. Oxidative lung damage resulting from repeated exposure to radiation and hyperoxia associated with space exploration. J Pulm Respir Med 2013;3:1–25. [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis ML, Reynolds JL, Cubano LA, et al. Spaceflight alters microtubules and increases apoptosis in human lymphocytes (Jurkat). FASEB J 1998;12:1007–18. [DOI] [PubMed] [Google Scholar]
- 18.Maier JA, Cialdai F, Monici M, et al. The impact of microgravity and hypergravity on endothelial cells. Biomed Res Int 2015;2015:434803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manti L. Does reduced gravity alter cellular response to ionizing radiation? Radiat Environ Biophys 2006;45:1–8. [DOI] [PubMed] [Google Scholar]
- 20.Mognato M, Girardi C, Fabris S, et al. DNA repair in modeled microgravity: double strand break rejoining activity in human lymphocytes irradiated with gamma-rays. Mutat Res 2009;663:32–9. [DOI] [PubMed] [Google Scholar]
- 21.Dang B, Yang Y, Zhang E, et al. Simulated microgravity increases heavy ion radiation-induced apoptosis in human B lymphoblasts. Life Sci 2014;97:123–8. [DOI] [PubMed] [Google Scholar]
- 22.Canova S, Fiorasi F, Mognato M, et al. “Modeled microgravity” affects cell response to ionizing radiation and increases genomic damage. Radiat Res 2005;163:191–9. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girardi C, De Pitta C, Casara S, et al. Analysis of miRNA and mRNA expression profiles highlights alterations in ionizing radiation response of human lymphocytes under modeled microgravity. PLoS One 2012;7:e31293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao A, Liu Y, Zhang H, et al. microRNA expression and biogenesis in cellular response to ionizing radiation. DNA Cell Biol 2014;33:667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva MF, Khokhar AR, Qureshi MZ, et al. Ionizing radiations induce apoptosis in TRAIL resistant cancer cells: in vivo and in vitro analysis. Asian Pac J Cancer Prev 2014;15:1905–7. [DOI] [PubMed] [Google Scholar]
- 27.Tessitore A, Cicciarelli G, Del Vecchio F, et al. MicroRNAs in the DNA damage/repair network and cancer. Int J Genomics 2014;2014:820248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girardi C, De Pitta C, Casara S, et al. Integration analysis of microRNA and mRNA expression profiles in human peripheral blood lymphocytes cultured in modeled microgravity. Biomed Res Int 2014;2014:296747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangala LS, Zhang Y, He Z, et al. Effects of simulated microgravity on expression profile of microRNA in human lymphoblastoid cells. J Biol Chem 2011;286:32483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson GA, Jones TA, Chesnut A, et al. Radiation-induced gene expression in the nematode Caenorhabditis elegans. J Radiat Res 2002;43 Suppl:S199–203. [DOI] [PubMed] [Google Scholar]
- 31.Higashitani A, Higashibata A, Sasagawa Y, et al. Checkpoint and physiological apoptosis in germ cells proceeds normally in spaceflown Caenorhabditis elegans. Apoptosis 2005;10:949–54. [DOI] [PubMed] [Google Scholar]
- 32.Adenle AA, Johnsen B, Szewczyk NJ. Review of the results from the International C . elegans first experiment (ICE-FIRST). Adv Space Res 2009;44:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selch F, Higashibata A, Imamizo-Sato M, et al. Genomic response of the nematode Caenorhabditis elegans to spaceflight. Adv Space Res 2008;41:807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adachi R, Takaya T, Kuriyama K, et al. Spaceflight results in increase of thick filament but not thin filament proteins in the paramyosin mutant of Caenorhabditis elegans. Adv Space Res 2008;41:816–23. [Google Scholar]
- 35.Higashibata A, Szewczyk NJ, Conley CA, et al. Decreased expression of myogenic transcription factors and myosin heavy chains in Caenorhabditis elegans muscles developed during spaceflight. J Exp Biol 2006;209:3209–18. [DOI] [PubMed] [Google Scholar]
- 36.Hartman PS, Hlavacek A, Wilde H, et al. A comparison of mutations induced by accelerated iron particles versus those induced by low earth orbit space radiation in the FEM-3 gene of Caenorhabditis elegans. Mutat Res 2001;474:47–55. [DOI] [PubMed] [Google Scholar]
- 37.Xu D, Gao Y, Huang L, et al. Changes in miRNA expression profile of space-flown Caenorhabditis elegans during Shenzhou-8 mission. Life Sci Space Res 2014;1:44–52. [DOI] [PubMed] [Google Scholar]
- 38.Gao Y, Xu D, Zhao L, et al. Effects of microgravity on DNA damage response in Caenorhabditis elegans during Shenzhou-8 spaceflight. Int J Radiat Biol 2015;91:531–9. [DOI] [PubMed] [Google Scholar]
- 39.Kato M, Paranjape T, Muller RU, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C . elegans and in vitro in human breast cancer cells. Oncogene 2009;28:2419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver BP, Zabinsky R, Weaver YM, et al. CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C . elegans. Elife 2014;3:e04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerisch B, Antebi A. Hormonal signals produced by DAF-9/cytochrome P450 regulate C . elegans dauer diapause in response to environmental cues. Development 2004;131:1765–76. [DOI] [PubMed] [Google Scholar]
- 42.Burnell AM, Houthoofd K, O'Hanlon K, et al. Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans. Exp Gerontol 2005;40:850–6. [DOI] [PubMed] [Google Scholar]
- 43.Onodera A, Yanase S, Ishii T, et al. Post-dauer life span of Caenorhabditis elegans dauer larvae can be modified by X-irradiation. J Radiat Res 2010;51:67–71. [DOI] [PubMed] [Google Scholar]
- 44.Yu X, Odera S, Chuang CH, et al. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell 2006;10:743–57. [DOI] [PubMed] [Google Scholar]
- 45.Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol 2008;6:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C . elegans. Cell 2001;104:43–56. [DOI] [PubMed] [Google Scholar]
- 47.Stiernagle T. Maintenance of C. elegans. In: WormBook: The Online Review of C. elegans Biology. Pasadena (CA): WormBook; 2006. Available from: http://www.ncbi.nlm.nih.gov/books/NBK19649/ (22 April 2015, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ibanez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS One 2008;3:e2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szewczyk NJ, Tillman J, Conley CA, et al. Description of International Caenorhabditis elegans Experiment first flight (ICE-FIRST). Adv Space Res 2008;42:1072–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gartner A, Milstein S, Ahmed S, et al. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C . elegans. Mol Cell 2000;5:435–43. [DOI] [PubMed] [Google Scholar]
- 51.Allen DL, Bandstra ER, Harrison BC, et al. Effects of spaceflight on murine skeletal muscle gene expression. J Appl Physiol (1985) 2009;106:582–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Claing A. beta-Arrestins: modulators of small GTPase activation and function. Prog Mol Biol Transl Sci 2013;118:149–74. [DOI] [PubMed] [Google Scholar]
- 53.Higashibata A, Imamizo-Sato M, Seki M, et al. Influence of simulated microgravity on the activation of the small GTPase Rho involved in cytoskeletal formation—molecular cloning and sequencing of bovine leukemia-associated guanine nucleotide exchange factor. BMC Biochem 2006;7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumei Y, Shimokawa H, Ohya K, et al. Small GTPase Ras and Rho expression in rat osteoblasts during spaceflight. Ann N Y Acad Sci 2007;1095:292–9. [DOI] [PubMed] [Google Scholar]
- 55.Rousseau M, Gaugler MH, Rodallec A, et al. RhoA GTPase regulates radiation-induced alterations in endothelial cell adhesion and migration. Biochem Biophys Res Commun 2011;414:750–5. [DOI] [PubMed] [Google Scholar]
- 56.Kidd AR, III, Snider JL, Martin TD, et al. Ras-related small GTPases RalA and RalB regulate cellular survival after ionizing radiation. Int J Radiat Oncol Biol Phys 2010;78:205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rutkowski R, Dickinson R, Stewart G, et al. Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling. PLoS Genet 2011;7:e1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kinchen JM, Cabello J, Klingele D, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C . elegans. Nature 2005;434:93–9. [DOI] [PubMed] [Google Scholar]
- 59.Pompeiano M, Blaschke AJ, Flavell RA, et al. Decreased apoptosis in proliferative and postmitotic regions of the Caspase 3-deficient embryonic central nervous system. J Comp Neurol 2000;423:1–12. [PubMed] [Google Scholar]
- 60.Weidhaas JB, Eisenmann DM, Holub JM, et al. A Caenorhabditis elegans tissue model of radiation-induced reproductive cell death. Proc Natl Acad Sci U S A 2006;103:9946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boulton SJ, Gartner A, Reboul J, et al. Combined functional genomic maps of the C. elegans DNA damage response. Science 2002;295:127–31. [DOI] [PubMed] [Google Scholar]
- 62.Liu J, Yang B, Ai J. Advance in research of microRNA in Caenorhabditis elegans. J Cell Biochem 2013;114:994–1000. [DOI] [PubMed] [Google Scholar]
- 63.Bostjancic E, Jerse M, Glavac D, et al. miR-1, miR-133a/b, and miR-208a in human fetal hearts correlate to the apoptotic and proliferation markers. Exp Biol Med (Maywood) 2015;240:211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weidhaas JB, Babar I, Nallur SM, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res 2007;67:11111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shimizu S, Takehara T, Hikita H, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol 2010;52:698–704. [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Shi ZM, Jiang CF, et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget 2014;5:5416–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen JZ, Zhang YY, Fu HY, et al. Overexpression of microRNA-143 inhibits growth and induces apoptosis in human leukemia cells. Oncol Rep 2014;31:2035–42. [DOI] [PubMed] [Google Scholar]
- 68.Xu C, Lu Y, Pan Z, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci 2007;120:3045–52. [DOI] [PubMed] [Google Scholar]
- 69.Wang J, Jia Z, Zhang C, et al. miR-499 protects cardiomyocytes from H2O2-induced apoptosis via its effects on Pdcd4 and Pacs2. RNA Biol 2014;11:339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Z, Chen Q, Li C, et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol 2014;16:1341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li XJ, Luo XQ, Han BW, et al. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J Cancer 2013;109:2189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korner C, Keklikoglou I, Bender C, et al. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J Biol Chem 2013;288:8750–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu X, Zhang Z, Sun L, et al. MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis 2011;32:1798–805. [DOI] [PubMed] [Google Scholar]
- 74.Breckenridge DG, Kang BH, Kokel D, et al. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol Cell 2008;31:586–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohnishi T, Tsuji K, Ohmura T, et al. Accumulation of stress protein 72 (HSP72) in muscle and spleen of goldfish taken into space. Adv Space Res 1998;21:1077–80. [DOI] [PubMed] [Google Scholar]
- 76.Yi S, Kim S, Song J. Analysis of the effect of space radiations on the nematode, Caenorhabditis elegans, through the simulated space radiation. Int J Astronom Astrophys 2013;3:291–302. [Google Scholar]
- 77.Kumari R, Singh KP, Dumond JW., Jr Simulated microgravity decreases DNA repair capacity and induces DNA damage in human lymphocytes. J Cell Biochem 2009;107:723–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.