Abstract
Background
Fusarium head blight (FHB) is a severe disease caused by different Fusarium species, which affects a wide range of cereal crops, including wheat. It determines from 10 to 30 % of yield loss in Europe. Chemical fungicides are mainly used to reduce the incidence of FHB, but low environmental impact solutions are looked forward. Applications of soil/rhizobacteria as biocontrol agents against FHB in wheat are described in literature, whereas the potential use of lactobacilli in agriculture has scarcely been explored.
Results
The aim of this work was to study the inhibitory effect of two bacterial strains, Lactobacillus plantarum SLG17 and Bacillus amyloliquefaciens FLN13, against Fusarium spp. in vitro and to assess their efficacy in field, coupled to the study of the microbial community profile of wheat seeds. Antimicrobial assays were performed on agar plates and showed that the two antagonistic strains possessed antimicrobial activity against Fusarium spp. In the field study, a mixture of the two strains was applied to durum wheat i) weekly from heading until anthesis and ii) at flowering, compared to untreated and fungicide treated plots. The FHB index, combining both disease incidence and disease severity, was used to evaluate the extent of the disease on wheat. A mixture of the two microorganisms, when applied in field from heading until anthesis, was capable of reducing the FHB index. Microbial community profile of seeds was studied via PCR-DGGE, showing the presence of L. plantarum SLG17 in wheat seeds and thus underlining an endophytic behavior of the strain.
Conclusions
L. plantarum SLG17 and B. amyloliquefaciens FLN13, applied as biocontrol agents starting from the heading period until anthesis of wheat plants, are promising agents for the reduction of FHB index.
Keywords: Biocontrol, Lattobacilli, Endophytes, Fusarium head blight
Background
Fusarium head blight (FHB) is a severe disease diffused worldwide affecting a wide range of cereal crops such as wheat (Triticum L.) and barley (Hordeum L.). Only in Europe the loss due to FHB in wheat harvest is estimated from 10 to 30 % [1]. The causal agents are mainly Fusarium graminearum Schwabe [teleomorph, Gibberella zeae (Schwabe) Petch] and Fusarium culmorum (WG Smith) Sacc. [2]. These Fusarium species are known to produce several mycotoxins, which can accumulate in the grains and are toxic to both animals and humans [1]. FHB epidemics in nature can strike suddenly, but its severity can vary depending on climate conditions, being favored by high humidity and rainfall during flowering [3]. Several authors reported different control measures for FHB [4–6], that include the use of agronomic control techniques, wheat genetic resistance and the use of chemical or biological antagonists. The limited time in which the heads are susceptible to FHB infection (during anthesis and for a short period afterwards) makes this disease a potential target for biological control.
The use of microorganisms against plant pathogens has increased in recent years due to consumers’ demand of a reduced chemical products use for the benefits of human health and the environment [7]. Some successful applications of soil/rhizobacteria as biocontrol agents against FHB, with a reduction of Fusarium incidence and mycotoxin level, have already been reported in in vitro assays, greenhouse experiments and field trials [6, 8–10]. These bacteria can also colonize root surface and enter root tissues, thus becoming endophytic. Endophytes are of special interest in the field of biocontrol, as they are harmless to plants and are able to inhabit specific ecological niches [11]. From the internal tissues, they can be a source of secondary metabolites acting as elicitors of plant defenses or as antimicrobial agents with potential use to control disease [12]. Typical examples of these molecules are cyclic lipopeptides (LPs) mainly synthesized by species of Bacillus and Pseudomonas [13]. Nevertheless the array of secondary metabolites produced by endophytic bacteria remains to be discovered [14].
In addition to these strains, Lactobacillus spp. members can be interesting as potential biocontrol agents. They are well known as probiotics and protective microorganisms in food and feed against spoilage bacteria or fungi [15]. However, in the agricultural field, their potential use is still sparse, although they can be found inside plant tissues [16, 17] and produce bioactive compounds (organic acids, bacteriocins, phenyllactic acid, cyclic dipeptides, fatty acids) with antimicrobial properties against a broad spectrum of phytopathogenic fungi. In particular, different molecules are active against Fusarium [18–20] and some Lactobacillus strains have been found to be capable of mycotoxin detoxification [21, 22].
In this work, the inhibitory effect of Lactobacillus plantarum SLG17 and Bacillus amyloliquefaciens FNL13 was evaluated against strains of F. culmorum and F. graminearum isolated from wheat, and the presence of genes responsible for antimicrobial peptide production was checked. Furthermore, the antagonistic efficacy in controlling FHB was studied in field conditions. PCR-DGGE analyses on wheat seeds were performed to investigate: a) the microbial community profile of wheat grains upon different treatments; b) the endophytic colonization feature of the microbial inoculants; c) Fusarium population in wheat.
Results
Antimicrobial activity against Fusarium spp
In vitro tests performed on agar plates showed that the two strains used in this work, L. plantarum SLG17 and B. amyloliquefaciens FLN13, possessed antimicrobial activity against all the five Fusarium strains isolated from diseased wheat (Table 1). Both antagonistic strains did not display reciprocal inhibition (data not shown). Antimicrobial activity of B. amyloliquefaciens FNL13 against F. culmorum Fc1 was also examined using scanning electron microscopy (SEM) since hyphae adjacent to the inhibition area in the plate tests revealed an unusual brown color. The adherence of B. amyloliquefaciens FNL13 cells on the hyphal surface of F. culmorum Fc1 is evident, and the contact zone showed shrinkage with loss of turgidness of hyphae (Fig. 1b) compared to the control grown hyphae (Fig. 1a). Conidia were also subjected to surface alteration (Fig. 1c). In addition, as shown in Fig. 1d, bacterial aggregates close to hyphae embedded in an extracellular matrix could be observed (Fig. 1c).
Table 1.
Source of isolation, species identity and fungal inhibition spectra of L. plantarum SLG17 and B. amyloliquefaciens FLN13; values are expressed as mean ± SD
| Antifungal activity: inhibition radius (mm) | ||||||
|---|---|---|---|---|---|---|
| Isolates | F. culmorum Fc1 | F. culmorum Fc2 | F. culmorum Fc3 | F. graminearum Fg1 | F. graminearum Fg2 | Source |
| L. plantarum SLG17 | 13.67 ± 0.58 | 12.67 ± 2.08 | 17.33 ± 2.31 | 15.67 ± 1.15 | 16.33 ± 3.21 | Silage |
| B amyloliquefaciens FLN13 | 10.87 ± 0.06 | 10.80 ± 0.10 | 10.67 ± 0.21 | 10.43 ± 0.06 | 10.67 ± 0.06 | Forest soil |
Fig. 1.
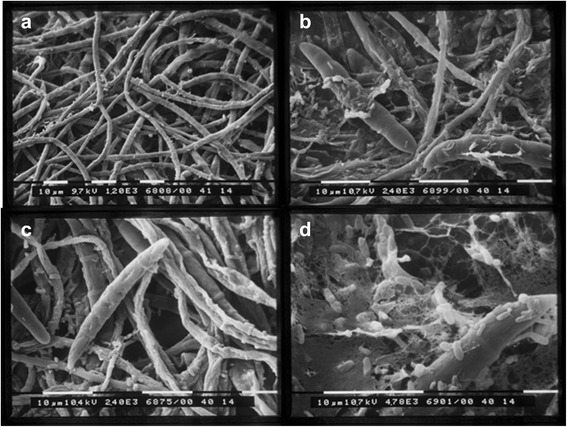
SEM analysis of antagonistic B. amyloliquefaciens FLN13 interacting with hyphae of F. culmorum Fc1 on PDA medium at 5th day after incubation. a normal hyphae of F. culmorum Fc1; b, c damaged hyphae and macroconidia of F. culmorum Fc1; d damaged hyphae with cells of B. amyloliquefaciens FLN13 embedded in a extracellular matrix
Detection of genes for plantaricin and LPs synthesis
L. plantarum SLG17 was positive for plnE/F, plnK, plnG, plnN genes, whereas the plnJ gene was not detected (data not shown). The PCR assay using degenerated primers followed by cloning and sequencing of the amplified bands allowed the detection of the genes related to the biosynthesis of surfactin, fengycin and mycosubtilin in the genome of B. amyloliquefaciens FLN13 (Table 2). On the other hand, the ituC fragment, involved in iturin A production, was not detected.
Table 2.
Blast results of the sequenced products obtained from PCR amplification using gene-specific degenerated primers from biosynthetic genes of Mycosubtilin synthase, Fengycin synthetase and Surfactin synthase in B. amyloliquefaciens FLN13
| Accession Number (GenBank) | Primer name | Size (bp) | GeneBank accession number | Identity (%) | E value | UniProt Accession number |
|---|---|---|---|---|---|---|
| KP944004 | Am1-F/Tm1-R | 405 | HG328254.1 | 99 % | 0.0 | BmyB protein, Mycosubtilin synthase subunit B (S6FI96) |
| KP944005 | Af2-F/Tf1-R | 441 | HF563562.1 | 99 % | 0.0 | FenD protein, Fengycin synthetase (M1XBL6) |
| KP944006 | As1-F/Ts2-R | 420 | CP003838.1 | 98 % | 0.0 | SrfAB protein, Surfactin synthase subunit 2 (L0BHY7) |
In field trial and FHB assessment in wheat
The isolation of FHB agents in the wheat grains resulted positive (75 % of kernel were positive to Fusarium spp. in the Ctr), confirming natural infection of the field.
DI was not significantly different among the four treatments at GS 73, whereas a few differences were observed in DS, with the highest values present in Ctr and Bio-2 (9.2 % and 11.5 %) and the lowest in Chem (3.3 %). Regarding the FHB index, the lowest value was obtained in Chem (1.6), being statistically different from all the other treatments (Table 3).
Table 3.
Mean values of FHB incidence (DI), disease severity (DS) and FHB index at Zadoks growth stage GS 73 and GS 87 in the different treatments (Ctr, Chem, Bio-1, Bio-2)
| Treatment* | GS 73 | GS 87 | ||||
|---|---|---|---|---|---|---|
| DI (%) | DS (%) | FHB index | DI (%) | DS (%) | FHB index | |
| Ctr | 30.0a | 9.2ab | 2.8b | 51.4b | 17.5b | 9.1c |
| Chem | 18.0a | 3.3a | 0.5a | 30.5a | 5.5a | 1.6a |
| Bio-1 | 23.0a | 7.2ab | 1.6b | 36.0a | 13.0ab | 4.6ab |
| Bio-2 | 29.0a | 11.5b | 3.5b | 54.0b | 17.4b | 9.0bc |
*Within columns, means followed by different letters differ significantly (Tukey’s test, P ≤ 0.05)
In the evaluation made at GS 87, the highest DI, DS and FHB index were observed in Ctr (51.4 %, 17.5 % and 9.1) and Bio-2 (54 %, 17.4 % and 9.0) and no significant differences were observed between them. On the other hand, a significant FHB index reduction was obtained in Bio-1 (49.5 % reduction) and Chem (82.4 % reduction) compared to Ctr; in both Bio-1 and Chem the DI is statistically different with respect to Ctr (36 % and 30.5 % vs 51.4 %), whereas DS showed a significant reduction only in Chem (5.5 % vs 17.5 %). The kernel weight was significantly increased (P ≤ 0.05) only in Chem (average weight: 2814 ± 99 g); on the other hand, the observed increase in BIO-1 was not significant compared to Ctr (2518 ± 99 g vs 2455 ± 102 g). Bio-2 had a kernel weight lower than Ctr (2410 ± 79) (Table 3).
PCR-DGGE analysis
Wheat seed bacterial community was firstly investigated in all treatment condition by PCR-DGGE analysis of 16S rRNA gene fragments targeting total eubacteria. Profiles of the bacterial communities and UPGMA dendrogram are shown in Fig. 2a and b. The cluster analysis showed a distinct division between Bio-2 and the group Ctr, Chem and Bio-1 (similarity less than 27 %), and a further division distinguishes Ctr from Chem and Bio-1 profiles.
Fig. 2.
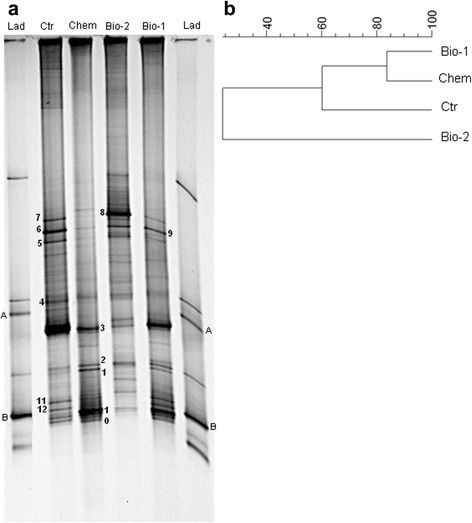
DGGE patterns of eubacterial 16S rDNA fragments amplified from wheat seeds (a) and cluster analysis (b); Ctr: untreated subplots; Chem: prothioconazole treated subplots; Bio-1: subplots treated with L. plantarum SLG17 and B. amyloliquefaciens FLN13 (weekly treatment-at Zadoks growth stage 50 to 67); Bio-2: subplots treated twice with L. plantarum SLG17 and B. amyloliquefaciens FLN13 at Zadoks growth stage 67 and 70; Lad: Ladder with known microorganism. A: L. plantarum SLG17; B: B. amyloliquefaciens FLN13
Sequencing of selected bands evidenced the presence of five different bacterial species (Table 4). Band 1, detected in Chem and Bio-1, showed 100 % similarity to Lysobacter soli, while bands 2, 3, and 12 (detected in all profiles, except for band 12 which is present only in Ctr and Bio-1) had 100 % similarity to Pantoea agglomerans. Bands 4, 5 and 7 showed 100 % identity with Erwinia rhapontici; bands 6, 9 (detected in all profiles) and 8 (Bio-2) were identified as Rahnella acquatilis (100 % identity). DGGE fingerprints targeting lactobacilli were rather simple, with a few well-defined bands (Fig. 3a), not detectable in all samples. Bands 1, 2, 3, 4 were particularly visible in the Ctr profile while their intensity decreased in the profiles Bio-1 and Bio-2. These bands and band 5 (Bio-1) had 100 % identity with Exiguobacterium sibiricum (Table 4). Bands 7 and 8 were detected only in Bio-1 and Bio-2 profile, and had the same migration distance of bands 6 and 9, which represent L. plantarum in the ladder profile. These four bands were also excised and sequenced, showing 100 % similarity with L. plantarum (Table 5). Unexpectedly, primers targeting lactobacilli allowed the amplification of B. amyloliquefaciens FLN13, which was thus present in this DGGE profile. An in silico evaluation of Lac1 and Lac2 primer sequences on 16S rDNA of Bacillus spp. evidenced a possible amplification of the ribosomal DNA.
Table 4.
Eubacteria sequence alignment with blast
| Band | Closest match | % similaritya | Accession number (GenBank) |
|---|---|---|---|
| 1 | Lysobacter soli - L. enzymogenes | 100 % | KP943989 |
| 2 | Pantoea agglomerans | 100 % | KP943990 |
| 3 | nd | - | - |
| 4 | Erwinia rhapontici strain | 100 % | KP943991 |
| 5 | Erwinia rhapontici strain | 100 % | KP943992 |
| 6 | Rahnella aquatilis strain | 100 % | KP943993 |
| 7 | Erwinia rhapontici strain | 100 % | KP943994 |
| 8 | Rahnella aquatilis strain | 100 % | KP943995 |
| 9 | nd. | - | - |
| 10 | nd | - | - |
| 11 | nd | - | - |
| 12 | Pantoea agglomerans | 100 % | KP943996 |
aSimilarity represents the % similarity shared with the sequences in the GenBank database. nd: not determined
Fig. 3.
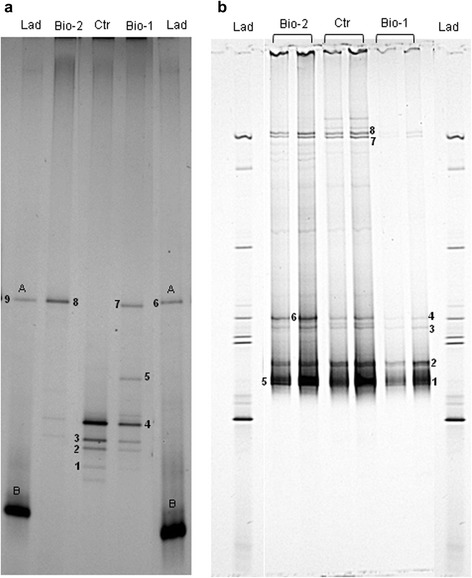
DGGE patterns of lactobacilli 16S rDNA fragments from wheat seeds (a) and DGGE patterns of partial region of the translation elongation factor1 alpha (EF-1 alpha) gene of Fusarium spp. from wheat seeds (b); Ctr: untreated subplots; Chem: prothioconazole treated subplots; Bio-1: subplots treated with L. plantarum SLG17 and B. amyloliquefaciens FLN13 (weekly treatment-at Zadoks growth stage 50 to 67); Bio-2: subplots treated twice with L. plantarum SLG17 and B. amyloliquefaciens FLN13 at Zadoks growth stage 67 and 70; Lad: Ladder with known microorganism. A: L. plantarum SLG17; B: B. amyloliquefaciens FLN13. Numbers indicate excised and sequenced DGGE bands
Table 5.
Lactobacilli sequence alignment with blast
| Band | Closest match | % similaritya | Accession number |
|---|---|---|---|
| 1-2-3 | Exiguobacterium sibiricum | 99 % | KP943997 |
| 4 | Exiguobacterium sibiricum | 100 % | KP943998 |
| 5 | nd | - | - |
| 6-9 | Lactobacillus plantarum | 100 % | KP943999 |
| 7–8 | Lactobacillus plantarum | 100 % | KP944000 |
aSimilarity represents the % similarity shared with the sequences in the GenBank database. nd: not determined
The DGGE fingerprints of the Fusarium community were also simple and not many differences were detected among plots treated with microorganisms and the control plots (Fig. 3b). A lower band intensity was shown in Bio-1 profile. Mainly, bands excised (1, 2, 3 and 4) had 100 % similarity to F. culmorum (Table 6) and were detected in all profiles. Bands 7 and 8, whose detection was weak in Bio-1, had sequence homology with F. graminearum.
Table 6.
Fusarium sequence alignment with blast
| Band | Closest match | % similaritya | Accession number |
|---|---|---|---|
| 1-2-5 | Fusarium culmorum | 100 % | KP944003 |
| 3-4-6 | Fusarium culmorum | 100 % | KP944002 |
| 7-8 | Fusarium graminearum strain | 100 % | KP944001 |
aSimilarity represents the % similarity shared with the sequences in the GenBank database
Discussion
The use of microorganisms for pathogen control in plants is largely studied and applied to several crops of economic value, with consequent reduction of the spread of chemical agents in the environment. Bacillus spp. are the most commercially successful biocontrol agents, with more than 10 Bacillus-based registered formulations [23], whereas Lactobacillus spp. strains, although included in multi-strain bio-fertilizers [17], have rarely been used for biocontrol intervention strategy [24]. In this work, two bacterial strains, B. amyloliquefaciens FLN13 and L. plantarum SLG17, previously selected for their strong inhibitory activity against several plant pathogenic fungi, were checked for their in vitro antifungal activity against strains of F. culmorum and F. graminearum and, as a mixture, they were tested for their potential to control FHB in durum wheat in field.
L. plantarum SLG17 exhibited antimicrobial activity against the tested Fusarium strains, and a consistent number of genes responsible for plantaricin production. Beside the role of organic acids which are known to be detrimental for pathogen growth [15], a possible action of plantaricins in the antagonist activity against FHB agents can be suggested. Moreover, results obtained from DGGE profiles of seeds deriving from wheat subjected to both biological treatments (Bio-1 and Bio-2) demonstrated that L. plantarum SLG17 was able to enter the internal tissues of spikelets to become an endophyte. Although no direct evidence (e.g. microscopic examination) was given to show the penetration of the sprayed microorganism, the absence of bands ascribed to L. plantarum in the Ctr profile strengthen the hyphotesis of the endophytic behavior of L. plantarum SLG17. Moreover, no data on Lactobacillus detection on wheat seeds are at present available in the literature, unless in silage fermentation [25]. In addition, B. amyloliquefaciens FLN13 was also able to inhibit in vitro all tested Fusarium strains and possessed the synthetase genes responsible for mycosubtilin, fengycin and surfactin production, which are known to be among the molecules (LPs, polyketides, antibiotics) involved in the antagonistic activity against plant pathogens [13]. In the in vitro antagonistic assay, morphological changes were observed by SEM analyses in the hyphae and macroconidia of F. culmorum Fc1, associated with the presence of an extracellular matrix (Fig. 1). These effects on fungi have already been described by several authors in the presence of different species of Bacillus, as a result of cell wall degradation, increased cell adhesion and lipopeptide excretion [8, 26, 27]. In addition, Gong et al. [28] clearly showed the destructive effect of secondary metabolites (iturin A and plipastatin A) from B. amyloliquefaciens S76-3 in F. graminearum conidia and hyphae. Unfortunately, no evidence on the presence of this strain inside wheat seeds was found in the PCR-DGGE profiles. As also reported by Crane et al. [29], following application of B. amyloliquefaciens strain TrigoCor on wheat, most Bacillus cells were present post-application as metabolically dormant spores, thus hindering potential internalization in the host plant. It should also be emphasised that the detection limit for DGGE is in the order of 103 cfu/ml [30]; as a consequence, microbial groups that are present and active, but whose population is lower than 103 cfu/ml, will not be evidenced.
Beside the clear efficacy of the fungicide treatment, the field trial showed, at GS 87, a significant FHB index reduction (49.6 %) in Bio-1 compared to the untreated subplots. Therefore, the efficacy of a preventive and repetitive treatment with the applied mixture of microorganisms was demonstrated, although this was not associated with a significant increase in kernel yield. On the other hand, the treatment applied twice at flowering (Bio-2) was not effective. This last result is different from what obtained by Schisler et al. [31] who showed a significant DS and DI reduction by using a B. subtilis/amyloliquefaciens strain at flowering in a field trial. However, as pointed out by the same author and by Crane et al. [29], several factors can contribute to the successful application of FHB antagonists in the field, such as the decline of antagonistic molecule levels on wheat surface within a few days, the deleterious effects of ultraviolet light, available nutrients, host variety and runoff from rainfall and irrigation. Last but not least, the intrinsic properties of the applied microorganisms and the inoculation technology should also be considered.
The identification of excised bands from the DGGE profiles evidenced the detection of different microbial species (P. agglomerans, E. raphontici), commonly isolated from plant tissues [32, 33] and the presence of F. culmorum and F. graminearum, confirming their endemic presence in wheat kernels. Finally, the PCR-DGGE, targeting lactobacilli, allowed the detection of L. plantarum in Bio-1 and Bio-2 seeds, indicating that the inoculated strain was able to become endophyte. This observation confirms that endophytes may not only derive from the rizosphere, but, as reviewed by Compant et al. [34], they may also originate from other sources, such as the phyllosphere, the anthosphere, or the spermosphere.
Conclusions
The obtained results suggest a possible use of the two microorganisms in controlling FHB spread throughout all the heading and ripening period, aimed at protecting the spikelets before and during anthesis. Moreover, the endophytic colonization of L. plantarum, which is one of the most characterized and used probiotic Lactobacillus species, can help to increase the nutritional properties of wheat and derived products (silages, flour and related bakery products). Further studies are necessary to deeply investigate the interaction with the host plant, the metabolite production of both strains and improve the inoculation technology.
Methods
Microorganisms and culture growth conditions
This work is based on the use of two microorganisms, L. plantarum SLG17, isolated from silages and B. amyloliquefaciens FNL13, isolated from a forest soil, both selected after an in vitro screening aimed at studying the capability of several Lactobacillus and Bacillus strains of inhibiting the growth of plant pathogenic fungi, including some Fusarium species [35]. These two strains, showing the highest antimicrobial activity, were therefore chosen to perform the current experiments with wheat-isolated strains of F. culmorum and F. graminearum, specifically associated to FHB.
L. plantarum SLG17 was grown on de Man, Rogosa and Sharpe (MRS) broth (BD, Difco, USA) for 24 h at 37 °C in anaerobic conditions (1.5 % agar for agar plates); while B. amyloliquefaciens FNL13 was grown in Nutrient Broth (NB, BD) for 24 h at 25–28 °C under gentle shaking. For long-term storage, microbial strains were stored at −80 °C.
F. culmorum Fc1, Fc2, Fc3 and F. graminearum Fg1, Fg2 strains, belonging to the main species involved in FHB, were isolated from naturally infected durum wheat kernels (cv Normanno) from fields at the University Farm of Bologna, located in Cadriano (Bologna, Italy) during 2010–2011 and identified by PCR using species-specific primers [36] (unpublished results). They were cultivated on Potato Dextrose agar plates (PDA, BD). For long-term storage the fungal strains were stored at 4 °C in the fungal collection of the Department of Agricultural Sciences, University of Bologna.
Assessment of the antimicrobial activity against Fusarium spp
Conidia production
Fusarium macroconidia inocula were obtained by growing the fungal strains on PDA plates for 7 days (or until sporulation). Then conidia were collected gently scraping the mycelium with a scalpel, filtered in cheese cloth, put into slants with sterile peptone water (0.2 % w/v) and the slants were vigorously shaked. Macroconidia concentrations were determined using a haemocytometer, and adjusted to 105 macroconidia per ml of sterile peptone water (0.2 % w/v).
Antimicrobial assay
The assay was performed according to Magnusson et al. [19]. 50 μl of L. plantarum and B. amyloliquefaciens overnight cultures were spotted on MRS and PDA plates, respectively, and allowed to grow for 24 h at 37 °C in anaerobic conditions the former and 25 °C in aerobiosis the latter. The plates were then overlaid with 10 ml of PD soft agar (0.8 %; Difco Laboratories) containing 105Fusarium spores (macroconidia) per ml. After 48–36 h of aerobic incubation at 25 °C, the inhibition radius (expressed in mm) was measured. Each assay was performed in triplicate and mean ± SD was recorded. The antimicrobial assay was also performed between the two antagonistic strains.
SEM analyses
An agar square (1x1 cm) plus mycelium was taken from the inhibition area of the dual cultures B. amyloliquefaciens FNL13—F. culmorum Fc1. Sample was immersed in 2.5 % glutaraldehyde for 4 h at room temperature, rinsed four times with phosphate buffer (0.1 M; pH 7.2), and dehydratated in a graded series of ethanol concentrations (10, 30, 50, 75, 95 and 100 % for 15 min each). It was then dried with a Critical Point Dryer unit (Balzers CPD 020), mounted on aluminium stub with double-sided tape and coated with gold-palladium film using an ion sputtering unit (Balzers MED 010). The sample was observed under a Philips 515 SEM at 7 KV and photographed with Nikon Coolpix 5400.
Detection of genes involved in antagonistic activity
The presence of genes belonging to pln locus (plnG, plnE/F, plnJ, plnK, plnN) and involved in plantaricin production was investigated according to Doulgeraki et al. [37] on L. plantarum SLG17, following DNA extraction from pure culture. In addition, the presence of four families of lipopeptides synthetase genes (surfactin synthetase, fengycin synthetase, mycosubtilin synthetase and IturinA) were also investigated in Bacillus amyloliquefaciens FLN13. PCR protocols and cloning procedure were performed according to Tapi et al. [38], while iturinA was analyzed according to Alvarez et al. [39] by amplifying the ItuC open reading frame. Primers used are listed in Table 7.
Table 7.
List of primers for detection of genes belongs to pln locus and LPs shyntetase
| Target gene | Primers | Annealing temperature (°C) | References |
|---|---|---|---|
| plnE/F | fw: 5′- GGCATAGTTAAAATTCCCCCC-3′ | 53.2 | Doulgeraki et al., 2013 [37] |
| rev: 5′- CAGGTTGCCGCAAAAAAAG-3′ | |||
| plnJ | fw: 5′-TAACGACGGATTGCTCTG-3′ | 51 | Doulgeraki et al., 2013 [37] |
| rev: 5′- AATCAAGAAATTATCACATTAGTC-3′ | |||
| plnK | fw: 5- CTGTAAGCATTGCTAACCAATC | 52.9 | Doulgeraki et al., 2013 [37] |
| rev: 5- ACTGCTGACGCTGAAAAG | |||
| plnG | fw: 5- TGCGGTTATCAGTATGTCAAAG | 52.8 | Doulgeraki et al., 2013 [37] |
| rev: 5-CCTCGAAACAATTTCCCCC | |||
| plnN | fw: 5- ATTGCCGGGTTAGGTATCG | 51.9 | Doulgeraki et al., 2013 [37] |
| rev: 5- CCTAAACCATGCCATGCAC | |||
| Surfactin synthetase | As1-f CGCGGMTACCGVATYGAGC | 43 °C | Tapi et al., 2010 [38] |
| Ts2-r ATBCCTTTBTWDGAATGTCCGCC | |||
| Fengycin synthetase | Af2-f GAATAYMTCGGMCGTMTKGA | 45 °C | Tapi et al., 2010 [38] |
| Tf1-r GCTTTWADKGAATSBCCGCC | |||
| Mycosubtilins syntetase | Am1-f CAKCARGTSAAAATYCGMGG | 45 °C | Tapi et al., 2010 [38] |
| Tm1-r CCDASATCAAARAADTTATC | |||
| Iturin A synthetase | ituC-f AAAGGATCCAAGCGTGCCTTTTACGGGAAA | 56 °C | Alvarez et al. 2011 [39] |
| ituC-r AAAAAGCTTAATGACGCCAGCTTTCTCTT |
Field trial
Experimental trial
The durum wheat cultivar Normanno, susceptible to FHB agents, was sown in autumn 2012 at the University Farm of Bologna, where durum wheat cultivation is repeated every year. The field was subdivided into small subplots (1 m x 2.2 m), one subplot represented one replication, and four replications were used for each treatment. Infection of heads by Fusarium spp. was dependent on naturally occurring levels of inoculum and rainfall (20 rainfall events for a total of 5.85 cm of rain, from flowering until ripening, were observed). The wheat stage of growth was measured using the Zadoks growth stage (GS) [40].
Four different treatments were designed: untreated subplots (Ctr), subplots treated with a prothioconazole-based fungicide (Proline®, Bayer Crop Science, Italy), according to the manufacturer instructions (Chem), subplots weekly treated with a suspension of the two microorganisms from heading (GS 50) to the beginning of flowering (GS 67) (Bio-1), subplots treated twice with the microorganisms suspension at GS 67 and at mid-flowering (GS 70) (Bio-2).
Production of antagonist inoculum
L. plantarum SLG17 and B. amyloliquefaciens FNL13 were grown in MRS broth and NB (under shaken), respectively, for 24 h as previously described. 10 ml of the overnight cultures were used as inoculum in flasks containing 400 ml of broth (MRS and NB) for 48 h to reach a concentration of approximately 107 cfu/ml. The cultures were then centrifuged (20 min at 12.000 rpm) and the pellets were combined in 400 ml of sterilized saline solution (0.8 % NaCl). The microbial inoculum was prepared before each treatment and sprayed immediately on the spikelets on late afternoon (100 ml/subplot).
Assessment of FHB
Five groups of ten spikes per subplot were chosen and marked with plastic labels for FHB assessment: disease incidence (DI) and disease severity (DS). The disease evaluation was assessed twice at GS 73 and 87. DI was calculated as the percentage of ears that are visibly diseased in relation to the total number assessed (50 spikes/subplot). For DS evaluation, the scale rating of Purahong et al. [41] was applied. This scale represents eight levels of percentage area infected on individual ears: 0 % (no infection), 2 %, 5 %, 10 %, 25 %, 50 %, 75 % and 90 % (90 % or more). DS was calculated as the weighted average of percentage area infected on individual ear in relation to the number of infected ears. Mean FHB Index was calculated as the product of DI and DS, divided by 100 [3]. Kernel yield per treatment was evaluated by calculating the average (±SD) of the kernel weight obtained from each subplot after harvest. The isolation of F. culmorum and F. gramineraum from 100 wheat grains, harvested from each plot, was performed to verify their presence.
Statistical analysis
The analysis of variance (ANOVA) was conducted on percentage values subjected to angular transformation for DI, DS and FHB index at both evaluation times. Significant differences among treatments were evaluated using the post hoc Tukey’s test with STATGRAPHICS PLUS software (STATPOINT TECHNOLOGIES, INC., Virginia). The level of significance was set at P ≤ 0.05.
DNA extraction
Wheat seeds from each treatment were collected two weeks before harvesting, and kept at 4 °C until further processing. Before DNA extraction, they were surface-sterilized with 70 % ethanol (3 min), treated with 2 % sodium hypochlorite (NaClO) (5 min), followed by repeated washing with sterile distilled water (3 times for 1 min). An aliquots of the final rinse was plated on Nutrient Agar and Luria-Bertani plates to ensure the surface sterilization efficiency. Later, they were snap frozen in liquid nitrogen and ground by mortar and pestle. Approximately 100 mg of powder was directly used for DNA extraction by applying the DNeasy plant mini kit (Qiagen GmbH, Milano, Italy), according to the manufacturer’s instructions. The purity and concentration of extracted DNA were determined by measuring the ratio of the absorbance at 260 and 280 nm (Infinite 200 PRO NanoQuant, Tecan, Mannedorf, Switzerland).
PCR amplification of 16S rDNA sequences and elongation factor1alpha
Three different PCR amplifications were performed, before DGGE, targeting eubacteria, lactobacilli and Fusarium spp. To avoid the amplification of the chloroplast or mitochondrial DNA, a nested PCR approach was used for the detection of eubacteria, according to Chelius and Triplett [42]. Firstly, a PCR was performed using universal primers 799f (5′-AACMGGATTAGATACCCKG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′). Following excision of the smaller PCR product (680 pb), a second PCR was then carried out in 30 μl volume, using GC-clamp 968f (5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGAACGCGAAGAACCTTAC-3′) and 1348r (5′-CGGTGTGTACAAGGCCCGGGAACG-3′). To deeply analyze possible differences between the treated plots with microorganisms and the Ctr plots, a PCR-DGGE targeting lactobacilli were also performed with primers Lac1 (5′-AGCAGTAGGGAATCTTCCA-3′) and Lac2-GC (5′-CGCCCGGGGCGCGCCCCGGGCGGCCCGGGGGCACCGGGGGATTYCACCGCTACACATG-3′) according to Gaggia et al. [17].
In addition, amplification of a partial region of the translation elongation factor1 alpha (EF-1α) gene was performed to investigate Fusarium diversity, according to Yergeau et al. [43], using the primer couple Alfie1-GC-f (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGTCGTCATCGGCCACGTCGACTC-3′) and Alfie2-r (5′-CCTTACCGAGCTCRGCGGCTTC-3′).
Electrophoretic conditions and identification of bands
The DGGE analysis was basically performed as first described by Muyzer et al. [44], using a DCode System apparatus (Bio-Rad, Richmond, CA, USA), employing 7 % polyacrylamide gels with a denaturing range of 35–55 % for total eubacteria, 30–55 % for group-specific lactobacilli and 40–55 % for fungi. Electrophoresis was performed at 65 V for 16 h at 60 °C. Gels were stained in a solution of 1X SYBR-Green (Sigma–Aldrich) in 1X TAE for 20 min and their images captured in UV transillumination with Gel DocTM 226 XR apparatus (Bio-Rad). Similarities between the DGGE profiles were determined by calculating similarity indices of the densitometric curves of the profiles using Pearson correlation coefficient with the aid of computer software (Gel Compare II, Applied Maths, Keistraat, Belgium). An unweighted pair group method using an arithmetic averages (UPGMA) algorithm was performed.
Well-defined bands in all DGGE experiments were excised from the gel with a sterile scalpel and DNA was eluted by incubating the gel fragments for 16 h in 50 μl of sterile deionised water at 4 °C. Two μl of the solutions were then used as template to re-amplify the band fragments using the couple primers 968f-GC/518r, Lac1-f/Lac2-GC andAlfie1-GC/Alfie2-r. Then, the positions of the excised bands in DGGE gel were confirmed with repeated DGGE. Bands showing the expected melting position in DGGE gels were amplified with the couple primers, without GC-clamp and the obtained amplicons were purified (Nucleospin gel and PCR clean-up kit; Macherey-Nagel GmbH & Co. KG, Germany). Finally, purified PCR products were sequenced by a commercial sequencing facility (EurofinsMWG Operon, Ebersberg, Germany). Sequence chromatograms were edited and analysed using the software programs Finch TV version 1.4.0 (Geospiza Inc., Seattle, WA, USA) and identity was then determined by the megablast algorithm in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/).
Acknowledgements
This work was done in the framework of the VII FP TRAFOON (Traditional food network to improve the transfer of knowledge for innovation), grant agreement no. 613912. Research on endophytes was performed within the EU COST FA1103 Action: “Endophytes in Biotechnology and Agriculture”. ND was the recipient of a fellowship within the JoinEU-SEEIII Action 2 Program.
Abbreviations
- FHB
Fusarium head blight
- SEM
Scanning electron microscopy
- PCR-DGGE
Polymerase chain reaction-denaturing gradient gel electrophoresis
- GS
Zadoks growth stage
- DI
Disease incidence
- DS
Disease severity
- LPs
lipopeptides
- MRS
de Man, Rogosa and Sharpe broth
- NB
Nutrient Broth
- PDA
Potato Dextrose agar
Footnotes
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
FG, LB and DDG designed the study. LB, FG and ND carried out microbiological analyses. AProdi and PN manipulated Fusarium strains. LB, FG and PN carried out the field study. AProdi and APisi performed SEM analyses. FG, LB and DDG wrote the manuscript, which was revised by BB. All Authors read and approved the final manuscript.
Contributor Information
Loredana Baffoni, Email: loredana.baffoni@unibo.it.
Francesca Gaggia, Email: francesca.gaggia@unibo.it.
Nereida Dalanaj, Email: nereida.dalanaj@gmail.com.
Antonio Prodi, Email: antonio.prodi@unibo.it.
Paola Nipoti, Email: paola.nipoti@unibo.it.
Annamaria Pisi, Email: annamaria.pisi@unibo.it.
Bruno Biavati, Email: bbiav01@um.edu.mt.
Diana Di Gioia, Phone: +39 051 2096269, Email: diana.digioia@unibo.it.
References
- 1.Bottalico A, Perrone G. Toxigenic Fusarium species and Mycotoxins Associated with Head Blight in Small-Grain Cereals in Europe. Eur J Plant Pathol. 2002;108:611–624
- 2.Goswami RS, Kistler HC. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol. 2004;5:515–525. doi: 10.1111/j.1364-3703.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 3.Xu X. Effects of Environmental Conditions on the Development of Fusarium Ear Blight. Eur J Plant Pathol. 2003;109:683–689
- 4.Parry DW, Jenkinson P, McLeod L. Fusarium ear blight (scab) in small grain cereals?a review. Plant Pathol. 1995;44:207–238. doi: 10.1111/j.1365-3059.1995.tb02773.x. [DOI] [Google Scholar]
- 5.Pancaldi D, Tonti S, Prodi A, Salomoni D, Pra’ MD, Nipoti P, et al. Survey of the main causal agents of fusarium head blight of durum wheat around Bologna, northern Italy. Phytopathologia Mediterranea. 2010;49:258-266.
- 6.Shi C, Yan P, Li J, Wu H, Li Q, Guan S. Biocontrol of Fusarium graminearum growth and deoxynivalenol production in wheat kernels with bacterial antagonists. Int J Environ Res Public Health. 2014;11:1094–105. doi: 10.3390/ijerph110101094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabak B, Dobson ADW, Var I. Strategies to prevent mycotoxin contamination of food and animal feed: a review. Crit Rev Food Sci Nutr. 2006;46:593–619. doi: 10.1080/10408390500436185. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Selvaraj JN, Xing F, Zhou L, Wang Y, Song H, et al. Antagonistic action of Bacillus subtilis strain SG6 on Fusarium graminearum. PLoS One. 2014;9:e92486. doi: 10.1371/journal.pone.0092486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J, Boland GJ, Zhou T. Concurrent selection for microbial suppression of Fusarium graminearum, Fusarium head blight and deoxynivalenol in wheat. J Appl Microbiol. 2009;106:1805–17. doi: 10.1111/j.1365-2672.2009.04147.x. [DOI] [PubMed] [Google Scholar]
- 10.Palazzini JM, Ramirez ML, Torres AM, Chulze SN. Potential biocontrol agents for Fusarium head blight and deoxynivalenol production in wheat. Crop Prot. 2007;26:1702–1710. doi: 10.1016/j.cropro.2007.03.004. [DOI] [Google Scholar]
- 11.Gray EJ, Smith DL. Intracellular and extracellular PGPR: commonalities and distinctions in the plant–bacterium signaling processes. Soil Biol Biochem. 2005;37:395–412. doi: 10.1016/j.soilbio.2004.08.030. [DOI] [Google Scholar]
- 12.Qin S, Xing K, Jiang J-H, Xu L-H, Li W-J. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl Microbiol Biotechnol. 2011;89:457–73. doi: 10.1007/s00253-010-2923-6. [DOI] [PubMed] [Google Scholar]
- 13.Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34:1037–62. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 14.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. Metabolic potential of endophytic bacteria. Curr Opin Biotechnol. 2014;27:30–7. doi: 10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaggia F, Di Gioia D, Baffoni L, Biavati B. The role of protective and probiotic cultures in food and feed and their impact in food safety. Trends Food Sci Technol. 2011;22:S58–S66. doi: 10.1016/j.tifs.2011.03.003. [DOI] [Google Scholar]
- 16.De Melo Pereira GV, Magalhães KT, Lorenzetii ER, Souza TP, Schwan RF. A multiphasic approach for the identification of endophytic bacterial in strawberry fruit and their potential for plant growth promotion. Microb Ecol. 2012;63:405–17. doi: 10.1007/s00248-011-9919-3. [DOI] [PubMed] [Google Scholar]
- 17.Gaggìa F, Baffoni L, Di Gioia D, Accorsi M, Bosi S, Marotti I, et al. Inoculation with microorganisms of Lolium perenne L.: evaluation of plant growth parameters and endophytic colonization of roots. N Biotechnol. 2013;30:695–704. doi: 10.1016/j.nbt.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Strom K, Sjogren J, Broberg A, Schnurer J. Lactobacillus plantarum MiLAB 393 Produces the Antifungal Cyclic Dipeptides Cyclo(L-Phe-L-Pro) and Cyclo(L-Phe-trans-4-OH-L-Pro) and 3-Phenyllactic Acid. Appl Environ Microbiol. 2002;68:4322–4327. doi: 10.1128/AEM.68.9.4322-4327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnusson J, Ström K, Roos S, Sjögren J, Schnürer J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol Lett. 2003;219:129–135. doi: 10.1016/S0378-1097(02)01207-7. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Yan Y, Wang J, Zhang H, Qi W. Production and characterization of antifungal compounds produced by Lactobacillus plantarum IMAU10014. PLoS One. 2012;7:e29452. doi: 10.1371/journal.pone.0029452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalié DKD, Deschamps AM, Richard-Forget F. Lactic acid bacteria – Potential for control of mould growth and mycotoxins: A review. Food Control. 2010;21:370–380. doi: 10.1016/j.foodcont.2009.07.011. [DOI] [Google Scholar]
- 22.Franco TS, Garcia S, Hirooka EY, Ono YS, dos Santos JS. Lactic acid bacteria in the inhibition of Fusarium graminearum and deoxynivalenol detoxification. J Appl Microbiol. 2011;111:739–48. doi: 10.1111/j.1365-2672.2011.05074.x. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-García A, Romero D, de Vicente A. Plant protection and growth stimulation by microorganisms: biotechnological applications of Bacilli in agriculture. Curr Opin Biotechnol. 2011;22:187–93. doi: 10.1016/j.copbio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Roselló G, Bonaterra A, Francés J, Montesinos L, Badosa E, Montesinos E. Biological control of fire blight of apple and pear with antagonistic Lactobacillus plantarum. Eur J Plant Pathol. 2013;137:621–633. doi: 10.1007/s10658-013-0275-7. [DOI] [Google Scholar]
- 25.Cai Y, Benno Y, Ogawa M, Kumai S. Effect of applying lactic acid bacteria isolated from forage crops on fermentation characteristics and aerobic deterioration of silage. J Dairy Sci. 1999;82:520–6. doi: 10.3168/jds.S0022-0302(99)75263-X. [DOI] [PubMed] [Google Scholar]
- 26.Vitullo D, Di Pietro A, Romano A, Lanzotti V, Lima G. Role of new bacterial surfactins in the antifungal interaction between Bacillus amyloliquefaciens and Fusarium oxysporum. Plant Pathol. 2012;61:689–699. doi: 10.1111/j.1365-3059.2011.02561.x. [DOI] [Google Scholar]
- 27.Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–25. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Gong A-D, Li H-P, Yuan Q-S, Song X-S, Yao W, He W-J, et al. Antagonistic mechanism of iturin A and plipastatin A from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS One. 2015;10:e0116871. doi: 10.1371/journal.pone.0116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crane JM, Gibson DM, Vaughan RH, Bergstrom GC. Iturin levels on wheat spikes linked to biological control of Fusarium head blight by Bacillus amyloliquefaciens. Phytopathology. 2013;103:146–55. doi: 10.1094/PHYTO-07-12-0154-R. [DOI] [PubMed] [Google Scholar]
- 30.Cocolin L, Bisson LF, Mills DA. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol Lett. 2000;189:81–87. doi: 10.1111/j.1574-6968.2000.tb09210.x. [DOI] [PubMed] [Google Scholar]
- 31.Schisler DA, Khan NI, Boehm MJ, Slininger PJ. Greenhouse and Field Evaluation of Biological Control of Fusarium Head Blight on Durum Wheat. Plant Dis. 2002;86:1350–1356. doi: 10.1094/PDIS.2002.86.12.1350. [DOI] [PubMed] [Google Scholar]
- 32.Ruppel S, Hecht-Buchholz C, Remus R, Ortmann U, Schmelzer R. Settlement of the diazotrophic, phytoeffective bacterial strain Pantoea agglomerans on and within winter wheat: An investigation using ELISA and transmission electron microscopy. Plant Soil. 1992;145:261–273. doi: 10.1007/BF00010355. [DOI] [Google Scholar]
- 33.Narula N, Remus R, Deubel A, Granse A, Dudeja SS, Behl RK, et al. Comparison of the effectiveness of wheat roots colonization by Azotobacter chroococcum and Pantoea agglomerans using serological techniques. Plant, Soil Environ. 2007;53:167–176. [Google Scholar]
- 34.Compant S, Duffy B, Nowak J, Clément C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol. 2005;71:4951–9. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaggia F, Dalanaj N, Baffoni L, Prodi A, Alkadri D, Biavati B, et al. Microorganisms as biological agents against pathogenic fungi of agricultural interest. In Schneider C, Leifert C, Feldman F, editors. Endophytes for Plant Protection: The State of the Art. 2013, p. 187. DPG Publisher; ISBN: 9783941261112.
- 36.Nicholson P, Simpson D, Weston G, Rezanoor H, Lees A, Parry D, et al. Detection and quantification ofFusarium culmorumandFusarium graminearumin cereals using PCR assays. Physiol Mol Plant Pathol. 1998;53:17–37. doi: 10.1006/pmpp.1998.0170. [DOI] [Google Scholar]
- 37.Doulgeraki AI, Paraskevopoulos N, Nychas GJE, Panagou EZ. An in vitro study of Lactobacillus plantarum strains for the presence of plantaricin genes and their potential control of the table olive microbiota. Antonie Van Leeuwenhoek. 2013;103:821–32. doi: 10.1007/s10482-012-9864-2. [DOI] [PubMed] [Google Scholar]
- 38.Tapi A, Chollet-Imbert M, Scherens B, Jacques P. New approach for the detection of non-ribosomal peptide synthetase genes in Bacillus strains by polymerase chain reaction. Appl Microbiol Biotechnol. 2010;85:1521–31. doi: 10.1007/s00253-009-2176-4. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez F, Castro M, Príncipe A, Borioli G, Fischer S, Mori G, et al. The plant-associated Bacillus amyloliquefaciens strains MEP2 18 and ARP2 3 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J Appl Microbiol. 2012;112:159–74. doi: 10.1111/j.1365-2672.2011.05182.x. [DOI] [PubMed] [Google Scholar]
- 40.Tottman DR. The decimal code for the growth stages of cereals, with illustrations. Ann Appl Biol. 1987;110:441–454. doi: 10.1111/j.1744-7348.1987.tb03275.x. [DOI] [Google Scholar]
- 41.Purahong W, Alkadri D, Nipoti P, Pisi A, Lemmens M, Prodi A. Validation of a modified Petri-dish test to quantify aggressiveness of Fusarium graminearum in durum wheat. Eur J Plant Pathol. 2012;132:381–391. doi: 10.1007/s10658-011-9883-2. [DOI] [Google Scholar]
- 42.Chelius MK, Triplett EW. The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L. Microb Ecol. 2001;41:252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- 43.Yergeau E, Filion M, Vujanovic V, St-Arnaud M. A PCR-denaturing gradient gel electrophoresis approach to assess Fusarium diversity in asparagus. J Microbiol Methods. 2005;60:143–54. doi: 10.1016/j.mimet.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


