Abstract
Proteomics has long been considered an ideal platform, and urine an ideal source for biomarker discovery in human autoimmune kidney diseases. A number of studies have examined the urine proteome to identify biomarkers of disease activity, kidney pathology, and response to therapy. Increasingly, proteomic studies of kidney disease have expanded to include blood, circulating cells and kidney tissue. Recently the clinical potential of renal proteomics has been realized through a handful of investigations whose results appear to be applicable to patient care. In this review, approaches to the proteomic evaluation of autoimmune kidney diseases will be considered in the context of developing clinically useful disease biomarkers.
Introduction
Several omic techniques are now available for the investigation of human disease. These include the broad categories of genomics, proteomics, metabolomics, and lipidomics. Within each category is the possibility of focusing down on more specific subgroups within an ome, such as peptides or phsophoproteins within the proteome. All of these techniques provide the tools to examine genes, proteins, metabolites or lipids in an unbiased fashion to uncover relationships of analytes to disease that may not be intuitive. Theoretically an agnostic omic approach may give a more complete understanding of disease than investigations in which analytes are chosen and studied on the basis of pre-conceived ideas of disease pathogenesis, which may be flawed or incomplete.
With respect to kidney diseases, and specifically autoimmune kidney diseases, the majority of studies thus far have focused on genomics and proteomics. Proteomics has been used to characterize proteins in the blood, urine, and renal tissue of patients with autoimmune kidney diseases. The goal of most of these studies has been to identify biomarkers of disease pathology, activity, and response to treatment, and to understand disease pathogenesis to uncover potential novel therapeutic approaches. This review will cover recent developments in the application of proteomics to autoimmune kidney disease with an emphasis on the translation of proteomics to the clinic.
Proteomic Techniques for the Investigation of Autoimmune Kidney Disease
The instrumentation and informatic tools used to study the proteome have evolved significantly over time. In the early days of proteomics two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) was used to separate proteins based on their molecular size and isoelectric point. Proteins from urine or the kidney were resolved and individual spots of differential intensity were cut from the gel and analyzed by mass spectrometry. In many studies, Matrix Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS) was used to identify proteins that distinguished normal from diseased tissue. Although several investigations applied 2D-PAGE to the urine proteome (1), it quickly became apparent that the resolving power of 2D-PAGE, especially for membrane proteins and highly basic proteins, was limited. Furthermore, MALDI-MS was ineffective for very low abundance proteins, and these early reports generally identified and quantitated only 30 to 40 urine proteins. The separation of proteins improved rapidly with the development of nanoscale reversed phase liquid chromatography (RPLC), and this rapidly replaced 2D-PAGE. When RPLC was coupled to mass spectrometers using nanoelectrospray ionization (ESI) sources, the number of proteins that could be resolved, identified and quantitated in urine and the kidney increased considerably, and several hundred proteins were detectable in urine (2, 3). Also essential to improved protein identification was the development of reliable protein search algorithms that permitted the design of high-throughput search engines. Subsequent advances in mass spectrometers using ion traps, and particularly orbitraps have dramatically increased the ability to detect proteins in tissue and biological fluids (4, 5). In conjunction with strategies to enrich individual sub-proteomes into fractions, these new approaches now permit the identification of several thousand proteins in human urine (6)
Proteomic Analysis of Biological Fluids in the Study of Kidney Disease
The earliest applications of proteomics focused on the urine proteome as urine is easily accessible and changes in its protein complement may reflect alterations in renal protein expression. In some studies, investigators focused on optimizing the number of proteins identified in normal urine so as to develop a database for future studies (1, 7). However, it quickly became apparent that the study of urine with proteomic techniques faced a number of barriers including variability in pH, solute content and concentration that may occur as a result of changes in hydration, diurnal variations and medication. In normal urine, the dynamic range of protein expression is several logs due to the abundance of uromodulin or Tamm-Horsfall protein (THP). Modern mass spectrometers use an approach termed data-dependent acquisition of ions that allows only a fixed number of precursor ions to be scanned in a single survey. In this setting, highly abundant proteins like THP compete for ionization with less abundant, and potentially interesting peptides so they are undetectable, thereby limit the sensitivity of the analysis. Affinity antibodies to highly abundant proteins have been used to “compress” the dynamic range of protein expression and allow detection and quantitation of low abundance proteins and peptides. Alternatively, THP can be removed by changing urine pH. The removal of highly abundant proteins with affinity antibodies is particularly necessary when studying urine with nephrotic range proteinuria.
The same broad dynamic range of protein expression is present in plasma and must be addressed in the study of renal disease. Removal of abundant plasma proteins by affinity antibodies is a common approach in this setting, as well (8). However, an alternative approach is to use mass spectrometry to study the low molecular weight peptides in plasma that represent intact protein degradation products. In that setting the peptides are separated from highly abundant proteins by size exclusion filters (9). The presence of these small molecular weight peptides in both plasma and urine, termed the peptidome, points out the complexity of biological fluids that must be appreciated in their proteomic analysis. Urine, in particular, has both peptidomic and proteomic components. Additionally, urine contains small microvesicles including exosomes and apoptotic bodies that are derived from all nephron segments (10). Exosomes contain proteins, peptides, RNA and metabolites that may reflect changes in renal function and pathology. Thus, these multiple constituents of urine may each serve as a source of information regarding how the kidney alters its behavior during normal physiologic changes and disease. In order to capture as much information as possible, a workflow to process urine and retain its multiple constituents is outlined in Figure 1. Even with this workflow some urine components will be lost during the separation steps.
Figure 1. Workflow for Investigating the Urine Proteome.
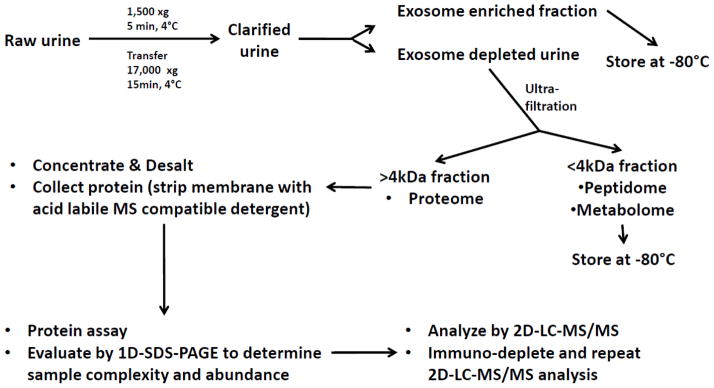
Urine is initially clarified of cells and debris by routine centrifugation. The acquired cellular sediments are stored for future studies. The clarified urine is then subjected to ultracentrifugation across sucrose density gradients to isolate exosomes and microparticles. The exosome and microparticle-depleted urine is then processed using size fractionating filters that partition the urine into fractions containing proteins and peptides either less than or greater than 4 kDa. The low molecular weight fraction is then stored for peptidomic and metabolomics studies. If proceeding to proteomic analysis the proteins >4kDa are stripped from the ultrafiltration membrane and the analyte is concentrated and de-salted before analysis by mass spectrometry. Immunodepletion can be used to remove the most highly abundant proteins from urine and improve resolution for low-abundance proteins.
Evaluation of the renal proteome can complement and extend the information obtained from the urine and serum proteomes. Differences between the proteomes of normal and affected renal parenchymal provide direct insights into disease-relevant mechanisms. Kidney injury in autoimmune disease is often initiated through glomerular immune complex accumulation however the ultimate fate of the kidneys is determined mainly by tubulointerstitial damage (11). The urine proteome is an integration of the whole kidney proteome; it cannot distinguish events occurring in the glomerular compartment from events occurring in the tubulointerstitial compartment. Using laser-capture microdissection (Fig. 2), the proteomes of the glomeruli and the tubulointerstitum can be evaluated separately (12). Additionally, differentially-expressed kidney proteins can focus urine biomarker searches to specific targets improving efficiency in developing non-invasive urine biomarkers of disease. Using only urine proteomics, such targets may be missed because of masking by more abundant, non-informative proteins. Parenchymal proteomics also identifies proteins that are not expressed or under-expressed in disease. It is much more difficult to be certain of the absence of a protein in a urine specimen given all the potential variables (e.g. osmolarity, pH, concentration, albumin) present in this heterogeneous fluid that could impair identification of unknown proteins.
Figure 2. Laser-capture microdissection of a kidney biopsy section.
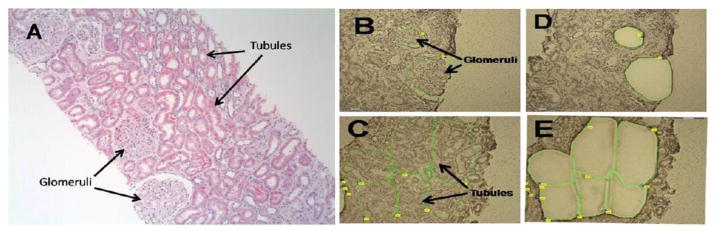
A. Hematoxylin and eosin-stained section of a kidney biopsy from a patient with lupus nephritis. B. Hematoxylin-stained section of the biopsy before laser dissection with the glomeruli to be captured outlined. C. Hematoxylin-stained section of the biopsy before laser dissection with the tubulointerstitial areas to be captured outlined. D. The same tissue section as in (B) after the glomeruli have been laser-dissected and captured. E. The same tissue section as in C after the glomeruli have been laser-dissected and captured.
Recently, we developed a proteomics workflow specifically for quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of laser-captured isolates from frozen and formalin-fixed paraffin-embedded (FFPE) tissue sections (13) (14). It is now feasible to perform proteomic analyses using the small amounts of tissue available from clinical kidney biopsies. An example of the data generated by this technique is shown in Figure 3. Two points of general importance may be taken from these data. First, despite labeling a patient as having Class IV or Class V LN based on clinical and pathological findings, examination of the kidney tissue at the molecular level shows different patterns of protein expression within an LN class. This is likely due to timing of the kidney biopsy during the disease course, and also to the intrinsic heterogeneity of all autoimmune kidney diseases. Proteomics and other omics offer platforms for understanding a disease in individuals and thereby personalizing therapies. Secondly, the tubulointerstitial compartment from some of the LN patients shows a marked decline in enzymes important for mitochondrial metabolism. It is intriguing to speculate that individuals with autoimmune kidney disease who develop chronic kidney injury do so because tubular metabolism is disrupted and energy cannot be generated properly for recovery. This mechanism could be tested, and may suggest a way to prevent chronic kidney disease therapeutically.
Figure 3. Differential kidney protein expression in lupus nephritis.
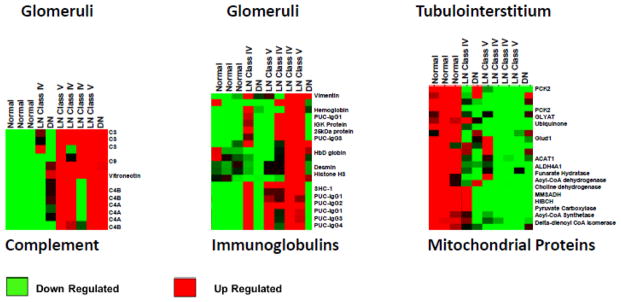
Kidney biopsies from normal kidney, lupus nephritis class IV or V kidneys, and diabetic nephropathy kidneys were laser-microdissected into glomerular and tubulointerstitial compartments. The kidney compartments were analyzed for global protein expression by LC-MS/MS and heat maps showing relative protein expression among the samples are shown. The glomerular heat maps are focused on complement proteins and immunoglobulins. It is not surprising that complement and immunoglobulins are generally increased in LN and not in normal kidney tissue. It is interesting that complement components are also increased in diabetic nephropathy. The interstitium shows a down-regulation of mitochondrial metabolic enzymes in most of the LN biopsies. It is notable that individuals with the same class of LN (compare the three Class IV LN biopsies) can have markedly different glomerular and interstitial proteomes.
FFPE biopsies have several advantages over frozen tissue, including storage and transportation logistics, and excellent preservation of kidney histology which is important for laser-capture microdissection. Approximately 8000–10,000 cells (roughly 2–3 μg of protein) are sufficient for proteomic analysis (14). Applying these requirements to clinical kidney biopsies translates into about 70 glomerular cross-sections and 1.5–2 million μm2 of TI from 10μm-thick tissue sections. Analyzing this quantity of kidney tissue on an early-generation mass spectrometer yielded 662 and 607 proteins from the glomerular and tubulointerstitial compartments, respectively (14). Using state-of-the-art technology (1 dimensional liquid chromatography coupled to a high-resolution LTQ-Orbitrap Elite mass spectrometer), 1399 proteins from the glomerular compartment and 1742 proteins from the tubulointerstitial compartment were identified from FFPE biopsies of patients with idiopathic immunoglobulin-A (IgA) nephropathy (unpublished data). In this analysis the false discovery rate was estimated at <1% using a decoy database generated from the UniprotKB Homo sapiens reference proteome. Many tissue proteins from autoimmune kidney diseases are also found in the urine, supporting the idea that tissue proteomics can focus the search for urine biomarkers. In a global proteomic screen of urine from 15 lupus nephritis (LN) patients we identified 1126 proteins and compared this list to the list of differentially-expressed proteins between control and Class IV LN tissue (n=3 each). This showed that 65% of proteins from the glomerular compartment and 54% of the proteins from the tubulointerstitial compartment were found in the LN urines (unpublished data).
Proteomic analysis of laser-captured glomeruli from kidney biopsies by LC-MS/MS is currently in use clinically to identify the composition of proteins in glomerular diseases with organized deposits, such as amyloidosis, fibrillary and immunotactoid glomerulonephritis, and membranoproliferative glomerulonephritis (15–17). Although these are not strictly autoimmune kidney diseases, proteomics has crossed the threshold into the clinical nephropathology laboratory as a diagnostic tool, suggesting it will not be long before proteomics will be applied to the diagnosis and evaluation of other types of kidney disease, including autoimmune diseases.
Proteomics in the Identification of Autoantibody Targets in Glomerulonephritis
One of the most successful and clinically relevant applications of proteomics to autoimmune kidney diseases has been in the identification of the target antigens of autoantibodies in a number of glomerular diseases. This is best exemplified by the discovery of the phospholipase A2 receptor as the dominant antigen in primary membranous nephropathy (IMN). In these studies, sera from patients with primary membranous nephropathy were used to immunoblot normal glomerular proteins separated by PAGE. The gel bands corresponding to bands of sera reactivity on the immunoblots were excised and analyzed by mass spectrometry. This identified eighteen proteins of relatively high abundance in the 185 kDa region corresponding to sera reactivity. Subsequent experiments identified one of these eighteen proteins, the M-type phospholipase A2 receptor, as the renal target antigen of antibodies present in 70% of primary membranous nephropathy patients (18). In recent follow-up experiments, the same proteomic approach was used to identify the thrombospondin type-1 domain-containing 7A protein as a membranous nephropathy target antigen in 10% of patients who did not produce antibodies to the phospholipase A2 receptor (19).
Similar approaches have been used to look for antigens in anti-neutrophil cytoplasmic antibody-associated vasculitis (AAV) and LN (17, 20). In AAV, the proteome of human umbilical vein endothelial cells (HUVEC) was separated into component proteins by 2D-PAGE and blotted with serum from patients with either microscopic polyangiitis (MPA) or granulomatosis with polyangiitis (GPA) to look for anti-endothelial cell reactivity. This study identified HUVEC antigens that distinguished MPA from GPA patients, including α-enolase, vimentin, lamin-A/C, far upstream binding protein 2, and protein disulfide-isomerase A3 precursor. Although these data provide evidence that the pathogenesis of MPA and GPA may be different, at present the data are not developed enough to suggest different approaches to therapy for these AAV subtypes. Furthermore, patients with AAV and renal involvement were not examined separately, so no conclusions can be drawn as to why only some patients have renal disease. Finally, HUVECs display relatively non-specific endothelial antigens. It would be interesting to repeat these experiments using cells from organs likely to be affected in AAV, such as glomerular capillary endothelial cells. Conceivably, auto-antibodies against glomerular endothelial cell antigens could be of use in predicting who will develop kidney injury. Some experiments were done using glomerular capillary endothelial cells and sera from patients with mixed connective tissue disease, and an antibody to human voltage-dependent anion selective channel 1 was identified, but its role/relationship in the glomerular injury of MCTD is not established (21).
The identification of autoantigens in LN kidneys was accomplished by eluting immunoglobulins from laser-captured glomeruli and assessing their reactivity to podocyte proteins separated by 2D-PAGE. This yielded several potential antigens, but antibodies to two antigens, annexin AI and α-enolase, were found in the glomeruli of all classes of LN and in all LN sera, indicating a potentially fundamental role in the pathogenesis of LN. These antibodies were isotyped as IgG2. Interestingly, anti-α-enolase was also found in glomeruli of patients with idiopathic membranous nephropathy, but was isotyped as IgG4 (20). Antibodies to annexin AI and α-enolase did not bind to nuclear material, and eluted anti-DNA antibodies did not bind to α-enolase. Intraperitoneal administration of hybridomas that produced anti-α-enolase in BALB/C mice led to the appearance of proteinuria in 25% of the mice; 66% of SCID mice injected with these hybridomas developed crescentic glomerulonephritis. These findings renew the controversy of the role of endogenous glomerular antigens versus planted nuclear antigens in the development of LN. However, although serum levels of anti-α-enolase were higher in LN patients then patients with non-renal SLE, there was moderate overlap, so it remains to be seen if these antibodies predict and/or mediate the development of LN in SLE.
Proteomics and Biomarkers of Autoimmune Kidney Diseases
Thus far, the main focus of proteomic investigations of autoimmune kidney diseases has been to identify disease biomarkers. In this context disease biomarkers have been sought that can be used diagnostically, prognostically, and to better understand the mechanisms of kidney injury. The approaches to biomarker discovery in autoimmune kidney disease will be reviewed with the intention of exemplifying important issues in this field. It should be pointed out that none of the biomarkers discussed below have been validated in independent, appropriately-sized patient cohorts, a ubiquitous problem that has limited the clinical qualification of most reported biomarkers.
Autoimmune kidney diseases are usually diagnosed by kidney biopsy. The possibility of having non-invasive diagnostic biomarkers that could substitute for the biopsy is appealing. The general approach to diagnostic biomarker development has been to identify patterns of differentially-expressed urine proteins characteristic of a particular autoimmune kidney disease. As an example, capillary-electrophoresis coupled to mass spectrometry was used to characterize the pattern of proteins/peptides in urine from patients with IgA and primary membranous nephropathy (22). A panel of 28 peptides based on the mass spectrometric pattern was sufficient to distinguish these two glomerular diseases with a sensitivity of 77% and a specificity of 100%. In other studies using capillary electrophoresis coupled to mass spectrometry, urine peptide panels were able to discriminate between IgAN and diabetic nephropathy, minimal change disease, or focal segmental glomerulosclerosis (FSGS) with high sensitivity and specificity. Importantly, the identity of many of the peptides within these individual panels was not determined. The mass spectrometric patterns were sufficient for the panels to serve as effective biomarkers. However to find novel therapeutic targets and to elucidate disease pathogenesis, knowing a protein’s identification is important.
A point to be emphasized from this IgAN study is that diagnostic accuracy was achieved with a panel of proteins/peptides, not a single biomarker. This concept was extended in another study that also used changes in the urine proteome to differentiate between LN, primary membranous nephropathy, diabetic nephropathy and FSGS. The relationship of the number of differentially-expressed proteins in a biomarker panel to the diagnostic sensitivity and accuracy of the panel was determined (23). Sensitivity was maintained above 70% until there were fewer than 5 proteins, but accuracy fell toward 50% below 20 proteins. It is therefore unlikely that single biomarkers will be able to reach sufficient discriminatory power for clinical use, so focusing on biomarker panels is appropriate.
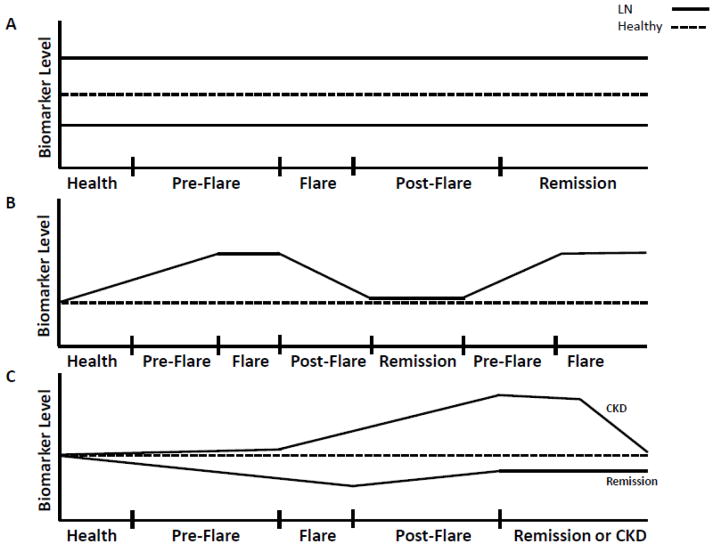
Another important diagnostic dilemma that has been addressed by biomarker studies is how to predict that a kidney disease will flare before the flare is clinically apparent. A biomarker of impending flare would open these diseases up to the possibility of flare prevention rather than flare treatment. Early intervention may limit inflammatory injury and prevent chronic kidney damage. To address this question we used SELDI-TOF mass spectrometry to analyze the low-molecular weight (< 30 kilo-daltons) urine proteome of urine samples collected from patients before, during and after LN flare (24). Among other differentially-expressed proteins during the LN flare cycle, these studies identified the 20 amino-acid form of hepcidin as a potential flare predictor, because it increased 4 months pre-flare and returned to baseline levels at flare. Given the dynamic changes of urine hepcidin expression over the LN flare cycle, it is unlikely that this potential biomarker would have been identified or correctly interpreted by examining only cross-sectional urine samples (Fig. 4A,B). This limitation probably applies to most other putative biomarkers of autoimmune kidney diseases, however to date the majority of proteomic studies have been cross-sectional.
Figure 4. Hypothetical urine biomarker patterns in lupus nephritis.
Several possible scenarios (not exhaustive) are presented to illustrate how a biomarker level may change over the course of an LN flare cycle. A. In this scenario a putative urine protein biomarker is found to be either higher or lower in LN than in healthy controls. If this protein was measured only at flare in a cross-sectional study the protein may be considered a biomarker of LN flare. However, when examined over time in serial urine samples it is seen that the biomarker does not change with the flare cycle, and levels were different even before LN flare. The biomarker may thus indicate an intrinsic abnormality of SLE as opposed to indicating active LN. B. In this scenario, the putative urine biomarker increases before flare, peaks during flare, and declines toward baseline as the patient is treated and remits. This pattern is repeated in the next flare cycle. If the urine was sampled only during flare, the biomarker may be interpreted as a marker of active LN. Following the biomarker levels in serial urine samples demonstrates a consistent increase before each flare, and the biomarker may thus be a predictor of impending LN flare. C. In this scenario the levels of a putative urine protein biomarker are shown for two separate patients. One patient remits after treatment and the other patient develops chronic kidney disease (CKD). During the evolution of CKD, the biomarker level increases during flare and treatment, whereas the biomarker level falls over time in the patient who responds to treatment. These biomarker patterns suggest that the biomarker may indicate and contribute to the development of chronic kidney injury.
The importance of longitudinal studies is emphasized by two recent investigations that identified potential biomarkers of chronic kidney damage in LN and IgAN (25, 26). Chronic kidney disease worsens prognosis, not only because patients can progress to end-stage kidney disease requiring renal replacement therapy, but also because there is increased risk of cardiovascular morbidity in patients with chronic kidney disease. Biomarkers of chronic kidney damage and progressive decline in kidney function are therefore high priority targets. Such biomarkers could identify patients in whom anti-fibrotic and kidney protective measures are needed.
Urine from patients with LN at or within 6 months of renal biopsy was evaluated with an antibody microarray that focused on cytokines (25). Angiostatin, an anti-angiogenic fragment of plasminogen was found to be greatly increased in LN urine compared to urine from healthy volunteers. Angiostatin correlated positively with serum creatinine and chronicity on the kidney biopsy, suggesting it may be a marker of progressive kidney failure.
The proteome of urine obtained from IgAN patients at kidney biopsy was investigated by 2D-PAGE and differentially-expressed proteins were identified by mass spectrometry (26). One interesting protein was significantly decreased in most of the IgA urines compared to urine from healthy individuals. This protein was identified as the LG3 fragment of endorepellin, which is a cleavage product of perlecan. Like angiostatin, LG3 has anti-angiogenic effects. Also like angiostatin, LG3 levels correlated inversely with glomerular filtration rate, and positively with increased fibrosis on kidney biopsy.
The observation that anti-angiogenic cytokines may be markers for, and/or involved in progressive parenchymal damage in two different autoimmune kidney diseases is compelling and suggests that angiogenesis may be important in preserving kidney tissue after inflammatory injury. However, before these cytokines can be accepted as biomarkers of prognosis, or used to develop targeted therapeutics, they need to be measured prospectively in patients to determine if they correlate with the development of chronic kidney disease (Fig. 4C).
Looking toward the Future
The use of proteomic approaches to identify auto-antibodies is now clearly established with the identification of phospholipase A2 receptor and the thrombospondin type-1 domain-containing 7A protein auto-antibodies in IMN. In a similar manner, proteomic analysis of kidney proteins isolated by laser capture micro-dissection appears to be established and has great potential to identify proteins relevant to renal pathology. Further advances in identifying pathology-related proteins and auto-antigens will likely require development of mass spectrometry methods that extend the ability of the instrument to identify more proteins in experimental specimens and to identify proteins with high confidence in the absence of validated antibodies for ELISA or immunoblot assays. Recent developments in mass spectometers and associated software permit a greater number of proteins to be identified in a specimen. As discussed previously, mass spectrometers have used an approach termed data-dependent acquisition of ions that allows only a fixed number of precursor ions to be scanned in a single survey. In this setting, highly abundant proteins displace less abundant and potentially interesting peptide ions from the scan, and thereby limit the sensitivity of the analysis. A new approach uses data-independent acquisition of ion characteristics that can, in theory, interrogate a specimen for the identity and quantity of any protein of interest. This approach uses extensive peptide fragment ion libraries and compares them to the observed peptide maps. This more “open-ended” approach to mass spectra analysis has been termed SWATH-MS after the swaths used to designate the ion scanning windows of the mass spectrometer. SWATH-MS depends upon the reproducibility of high-resolution spectra and the recent availability of proteome-wide spectral libraries (27).
A second recent advance in proteomics is the use of targeted analysis termed selected reaction monitoring (SRM), also known as multiple reaction monitoring (MRM). In SRM, selected precursor fragments of peptides of interest are monitored. The selected peptide fragments and their resultant ions are determined by either ion scanning, repository data or rules that govern enzymatic proteolysis of protein amino acid sequences. When coupled with isotopic labeling of peptides, SRM can be quantitative for individual proteins down to the sub-femtomole level. Thus, SRM assays can be developed that are quantitative for proteins for which validated antibodies have not been developed and can serve as an alternative to antibody based assays such as ELISA. SRM assays are currently being used to quantitate small molecular weight peptides that may reflect flares of autoimmune renal disease.
These techniques are expected to accelerate the pace at which proteomic findings achieve clinical applicability in the diagnosis and management of autoimmune kidney diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thongboonkerd V, McLeish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62(4):1461–9. doi: 10.1111/j.1523-1755.2002.kid565.x. [DOI] [PubMed] [Google Scholar]
- 2.Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High–efficiency nanoscale liquid chromatography coupled on-line with mass spectrometry using nanoelectrospray ionization for proteomics. Anal Chem. 2002;74(16):4235–49. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- 3.Cutillas PR, Norden AG, Cramer R, Burlingame AL, Unwin RJ. Detection and analysis of urinary peptides by on-line liquid chromatography and mass spectrometry: application to patients with renal Fanconi syndrome. ClinSci(Lond) 2003;104(5):483–90. doi: 10.1042/CS20020342. [DOI] [PubMed] [Google Scholar]
- 4.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7(9):R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Muller M, et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11(3):O111 013698. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santucci L, Candiano G, Petretto A, Bruschi M, Lavarello C, Inglese E, et al. From hundreds to thousands: Widening the normal human Urinome (1) J Proteomics. 2015;112:53–62. doi: 10.1016/j.jprot.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 7.Thongboonkerd V, Klein JB, Arthur JM. Proteomic identification of a large complement of rat urinary proteins. Nephron Exp Nephrol. 2003;95(2):e69–e78. doi: 10.1159/000073674. [DOI] [PubMed] [Google Scholar]
- 8.Kullolli M, Warren J, Arampatzidou M, Pitteri SJ. Performance evaluation of affinity ligands for depletion of abundant plasma proteins. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;939:10–6. doi: 10.1016/j.jchromb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Merchant ML, Niewczas MA, Ficociello LH, Lukenbill JA, Wilkey DW, Li M, et al. Plasma kininogen and kininogen fragments are biomarkers of progressive renal decline in type 1 diabetes. Kidney Int. 2013;83(6):1177–84. doi: 10.1038/ki.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101(36):13368–73. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh C, Chang A, Brandt D, Guttikonda R, Utset TO, Clark MR. Predicting outcomes of lupus nephritis with tubulointerstitial inflammation and scarring. Arthritis care & research. 2011;63(6):865–74. doi: 10.1002/acr.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, et al. Laser capture microdissection. Science. 1996;274(5289):998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 13.Satoskar AA, Shapiro JP, Bott CN, Song H, Nadasdy GM, Brodsky SV, et al. Characterization of glomerular diseases using proteomic analysis of laser capture microdissected glomeruli. Mod Pathol. 2012;25(5):709–21. doi: 10.1038/modpathol.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro JP, Biwas S, Merchant AS, Satoskar A, Taslim C, Lin S, et al. A quantitative proteomic workflow for characterization of frozen clinical biopsies: Laser capture microdissection coupled with label-free mass spectrometry. J Prot. 2012;77:433–40. doi: 10.1016/j.jprot.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sethi S, Vrana JA, Theis JD, Dogan A. Mass spectrometry based proteomics in the diagnosis of kidney disease. Curr Opin Nephrol Hypertens. 2013;22(3):273–80. doi: 10.1097/MNH.0b013e32835fe37c. [DOI] [PubMed] [Google Scholar]
- 16.Jain D, Green JA, Bastacky S, Theis JD, Sethi S. Membranoproliferative glomerulonephritis: the role for laser microdissection and mass spectrometry. Am J Kidney Dis. 2014;63(2):324–8. doi: 10.1053/j.ajkd.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Regent A, Lofek S, Dib H, Bussone G, Tamas N, Federici C, et al. Identification of target antigens of anti-endothelial cell antibodies in patients with anti-neutrophil cytoplasmic antibody-associated vasculitides: a proteomic approach. Clin Immunol. 2014;153(1):123–35. doi: 10.1016/j.clim.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Beck LH, Jr, Bonegio RGB, Lambeau G, Beck DM, Powell DW, Cummins TD, et al. M-Type Phospholipase A(sub 2) Receptor as Target Antigen in Idiopathic Membranous Nephropathy. New England Journal of Medicine. 2009;361(1):11–21. doi: 10.1056/NEJMoa0810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomas NM, Beck LH, Jr, Meyer-Schwesinger C, Seitz-Polski B, Ma H, Zahner G, et al. Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med. 2014;371(24):2277–87. doi: 10.1056/NEJMoa1409354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruschi M, Sinico RA, Moroni G, Pratesi F, Migliorini P, Galetti M, et al. Glomerular autoimmune multicomponents of human lupus nephritis in vivo: alpha-enolase and annexin AI. J Am Soc Nephrol. 2014;25(11):2483–98. doi: 10.1681/ASN.2013090987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikuchi T, Yoshida Y, Morioka T, Gejyo F, Oite T. Human voltage-dependent anion selective channel 1 is a target antigen for antiglomerular endothelial cell antibody in mixed connective tissue disease. Mod Rheumatol. 2008;18(6):570–7. doi: 10.1007/s10165-008-0094-4. [DOI] [PubMed] [Google Scholar]
- 22.Haubitz M, Wittke S, Weissinger EM, Walden M, Rupprecht HD, Floege J, et al. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney international. 2005;67(6):2313–20. doi: 10.1111/j.1523-1755.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 23.Varghese SA, Powell TB, Budisavljevic MN, Oates JC, Raymond JR, Almeida JS, et al. Urine biomarkers predict the cause of glomerular disease. J Am Soc Nephrol. 2007;18:913–22. doi: 10.1681/ASN.2006070767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Jin M, Wu H, Nadasdy T, Nadasdy G, Harris N, et al. Biomarkers of lupus nephritis determined by serial urine proteomics. Kidney international. 2008;74(6):799–807. doi: 10.1038/ki.2008.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu T, Du Y, Han J, Singh S, Xie C, Guo Y, et al. Urinary angiostatin--a novel putative marker of renal pathology chronicity in lupus nephritis. Mol Cell Proteomics. 2013;12(5):1170–9. doi: 10.1074/mcp.M112.021667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surin B, Sachon E, Rougier JP, Steverlynck C, Garreau C, Lelongt B, et al. LG3 fragment of endorepellin is a possible biomarker of severity in IgA nephropathy. Proteomics. 2013;13(1):142–52. doi: 10.1002/pmic.201200267. [DOI] [PubMed] [Google Scholar]
- 27.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11(6):O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]