Abstract
Context
Head and neck squamous cell carcinoma (HNSCC) is a highly invasive cancer with an association with locoregional recurrence and lymph node metastasis. We have previously reported that low microRNA-375 (miR-375) expression levels correlate with poor patient survival, increased locoregional recurrence, and distant metastasis. Increasing miR-375 expression in HNSCC cell lines to levels found in normal cells results in suppressed invasive properties. HNSCC invasion is mediated in part by invadopodia-associated degradation of the extracellular matrix.
Objective
To determine whether elevated miR-375 expression in HNSCC cell lines also affects invadopodia formation and activity.
Design
For evaluation of the matrix degradation properties of the HNSCC lines, an invadopodial matrix degradation assay was used. The total protein levels of invadopodia-associated proteins were measured by Western blot analyses. Immunoprecipitation experiments were conducted to evaluate the tyrosine phosphorylation state of cortactin. Human Protease Arrays were used for the detection of the secreted proteases. Quantitative real time–polymerase chain reaction measurements were used to evaluate the messenger RNA (mRNA) expression of the commonly regulated proteases.
Results
Increased miR-375 expression in HNSCC cells suppresses extracellular matrix degradation and reduces the number of mature invadopodia. Higher miR-375 expression does not reduce cellular levels of selected invadopodia-associated proteins, nor is tyrosine phosphorylation of cortactin altered. However, HNSCC cells with higher miR-375 expression had significant reductions in the mRNA expression levels and secreted levels of specific proteases.
Conclusions
MicroRNA-375 regulates invadopodia maturation and function potentially by suppressing the expression and secretion of proteases.
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide with roughly 50 000 new cases and 11 000 cancer deaths in the United States every year.1 HNSCC develops in the mucosal layer of the upper aerodigestive tract. Owing to the highly invasive nature of HNSCC, it is often associated with locoregional recurrence and metastasis to cervical lymph nodes.2–5 Early-stage tumors are often treated with radiation or surgery, while later-stage tumors are treated with the combination of surgery, radiotherapy, and/or chemotherapy.6 Despite attempts to develop new and improved treatment strategies, the 5-year survival rate associated with HNSCC has not significantly changed and still remains approximately 40% to 50%.1 Targeting molecules driving HNSCC invasion and understanding the mechanisms of invasion may lead to improvements in patient outcome.
MicroRNAs (miRNAs) are small noncoding RNAs that are approximately 22 nucleotides in length and function as posttranscriptional regulators of gene expression.7–9 Micro-RNAs are thought to regulate roughly 30% of protein-coding genes.9 Some miRNAs, including miR-375, have been identified as diagnostic and prognostic markers in human cancers.8,10–12 MicroRNAs regulate the proteome directly and indirectly by controlling many processes of cancer cells, including cellular proliferation, differentiation, apoptosis, invasion/metastasis, and angiogenesis.8,12–14 Multiple studies have demonstrated lower expression of miR-375 in HNSCC and other tumors compared with normal cells.11,15–19 The ectopic overexpression of miR-375 in HNSCC cell lines and other cancer cells results in the suppression of cell proliferation, migration, and invasion.11,15,19–23
The ability of cells to degrade extracellular matrix (ECM) through secretion of proteases allows local invasion into the surrounding stroma, as well as possible metastasis.24 Invadopodia are specialized actin-rich structures, which provide cancer cells with ECM degradation capacity.25–28 Invadopodium formation involves the assembly of actin regulators, including cortactin, cofilin, neural Wiskott-Aldrich syndrome protein (N-WASP), and tyrosine kinase substrate with 5 SH3 domains (Tks5/Fish).29,30 These invadopodium precursor structures lack the ability to degrade the ECM.31,32 Invadopodium precursors are stabilized by the association of Tks5 with phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2).30 Fascin, an actin-bundling protein, stabilizes actin in invadopodia.33 In HNSCC cells, increased epithelial growth factor receptor signaling results in elevated Src and extracellular-signal–regulated kinase (Erk) 1/2 activity, which induces tyrosine and serine phosphorylation of cortactin.34 Tks5/Fish is tyrosine phosphorylated by Src.35–37 The tyrosine phosphorylation of cortactin and Tks5 enhances binding to noncatalytic region of tyrosine kinase (Nck) family proteins in invadopodia.37–39 Intracellular pH changes reverse the inhibitory interaction of cofilin and cortactin, allowing cofilin to sever F-actin, which induces actin polymerization, leading to membrane protrusion at invadopodia.37–39 The final step of the invadopodia maturation process involves the secretion of proteases, such as matrix metalloproteinase 2 (MMP-2) and matrix metal-loproteinase 9 (MMP-9), and presentation of membrane type 1 metalloproteinase (MT1-MMP) at the protruding invadopodia.40
We previously reported that miR-375 was identified as a potential prognostic marker of poor outcome and metastasis in HNSCC.11 Also in that study, we observed that diminished in vitro invasive properties correlate with increased miR-375 expression in HNSCC cell lines. HNSCC invasion has been previously associated with invadopodia activity.34,40–44 Here we report that increased miR-375 expression in HNSCC cell lines suppresses the number of mature invadopodia as well as mRNA expression and secreted levels of proteases, resulting in markedly reduced ECM degradation.
MATERIALS AND METHODS
Generation of OSC19 Transductant Lines and Cell Culture Conditions
The generation of stable UMSCC1 control and precursor miR-375–expressing (Pre375) lentiviral transductant cell lines was previously described.11 Another oral cavity human papillomavirus–negative squamous cell carcinoma cell line, OSC19, was kindly provided by Jeffrey Myers, PhD, from University of Texas MD Anderson Cancer Center, Houston, Texas. For the generation of stable OSC19 lentiviral transductant cell lines, empty vector control or precursor miR-375 pseudoviral particle-containing medium supplemented with polybrene (8 µg/mL) was applied on OSC19 cells. Stable OSC19 empty vector control and Pre375 transductant lines were sorted for similar green fluorescent protein levels. Cells were grown in Dulbecco modified Eagle medium (DMEM)/High Glucose (Catalog No. [Cat.] SH30243, Hyclone/Thermo Fisher, Waltham, Massachusetts), 10% fetal bovine serum (Cat. 10082-147, Gibco/Life Technologies, Norwalk, Connecticut), penicillin-streptomycin (100 units/mL penicillin, 100 µg/mL streptomycin; Cat. 15140-122, Gibco/Life Technologies), 100 µM nonessential amino acids (Cat. 11140-050, Gibco/Life Technologies), and 2 mM l-glutamine (Cat. 25030-081, Gibco/Life Technologies). Both the UMSCC1 and OSC19 transductant cell lines were treated with 50 µg/mL Plasmocure (Cat. ant-pc, InvivoGen, San Diego, California) for more than 2 weeks. The transductant cell lines were maintained in a 37°C incubator with 5% CO2.
Antibodies
Total cortactin (Cat. 05–180) and phosphorylated tyrosine 421 (pY421) cortactin (Cat. AB3852) antibodies were purchased from EMD Millipore, Billerica, Massachusetts. Total Tks5 (Cat. sc-30122) antibody was purchased from Santa Cruz Biotechnology, Inc, Santa Cruz, California. β-tubulin (Cat. T4026), β-actin (Cat. A5441), and pY421 cortactin (Cat. C0739) antibodies were purchased from Sigma-Aldrich, St Louis, Missouri. Fascin (Cat. 9269) and total phosphorylated tyrosine proteins (P-Tyr-1000) (Cat. 8954) antibody were purchased from Cell Signaling Technology, Danvers, Massachusetts. Rabbit immunoglobulin G (IgG) antibody (Cat. 02– 6102) was purchased from Invitrogen/Life Technologies, Norwalk, Connecticut. IRDye 680RD anti-mouse (Cat. 926–68072) and IRDye 800CW anti-rabbit (Cat. 926–32213) secondary antibodies were purchased from LI-COR Biosciences, Lincoln, Nebraska. Mouse IgG (Cat. 015-000-003), cyanine (Cy) 5 anti-rabbit (Cat. 711-175-152), and Cy3 anti-mouse (Cat. 715-165-161) antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc, West Grave, Pennsylvania.
Matrix Degradation Assay and Immunofluorescence
Labeling of gelatin with Alexa Fluor 405 dye (Life Technologies, Norwalk, Connecticut) and preparation of gelatin-coated MatTek dishes (MatTek Corporation, Ashland, Massachusetts) were performed as previously described.45 For the matrix degradation assay, 1 × 105 UMSCC1 transductant cells or 0.5 × 105 OSC19 transductant cells were plated in DMEM culture media on Alexa Fluor 405 gelatin-coated MatTek dishes and cultured for 4 hours. Cells were fixed in 4% paraformaldehyde for 20 minutes. Foci of degraded matrix were identified as dark spots that lacked Alexa Fluor 405 fluorescence. For immunofluorescence with invadopodium markers, cells were permeabilized in 0.05% Triton X-100 in 1× phosphate-buffered saline for 5 minutes after fixation. Cells were blocked in 1% bovine serum albumin/1% fetal bovine serum in 1× phosphate-buffered saline for 30 minutes and incubated with total cortactin (1:100) and Tks5 (1:50) or phosphocortactin (pY421) (1:200; Sigma) antibodies for 1 hour at room temperature. Cells were then incubated with Cy5 anti-rabbit and Cy3 anti-mouse (both 1:400) secondary antibodies for 1 hour at room temperature. Cells were imaged on an Inverted Olympus IX71 microscope (Olympus, Center Valley, Pennsylvania). Ten fields per cell line were analyzed in 3 independent experiments. Matrix degradation area (µm2) per field was quantified by using the FIJI image processing program46 and normalized to the number of cells in the field. Invadopodium precursors were identified as Tks5 and cortactin colocalized puncta lacking gelatin degradation; mature invadopodia were defined as Tks5 and cortactin colocalized puncta that were overlying Alexa Fluor 405 matrix degradation holes.29,31,32,45 For the assessment of cortactin activation at sites of matrix degradation, regions of interest were drawn in FIJI around puncta labeled for both total cortactin and phosphocortactin (pY421), which were overlying gelatin degradation holes. The signal intensities of total cortactin and phosphocortactin were measured and ratio of phosphocortactin to total cortactin was determined.
Western Blot
UMSCC1 transductant cells were lysed in radioimmunoprecipitation assay buffer containing 1× protease inhibitors (Cat. 11836170-001 [Roche, Nutley, New Jersey]). Cell lysates were sonicated for 15 seconds at 4°C. Quantitation of total protein concentrations was performed with the Pierce Bicinchoninic Acid Protein Assay (Cat. 23225 [Thermo Scientific, Rockford, Illinois]). Sodium dodecyl sulfate–polyacrylamide gel electrophoresis sample loading buffer was added to 40 µg of total protein and heated at 95°C for 5 minutes. Samples were loaded in polyacrylamide gels and run at 120 volts for approximately 1.5 hours. Gels were transferred to nitrocellulose membranes at 100 volts for 1 hour. Membranes were blocked in LI-COR Odyssey blocking buffer (Cat. 927–40000 [LI-COR Biosciences]) for 1 hour at room temperature and incubated with primary antibodies (β-tubulin [1:1000] and Tks5 [1:1000] or cortactin [1:1000]; β-actin [1:20 000] and fascin [1:1000]) overnight at 4°C. The membranes were washed 3 times with 1× Tris Buffered Saline-Tween (TBS-T) and incubated with secondary antibodies (IRDye 680RD anti-mouse and IRDye 800CW anti-rabbit [1:5000]) for 1 hour at room temperature. The membranes were then washed again 3 times with 1× TBS-T and scanned with the LI-COR Odyssey Infrared Imaging System. The Western blot images were analyzed by using ImageJ.47 β-tubulin or β-actin was used as a loading control. The mean intensities of the Tks5, cortactin, fascin, β-actin, and β-tubulin bands were measured and the corresponding background mean intensities were subtracted. For Tks5 and cortactin, the background subtracted mean intensities of the Tks5 and cortactin bands were normalized to the background subtracted mean intensity of the β-tubulin band, and ratios of the value for the miR-375 line to its respective control line were calculated. For fascin, the background subtracted mean intensities of the fascin band were normalized to the background subtracted mean intensities of the β-actin bands, and ratio of the value for the miR-375 line to its respective control line was calculated.
Immunoprecipitation
Immunoprecipitation (IP) experiments were conducted with the Pierce Classic IP Kit (Cat. 26146). UMSCC1 transductant cells were lysed with cold IP Lysis/Wash Buffer containing phosphatase inhibitor cocktails (set 1: Cat. 524624 [Chemicon, Billerica, Massachusetts] and set 2: Cat. P5726 [Sigma]) and protease inhibitors (Cat. 11836170-001; Roche). Cell lysates were centrifuged at 13 000g for 10 minutes and protein concentrations of lysates were determined by bicinchoninic acid protein assay. One milligram of lysate was incubated with 10 µg of cortactin antibody, mouse IgG, or rabbit IgG, or phosphotyrosine antibody diluted 1:200 overnight at 4°C to form the immune complex. Pierce Protein A/G beads were added to Pierce Spin Columns and rinsed twice with ice-cold IP Lysis/Wash Buffer. Immune complexes were captured by incubation with Protein A/G beads in spin columns for 6 hours. Columns were centrifuged to remove the immunodepleted lysate and were rinsed 3 times with ice-cold IP Lysis/Wash Buffer. Samples were eluted with 23 reducing sample buffer containing 20 mM dithiothreitol and analyzed by Western blot. Immunoprecipitated phosphotyrosine containing proteins were probed for cortactin levels. The intensities of the cortactin in the phosphotyrosine immunoprecipitate and total cortactin in the input were measured in ImageJ, background subtracted, and the ratio determined. Conversely, total immunoprecipitated cortactin was probed by using total cortactin and phosphocortactin (pY421) (Millipore) antibodies. The intensities of the total cortactin and phosphocortactin bands were measured in ImageJ, background subtracted, and the ratio of phosphocortactin to total cortactin was determined.
Protease Array
Secreted protease profiling was performed with Human Proteome Profiler Protease Array from R & D Systems, Minneapolis, Minnesota (Cat. ARY021). To generate conditioned cell culture medium for the protease arrays, 1 × 106 cells were cultured in a 6-cm dish in 3 mL of cell culture medium for 28 hours, followed by collection of the conditioned culture medium. The protease array membranes were incubated for 1 hour in Assay Buffer 6. Five hundred microliters of Assay Buffer 4 and 1 mL of conditioned cell culture medium from the HNSCC cells were combined with 15 µL of Detection Antibody Cocktail and incubated for 1 hour. The Assay Buffer 6 was aspirated from the wells containing the membranes. The prepared sample/antibody mixtures were added to the membranes and incubated overnight. Membranes were then washed with 1× Wash Buffer 3 times. Two milliliters of diluted Streptavidin-HRP was added to each membrane and incubated for 30 minutes. Membranes were washed 3 times with 1×Wash Buffer. One milliliter of the prepared Chemi Reagent Mix was added onto each membrane and incubated for 1 minute. Two methods were used for the determination of the signal intensities of the proteases. In the case of the first 2 UMSCC1 experiments, the membranes were exposed to x-ray film for varying amounts of time. The x-ray films were scanned and the intensities of the proteases were determined from scanned images in ImageJ. For all other experiments, the chemilluminescence signals of proteases were measured with a LI-COR Odyssey Fc scanner. The intensities of proteases were determined in ImageStudio Lite program and the protease signal intensities from the cell line with increased expression were compared to its respective control line.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from HNSCC cells by using the RNeasy Mini Kit from QIAGEN, (Cat. 74104, Germantown, Maryland) following manufacturer’s protocol. Two hundred nanograms of RNA from each cell line was used for the quantitative real-time polymerase chain reaction (qPCR) measurements. The measurements were made with the TaqMan RNA to Ct One Step Kit (Cat. 4392938) from Life Technologies, by using their recommended parameters. TaqMan Gene Expression assays used were Hs00160519_m1 (kallikrein 6, KLK6), Hs00173611_m1 (kallikrein 10, KLK10), Hs00234579_m1 (matrix metalloproteinase 9, MMP9) and Hs99999905_m1 (glyceraldehyde 3-phosphate dehydrogenase, GAPDH). For each cell line, a mean cycle threshold (CT) value was calculated from the 3 replicates. For each cell line, the changes in the mean CT values (ΔCT) were computed by subtracting the mean CT value of GAPDH from the mean CT value of KLK6, KLK10, or MMP9. To calculate the change in the ΔCT value in the UMSCC1 and OSC19 Pre375 cell lines relative to their respective control lines, ΔΔCT values were computed and used to calculate the fold change as 2−ΔΔCT.
Ingenuity Pathway Analysis
The predicted miR-375 target list was obtained from Target-Scan.48 A functional analysis was performed with the use of QIAGEN’S Ingenuity Pathway Analysis (QIAGEN Redwood City, California; www.qiagen.com/ingenuity). In the Diseases and Disorders category, Invasion was filtered for in the Cancer subcategory. A list of putative miR-375 targets associated with cell movement was obtained. In the Molecular and Cellular Functions category, putative miR-375 targets involved in formation of cellular protrusions and organization of actin cytoskeleton were identified in the Cellular Assembly and Organization subcategory. These putative miR-375 targets were then compared to an updated list of proteins in the invadopodia network.49 A modified version of the invadopodia network was generated in Ingenuity Pathway Analysis, showing some of the interactions between the invado-podia-associated proteins and highlighting the theoretical miR-375 targets, which are invadopodia-associated proteins.
Statistical Analysis
All of the results shown are based on at least 3 independent experiments. Statistical analyses were assessed by using 2-tailed unpaired t tests or z tests.
RESULTS
Increased miR-375 Expression in HNSCC Cell Lines Suppresses Matrix Degradation and Mature Invadopodia
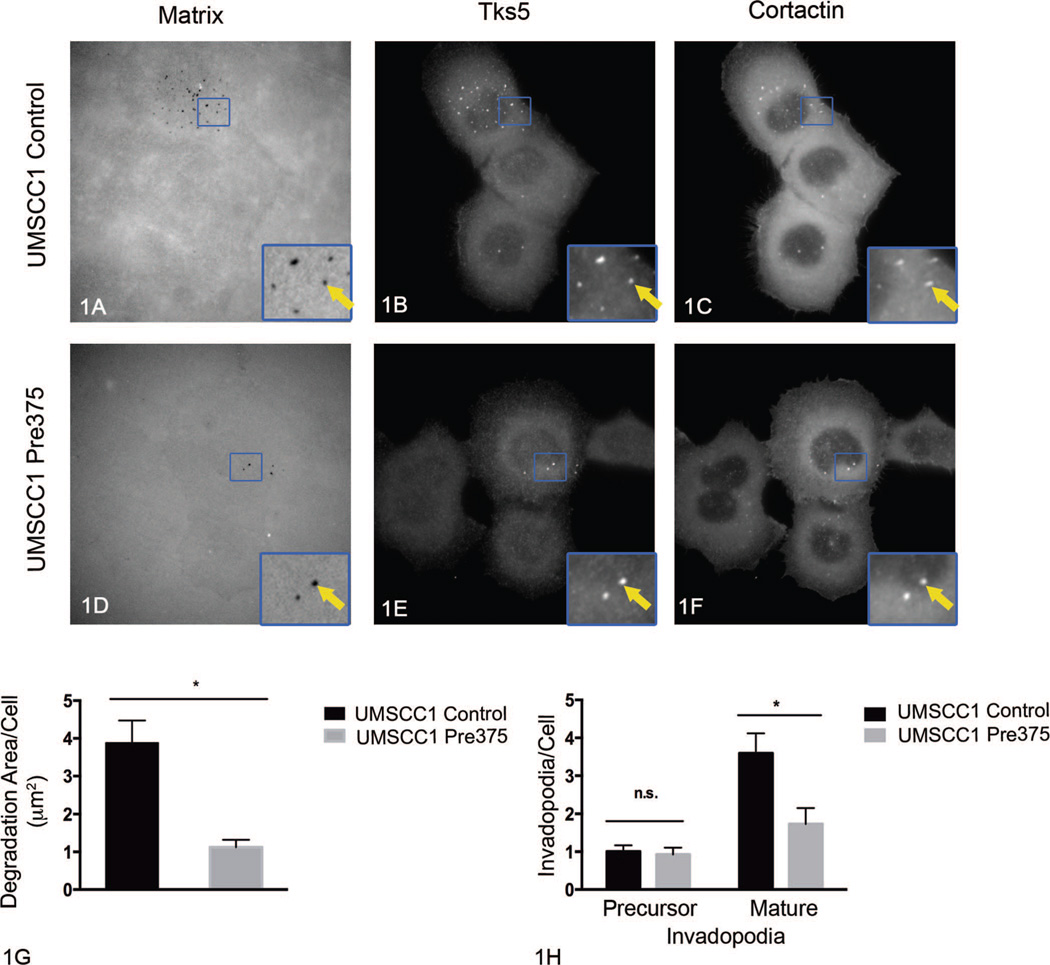
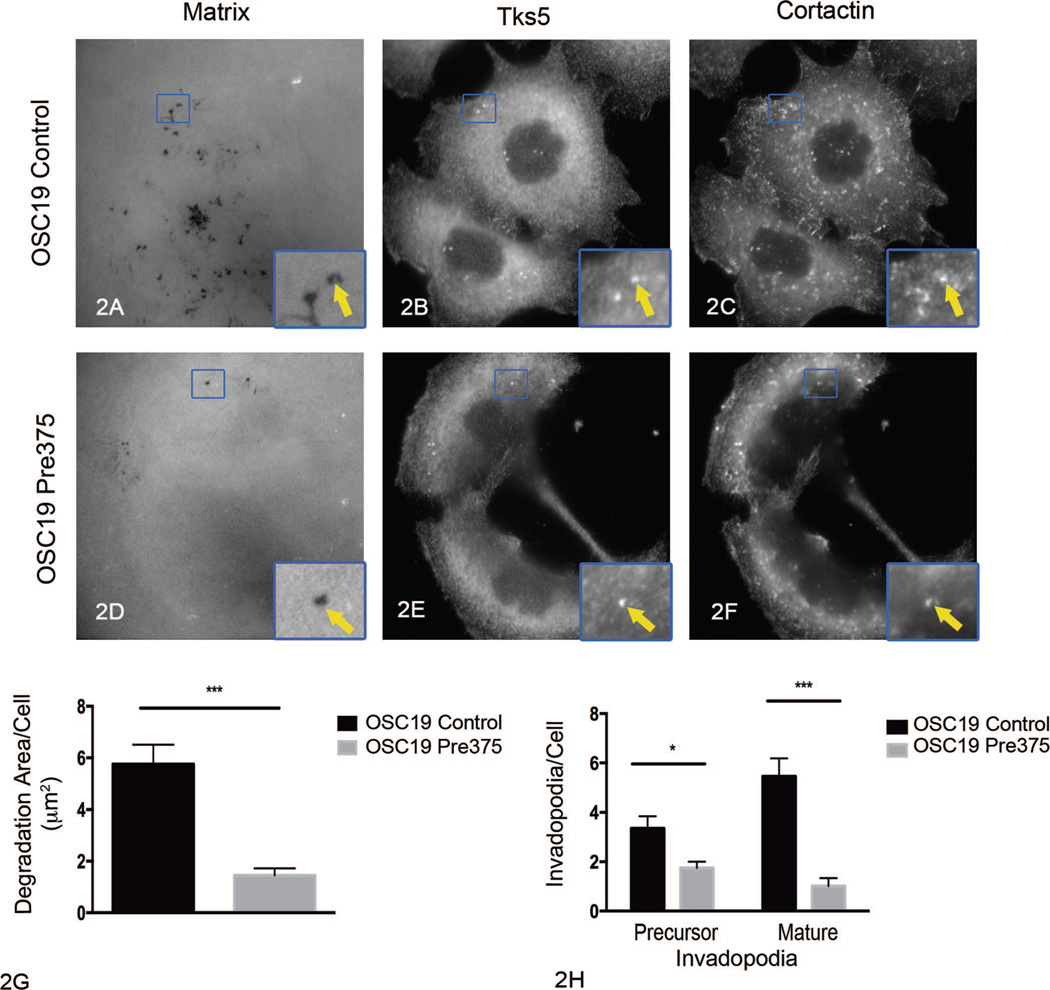
We previously reported that stable increased miR-375 expression in the UMSCC1 cell line results in diminished invasion in vitro.11 One major determining feature of the ability of cells to invade is their capability to degrade matrix barriers.24 Invadopodia are cellular structures, which can mediate degradation of extracellular matrix barriers.25–28,45,50 Since invadopodia mediate invasion and matrix degradation, we asked if elevated miR-375 expression in HNSCC cell lines affects invadopodium formation and maturation. To do that, we used the fluorescent matrix degradation assay in combination with the immunolocalization of cortactin and Tks5 (2 invadopodia markers). From this assessment, the area of Alexa Fluor 405 gelatin matrix degradation and numbers of precursor and mature invadopodia were determined. Invadopodium precursors were defined as puncta of Tks5 and cortactin colocalization that were not associated with gelatin degradation; mature invadopodia were defined as Tks5 and cortactin colocalized puncta that were associated with gelatin degradation holes.29,31,32,45 After incubation on the gelatin matrix for 4 hours, UMSCC1 cells expressing increased miR-375 expression (UMSCC1 Pre375) showed a 71% reduction in matrix degradation area per cell and a 52% reduction in the number of mature invadopodia per cell, compared to empty vector UMSCC1 control cells (Figure 1, A through H). In contrast to measurements of mature invadopodia, increased miR-375 expression in UMSCC1 cells did not significantly change the numbers of invadopodium precursors per cell (Figure 1, H). OSC19 cells expressing higher levels of miR-375 (OSC19 Pre375) showed a 73% reduction in matrix degradation area per cell when compared to OSC19 control cells (Figure 2, A through H). OSC19 cells with increased miR-375 expression showed an 81% reduction in the number of mature invadopodia per cell, as well as a 48% reduction in invadopodium precursors, compared to OSC19 control cells (Figure 2, A through H). The major effect of increased miR-375 expression in the UMSCC1 and OSC19 cells is on matrix degradation and mature invadopodia numbers.
Figure 1.
Increased microRNA-375 expression impairs extracellular matrix degradation and number of mature invadopodia in UMSCC1 cells. Representative images of fluorescent matrix (A, D), tyrosine kinase substrate with 5 SH3 domains (Tks5) (B, E), and cortactin (C, F) immunostaining for UMSCC1 transductant cells. Arrows in zoomed areas show matrix degradation holes colocalized with Tks5 and cortactin puncta. Quantitation of matrix degradation per cell for UMSCC1 transductant cells (G) shown as mean and standard error of the mean (SEM) of 4 independent experiments; **P=.002. Quantitation of invadopodium precursors and mature invadopodia per cell for UMSCC1 transductant cells (H) shown as mean and SEM of 4 independent experiments; *P=.02 (original magnification × 413 [A through F]; original magnification × 1270 [insets A through F]). Abbreviation: n.s., not significant.
Figure 2.
Increased microRNA-375 expression impairs extracellular matrix degradation and invadopodia numbers in OSC19 cells. Representative images of fluorescent matrix (A, D), tyrosine kinase substrate with 5 SH3 domains (Tks5) (B, E), and cortactin (C, F) immunostaining for OSC19 transductant cells. Arrows in zoomed areas show matrix degradation holes colocalized with Tks5 and cortactin puncta. Quantitation of matrix degradation per cell for OSC19 transductant cells (G) shown as mean and standard error of the mean (SEM) of 5 independent experiments; ***P< .001. Quantitation of invadopodium precursors and mature invadopodia per cell for OSC19 transductant cells (H) shown as mean and SEM of 5 independent experiments; *P=.01, ***P <.001 (original magnification ×413 [A through F]; original magnification ×1191 [insets A through F]).
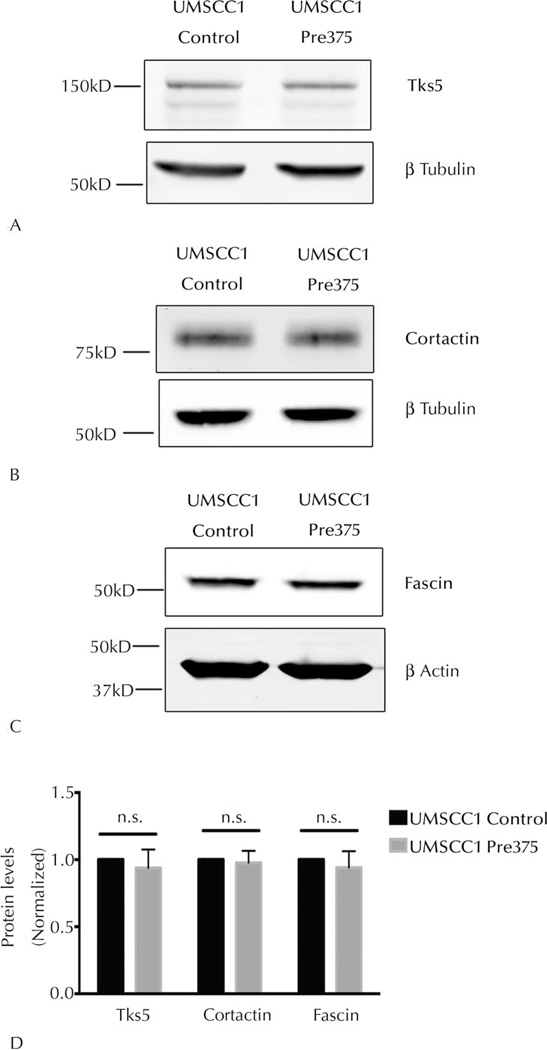
Elevated Levels of miR-375 in UMSCC1 Did Not Significantly Alter Total Protein Levels of Tks5, Cortactin, and Fascin
One possible mechanism for having fewer invadopodia is the suppression of the levels of key invadopodia-associated proteins. Total levels of Tks5, cortactin, and fascin proteins in the UMSCC1 transductant cells were assessed by Western blot analyses. Tks5 has been identified as a theoretical miR-375 direct-target regulated gene by various miRNA target-prediction algorithms, including TargetScan,48 Mirwalk,51 DIANAmT,52 and Miranda,53 and was therefore of particular interest. Quantitative Western blot analysis showed that there were no significant reductions in the total Tks5, cortactin, or fascin protein levels in UMSCC1 Pre375 cells compared to the control line (Figure 3, A through D). Tks5 therefore does not appear to be a direct target of miR-375, nor are the other measured proteins regulated by this miRNA.
Figure 3.
Elevated microRNA-375 expression does not significantly alter total Tks5 (tyrosine kinase substrate with 5 SH3 domains), cortactin, and fascin protein levels in UMSCC1 cells. Representative Western blots of total Tks5 (A), cortactin (B), and fascin (C) protein levels in UMSCC1 transductant lines. β-Tubulin or β-actin was used as a loading control for Western blots. Quantitation of total Tks5, cortactin, and fascin levels in UMSCC1 transductant cells (D) shown as means and standard error of the mean of 5 independent experiments. Abbreviation: n.s., not significant.
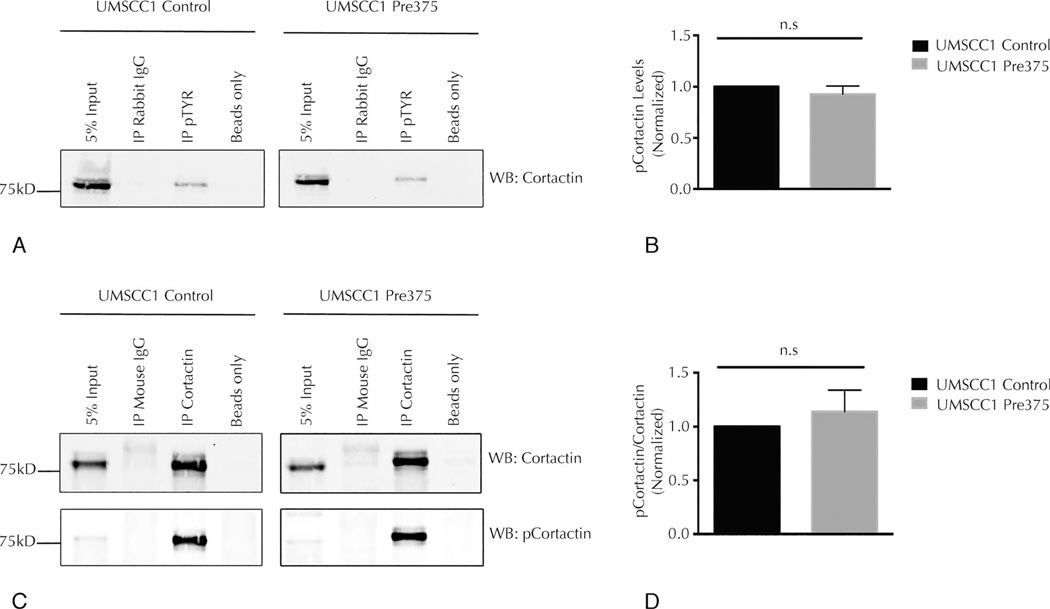
Higher miR-375 Expression in UMSCC1 Cells Does Not Diminish the Phosphorylation State of Cortactin
Given the unchanged levels of cortactin and other invadopodia proteins, we looked for tyrosine phosphorylation of cortactin as a way to regulate invadopodial maturation.54 Immunoprecipitation experiments of total tyrosine phosphorylated proteins in the UMSCC1 trans-ductant cells were conducted to evaluate the overall tyrosine phosphorylation state of cortactin. The levels of tyrosine phosphorylation of cortactin measured by this assay were not significantly changed in UMSCC1 cells with increased miR-375 expression compared to its empty vector control line (Figure 4, A and B). We then evaluated the phosphorylation levels of a specific key tyrosine residue (Y421) by immunoprecipitation of cortactin protein in UMSCC1 transductant cells, followed by Western blots for cortactin and phosphocortactin (pY421). The level of phosphocortactin (pY421) was not consistently changed in UMSCC1 cells containing higher levels of miR-375 compared to its control line (Figure 4, C and D). Taken together, the data presented in Figures 3 and 4 indicate that altered cellular levels of total invadopodia-associated proteins or levels of phosphocortactin are not responsible for the reduction in mature invadopodia seen in the cell lines with higher miR-375 expression.
Figure 4.
Higher microRNA-375 expression does not reduce tyrosine phosphorylation of cortactin in UMSCC1 cells. A, Representative Western blots (WBs) for cortactin in total phosphotyrosine immunoprecipitates (IP pTYR) compared to the total cell lysate (5% input) and nonspecific rabbit immunoglobin G (IP Rabbit IgG) and beads (Beads only) control. B, Quantitation of cortactin levels in immunoprecipated phosphotyrosine proteins, shown as mean and standard error of the mean (SEM) of 4 independent experiments. C, Representative WBs for cortactin and phosphocortactin (pY421) in cortactin immunoprecipitates (IP Cortactin) compared to the total cell lysate (5% input) and nonspecific mouse IgG (IP Mouse IgG) and beads (Beads only) controls. D, Quantitation of phosphocortactin levels compared to total cortactin in cortactin immunoprecipitates, shown as mean and SEM of 4 independent experiments. Abbreviation: n.s., not significant.
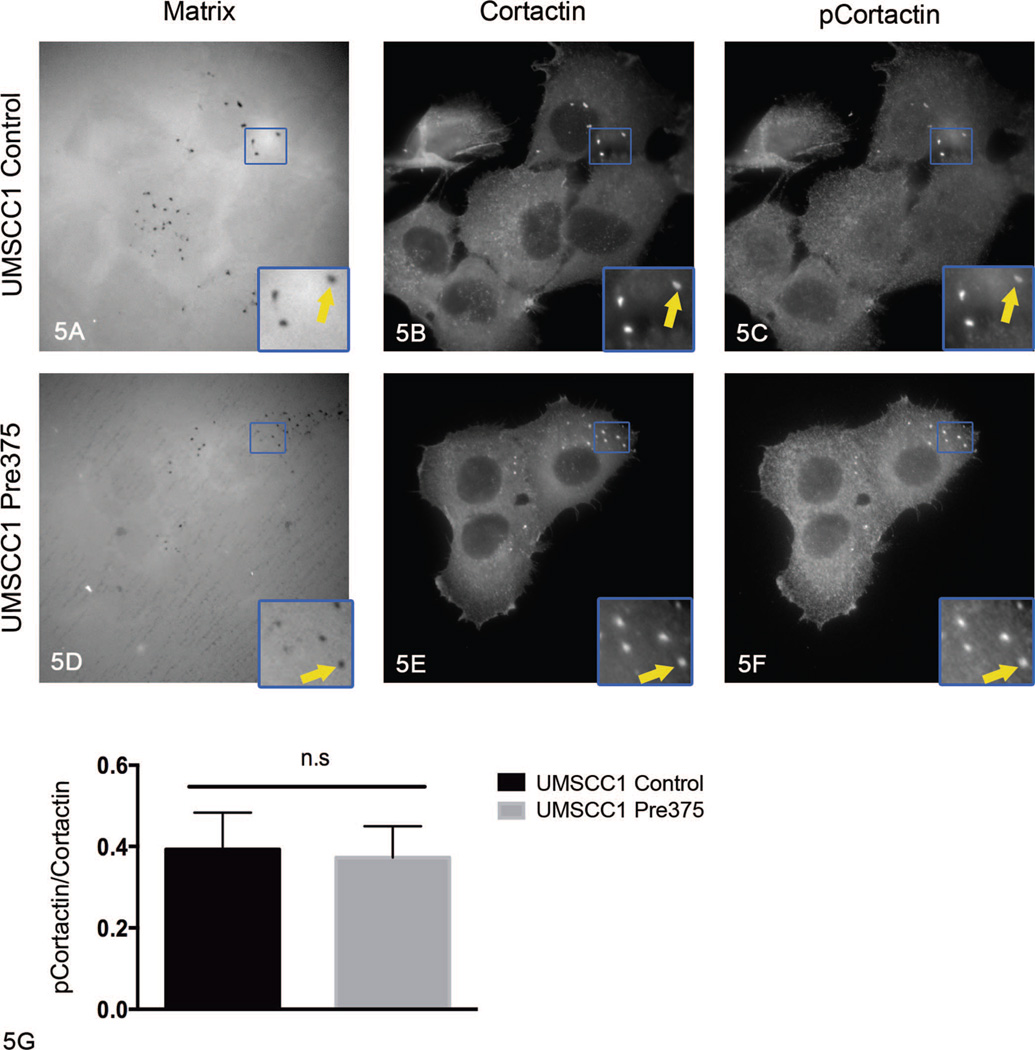
The experiments presented above did not evaluate the possibility that the phosphorylation state of cortactin was selectively altered in the mature invadopodia. To assess whether there is reduced tyrosine phosphorylation of cortactin specifically at sites of matrix degradation, the fluorescent matrix degradation assay was combined with immunofluorescence for cortactin and phosphocortactin (pY421).54 For this assessment, UMSCC1 transductant cells were plated on the Alexa Fluor 405 gelatin-coated MatTek dishes, cultured for 4 hours, fixed, and stained by using total anti-cortactin and anti-phosphocortactin (pY421) antibodies. The signal intensity of total cortactin and phosphocortactin puncta, which were colocalized with matrix degradation holes, were measured and the ratio of phosphocortactin to total cortactin was determined. The ratio of phosphocortactin to total cortactin at matrix degradation holes was not significantly changed in UMSCC1 Pre375 cells compared to UMSCC1 control cells (Figure 5, A through G). This result suggests that the decreases in mature invadopodia formed in the cells with elevated miR-375 expression are not due to reduced cortactin pY421 phosphorylation in invadopodia.
Figure 5.
Increased microRNA-375 expression in UMSCC1 cells did not diminish cortactin tyrosine phosphorylation at matrix degradation holes. Representative images of fluorescent matrix (A, D), cortactin (B, E), and phosphocortactin (pY421) (C, F) immunostaining for UMSCC1 transductant cells. Arrows in zoomed areas show matrix degradation holes colocalized with cortactin and phosphocortactin puncta. G, Quantitation of the ratio of signal intensity of total cortactin and phosphocortactin at matrix degradation holes, shown as mean and standard error of the mean of 3 independent experiments (original magnification ×413 [A through F]; original magnification ×953 [insets A through F]).
Increased miR-375 Expression in HNSCC Cells Causes Reductions in Numerous Secreted Proteases
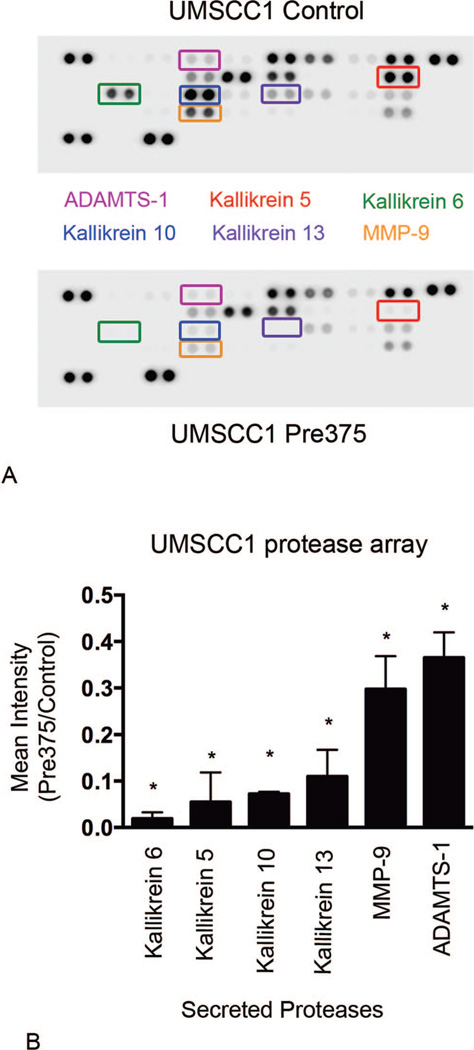
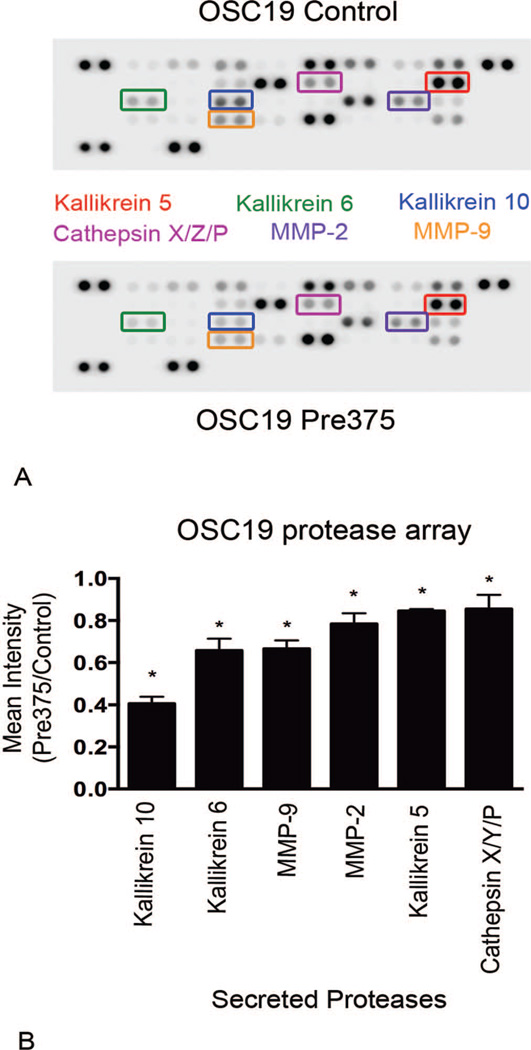
Extracellular matrix degradation is caused by the action of proteases acting on their substrates within the matrix.26,28,55 For the detection of possible effects of increased miR-375 expression in HNSCC cells on the secretion of proteases from the cells, Proteome Profile Human Protease Arrays were used. The protease arrays allow for quantitative measurements of proteases secreted into the cell culture medium. Increased levels of miR-375 expression in UMSCC1 cells resulted in dramatic reductions in secreted proteases. In the conditioned cell culture medium from UMSCC1 cells with higher expression of miR-375, the most significantly reduced secreted proteases were a group of serine proteases known as kallikreins: kallikrein 5, kallikrein 6, kallikrein 10, and kallikrein 13. The secreted levels of these proteases were significantly suppressed by 95%, 98%, 93%, 89%, respectively, compared to the empty vector control line (Figure 6, A and B). In the conditioned medium from UMSCC1 cells with increased expression of miR-375 there was also significant reductions in the secreted levels of MMP-9 and ADAMTS-1, which were diminished by 70% and 64%, respectively (Figure 6, A and B). In OSC19 cells, higher miR-375 expression resulted in less dramatic, but significant reductions in secreted levels of kallikrein 6, kallikrein 10, MMP-9, and kallikrein 5. These proteases were reduced by 59%, 34%, 33%, and 15%, respectively, compared to the empty vector control line (Figure 7, A and B). OSC19 cells with increased miR-375 expression also showed MMP-2 and cathepsin X/Z/P secreted levels significantly diminished by 22% and 15%, respectively. Our results revealed reproducible reductions in some proteases with additional decreases in other proteases unique to each cell line. The most strongly diminished proteases in the lines with higher miR-375 expression were kallikrein 6, kallikrein 10, and MMP-9.
Figure 6.
UMSCC1 cells expressing increased levels of microRNA-375 show dramatic reductions in protease secretion levels. Conditioned cell culture supernatants were harvested and assayed for protease secretion levels by using the Proteome Profiler Human Protease Arrays. A, Representative images of protease arrays incubated with cell culture supernatants from UMSCC1 transductant cells. B, Ratio of the 6 proteases that were most diminished in the protease array analyses, shown as means and standard error of the mean of 3 independent experiments; *P < .001. Abbreviations: ADAMTS-1, ADAM metal-lopeptidase with thrombospondin type 1 motif, 1; MMP-9, matrix metalloproteinase 9.
Figure 7.
OSC19 cells expressing higher levels of microRNA-375 exhibit declines in secreted levels of proteases. Conditioned cell culture supernatants were harvested and assayed for protease secretion by using the Proteome Profiler Human Protease Arrays. A, Representative images of protease arrays incubated with cell culture supernatant from OSC19 transductant cells. B, Ratio of the 6 proteases that were most diminished in the protease array analyses, shown as means and standard error of the mean of 3 independent experiments; *P <.001. Abbreviation: MMP, matrix metalloproteinase.
Elevated miR-375 Expression in HNSCC Cells Regulates Kallikrein 6, Kallikrein 10, and MMP-9 mRNA Expression
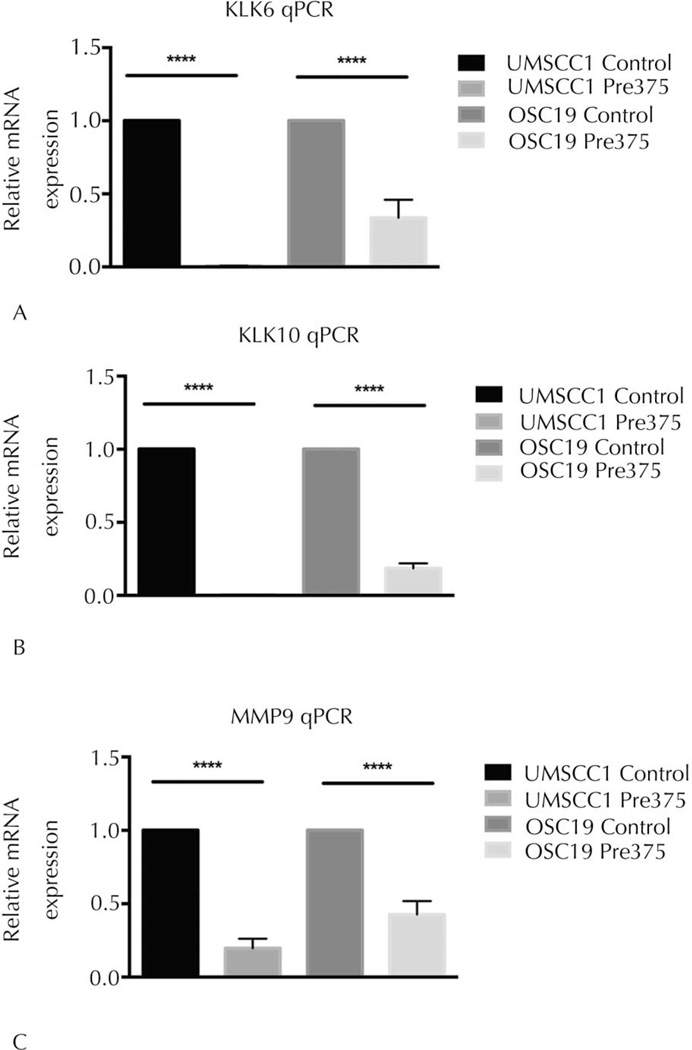
The qPCR measurements were used to evaluate the impact of elevated miR-375 expression on the mRNA expression of the commonly regulated proteases, kallikrein 6 (KLK6), kallikrein 10 (KLK10), and matrix metalloproteinase 9 (MMP9). We observed that the mRNA expression levels of KLK6 were reduced by 99% and 64% in the UMSCC1 and OSC19 cells with increased miR-375 expression, respectively, compared to their control lines (Figure 8, A). We determined that the UMSCC1 and OSC19 Pre375 cells had reductions in the mRNA expression levels of KLK10, by 99% and 81%, respectively (Figure 8, B). Likewise, we observed that the MMP9 mRNA expression levels were reduced by 80% and 53% in the UMSCC1 and OSC19 Pre375 cells, respectively, compared to their control lines (Figure 8, C). All of these mRNA reductions were significant (P < .001). These results validated the protease array measurements and demonstrate that the reductions in secreted levels of these proteases correlate with diminished mRNA levels.
Figure 8.
Elevated expression of microRNA-375 in HNSCC cells significantly reduces kallikrein 6, kallikrein 10, and MMP-9 messenger RNA (mRNA) expression. The mRNA expression levels in the transductant lines were measured by TaqMan quantitative real-time polymerase chain reaction (qPCR). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal control per cell line. Relative kallikrein 6 (KLK6), kallikrein 10 (KLK10), and matrix metal-loproteinase 9 (MMP9) mRNA expression levels in UMSCC1 and OSC19 transductant lines, shown in A through C, respectively. Data shown as mean and standard error of the mean of the fold change (2−ΔΔCT) from 5 independent experiments; ****P<.001.
COMMENT
This study extends our previous observation that the invasive properties of cells are regulated by miR-375 by demonstrating that increased miR-375 expression results in reduced matrix degradation and invadopodial activity. This was shown by identifying the invadopodia as sites of colocalization of cortactin and Tks5 in discrete puncta of matrix degradation. In particular, the number of mature invadopodia per cell was significantly reduced by miR-375 expression in parallel with the suppression of matrix degradation. The reduction in mature invadopodia was not accompanied by decreases in expression of selected invadopodia-associated proteins or changes in tyrosine phosphorylation of cortactin. In contrast, we found that there was a reduction in the levels of specific proteases, including kallikrein 6, kallikrein 10, and MMP-9.
MicroRNA-375 affects a variety of tumor phenotypes. The expression level of miR-375 is a potential biomarker for HNSCC and other cancers. Low expression of miR-375 in solid tumors, such as esophageal adenocarcinoma, glioma, and HNSCC, is associated with diminished patient survival and poor prognosis, whereas low expression of miR-375 in pediatric acute myeloid leukemia has been associated with better patient prognosis.11,17,56–58 In addition, although ectopic expression of miR-375 by transient transfection causes reduction in the proliferation of cervical, hepatocellular, gastric, colorectal, oral, and hypopharyngeal cancer cells, it stimulates proliferation of estrogen receptor– positive breast cancer cells.19,23,59–64 As we reported previously, the proliferation rates of stable transductant lines were not impaired by increased miR-375 expression, compared to the empty vector control lines.11 These findings suggest that the reduced survival of patients with HNSCC in low miR-375–expressing tumors is not due to altered proliferation. Ectopic overexpression of miR-375 impairs cell invasion of oral cavity, hypopharyngeal, nasopharyngeal, esophageal, breast, hepatocellular, cervical, and gastric cancer cells.15,20,22,23,65–69 Our previous study11 reported that stable increased miR-375 expression in both UMSCC1 and UMSCC47 cell lines results in diminished invasion in vitro. The invasive nature of HNSCC is often associated with locoregional invasion and metastasis to cervical lymph nodes. Identifying the drivers of HNSCC invasion is pivotal for new and improved treatment strategies for treating this disease.
Cancer cell invasion is mediated in part by invadopodia, which facilitate ECM degradation.25–28 The core structure of invadopodia consists of actin, cortactin, Tks5, cofilin, N-WASP, and actin-related protein-2/3 (Arp2/3) complex.70 Tks5 and cortactin are essential for invadopodium precursor formation.31,45 Tks5 is needed for invadopodium precursor stabilization, but it is not essential for the secretion of proteases leading to ECM degradation.30,32 Our data have shown that higher miR-375 expression in HNSCC cells did not correlate with changes in total Tks5 or cortactin. Tyrosine phosphorylation of cortactin contributes to actin polymerization responsible for membrane protrusion at invadopodia and efficient ECM degradation.31,54 Increased miR-375 expression did not affect tyrosine phosphorylation of cortactin. However, we did find that mature invadopodia were decreased in the cells expressing higher levels of miR-375, suggesting that a step subsequent to formation of precursor invadopodia is affected by miR-375 expression.
An Ingenuity Pathway Analysis of theoretical targets of miR-375 from TargetScan prediction database identified target candidates that have involvement in cell movement, formation of cellular protrusions, and organization of actin cytoskeleton (see Supplemental Table 1; see supplemental digital content). Some of these theoretical targets have been reported to be invadopodia-associated proteins, including tyrosine kinase Arg (ABL2), integrin α3 (ITGA3), podocalyxin-like protein (PODXL), protein kinase C α (PRKCA), podoplanin (PDPN), Tks4 (SH3PXD2B), and ADAM metallopeptidase with thrombospondin type 1 motif, 1 (ADAMTS1). Studies using breast cancer cell lines have shown that β-integrin 1 associates with tyrosine kinase Arg, which subsequently leads to the phosphorylation of cortactin at tyrosine 421.71,72 Additional work with HNSCC cell lines has identified that the Abelson kinases, Abl and Arg, are not essential for cortactin phosphorylation when there is elevated Src activity.34 Integrins α3 and β1 border the heads and tails of invadopodia73 and are needed for the association of seprase, a transmembrane serine protease, at invadopodia.74 In studies conducted with oral squamous cell carcinoma (OSCC) cells, podocalyxin-like 1 protein, a sialomucin, is needed for OSCC migratory, invasive, and proliferative properties.75 Podocalyxin-like 1 protein promotes invadopodia formation by regulating cortactin phosphorylation and Ras-related C3 botulinum toxin substrate 1 (Rac1)/cell division cycle 42 (CDC42) activity.76 In squamous cell carcinoma cells, podoplanin, a type 1 transmembrane glycoprotein, localizes to invadopodia-associated adhesion rings and serves an important role in the stabilization of invadopodia.77 Podoplanin controls the maturation of invadopodia of squamous cell carcinoma cells by regulating RhoC activity, affecting the Rho-associated protein (ROCK)/LIM-domain kinase (LIMK) pathway, which subsequently controls the phosphorylation of cofilin.77 In HNSCC cells with regulated (wild-type) PI3 kinase activity, protein kinase C α stimulates invadopodium formation.49 Tyrosine kinase substrate with 4 SH3 domains (Tks4), a Tks5-related protein, is important for the localization and stabilization of the transmembrane protease, MT1-MMP, in invadopodia.78 Tks4 and Tks5 are also needed for extravasation and lung metastasis formation of breast cancer cells.79 Depletion of ADAMTS-1, a secreted protease, increases migration, invasion, and invadopodia formation in breast cancer cells.80 On the other hand, ADAMTS-1 also facilitates cell invasion by cleaving heparin-binding epithelial growth factor–like growth factor and amphiregulin.81,82 Interestingly, in our data from the protease arrays of the UMSCC1 cells, the secreted levels of ADAMTS-1 was identified as being significantly diminished in conditioned cell culture medium from UMSCC1 cells with increased miR-375 expression compared to its control line. The interactions of the theoretical miR-375 targets mentioned above with other invadopodia-associated proteins are shown in Supplemental Figure 1 (see supplemental digital content). These target candidates may warrant further investigation of a potential role in defective matrix degradation of HNSCC cells as a result of increased miR-375 expression.
Proteolysis of matrix components at invadopodia is mediated in part by transmembrane and secreted proteases. Matrix metalloproteinases, membrane-bound MT1-MMP, and secreted MMP-2 and MMP-9 have been reported to play key roles in invadopodia-mediated ECM degradation.29,40 Our data showed that increased miR-375 expression levels regulate MMP-9 mRNA and secreted MMP-9 in both oral cavity human papillomavirus-negative HNSCC cell lines, indicating possible defects in transcription of MMP-9. MMP-9 is not predicted to be a direct target of miR-375; thus, miR-375 may be targeting a transcriptional regulator of MMP-9. Activator protein 1 (AP-1) is important for the expression of both MMP2 and MMP9 genes.83 Specificity protein 1 (Sp1) has been shown to bind to the promoter of MMP9 and to induce its transcription.84 Metadherin (MTDH), also known as astrocyte elevated gene-1 (AEG-1), activates the transcription of MMP9 by directly binding to the MMP9 promoter.85 Both Sp1 and MTDH are validated direct targets of miR-375 and therefore are candidates for playing a part in this phenotype.
Transmembrane type II serine proteases, fibroblast activation protein (or seprase), and dipeptidyl peptidase IV (DPPIV) localize to invadopodia, and DPPIV-seprase complex has been reported to play a vital role in the gelatin ECM degradation in fibroblasts.86–88 Our protease arrays revealed another family of serine proteases regulated by elevated miR-375 expression. Kallikreins, a family of secreted serine proteases, have been identified as diagnostic and biomarkers for various malignancies.89,90 Kallikreins have several physiological roles, including regulating cancer cell growth, angiogenesis, invasion, and metastasis.89 Kallikreins can have direct and indirect effects on the degradation of ECM components. Kallikrein 5, kallikrein 6, kallikrein 10, and kallikrein 13 degrade fibronectin, laminin, and collagen type I–IV.91–94 Kallikreins facilitate ECM degradation by activating other proteases. Kallikrein 1 activates pro–MMP-2 and pro–MMP-989 and kallikrein 7 activates pro–MMP-9 by cleaving its C-terminus.95
Our data show that increased miR-375 expression results in reductions in the secretion of kallikrein 6 and kallikrein 10 in both UMSCC1 and OSC19 cells. The mRNA expression levels of these proteases were also greatly diminished in the cells with higher miR-375 expression, indicating that miR-375 may be directly or indirectly affecting the transcription of kallikrein 6 and kallikrein 10. One possible mechanism for the regulation of the expression of the kallikreins is through hypermethylation of CpG islands in their promoters. The promoters of kallikrein 5, kallikrein 6, kallikrein 10, kallikrein 11, and kallikrein 12 are epigenetically regulated by DNA methylation.96–99 The promoters of kallikrein 6 and kallikrein 10 have Sp1 consensus sequences, as well as an AP-1 binding site.99,100 Perhaps in the proper cellular context, these factors could contribute to a somewhat widespread change in protease expression patterns. Since we have observed that 2 classes of proteases are affected by miR-375 on the mRNA level, this suggests that miR-375 might be targeting a common transcriptional regulator needed to activate the transcription of the proteases.
In summary, miR-375 was shown to suppress invadopodial function. The miR-375 gene is significantly repressed in head and neck cancers, and our data suggest that reduced miR-375 expression in patients with HNSCC can contribute to the invasive properties of head and neck cancer through increased invadopodial activity. Our data indicate that increased miR-375 expression does not prevent invadopodium formation but affects a step in the invadopodia maturation process. We have provided evidence that increased miR-375 expression affects the expression of secreted proteases, which potentially directly impact ECM degradation properties of HNSCC cells.
Supplementary Material
Acknowledgments
This work was supported by a predoctoral fellowship from National Institutes of Health (NIH) (CA168337) to L.J., a postdoctoral fellowship from Susan G. Komen for the Cure (KG111405) to V.P.S., NIH grant (CA150344) to J.C. and V.P.S., NIH grants (CA77522 and CA100324) to J.E.S., and by the Department of Pathology, Albert Einstein College of Medicine/ Montefiore Medical Center.
Footnotes
Supplemental digital content is available for this article. See text for hyperlink
The authors have no relevant financial interest in the products or companies described in this article.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Url C, Schartinger VH, Riechelmann H, et al. Radiological detection of extracapsular spread in head and neck squamous cell carcinoma (HNSCC) cervical metastases. Eur J Radiol. 2013;82(10):1783–1787. doi: 10.1016/j.ejrad.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 3.Oksuz DC, Prestwich RJ, Carey B, et al. Recurrence patterns of locally advanced head and neck squamous cell carcinoma after 3D conformal (chemo)-radiotherapy. Radiat Oncol. 2011;6:54. doi: 10.1186/1748-717X-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabor MP, Brakenhoff RH, Ruijter-Schippers HJ, Kummer JA, Leemans CR, Braakhuis BJ. Genetically altered fields as origin of locally recurrent head and neck cancer: a retrospective study. Clin Cancer Res. 2004;10(11):3607–3613. doi: 10.1158/1078-0432.CCR-03-0632. [DOI] [PubMed] [Google Scholar]
- 5.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63(8):1727–1730. [PubMed] [Google Scholar]
- 6.Haddad RI, Shin DM. Recent advances in head and neck cancer. New Engl J Med. 2008;359(11):1143–1154. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 8.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 10.Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174(3):736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris T, Jimenez L, Kawachi N, et al. Low-level expression of miR-375 correlates with poor outcome and metastasis while altering the invasive properties of head and neck squamous cell carcinomas. Am J Pathol. 2012;180(3):917–928. doi: 10.1016/j.ajpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu HF, Lin SC, Chang KW. MicroRNA aberrances in head and neck cancer: pathogenetic and clinical significance. Curr Opin Otolaryngol Head Neck Surg. 2013;21(2):104–111. doi: 10.1097/MOO.0b013e32835e1d6e. [DOI] [PubMed] [Google Scholar]
- 13.Singh R, Mo YY. Role of microRNAs in breast cancer. Cancer Biol Ther. 2013;14(3):201–212. doi: 10.4161/cbt.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Li Y, Zhou J, et al. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179(5):2580–2588. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siow MY, Ng LP, Chong VK, et al. Dysregulation of miR-31 and miR-375 expression is associated with clinical outcomes in oral carcinoma. Oral Dis. 2014;20(4):345–351. doi: 10.1111/odi.12118. [DOI] [PubMed] [Google Scholar]
- 17.Chang C, Shi H, Wang C, et al. Correlation of microRNA-375 downregulation with unfavorable clinical outcome of patients with glioma. Neurosci Lett. 2012;531(2):204–208. doi: 10.1016/j.neulet.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu S, Ichikawa D, Takeshita H, et al. Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin Biol Ther. 2012;12(suppl 1):S53–S59. doi: 10.1517/14712598.2012.681373. [DOI] [PubMed] [Google Scholar]
- 19.Ding L, Xu Y, Zhang W, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20(7):784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 20.Jung HM, Patel RS, Phillips BL, et al. Tumor suppressor miR-375 regulates MYC expression via repression of CIP2A coding sequence through multiple miRNA-mRNA interactions. Mol Biol Cell. 2013;24(11):1638–1648. S1631–S1637. doi: 10.1091/mbc.E12-12-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui AB, Bruce JP, Alajez NM, et al. Significance of dysregulated metadherin and microRNA-375 in head and neck cancer. Clin Cancer Res. 2011;17(24):7539–7550. doi: 10.1158/1078-0432.CCR-11-2102. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita T, Nohata N, Yoshino H, et al. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. Int J Oncol. 2012;40(1):185–193. doi: 10.3892/ijo.2011.1196. [DOI] [PubMed] [Google Scholar]
- 23.Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394(3):623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Weaver AM. Invadopodia: specialized cell structures for cancer invasion. Clin Exp Metastasis. 2006;23(2):97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- 26.Linder S, Wiesner C, Himmel M. Degrading devices: invadosomes in proteolytic cell invasion. Annu Rev Cell Dev Biol. 2011;27:185–211. doi: 10.1146/annurev-cellbio-092910-154216. [DOI] [PubMed] [Google Scholar]
- 27.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol. 2012;24(2):277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nature Rev Mol Cell Biol. 2011;12(7):413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66(6):3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- 30.Sharma VP, Eddy R, Entenberg D, Kai M, Gertler FB, Condeelis J. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr Biol. 2013;23(21):2079–2089. doi: 10.1016/j.cub.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oser M, Yamaguchi H, Mader CC, et al. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol. 2009;186(4):571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seals DF, Azucena EF, Jr, Pass I, et al. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 2005;7(2):155–165. doi: 10.1016/j.ccr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Li A, Dawson JC, Forero-Vargas M, et al. The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr Biol. 2010;20(4):339–345. doi: 10.1016/j.cub.2009.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayes KE, Walk EL, Ammer AG, Kelley LC, Martin KH, Weed SA. Ableson kinases negatively regulate invadopodia function and invasion in head and neck squamous cell carcinoma by inhibiting an HB-EGF autocrine loop. Oncogene. 2013;32(40):4766–4777. doi: 10.1038/onc.2012.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fekete A, Bögel G, Pesti S, Peterfi Z, Geiszt M, Buday L. EGF regulates tyrosine phosphorylation and membrane-translocation of the scaffold protein Tks5. J Mol Signal. 2013;8(1):8. doi: 10.1186/1750-2187-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burger KL, Learman BS, Boucherle AK, et al. Src-dependent Tks5 phosphorylation regulates invadopodia-associated invasion in prostate cancer cells. Prostate. 2014;74(2):134–148. doi: 10.1002/pros.22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stylli SS, Stacey TT, Verhagen AM, et al. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J Cell Sci. 2009;122(pt 15):2727–2740. doi: 10.1242/jcs.046680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abram CL, Seals DF, Pass I, et al. The adaptor protein fish associates with members of the ADAMs family and localizes to podosomes of Src-transformed cells. J Biol Chem. 2003;278(19):16844–16851. doi: 10.1074/jbc.M300267200. [DOI] [PubMed] [Google Scholar]
- 39.Oser M, Dovas A, Cox D, Condeelis J. Nck1 and Grb2 localization patterns can distinguish invadopodia from podosomes. Eur J Cell Biol. 2011;90(2–3):181–188. doi: 10.1016/j.ejcb.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark ES, Whigham AS, Yarbrough WG, Weaver AM. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 2007;67(9):4227–4235. doi: 10.1158/0008-5472.CAN-06-3928. [DOI] [PubMed] [Google Scholar]
- 41.Ammer AG, Kelley LC, Hayes KE, et al. Saracatinib impairs head and neck squamous cell carcinoma invasion by disrupting invadopodia function. J Cancer Sci Ther. 2009;1(2):52–61. doi: 10.4172/1948-5956.1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang YS, Park KK, Chung WY. Invadopodia formation in oral squamous cell carcinoma: the role of epidermal growth factor receptor signalling. Arch Oral Biol. 2012;57(4):335–343. doi: 10.1016/j.archoralbio.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Hwang YS, Park KK, Cha IH, Kim J, Chung WY. Role of insulin-like growth factor-II mRNA-binding protein-3 in invadopodia formation and the growth of oral squamous cell carcinoma in athymic nude mice. Head Neck. 2012;34(9):1329–1339. doi: 10.1002/hed.21929. [DOI] [PubMed] [Google Scholar]
- 44.Hwang YS, Park KK, Chung WY. Epigallocatechin-3 gallate inhibits cancer invasion by repressing functional invadopodia formation in oral squamous cell carcinoma. Eur J Pharmacol. 2013;715(1–3):286–295. doi: 10.1016/j.ejphar.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Sharma VP, Entenberg D, Condeelis J. High-resolution live-cell imaging and time-lapse microscopy of invadopodium dynamics and tracking analysis. Methods Mol Biol. 2013;1046:343–357. doi: 10.1007/978-1-62703-538-5_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 49.Hoshino D, Jourquin J, Emmons SW, et al. Network analysis of the focal adhesion to invadopodia transition identifies a PI3K-PKCalpha invasive signaling axis. Sci Signal. 2012;5(241):ra66. doi: 10.1126/scisignal.2002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parekh A, Weaver AM. Regulation of cancer invasiveness by the physical extracellular matrix environment. Cell Adh Migr. 2009;3(3):288–292. doi: 10.4161/cam.3.3.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Vlachos IS, Kostoulas N, Vergoulis T, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40(Web server issue):W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Research. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oser M, Mader CC, Gil-Henn H, et al. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J Cell Sci. 2010;123(pt 21):3662–3673. doi: 10.1242/jcs.068163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poincloux R, Lizarraga F, Chavrier P. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J Cell Sci. 2009;122(pt 17):3015–3024. doi: 10.1242/jcs.034561. [DOI] [PubMed] [Google Scholar]
- 56.Leidner RS, Ravi L, Leahy P, et al. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett’s esophageal carcinogenesis. Genes Chromosomes Cancer. 2012;51(5):473–479. doi: 10.1002/gcc.21934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Hong Z, Gao F, Feng W. Upregulation of microRNA-375 is associated with poor prognosis in pediatric acute myeloid leukemia. Mol Cell Biochem. 2013;383(1–2):59–65. doi: 10.1007/s11010-013-1754-z. [DOI] [PubMed] [Google Scholar]
- 58.Mathe EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15(19):6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Souza Rocha Simonini P, Breiling A, Gupta N, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor {alpha} in breast cancer cells. Cancer Res. 2010;70(22):9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 60.Hui AB, Lenarduzzi M, Krushel T, et al. Comprehensive microRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16(4):1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 61.Nohata N, Hanazawa T, Kikkawa N, et al. Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC) J Hum Genetics. 2011;56(8):595–601. doi: 10.1038/jhg.2011.66. [DOI] [PubMed] [Google Scholar]
- 62.Shen ZY, Zhang ZZ, Liu H, Zhao EH, Cao H. miR-375 inhibits the proliferation of gastric cancer cells by repressing ERBB2 expression. Exp Ther Med. 2014;7(6):1757–1761. doi: 10.3892/etm.2014.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miao L, Liu K, Xie M, Xing Y, Xi T. miR-375 inhibits Helicobacter pylori-induced gastric carcinogenesis by blocking JAK2-STAT3 signaling. Cancer Immunol Immunother. 2014;63(7):699–711. doi: 10.1007/s00262-014-1550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Tang Q, Li M, Jiang S, Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun. 2014;444(2):199–204. doi: 10.1016/j.bbrc.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 65.He XX, Chang Y, Meng FY, et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31(28):3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 66.Mazar J, DeBlasio D, Govindarajan SS, Zhang S, Perera RJ. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 2011;585(15):2467–2476. doi: 10.1016/j.febslet.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y, Jin J, Liu Y, et al. Snail-regulated MiR-375 inhibits migration and invasion of gastric cancer cells by targeting JAK2. PloS One. 2014;9(7):e99516. doi: 10.1371/journal.pone.0099516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong S, Noh H, Teng Y, et al. SHOX2 is a direct miR-375 target and a novel epithelial-to-mesenchymal transition inducer in breast cancer cells. Neoplasia. 2014;16(4):279–290. e271–e275. doi: 10.1016/j.neo.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo D, Wilson JM, Harvel N, et al. A systematic evaluation of miRNA: mRNA interactions involved in the migration and invasion of breast cancer cells. J Transl Med. 2013;11:57. doi: 10.1186/1479-5876-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beaty BT, Condeelis J. Digging a little deeper: the stages of invadopodium formation and maturation. Eur J Cell Biol. 2014;93(10–12):438–444. doi: 10.1016/j.ejcb.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beaty BT, Sharma VP, Bravo-Cordero JJ, et al. Beta1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Mol Biol Cell. 2013;24(11):1661–1675. S1–S11. doi: 10.1091/mbc.E12-12-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mader CC, Oser M, Magalhaes MA, et al. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71(5):1730–1741. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takkunen M, Hukkanen M, Liljestrom M, Grenman R, Virtanen I. Podosome-like structures of non-invasive carcinoma cells are replaced in epithelial-mesenchymal transition by actin comet-embedded invadopodia. J Cell Mol Med. 2010;14(6B):1569–1593. doi: 10.1111/j.1582-4934.2009.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mueller SC, Ghersi G, Akiyama SK, et al. A novel protease-docking function of integrin at invadopodia. J Biol Chem. 1999;274(35):24947–24952. doi: 10.1074/jbc.274.35.24947. [DOI] [PubMed] [Google Scholar]
- 75.Lin CW, Sun MS, Wu HC. Podocalyxin-like 1 is associated with tumor aggressiveness and metastatic gene expression in human oral squamous cell carcinoma. Int J Oncol. 2014;45(2):710–718. doi: 10.3892/ijo.2014.2427. [DOI] [PubMed] [Google Scholar]
- 76.Lin CW, Sun MS, Liao MY, et al. Podocalyxin-like 1 promotes invadopodia formation and metastasis through activation of Rac1/Cdc42/ cortactin signaling in breast cancer cells. Carcinogenesis. 2014;35(11):2425–2435. doi: 10.1093/carcin/bgu139. [DOI] [PubMed] [Google Scholar]
- 77.Martin-Villar E, Borda-d’Agua B, Carrasco-Ramirez P, et al. Podoplanin mediates ECM degradation by squamous carcinoma cells through control of invadopodia stability [published online ahead of print December 8, 2014] Oncogene. doi: 10.1038/onc.2014.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buschman MD, Bromann PA, Cejudo-Martin P, Wen F, Pass I, Courtneidge SA. The novel adaptor protein Tks4 (SH3PXD2B) is required for functional podosome formation. Mol Biol Cell. 2009;20(5):1302–1311. doi: 10.1091/mbc.E08-09-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leong HS, Robertson AE, Stoletov K, et al. Invadopodia are required for cancer cell extravasation and are a therapeutic target for metastasis. Cell Rep. 2014;8(5):1558–1570. doi: 10.1016/j.celrep.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 80.Freitas VM, do Amaral JB, Silva TA, et al. Decreased expression of ADAMTS-1 in human breast tumors stimulates migration and invasion. Mol Cancer. 2013;12:2. doi: 10.1186/1476-4598-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu X, Wang Q, Hu G, et al. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009;23(16):1882–1894. doi: 10.1101/gad.1824809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le Bras GF, Taylor C, Koumangoye RB, Revetta F, Loomans HA, Andl CD. TGFbeta loss activates ADAMTS-1-mediated EGF-dependent invasion in a model of esophageal cell invasion. Exp Cell Res. 2015;330(1):29–42. doi: 10.1016/j.yexcr.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hasegawa H, Senga T, Ito S, Iwamoto T, Hamaguchi M. A role for AP-1 in matrix metalloproteinase production and invadopodia formation of v-Crk-transformed cells. Exp Cell Res. 2009;315(8):1384–1392. doi: 10.1016/j.yexcr.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 84.Sato H, Seiki M. Regulatory mechanism of 92 kDa type IV collagenase gene expression which is associated with invasiveness of tumor cells. Oncogene. 1993;8(2):395–405. [PubMed] [Google Scholar]
- 85.Liu L, Wu J, Ying Z, et al. Astrocyte elevated gene-1 upregulates matrix metalloproteinase-9 and induces human glioma invasion. Cancer Res. 2010;70(9):3750–3759. doi: 10.1158/0008-5472.CAN-09-3838. [DOI] [PubMed] [Google Scholar]
- 86.Monsky WL, Lin CY, Aoyama A, et al. A potential marker protease of invasiveness, seprase, is localized on invadopodia of human malignant melanoma cells. Cancer Res. 1994;54(21):5702–5710. [PubMed] [Google Scholar]
- 87.O’Brien P, O’Connor BF. Seprase: an overview of an important matrix serine protease. Biochim Biophys Acta. 2008;1784(9):1130–1145. doi: 10.1016/j.bbapap.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 88.Ghersi G, Dong H, Goldstein LA, et al. Regulation of fibroblast migration on collagenous matrix by a cell surface peptidase complex. J Biol Chem. 2002;277(32):29231–29241. doi: 10.1074/jbc.M202770200. [DOI] [PubMed] [Google Scholar]
- 89.Borgono CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nature Rev Cancer. 2004;4(11):876–890. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- 90.Paliouras M, Borgono C, Diamandis EP. Human tissue kallikreins: the cancer biomarker family. Cancer Lett. 2007;249(1):61–79. doi: 10.1016/j.canlet.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 91.Ghosh MC, Grass L, Soosaipillai A, Sotiropoulou G, Diamandis EP. Human kallikrein 6 degrades extracellular matrix proteins and may enhance the metastatic potential of tumour cells. Tumor Biol. 2004;25(4):193–199. doi: 10.1159/000081102. [DOI] [PubMed] [Google Scholar]
- 92.Magklara A, Mellati AA, Wasney GA, et al. Characterization of the enzymatic activity of human kallikrein 6: autoactivation, substrate specificity, and regulation by inhibitors. Biochem Biophys Res Commun. 2003;307(4):948–955. doi: 10.1016/s0006-291x(03)01271-3. [DOI] [PubMed] [Google Scholar]
- 93.Kapadia C, Ghosh MC, Grass L, Diamandis EP. Human kallikrein 13 involvement in extracellular matrix degradation. Biochem Biophys Res Commun. 2004;323(3):1084–1090. doi: 10.1016/j.bbrc.2004.08.206. [DOI] [PubMed] [Google Scholar]
- 94.Michael IP, Sotiropoulou G, Pampalakis G, et al. Biochemical and enzymatic characterization of human kallikrein 5 (hK5), a novel serine protease potentially involved in cancer progression. J Biol Chem. 2005;280(15):14628–14635. doi: 10.1074/jbc.M408132200. [DOI] [PubMed] [Google Scholar]
- 95.Ramani VC, Kaushal GP, Haun RS. Proteolytic action of kallikrein-related peptidase 7 produces unique active matrix metalloproteinase-9 lacking the C-terminal hemopexin domains. Biochim Biophys Acta. 2011;1813(8):1525–1531. doi: 10.1016/j.bbamcr.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang W, Zhong J, Wu LY, et al. Downregulation and CpG island hypermethylation of NES1/hK10 gene in the pathogenesis of human gastric cancer. Cancer Lett. 2007;251(1):78–85. doi: 10.1016/j.canlet.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y, Song H, Miao Y, Wang R, Chen L. Frequent transcriptional inactivation of Kallikrein 10 gene by CpG island hypermethylation in non-small cell lung cancer. Cancer Sci. 2010;101(4):934–940. doi: 10.1111/j.1349-7006.2009.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pampalakis G, Diamandis EP, Sotiropoulou G. The epigenetic basis for the aberrant expression of kallikreins in human cancers. Biol Chem. 2006;387(6):795–799. doi: 10.1515/BC.2006.100. [DOI] [PubMed] [Google Scholar]
- 99.Pampalakis G, Sotiropoulou G. Multiple mechanisms underlie the aberrant expression of the human kallikrein 6 gene in breast cancer. Biol Chem. 2006;387(6):773–782. doi: 10.1515/BC.2006.097. [DOI] [PubMed] [Google Scholar]
- 100.Borgono CA, Michael IP, Diamandis EP. Human tissue kallikreins: physiologic roles and applications in cancer. Mol Cancer Res. 2004;2(5):257–280. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.