Abstract
Allergy and type 1 diabetes are immune mediated diseases that, despite being etiologically distinct, have inappropriate activation and effector function of antigen-specific T cells in the pathogenic process. Understanding changes in frequency and phenotype of these cells is critical to improve assessment of disease diagnosis and prognosis and effectively assess immunological response to therapy. In the setting of antigen-specific therapy in allergy and type 1 diabetes, assays to monitor the immunological mechanisms of disease have been improving in recent years, and we are getting closer to an accurate understanding of how the cellular immune response is modulated during treatment. In this review, we summarize the current state of cell-based immune monitoring of antigen therapy trials. We then discuss emerging advances in antigen-specific biomarkers that are transforming our knowledge about allergy and that have the potential to dramatically impact our understanding of T cell-mediated autoimmune diseases, such as type 1 diabetes.
Keywords: Allergy, type 1 diabetes, biomarkers, pathogenesis, immunotherapy
1. Introduction
Allergy and type 1 diabetes (T1D) are complex immunological disorders with multiple cellular and molecular alterations in pathways involving both activation and effector function. To rationally evaluate the mechanistic impact of candidate therapies in these diseases, therefore, it will be essential to illuminate stages of pathogenesis with the help of informative biomarkers. Biomarkers have potential applicability in multiple phases of drug development and clinical practice (Table 1). Specific opportunities to develop correlates of immune mediated disease outcome include inappropriate expansion of cells specific for intrinsic or extrinsic antigens, increased or unregulated effector functions of pro-inflammatory cells as a whole, and alterations in gene expression pathways reflecting defective homeostasis. Biomarkers directly related to pathogenesis are currently employed in both allergy (IgE and basophil activation) and T1D (hyperglycemia measures and c-peptide levels), but in both cases there remains a need to accurately measure the cellular immunopathogenic process, as well as pharmacological responses in the context of a therapeutic intervention. In other words, clinical utilization of biomarkers that directly assess pathogenic mechanisms is fundamental both for improving clinical trials and for generating new concepts for intervention. Biomarker assays used to assess clinical trial outcomes must have appropriate performance characteristics (Table 2), so it is important to consider early on whether a candidate assay will be amenable to validation.
Table 1.
Types of biomarkers
| Biomarker Class | Clinical Use |
|---|---|
| Diagnostic | Indicate presence of a disease |
| Prognostic | Indicate likely disease course if untreated |
| Companion Diagnostic | Identify patients likely to respond to a specific therapy |
| Pharmacodynamic | Identify pharmacological response to treatment |
| Screening | Identify patients at risk for disease development |
| Surrogate | Substitute for clinical outcome in efficacy trials |
Table 2.
Performance characteristics for analytical methods
| Characteristic | Definition |
|---|---|
| Feasibility | Utilizes accessible biological tissue and possible to scale up for routine use |
| Precision | Closeness of test values to one another when analyzing the same specimen |
| Accuracy | Closeness of test values to the true value |
| Specificity | Ability to measure only the intended analyte, particularly in the presence of other entities |
| Sensitivity | Lower limit of quantitation within a defined degree of confidence |
| Robustness | Assay performance maintained across sites, operators, instruments, and reagents |
Both allergy and T1D offer the opportunity to focus on a key step in disease pathogenesis — namely, the antigen-specific T cell response. In both cases, therapy with specific antigen is designed to perturb the pathogenic response and restore homeostatic balance, but in neither case is there yet a qualified immunological measure that predicts clinical outcome. Clinical trials designed to achieve antigen-specific tolerance, whether desensitization protocols with allergens or tolerogenic immunization with T1D autoantigens, create opportunities to develop such biomarkers by specifically measuring cellular immune responses to individual disease-associated antigens. Technical advances over the last decade with major histocompatibility complex (MHC) tetramers, multiparameter flow cytometry, and gene expression profiling have dramatically enhanced the quality and quantity of information that can be obtained regarding specific T cell immunobiology. Although still technically challenging, it is now possible to interrogate antigen-specific responses in the peripheral blood of patients in antigen therapy trials, revealing new insights into disease pathogenesis and creating new biomarkers for evaluation.
2. Antigen-specific biomarkers in allergy
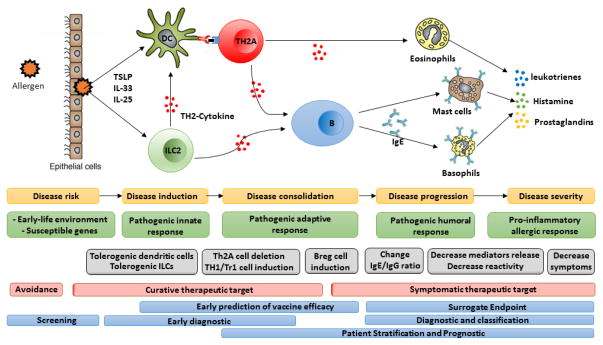
Figure 1 presents a schematic view of allergic responses, highlighting measurable checkpoints for disease progression. The core features of this model apply to many immune mediated and autoimmune diseases, in which individuals at increased risk undergo a step-wise sequence of cellular activation and maturation events that lead to effector responses directly implicated in pathogenesis. Existing clinical markers of allergic inflammation focus on IgE antibodies and histamine release by mast cells and basophils [1,2], which are the downstream effectors of allergic symptoms. The use of circulating allergen-specific immunoglobulin and basophils as surrogate biomarkers for clinical efficacy has been intensively investigated [3–5]. Specific IgE is not generally considered appropriate for monitoring immunotherapy in allergy, as the decrease in serum IgE level is modest and occurs late in treatment. It has been suggested that the presence of specific IgG4 may be associated with successful therapy [5], but this correlation is not always present, suggesting that IgG4 levels may merely reflect high allergen exposure rather than a tolerant status [6]. As clinical symptoms are mediated by allergen and IgE-dependent histamine release by basophils, assays assessing antigen-dependent activation of basophils, such as through measurement of CD203c [7] or diamine oxidase [8], have also been suggested to correlate with treatment efficacy. In allergic rhinitis, blood eosinophil counts [9,10] and serum levels of tryptase, eosinophil cationic protein [11], and osteopontin [12] have also been proposed as surrogate endpoints for therapy. While each of these candidate biomarkers merits further investigation, they all relate to the end stage of the allergic chain reaction and therefore lack sufficient sensitivity to predict the onset of atopic disease and to predict early efficacy in therapeutic trials.
Figure 1. The process of type 1 allergic disease pathogenesis, showing opportunities for identifying biomarkers.
Upon allergen recognition, epithelial cells release cytokines, such as IL-25, IL-33, and TSLP. Activation of ILC2 amplify and coordinate local immune responses. Allergens are captured by antigen-presenting dendritic cells or macrophages, and allergen-derived antigens presented by these cells are recognized by CD4+ T cells, which proliferate and differentiate. Primed allergen-specific Th2 cells and release of Th2 cytokines activate eosinophils and trigger the maturation of antigen-specific B cell populations into plasma cells. Plasma cells release antigen-specific IgE, which binds to IgE receptors on mast cells and basophils, initiating downstream histamine release when cross-linked with antigen.
Key: Yellow and green boxes show the process of type 1 allergic disease pathogenesis. Grey boxes show potential treatment response markers on allergic disease process. Red boxes show principal targeted cells during current potential therapy. Blue boxes show potential types of biomarkers in an allergic disease process.
New technologies are emerging, which are capable of comprehensive analysis of genes, transcripts, proteins, immune cells, and other significant biological molecules designed to discover biomarkers upstream in the allergic disease process (Figure 1). For instance, molecular changes at the level of dendritic cells (DC) have been recently described for response-monitoring at the early stages of allergen immunotherapy with the use of label-free mass spectrometry approaches [13]. In this elegant study, Zimmer and colleagues demonstrate that expression of complement component 1 and Stabilin-1 may represent an early signature predictive of clinical tolerance during therapy and suggest that these proteins themselves may play a role in the desensitization process. Similarly, technical advances in polychromatic flow cytometry have now enabled a more detailed phenotypic evaluation of a multitude of immune cell subsets that may causally correlate with treatment efficacy. Innate lymphoid type 2 cells (ILC2), for example, have been recently reported to decrease dramatically after successful grass pollen subcutaneous immunotherapy in patients with seasonal allergic rhinitis [14].
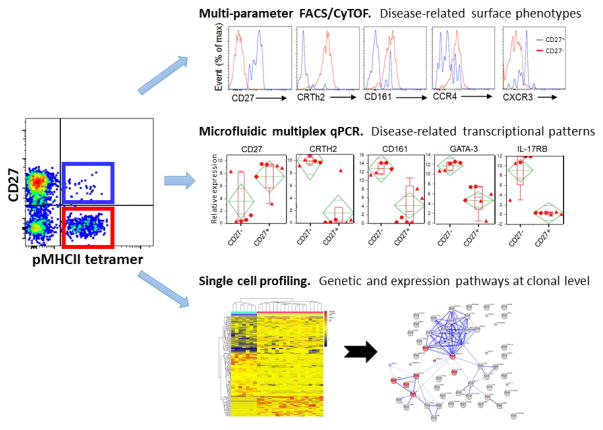
An additional approach focused on pivotal early stages of disease is to use the frequency and phenotype of allergen-specific CD4 cells as a clinically meaningful biomarker in allergy. T lymphocytes drive allergic sensitization, primarily through a Th2-biased response pathway initiated by soluble mediators, such as thymic stromal lymphopoietin (TSLP) and IL-33. As shown in Figure 1, there is a pivotal role for T cell-derived cytokines, such as IL-4 and IL-13, in driving downstream effector responses, including eosinophil activation and B cell production of allergen-specific Ig. Since T cell activation and commitment to the Th2 lineage precede the main effector phases of allergy, biomarkers that detect allergen-specific early Th2 induction have the potential to offer both early, potentially pre-symptomatic, diagnosis and improved assessment of prognosis, particularly in the context of specific immunotherapy. We have focused on this objective, based on the notion that such cells are highly specific for the antigen and are therefore more likely to directly reflect the causative events of disease compared with biomarkers of more downstream events. A major impediment to the use of allergen-specific T cells as a clinically useful biomarker is their low frequency in peripheral blood and the lack of a validated method for their identification and discrimination from overall non-pathogenic Th2 cell types. However, the recent advances in peptide-MHC class II (pMHCII) tetramer staining has now allowed reliable and direct ex vivo visualization of antigen-specific CD4 T cells [15,16]. This analysis can be combined with large panels of phenotypic markers that now allow more biological information to be extracted from such rare cells (Figure 2). Additionally, isolation of pMHCII-binding cells provides the capacity to search for molecular biomarkers in an unbiased manner by using single cell transcriptome analysis. Using this approach, we recently demonstrated that allergen-specific Th2 cells are confined to allergic individuals and their disappearance is indicative of clinical responses induced by allergen-specific immunotherapy[16,17]. In addition, we have shown that CD27 expression on allergen-specific T cells predicts successful clinical outcome in allergen-specific immunotherapy [18,19]. Some challenges to the broad application of pMHCII tetramer technology in clinical development continue to be low target cell frequency, the presence of multiple allergic protein components, and the difficulty in producing class II MHC-based reagents. With these limitations in mind, we recently demonstrated an alternative approach based on characterization of an allergic disease-related phenotype shared among all allergen-specific Th2 cells (CD4+, CRTh2+, CD161+, CD49d+, CD27-, and CD45RB-) [20] (and manuscript submitted). Remarkably, the proportion of these allergy-prone Th2 cells was extremely low in non-atopic individuals compared with allergic individuals, and these cells were preferentially deleted during successful allergen-specific immunotherapy, suggesting a possible role in the pathogenesis of the disease and in disease severity. As such, we have denoted the pathogenic subpopulation of Th2 effector cells, virtually unique to atopic individuals, as the Th2A cell subset [20]. The application of this technique has the potential to transform our ability to profile allergen-specific Th2 cells with the goal of illuminating biology and utilizing clinical biomarkers in allergy (Figure 3). Therapeutic strategies directed against molecular targets found in the Th2A subset will shift the focus of treatment to pathogenic steps earlier in the disease process, and these studies are now supported by assays specific for the Th2A biomarker.
Figure 2. Strategies for discovering allergen-specific CD4+ T cell biomarkers.
MHC class II tetramer technology enables technological advances in immunological research that foster biomarker discovery. Multi-parameter flow cytometry and mass cytometry provide us the ability to search for cell-surface marker signatures within pMHCII tetramer-positive CD4+ T cells. Simultaneously, we can use isolated antigen-specific CD4+ T cells to search for molecular biomarkers by micro-scaled RNA sequencing or single cell RNA sequencing.
Figure 3. Illustration of potential applications of cell surface marker-based allergen-specific Th2 cells.
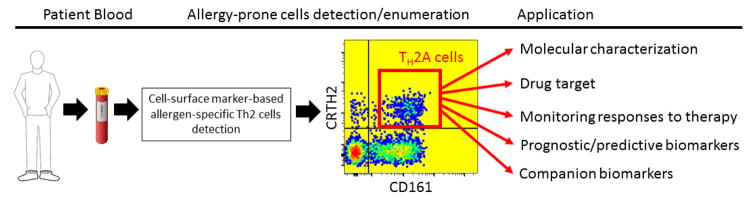
Blood samples are collected from patients and processed to allow detection of whole allergen-specific Th2 cells (Th2A subset), based on an allergic disease-related phenotype shared among these cells. Enumeration of Th2A cells can then be applied as a clinically useful biomarker in allergy, while these cells can be purified for more molecular profiling and analysis in cell culture.
3. Antigen-specific cellular immunophenotyping in T1D
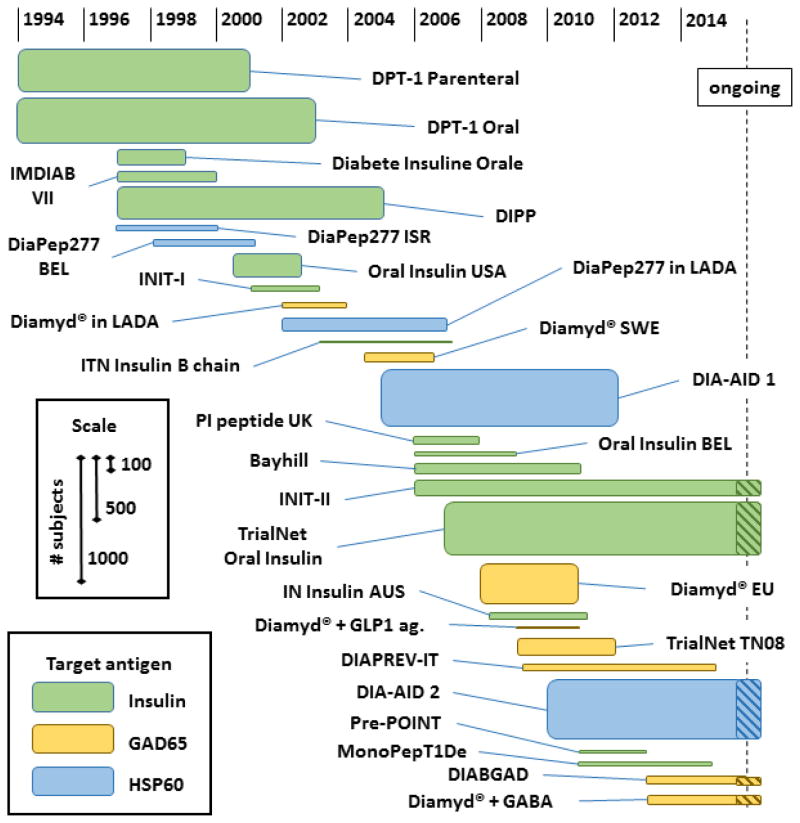
There are several parallel elements and corresponding lessons to be learned when comparing immune monitoring approaches in allergy and T1D. In T1D current surrogate markers for disease progression include metabolic parameters associated with hyperglycemia, insulin-secretory capacity of islet cells as measured by circulating c-peptide, and the presence of autoantibodies specific for beta cell antigens. As in allergy, these assessments predominantly measure late stage outcomes and are poor representations of the immunological status that directly drives the autoimmune response against islet cells. This issue is particularly vexing because the goal of therapeutic intervention in autoimmune disease, such as in T1D, is to restore tolerance to autoantigens while sparing the function of T cells specific for pathogens or cancer antigens. Unlike standard immunosuppressive interventions, this goal is likely feasible only by targeting specific cells directly through antigen immunotherapy. There are at least five beta cell antigens that are commonly targeted in T1D patients: insulin, GAD65, IA2, IGRP, and ZnT8. Since 1994 to the present, various forms of two of these antigens, insulin and GAD65, have been used in clinical trials, as summarized in Figure 4. These trials have been evaluated in the treatment mode, with the goal of maintaining residual endogenous beta cell function, or in the secondary prevention mode, with the goal of preventing the progression of disease in individuals with early autoimmunity prior to complete loss of beta cells. While there have been interesting post hoc findings in subgroups of these trials, none has consistently met its primary efficacy endpoints. Of even more concern, few of these trials utilized immunological biomarkers that effectively characterized the T cell response, so our ability to understand the failure to achieve therapeutic benefit is lacking. Many variables in the therapeutic approach remain untested, and the mechanistic rationale for antigen administration in any particular formulation or regimen is weak. Thus, it is evident that we cannot advance rationally toward an effective autoantigen therapy via large efficacy trials and clinical outcome endpoints alone. A set of validated biomarker assays focused on the quantity and quality of islet antigen-specific T cells is needed to assess directly whether candidate therapies are achieving their mechanistic goals.
Figure 4. Antigen immunotherapy trials in T1D.
Trial data, including enrollment number, start date, and primary endpoint stop date, are determined from literature references, the website www.ClinicalTrials.gov, and EudraCT. Estimates are made when specific data were unavailable.
Several assays have been employed in an attempt to measure islet-responsive T cells in the context of antigen-specific therapeutic clinical trials, as summarized in Table 3. The first, T cell proliferation based on incorporation of 3H-thymidine, was one of the earliest techniques employed in cellular immunology. The findings from this assay can essentially be classified into two results: enhanced or suppressed proliferative responses as a consequence of antigen therapy. In trials of intranasal insulin [21] and subcutaneous insulin B-chain treatment [22], reduced insulin-specific proliferation was observed in the active arms, which could be evidence of deletion, margination, or alteration of the responsiveness of insulin-specific cells. Interestingly, in a study of a subset of diabetes prevention trial-type 1 participants, treatment with parenteral insulin was also associated with reduced proliferation in response to a range of antigenic islet cell extract fractions [23]. This result may suggest the occurrence of tolerogenic epitope spreading, though the specificity of these responses remains to be defined. In clear contrast to these trials, treatment with Diamyd® GAD-alum clearly enhances GAD-specific proliferative response, likely due to the presence of adjuvant [24]. Other methods for assessing T cell proliferation, particularly CFSE dilution, have also been studied in the context of T1D [25], but it remains to be determined whether CFSE-based methods will be consistently sensitive enough for islet antigen trial monitoring.
Table 3.
Cellular biomarker assays in antigen immunotherapy for T1D
| Assay | Trial | Agent | Treatment effect |
|---|---|---|---|
| 3H-thymidine incorporation | INIT-I | Insulin | ↓ insulin response vs. placebo [21] |
| DiaPep277 BEL | HsP60 peptide | Negative change in HSP60 response associated with better outcome [39] | |
| ITN Insulin B chain | Insulin B chain | ↑ insulin B chain response vs. placebo [22] | |
| DPT-1 Parenteral | Insulin | ↓ response to islet extracts vs. placebo [23] | |
| Diamyd® EU | GAD-alum | ↑ GAD65-specific response vs. placebo [24] | |
| Stimulation phenotyping | Diamyd® SWE | GAD-alum | ↑ GAD65-induced CD25 and FOXP3 protein, multiple cytokine transcripts including TGFβ [28] |
| Diamyd® SWE | GAD-alum | ↑ GAD65-induced blasting cells with activated/TEM phenotype [29] | |
| Diamyd® SWE | GAD-alum | ↑ GAD65-induced CD25+ CD127-cells [30] | |
| ELISPOT | DiaPep277 ISR | HSP60 peptide | ↑ HSP60-specific Th2/Th1 ratio in active vs placebo [40] |
| PI peptide UK | Proinsulin peptide | Transient ↑ PI-specific IL-10+ cells [27] | |
| IN Insulin AUS | Insulin | ↓ PI-specific IFN-γ+ cells [26] | |
| Tetramer | Bayhill | Proinsulin plasmid | ↓ PI-specific CD8 frequency in subset with positive outcome [35] |
| Cytokine secretion | DiaPep277 BEL | HSP60 peptide | ↑ HSP60-specific production of multiple cytokines [39] |
| ITN Insulin B chain | Insulin B chain | ↑ insulin-specific TGF-β production [22] | |
| Diamyd® SWE | GAD-alum | ↑ GAD65-specific cytokines, mostly Th2 [29] | |
| Diamyd® SWE | GAD-alum | ↑ GAD65-specific cytokines, mostly Th2 [28] | |
| Diamyd® EU | GAD-alum | ↑ GAD65-specific cytokines, ↑ Th2 skewing with additional doses [24] |
A major limitation of proliferation assays is that they do not provide information about the functional characteristics of the responding cells. Among antigen-specific assays that assess function, ELISPOT is by far the most validated and widely used in the infectious disease vaccine and cancer immunotherapy fields. This assay was also used to show evidence of induced tolerance to insulin, with a reduction in specific IFN-γ secreting cells after three months of intranasal insulin treatment [26]. An earlier trial by the same group, however, did not achieve the same effect [21]. In a small study of intradermal injection of a single proinsulin peptide, there was no change in peptide-specific IFN-γ, IL-4, IL-5, or IL-13 by ELISPOT, although there was anecdotal evidence of transient increases in IL-10+ cells in the lower dose arm [27]. A limitation of ELISPOT is the number of cytokines, one or two, that is measurable in a single assay, which limits the amount of information gained per sample. Other T cell cytokine assays, such as intracellular cytokine staining or cytokine capture assays, have not been reported for antigen therapy trials in T1D. Flow cytometry-based assays generally have lower sensitivity than ELISPOT, putting them at a disadvantage for detection of rare T cells with anti-islet specificities. A complementary approach that has been effective in quantifying multiple cytokines at once is multiplex cytokine analysis, such as Luminex, on stimulated cell supernatants. While this approach assesses the stimulated population as a whole, rather than quantifying the number of responding cells, it has been effective in generating qualitative information about the T cell response induced by GAD-alum therapy. In a Swedish Phase II study, a subset of subjects receiving two GAD-alum treatments showed highly significant increases in a broad range of GAD-specific cytokine responses, including IL-4, IL-5, IL-6, IL-10, IL-13, IL-17, IFN-γ, and TNF-α [28]. In long term follow-up of these subjects four years later, another Luminex analysis showed similar results with the addition of elevated IL-1β and IL-2, but not IL-6 [29]. In a subsequent Phase III trial, the investigators employed a modified assay with an extended 7-day culture period. A GAD-alum-treated group showed dose-dependent increases in the same array of GAD-induced cytokines, with the Th2-associated cytokines notably increasing in prevalence with the third and fourth doses [24]. Unfortunately, as these trials have not shown consistent evidence of efficacy, it is not clear whether an islet-specific Th2 response in this setting is clinically beneficial.
An alternative approach taken to assess the phenotype of autoantigen-specific T cells has been to measure antigen-induced changes in surface markers or gene expression of the entire lymphocyte population as a reflection of the characteristics of the responding cells. This strategy was also used in context of GAD-alum, where findings included treatment-associated gene upregulation (e.g., PD-L1, CD25, IL-2, Foxp3, IL-15R, and TGF-β) [28,29] and induction of CD25+ CD127− and FOXP3+ cells after GAD stimulation [28,30]. These data suggest that GAD-alum therapy, in addition to promoting the activation of effector T cells, particularly those with a Th2 phenotype, may also expand GAD-specific regulatory T cells. Confirming these findings with more direct methods will be crucially important.
A more direct method for characterizing T cells with a defined specificity in T1D, analogous to the methods discussed above in allergy, is using MHC-peptide multimers [31–33]. A recent innovation in this area, the Diab-Q Kit, uses a combinatorial labelling approach to allow monitoring of multiple CD8 islet-antigen specificities simultaneously [34]. This assay was used to analyze patients receiving Bayhill BHT-3021, a plasmid vector encoding proinsulin. Reductions in proinsulin-specific CD8 T cells were observed in a subset of plasmid-, but not placebo-, treated patients that were tetramer positive at baseline, and this change in specific CD8 T cell frequency appeared to correlate with maintenance of c-peptide [35]. Though they are approximately 10-fold less frequent than in the CD8 compartment, islet-specific CD4 T cells can also be directly measured by pMHC multimers. For specificities with frequencies below approximately 50 per one million CD4 T cells, the range in which islet antigen reactivity is typically found, it is critical to use magnetic enrichment to allow reliable detection above background [36]. The combination of fine specific frequency and phenotypic data provided by pMHC multimer assays make an attractive approach to monitor antigen-specific immunotherapy in T1D, using the same strategies outlined in Figure 2. One limitation to the pMHC multimer strategy is the requirement for known immunodominant peptide responses mapped for each relevant HLA type. While this limitation is not a barrier to effective use in a proof-of-mechanism setting focused on HLA-restricted populations, an HLA-independent assay would be ideal for downstream use as a surrogate endpoint. Assays based on the induction of surface activation markers, such as CD137 [37] or CD154 [38], on antigen-specific cells are also compatible with magnetic enrichment and could employ stimulation with overlapping peptide libraries or whole antigen. Outcomes from this type of assay have not been reported yet in the context of clinical antigen-specific therapy in T1D, but we expect these methods to be improved and highly informative in future trials.
4. Concluding remarks
We are now in an era with an ever expanding number of potential therapeutic approaches to specifically modulate antigen-specific T cells in allergy and autoimmune diseases, such as T1D. More than ever, there is a critical need to develop and validate biomarker assays that will guide target selection, streamline the evaluation process, and expedite the path to regulatory approval. With antigen-specific T cells at the core of the pathologic process in these diseases, development and validation of assays that reliably assess both the quantity and quality of these cells will provide the key data to move rationally from antigen therapy concepts to clinical application.
Highlights.
Discovering and understanding biomarkers informs multiple phases of drug development and clinical practice.
Biomarker assays used to assess clinical trial outcomes must have appropriate performance characteristics and require validation.
Clinical trials designed to achieve antigen-specific tolerance create opportunities to develop biomarkers by specifically measuring cellular immune responses to individual disease-associated antigens.
Acknowledgments
The authors acknowledge the following sources of support for immune mediated disease research: JDRF (T1D Biomarker Core Assay and Validation Center to JMO, GTN) and NIH/NIAID (1 R01 AI108839 to EW). The authors thank their colleagues and clinical collaborators at Benaroya Research Institute, who contributed to the work described in this review.
Abbreviations
- T1D
type 1 diabetes
- MHC
major histocompatibility complex
- DC
dendritic cells
- ILC2
innate lymphoid type 2 cells
- pMHCII
peptide-MHC class II
- TSLP
thymic stromal lymphopoietin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jared M. Odegard, Email: jodegard@benaroyaresearch.org.
Gerald T. Nepom, Email: nepom@benaroyaresearch.org.
Erik Wambre, Email: ewambre@benaroyaresearch.org.
Reference List
- 1.Ciprandi G, Alesina R, De Amici M. Serum specific IgE: biomarker for specific immunotherapy responsiveness? Allergol Immunopathol (Madr) 2014;42:369–371. doi: 10.1016/j.aller.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Özdemir SK, Güloğlu D, Sin BA, Elhan AH, Ikinciogullari A, Misirligil Z. Reliability of basophil activation test using CD203c expression in diagnosis of pollen allergy. Am J Rhinol Allergy. 2011;25:e225–e231. doi: 10.2500/ajra.2011.25.3723. [DOI] [PubMed] [Google Scholar]
- 3.Santos AF, Douiri A, Becares N, Wu SY, Stephens A, Radulovic S, Chan SM, Fox AT, Du TG, Turcanu V, Lack G. Basophil activation test discriminates between allergy and tolerance in peanut-sensitized children. J Allergy Clin Immunol. 2014;134:645–652. doi: 10.1016/j.jaci.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patil SU, Shreffler WG. Immunology in the Clinic Review Series; focus on allergies: basophils as biomarkers for assessing immune modulation. Clin Exp Immunol. 2012;167:59–66. doi: 10.1111/j.1365-2249.2011.04503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nouri-Aria KT, Wachholz PA, Francis JN, Jacobson MR, Walker SM, Wilcock LK, Staple SQ, Aalberse RC, Till SJ, Durham SR. Grass pollen immunotherapy induces mucosal and peripheral IL-10 responses and blocking IgG activity. J Immunol. 2004;172:3252–3259. doi: 10.4049/jimmunol.172.5.3252. [DOI] [PubMed] [Google Scholar]
- 6.Bodtger U, Ejrnaes AM, Hummelshoj L, Jacobi HH, Poulsen LK, Svenson M. Is immunotherapy-induced birch-pollen-specific IgG4 a marker for decreased allergen-specific sensitivity? Int Arch Allergy Immunol. 2005;136:340–346. doi: 10.1159/000084227. [DOI] [PubMed] [Google Scholar]
- 7.Nagao M, Hiraguchi Y, Hosoki K, Tokuda R, Usui T, Masuda S, Yamaguchi M, Fujisawa T. Allergen-induced basophil CD203c expression as a biomarker for rush immunotherapy in patients with Japanese cedar pollinosis. Int Arch Allergy Immunol. 2008;146(Suppl 1):47–53. doi: 10.1159/000126061. [DOI] [PubMed] [Google Scholar]
- 8.Shamji MH, Layhadi JA, Scadding GW, Cheung DK, Calderon MA, Turka LA, Phippard D, Durham SR. Basophil expression of diamine oxidase: A novel biomarker of allergen immunotherapy response. J Allergy Clin Immunol. 2014;6749:1477–1478. doi: 10.1016/j.jaci.2014.09.049. [DOI] [PubMed] [Google Scholar]
- 9.Di Lorenzo G, Mansueto P, Pacor ML, Martinelli N, Rizzo M, Ditta V, Leto-Barone MS, D’Alcamo A, Politi D, Pepe I, Rotolo G, Di Fede G, Caruso C, Rini GB, Corrocher R. Clinical importance of eosinophil count in nasal fluid in patients with allergic and non-allergic rhinitis. Int J Immunopathol Pharmacol. 2009;22:1077–1087. doi: 10.1177/039463200902200424. [DOI] [PubMed] [Google Scholar]
- 10.Busse W, Spector S, Rosen K, Wang Y, Alpan O. High eosinophil count: a potential biomarker for assessing successful omalizumab treatment effects. J Allergy Clin Immunol. 2013;132:485–486. doi: 10.1016/j.jaci.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 11.Scadding GW, Calderon MA, Bellido V, Koed GK, Nielsen NC, Lund K, Togias A, Phippard D, Turka LA, Hansel TT, Durham SR, Wurtzen PA. Optimisation of grass pollen nasal allergen challenge for assessment of clinical and immunological outcomes. J Immunol Methods. 2012;384:25–32. doi: 10.1016/j.jim.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Konno S, Golden DB, Schroeder J, Hamilton RG, Lichtenstein LM, Huang SK. Level of osteopontin is increased after bee venom immunotherapy. J Allergy Clin Immunol. 2005;115:1317–1318. doi: 10.1016/j.jaci.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Zimmer A, Bouley J, Le Mignon M, Pliquet E, Horiot S, Turfkruyer M, Baron-Bodo V, Horak F, Nony E, Louise A, Moussu H, Mascarell L, Moingeon P. A regulatory dendritic cell signature correlates with the clinical efficacy of allergen-specific sublingual immunotherapy. J Allergy Clin Immunol. 2012;129:1020–1030. doi: 10.1016/j.jaci.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Lao-Araya M, Steveling E, Scadding GW, Durham SR, Shamji MH. Seasonal increases in peripheral innate lymphoid type 2 cells are inhibited by subcutaneous grass pollen immunotherapy. J Allergy Clin Immunol. 2014;134:1193–1195. doi: 10.1016/j.jaci.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 15.Kwok WW, Roti M, DeLong JH, Tan V, Wambre E, James EA, Robinson D. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol. 2010;125:1407–1409. doi: 10.1016/j.jaci.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wambre E, James EA, Kwok WW. Characterization of CD4+ T cell subsets in allergy. Curr Opin Immunol. 2012;24:700–706. doi: 10.1016/j.coi.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wambre E, DeLong JH, James EA, Torres-Chinn N, Pfutzner W, Mobs C, Durham SR, Till SJ, Robinson D, Kwok WW. Specific immunotherapy modifies allergen-specific CD4(+) T-cell responses in an epitope-dependent manner. J Allergy Clin Immunol. 2014;133:872–879. doi: 10.1016/j.jaci.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wambre E, DeLong JH, James EA, Lafond RE, Robinson D, Kwok WW. Differentiation stage determines pathologic and protective allergen-specific CD4+ T-cell outcomes during specific immunotherapy. J Allergy Clin Immunol. 2012;129:544–551. doi: 10.1016/j.jaci.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilles S, Traidl-Hoffmann C. CD27 expression on allergen-specific T cells: a new surrogate for successful allergen-specific immunotherapy? J Allergy Clin Immunol. 2012;129:552–554. doi: 10.1016/j.jaci.2011.12.967. [DOI] [PubMed] [Google Scholar]
- 20.Wambre E, DeLong JH, James EA, Robinson DM, Kwok WW. TH2A cells as a unique TH2 cell subset in allergic individuals: Steps toward a T cell biomarker for allergy. J Allergy Clin Immunol. 2012;129:AB129. [Google Scholar]
- 21.Harrison LC, Honeyman MC, Steele CE, Stone NL, Sarugeri E, Bonifacio E, Couper JJ, Colman PG. Pancreatic beta-cell function and immune responses to insulin after administration of intranasal insulin to humans at risk for type 1 diabetes. Diabetes Care. 2004;27:2348–2355. doi: 10.2337/diacare.27.10.2348. [DOI] [PubMed] [Google Scholar]
- 22.Orban T, Farkas K, Jalahej H, Kis J, Treszl A, Falk B, Reijonen H, Wolfsdorf J, Ricker A, Matthews JB, Tchao N, Sayre P, Bianchine P. Autoantigen-specific regulatory T cells induced in patients with type 1 diabetes mellitus by insulin B-chain immunotherapy. J Autoimmun. 2010;34:408–415. doi: 10.1016/j.jaut.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenbaum CJ, McCulloch-Olson M, Chiu HK, Palmer JP, Brooks-Worrell B. Parenteral insulin suppresses T cell proliferation to islet antigens. Pediatr Diabetes. 2011;12:150–155. doi: 10.1111/j.1399-5448.2010.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Axelsson S, Cheramy M, Akerman L, Pihl M, Ludvigsson JR. Casas, Cellular and humoral immune responses in type 1 diabetic patients participating in a phase III GAD-alum intervention trial. Diabetes Care. 2013;36:3418–3424. doi: 10.2337/dc12-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monti P, Scirpoli M, Rigamonti A, Mayr A, Jaeger A, Bonfanti R, Chiumello G, Ziegler AG, Bonifacio E. Evidence for in vivo primed and expanded autoreactive T cells as a specific feature of patients with type 1 diabetes. J Immunol. 2007;179:5785–5792. doi: 10.4049/jimmunol.179.9.5785. [DOI] [PubMed] [Google Scholar]
- 26.Fourlanos S, Perry C, Gellert SA, Martinuzzi E, Mallone R, Butler J, Colman PG, Harrison LC. Evidence that nasal insulin induces immune tolerance to insulin in adults with autoimmune diabetes. Diabetes. 2011;60:1237–1245. doi: 10.2337/db10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thrower SL, James L, Hall W, Green KM, Arif S, Allen JS, Van-Krinks C, Lozanoska-Ochser B, Marquesini L, Brown S, Wong FS, Dayan CM, Peakman M. Proinsulin peptide immunotherapy in type 1 diabetes: report of a first-in-man Phase I safety study. Clin Exp Immunol. 2009;155:156–165. doi: 10.1111/j.1365-2249.2008.03814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hjorth M, Axelsson S, Ryden A, Faresjo M, Ludvigsson J, Casas R. GAD-alum treatment induces GAD65-specific CD4+CD25highFOXP3+ cells in type 1 diabetic patients. Clin Immunol. 2011;138:117–126. doi: 10.1016/j.clim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Axelsson S, Chéramy M, Hjorth M, Pihl M, Akerman L, Martinuzzi E, Mallone R, Ludvigsson J, Casas R. Long-lasting immune responses 4 years after GAD-alum treatment in children with type 1 diabetes. PLoS ONE. 2011;6:e29008. doi: 10.1371/journal.pone.0029008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pihl M, Akerman L, Axelsson S, Chéramy M, Hjorth M, Mallone R, Ludvigsson J, Casas R. Regulatory T cell phenotype and function 4 years after GAD-alum treatment in children with type 1 diabetes. Clin Exp Immunol. 2013;172:394–402. doi: 10.1111/cei.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reijonen H, Kwok WW, Nepom GT. Detection of CD4+ autoreactive T cells in T1D using HLA class II tetramers. Ann NY Acad Sci. 2003;1005:82–87. doi: 10.1196/annals.1288.009. [DOI] [PubMed] [Google Scholar]
- 32.Pinkse GG, Tysma OH, Bergen CA, Kester MG, Ossendorp F, van Veelen PA, Keymeulen B, Pipeleers D, Drijfhout JW, Roep BO. Autoreactive CD8 T cells associated with beta cell destruction in type 1 diabetes. Proc Natl Acad Sci USA. 2005;102:18425–18430. doi: 10.1073/pnas.0508621102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nepom GT. MHC class II tetramers. J Immunol. 2012;188:2477–2482. doi: 10.4049/jimmunol.1102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velthuis JH, Unger WW, Abreu JR, Duinkerken G, Franken K, Peakman M, Bakker AH, Reker-Hadrup S, Keymeulen B, Drijfhout JW, Schumacher TN, Roep BO. Simultaneous detection of circulating autoreactive CD8+ T-cells specific for different islet cell-associated epitopes using combinatorial MHC multimers. Diabetes. 2010;59:1721–1730. doi: 10.2337/db09-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roep BO, Solvason N, Gottlieb PA, Abreu JR, Harrison LC, Eisenbarth GS, Yu L, Leviten M, Hagopian WA, Buse JB, von Herrath M, Quan J, King RS, Robinson WH, Utz PJ, Garren H, Steinman L. Plasmid-encoded proinsulin preserves C-peptide while specifically reducing proinsulin-specific CD8(+) T cells in type 1 diabetes. Sci Transl Med. 2013;5:191ra82. doi: 10.1126/scitranslmed.3006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scriba TJ, Purbhoo M, Day CL, Robinson N, Fidler S, Fox J, Weber JN, Klenerman P, Sewell AK, Phillips RE. Ultrasensitive detection and phenotyping of CD4+ T cells with optimized HLA class II tetramer staining. J Immunol. 2005;175:6334–6343. doi: 10.4049/jimmunol.175.10.6334. [DOI] [PubMed] [Google Scholar]
- 37.Wolfl M, Kuball J, Ho WY, Nguyen H, Manley TJ, Bleakley M, Greenberg PD. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110:201–210. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacher P, Schink C, Teutschbein J, Kniemeyer O, Assenmacher M, Brakhage AA, Scheffold A. Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naive and memory Th cell repertoire. J Immunol. 2013;190:3967–3976. doi: 10.4049/jimmunol.1202221. [DOI] [PubMed] [Google Scholar]
- 39.Huurman VA, van der Meide PE, Duinkerken G, Willemen S, Cohen IR, Elias D, Roep BO. Immunological efficacy of heat shock protein 60 peptide DiaPep277 therapy in clinical type I diabetes. Clin Exp Immunol. 2008;152:488–497. doi: 10.1111/j.1365-2249.2008.03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raz I, Elias D, Avron A, Tamir M, Metzger M, Cohen IR. Beta-cell function in new-onset type 1 diabetes and immunomodulation with a heat-shock protein peptide (DiaPep277): a randomised, double-blind, phase II trial. Lancet. 2001;358:1749–1753. doi: 10.1016/S0140-6736(01)06801-5. [DOI] [PubMed] [Google Scholar]