Abstract
Oncocytomas are mostly benign tumors characterized by accumulation of defective mitochondria, and in sporadic cases, are associated with disruptive mitochondrial DNA (mtDNA) mutations. However, the role mtDNA mutations play in renal tumors of Birt-Hogg-Dubé (BHD) patients and other renal oncocytomas with an apparent genetic component has not been investigated to date. Here we characterize the mitochondrial genome in different renal tumors and investigate the possibility of employing mtDNA sequencing analyses of biopsy specimens to aid in the differential diagnosis of oncocytomas. The entire mitochondrial genome was sequenced in 25 samples of bilateral and multifocal (BMF) renal oncocytomas, 30 renal tumors from BHD patients and 36 non-oncocytic renal tumors of different histologies as well as in biopsy samples of kidney tumors. mtDNA sequencing in BMF oncocytomas revealed that all tumors carry disruptive mutations, which impair the assembly of the NADH-ubiquinone oxidoreductase. Multiple tumors from a given BMF oncocytoma patient mainly harbor the same somatic mutation and the kidneys of these patients display diffuse oncocytosis. In contrast, renal oncocytomas of patients with BHD syndrome and renal tumors with different histologies do not show disruptive mtDNA mutations. Moreover, we demonstrate that it is feasible to amplify and sequence the entire mtDNA in biopsy specimens, and that these sequences are representative of the tumor DNA. These results show that pathogenic mtDNA mutations affecting complex I of the respiratory chain are strongly correlated with the oncocytoma phenotype in non-BHD-related renal tumors and that mtDNA sequences from biopsies are predictive of the tumor genotype. This work supports a role for mtDNA mutations in respiratory chain complexes as diagnostic markers for renal oncocytomas.
Introduction
In recent years, the mitochondrial genome (mtDNA) has been attributed an important role in many pathological conditions, including cancer (1). Human mtDNA is a maternally inherited circular chromosome of ca. 16.6 kb that encodes 37 genes, including tRNAs, rRNAs and 13 proteins that are assembled into the respiratory chain complexes. Mitochondria contain multiple copies of mtDNA and each cell may contain several thousands of mtDNA copies (1, 2). The presence of multiple mtDNA copies within a cell implies the possibility of a coexistence of mutant and wild-type alleles within a single cell or tissue, a phenomenon known as heteroplasmy. When mtDNA mutations are present, the level of heteroplasmy will determine the phenotypic effect (1). Replication and distribution of mtDNA copies are stochastic processes and their allocation is influenced by fission and fusion events of mitochondria. Yet, mitochondrial genome replication and transcription are tightly regulated and carried out by nuclear-encoded genes. One of the master regulators of mitochondrial energy metabolism is the Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1 alpha (PGC1alpha), a nuclear transcriptional coactivator that coordinates mitochondrial biogenesis with metabolic needs of the cell or tissue (3).
Kidney cancer is not a single entity, but is made up of a number of tumor types that are caused by mutations in different genes and present different histologies and clinical courses. Therefore, clinical treatment needs to be optimized for each type of renal tumor (4). In particular, patient management of benign tumors will differ markedly from the management of aggressive tumors, and to date, imaging techniques and biopsy sampling are not always sufficient to distinguish between benign and malignant tumors (5). Renal oncocytomas behave mostly in a benign manner with no recurrence, metastasis or mortality (6). A non-invasive assay to differentiate oncocytomas from more aggressive renal cell carcinoma (RCC) could better facilitate clinical management. Oncocytomas are tumors of epithelial origin, characterized by dense accumulation of defective mitochondria that confers a granular eosinophilic cytoplasm (7). To date, no nuclear gene has been found to be responsible for these tumors, which often have a stable karyotype (7, 8). Notably, a correlation of the oncocytoma phenotype with the occurrence of disruptive mitochondrial DNA (mtDNA) mutations has been demonstrated in sporadic oncocytomas from different organs (9–11). In particular, sporadic renal oncocytomas have been found to lose activity of the electron transport chain complex I (NADH-ubiquinone oxidoreductase, CI) due to mutations in the mtDNA (11–13). Accordingly, complex I proteins are down-regulated in renal oncocytoma, as compared to chromophobe RCC, while proteins assembled into other respiratory chain complexes are up-regulated (14). Previous findings also show that the presence of homoplasmic mtDNA mutations in complex I genes trigger the benign behavior of these tumors (15), while somatic mtDNA nucleotide changes have been found in other renal tumors, but mostly with a low impact on respiratory chain function (16).
Sequencing analyses of the mtDNA have been carried out in renal tumors (17, 18) and several other types of tumors [e.g. (36) (colorectal cancer); (20) (breast cancer), (21)] and heteroplasmic mutations have frequently been found in neoplastic tissues. However, the pathogenic potential of these mutations has rarely been evaluated, even though a role of mtDNA mutations in oncogenesis has been suggested (22).
Renal oncocytomas can be found as single sporadic neoplasms, or can occur as bilateral and multifocal (BMF) tumors. First described as “oncocytomatosis” (23), the presence of numerous concurrent oncocytic nodules scattered throughout the renal parenchyma is now known as renal oncocytosis (24) and mainly causes reduced renal function in affected patients (25). It has been described in sporadic cases, and in familial cases related to Birt-Hogg-Dubé (BHD) syndrome (19, 25, 26). The most common histologies in the setting of BHD syndrome are hybrid oncocytic tumors with features of oncocytoma and chromophobe RCC, as well as pure chromophobe or oncocytic renal masses (25). In cases where renal oncocytosis occurs in a non-BHD setting without FLCN mutations, the responsible gene remains unknown (25). On the other hand, patients affected with BHD syndrome typically present with cutaneous lesions and pulmonary cysts in addition to an increased risk for multifocal renal neoplasms with oncocytic features. While sporadic renal oncocytomas are characterized by dysfunctional mitochondria, tumors with FLCN deficiency have been shown to carry functional mitochondria (27).
In the present work we investigate the involvement of mtDNA mutations in renal tumors with oncocytic histology. We show that BMF oncocytomas, similar to sporadic oncocytomas, carry disruptive mtDNA mutations affecting respiratory chain complex I genes, while BHD-related renal tumors have mostly intact mitochondrial genomes similar to non-oncocytic RCCs. We also provide evidence that renal oncocytosis is related to the presence of disruptive mtDNA mutations in affected cells. Hence, the present work demonstrates a role for mtDNA mutations in the development of renal oncocytomas with wild-type FLCN.
Materials and Methods
Samples
Twenty-five samples from 7 patients with sporadic bilateral and multifocal (BMF) renal oncocytoma were selected for this study. Five oncocytomas from patient BMF2 were derived from one kidney, and an additional six samples were derived from the contralateral kidney. Patients had no family history of renal tumors and were not affected by any genetic syndrome, including BHD. Detailed information about BMF patients and tumors are presented in Supplementary Table S1. Thirty renal tumor samples from 13 different patients, affected with BHD syndrome, were included in the analysis. Seven of these tumors displayed an oncocytic histology, 4 samples had chromophobe histology, and 18 tumors were hybrid oncocytic tumors, presenting features of both chromophobe RCC and oncocytoma, while one tumor was poorly differentiated. A panel of 36 non-oncocytic renal cell carcinomas with different histologies and genetic backgrounds were used as controls. Sporadic clear cell renal cell carcinomas (ccRCC) (n=6), BMF ccRCCs (n=4) and VHL-related ccRCC (n=5), as well as sporadic papillary type 1 RCC (n=5) and FH-mutated RCC (HLRCC) (n=4; 3 cell lines and 1 tumor) were included in the study. Chromophobe RCCs were selected from patients without family history of kidney tumors or BHD-related manifestations and without FLCN mutations (n=12).
Blood DNA was collected from each patient to evaluate the somatic versus germline status of mutations. The histology of all tumors was reviewed by one pathologist (MM). Informed consent was obtained from all patients prior to surgery, in accordance with the National Institutes of Health Review Board.
mtDNA sequencing
DNA was extracted from frozen tumor tissue and blood with Maxwell 16 Tissue and Blood DNA purification kits (Promega, Madison, WI, USA). Microdissection of tumor and normal tissue was performed manually on formalin fixed and paraffin embedded tissue slides and DNA was extracted with Maxwell 16 FFPE Tissue LEV DNA purification kit (Promega) after 16 h incubation with proteinase K at 65°C.
The entire mitochondrial genome was sequenced with the MitoAll kit (Applied Biosystems, Foster City, CA, USA) (10). PCR reactions were performed in 10 µl with KAPA2G Fast Readymix on 10 ng of DNA. PCR amplicons were purified by adding 1U of FastAP Thermosensitive Alkaline Phosphatase and 0.5 U of Exonuclease I to each PCR reaction and incubation at 37°C for 30 min, followed by heat inactivation at 80°C for 15 min. Sequencing was performed with BigDye v1.1 (Applied Biosystems) according to the manufacturer’s instructions. The reactions were cleaned up with Performa DTR Plates (Edge Bio) and run in an ABI 3730 Genetic Analyzer automated sequencing machine (Applied Biosystems). Electropherograms were inspected and aligned to the human mitochondrial reference sequence (NC_012920) using Sequencher 4.10.4 (Genecodes, Ann Arbor, MI, USA). All sequences were submitted to the NCBI database (KJ801401 – KJ801483).
Mutation analysis
The polymorphic status of mtDNA nucleotide changes was verified by assigning variability scores according to the Human mitochondrial Database [hmtdb; www.hmtdb.uniba.it; (28)] as well as by inspecting the nucleotide changes in MITOMAP, a human mitochondrial genome database [mitomap.org; (29)]. The pathogenic potential of novel mtDNA mutations was assessed using PolyPhen-2 (30).
Heteroplasmy evaluation
Heteroplasmy of indel mutations was evaluated by Fluorescent PCR as previously described (31). Heteroplasmy levels of the m.G4036A mutation were evaluated by application of a custom castPCR (Applied Biosystems), using a wild-type and mutant assay. The amount of amplifiable DNA (mitochondrial DNA) was adjusted by real-time PCR performed with Maxima SYBR Green/ROX qPCR Master Mix (Fermentas, Waltham, MA, USA) in 20-µl reactions using primers specific for MT-CYB and starting with 1 ng of DNA per reaction (FW primer: TCCCACCCTCACACGATTCTTTACC, RV primer: GTGCAAGAATAGGAGGTGGAGTGCT). ΔCt cutoff values for castPCR were detected through castPCR assay on plasmid DNA, containing cloned wild-type or mutant alleles (Topo-TA cloning kit, Invitrogen, Carlsbad, CA, USA). CastPCR assays were performed in 10-µl reactions with TaqMan Universal PCR Master Mix (Applied Biosystems) on a Viia7 real-time PCR machine (Applied Biosystems) following the instructions given by the manufacturer. Data analysis was performed with Mutation Detector Software (Applied Biosystems).
Complex I activity assay
Complex I activity assay was performed on protein lysates from tumor and normal kidney tissue with Complex I Enzyme Activity Microplate Assay Kit (MitoSciences, Eugene, OR, USA) according to manufacturer’s instructions.
Immunofluorescence
Immunofluorescence staining was performed on 5-µm sections of frozen tissue, mounted on positively charged slides, as previously described (32). Staining for complex I (NDUFB8; 1:200, Invitrogen), complex II (SDHA; 1:200, Abcam; Cambridge, UK) and complex IV (COXIV; 1:400, Cell Signaling; Beverly, MA, USA) of the respiratory chain was performed by incubation with the primary antibody at 4°C overnight followed by 1 hour incubation at room temperature with secondary antibodies Alexa Fluor 488 goat anti-rabbit IgG (1:400) and/or Alexa Fluor 594 goat anti-mouse IgG (1:400) (Invitrogen). Slides were counterstained and mounted with Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA, USA) and analyzed with a fluorescent microscope (Leica DM RXA).
Analysis of tumor biopsies
Three biopsy cores were obtained from two different tumor specimens immediately after their surgical removal with 18G × 6 cm Biopsy Systems (Temno, San Diego, CA, USA) and kept in sterile PBS until further processing. DNA extraction from single biopsy cores and mtDNA sequencing were performed as described above.
Results
mtDNA sequencing in bilateral and multifocal (BMF) renal oncocytomas identifies complex I mutations
Pathogenic mtDNA mutations are hallmarks of sporadic oncocytomas (33). To investigate whether mtDNA mutations also play a role in multifocal renal oncocytomas, we sequenced the entire mitochondrial genome in 15 renal oncocytomas, resected in 8 surgeries from 7 different patients, who were diagnosed to have bilateral and multifocal renal oncocytomas. MtDNA sequence analysis showed that all of these samples carried pathogenic mutations in the mitochondrial genome. All initially sequenced tumors from patients BMF2 and BMF4 showed a distinct mutation (11038delA and 3571insC, respectively). Therefore, five additional tumors from each of these two patients were analyzed to determine their mutation status. In total, we analyzed 25 samples and found that all samples but one (patient BMF7) carried disruptive (frameshift or nonsense) mutations in genes coding for proteins assembled into complex I (CI) of the respiratory chain (MT-ND1, MT-ND4, MT-ND5) (Table 1). The tumor (patient BMF7) that did not carry a mutation in a CI gene had a heteroplasmic mutation in a tRNA (m.C7483T), affecting the anticodon of MT-TS1 and changing the anticodon triplet from UCA, coding for Serine, into UUA, the anticodon normally displayed by the tRNA for Leucine.
Table 1.
Somatic mtDNA mutations in BMF oncocytomas.
| Patient # | Surgery/Tumor | Mutation nucleotide | Mutation protein | Gene | Heteroplasmy |
|---|---|---|---|---|---|
| BMF1 | 1 / 1 | m.12425delA | p.N30Lfs*26 | MT-ND5 | 71% |
| 1 / 2 | m.3571insC | p.L89Pfs*13 | MT-ND1 | 88% | |
| m.12425delA | p.N30Lfs*26 | MT-ND5 | 27% | ||
| BMF2 | 1 / 1 | m.11038delA | p.K93Dfs*7 | MT-ND4 | 99% |
| 1 / 2 | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| 1 / 3 | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| 1 / 4 | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| 1 / 5 | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| 2 / 1a | m.11038delA | p.K93Dfs*7 | MT-ND4 | 98% | |
| 2 / 2a,b | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| 2 / 3a,b | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| 2 / 4 a,b | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| 2 / 5a,b | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| 2 / 6a,b | m.11038delA | p.K93Dfs*7 | MT-ND4 | hom | |
| BMF3 | 1 / 1 | m.G13915A | p.G524X | MT-ND5 | hom |
| BMF4 | 1 / 1 | m.3571insC | p.L89Pfs*13 | MT-ND1 | 93% |
| 1 / 2 | m.3571insC | p.L89Pfs*13 | MT-ND1 | 93% | |
| 1 / 3b | m.3571insC | p.L89Pfs*13 | MT-ND1 | 12% | |
| 1 / 4b | m.3571insC | p.L89Pfs*13 | MT-ND1 | 88% | |
| 1 / 5b | m.3571insC | p.L89Pfs*13 | MT-ND1 | 63% | |
| 1 / 6b | m.3571insC | p.L89Pfs*13 | MT-ND1 | 90% | |
| 1 / 7b | m.3571insC | p.L89Pfs*13 | MT-ND1 | 16% | |
| BMF5 | 1 / 1 | m.G4036A | p.G244X | MT-ND1 | 99% |
| 1 / 2 | m.G4036A | p.G244X | MT-ND1 | 98% | |
| BMF6 | 1 / 1 | m.10952insC | p.L65Pfs*45 | MT-ND4 | hom |
| BMF7 | 1 / 1 | m.C7483T | Anticodon UCA -> UUA | MT-TS1 | ca. 50% |
Samples from surgery 2 of patient BMF2 derived from the contralateral kidney.
Approximately 600 bp of the mitochondrial genome, spanning the mutation, have been sequenced in these samples.
hom: homoplasmic mutation as detected by electropherogram
In addition to these clearly pathogenic mutations, 10 samples carried additional somatic base changes, including known polymorphisms [m.309insC (BMF2-1/3, BMF4-1/1, BMF7), m.C16188T (BMF1-1/1) and m.524insAC (BMF7, BMF5-1/2)] in the variable D-Loop region, tRNA mutations of unclear pathogenic potential [m.G15961A – MT-TP (BMF2-2/1); m.G1664A MT-TV (BMF5-1/1)], a frameshift mutation and a nonsense mutation at a low heteroplasmic level [m.5761insC – MT-ND2 (BMF4-1/1); m.G12923A-MT-ND5 (BMF6)].
mtDNA mutations in BMF oncocytomas are somatic and homoplasmic
In order to determine the mutation load of the tumor samples and to test if the mutations are somatic and exclusive to the tumor tissue, we analyzed the heteroplasmy levels of the mutations found in the BMF oncocytomas and assessed the somatic status of the mutations by analyzing blood DNA and normal renal tissue. We evaluated the heteroplasmy level of frameshift mutations through Fluorescent-PCR (F-PCR) and the data are summarized in Table 1. Tumor 1 of patient BMF1 carried an m.12425delA mutation at 71% heteroplasmy level (71% mutant allele), whereas tumor 2 from the same patient had the same mutation at 27% heteroplasmy level, but carried an additional mutation (m.3571insC) at a high heteroplasmy level (88% mutant allele). Neither of these mutations was detected in the blood DNA of this patient.
F-PCR analysis of an m.11038delA mutation that occurred in two tumor samples from different surgeries and contralateral kidneys of patient BMF2 revealed mutation loads of 99% and 98.5%, whereas no mutation was detectable in the blood DNA of the same patient. We also analyzed all 7 samples from patient BMF4 for the m.3571insC mutation. Samples that had a mutation load of 88% or higher were of high tumor purity (Table 1) whereas the three samples with lower mutation load were noted by histological evaluation to have a corresponding amount of normal tissue contamination. Again, analysis of the blood DNA confirmed the somatic status of this mutation.
Mutation m.G4036A (patient BMF5) was analyzed by real-time PCR. Both tumors had a high mutation load (>98%), and the mutation was not detectable in the blood DNA of the respective patient (Table 1).
To investigate whether the mutations found in the tumors, and absent from the blood DNA, were present in the normal kidney parenchyma, we microdissected kidney tissue adjacent to the tumors, and the tumor tissue itself, in two patients (BMF1 and BMF4). Tissue adjacent to tumor 1/1 of patient BMF1 (Table 1) was analyzed for mutations m.12425insA and m.3571delC, but the mutations could not be detected. While the microdissected tumor of patient BMF4 had a high mutation load (>94%), the mutation was not detectable in the corresponding microdissected normal tissues (Figure 1D). These data confirm that the mtDNA mutations found in BMF oncocytomas are somatic and only present in the tumor tissue.
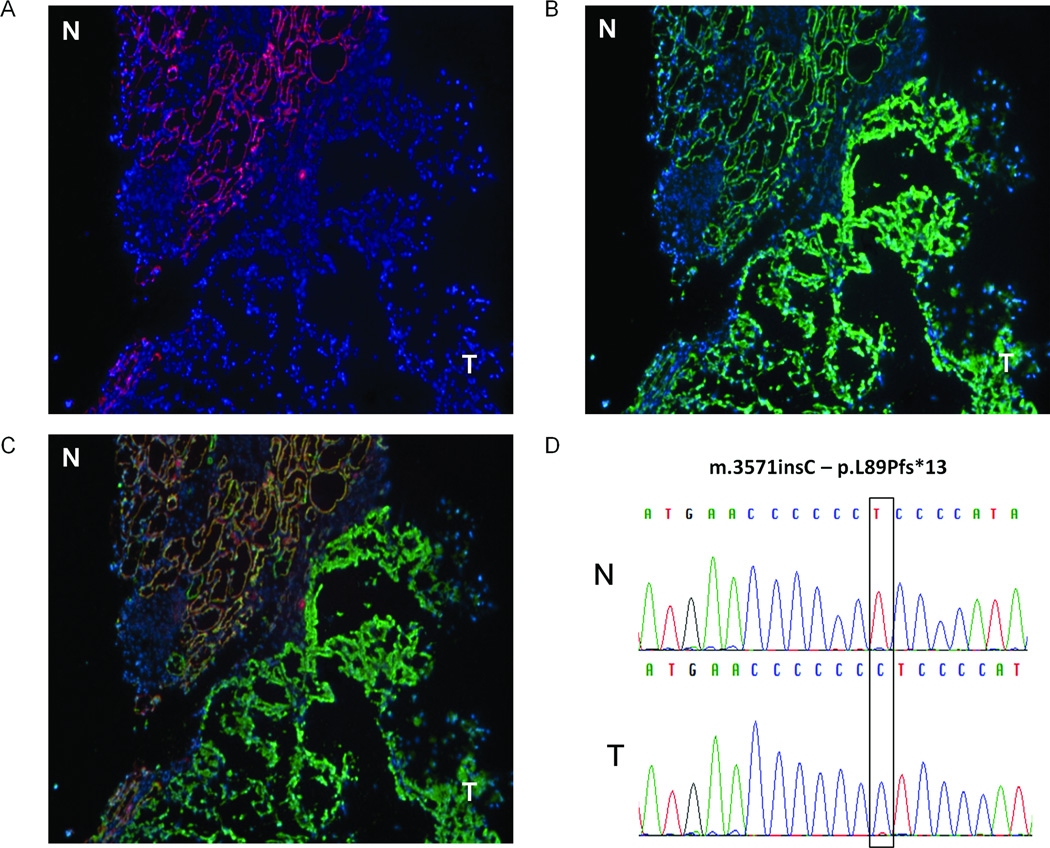
Figure 1.
BMF oncocytomas lack complex I. Representative immunohistochemistry of tissue sections containing normal (N) and oncocytic tissue (T) for subunits of respiratory chain complex I (NDUFB8) (A) and complex IV (COXIV) (B). (C) Overlap of A and B. (50× magnification) (D) Sequence electropherogram showing frameshift mutation m.3571insC in sample BMF4.
Immunohistochemistry of BMF oncocytomas reveals absence of respiratory chain complex I
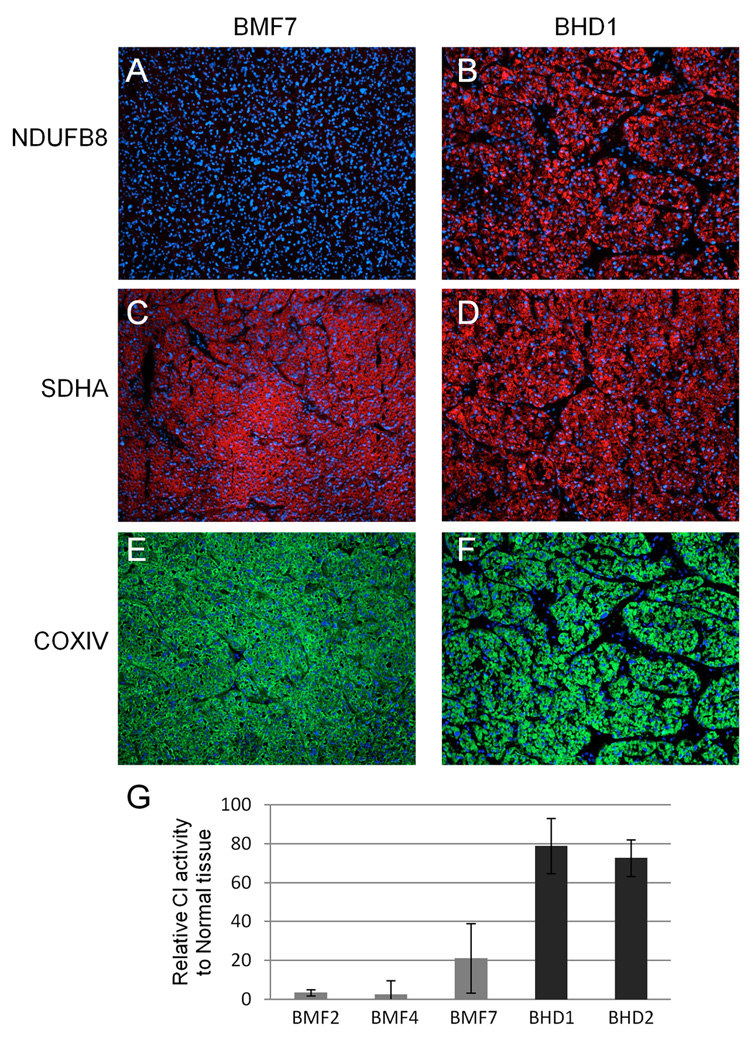
Disruptive mutations of mitochondria-encoded subunits of CI are expected to impair the proper assembly of the enzyme complex. To test this assumption, we stained frozen tissue sections of BMF oncocytomas with antibodies for different subunits of respiratory chain complexes (CI, CII and CIV) and found that all tumors yielded strong staining for CIV (COXIV) (Figure 1B, C) and CII (SDHA) (Figure 2C) proteins, which is in agreement with a high mitochondrial content, a hallmark of oncocytomas. Conversely, a nuclear encoded subunit of CI (NDUFB8) did not stain in BMF oncocytomas, although it was clearly present in adjacent normal renal tissue (Figure 1A,C). This staining pattern was consistent for all samples with mutations in CI genes, as well as for the case with the heteroplasmic mutation in the tRNA MT-TS1 (patient BMF7, mutation m.C7483T; Figure 2A,C,E). In addition, an in vitro assay confirmed that the activity of CI was seriously impaired in BMF oncocytomas (Figure 2G).
Figure 2.
A tRNA mutation disrupts respiratory chain complex I in renal oncocytoma while BHD oncocytomas have intact mitochondria. Immunohistochemistry for complex I (NDUFB8) (A, B), complex II (SHDA) (C, D), and complex IV (COXIV) (E, F) in renal oncocytoma sample with heteroplasmic tRNA mutation compared to BHD oncocytoma shows absence of CI staining in oncocytoma with tRNA mutation (A), but presence of CI staining in BHD oncocytoma (B). CII and CIV staining is present in both tumors (100× magnification). G, Complex I activity assay confirms very low activity in BMF oncocytomas, compared to normal kidney tissue activity, but similar CI activity between BHD tumors and normal kidney tissue.
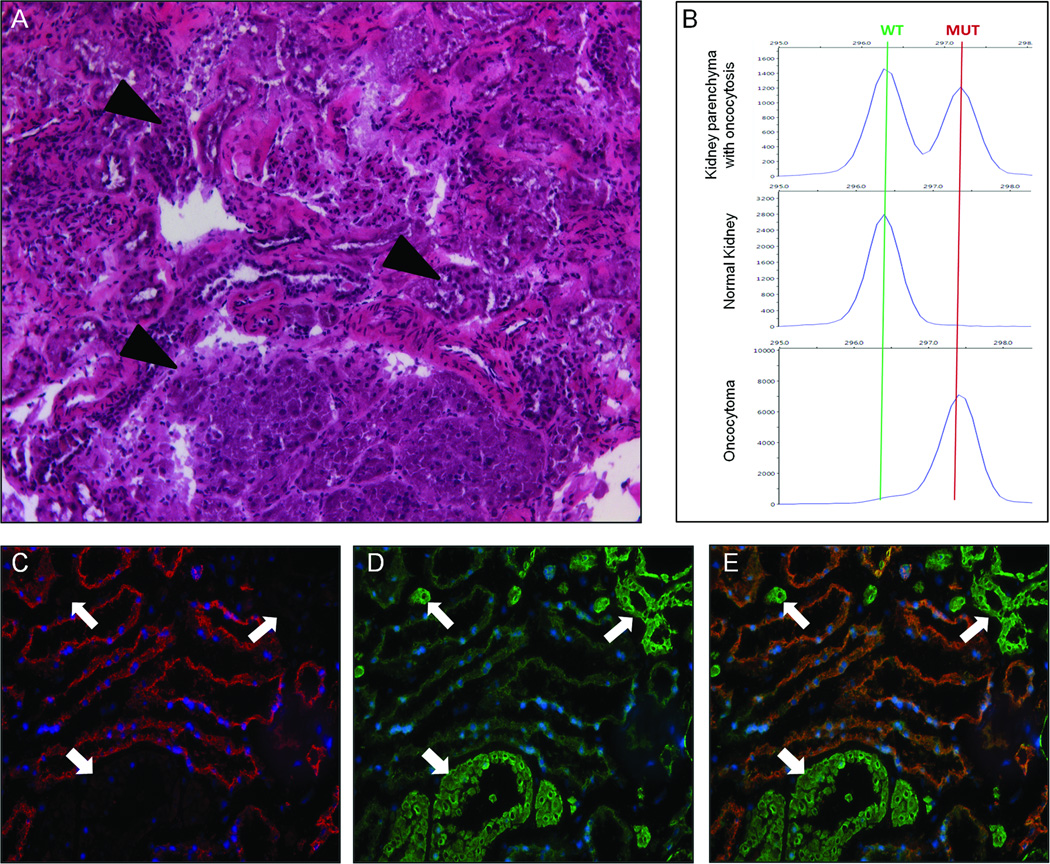
BMF oncocytoma patients can present with oncocytosis in their kidneys
BMF oncocytomas have been described to occur in the context of renal oncocytosis and the incidence of numerous oncocytomas in both kidneys is suggestive of this diagnosis (24). For example, patient BMF4 underwent surgery on both kidneys and a total of 10 tumors were removed. For this patient, the analysis of tissue adjacent to the resected tumors allowed us to investigate the presence of oncocytosis in the “normal” kidney parenchyma. Hematoxylin and eosin staining in fact revealed the presence of numerous cells and cell foci with an oncocytic appearance (Figure 3A). Immunohistochemistry (IHC) staining for subunits of respiratory chain complexes revealed that all oncocytic cells had the same staining pattern seen in the oncocytoma: high abundance of CIV (COXIV) proteins (Figure 3D,E), and absence of CI subunits (NDUFB8) (Figure 3C, E). In agreement with this finding, genetic analysis of microdissected tissue with oncocytosis confirmed that oncocytic cells carried a disruptive mtDNA mutation (in this case m.3571insC), while non-affected cells presented with a wild-type mitochondrial genome (Figure 3B).
Figure 3.
H&E staining, IHC and Fluorescent PCR (F-PCR) of kidney samples from patient BMF4 showing oncocytosis. A, H&E staining of “normal” renal tissue showing numerous oncocytic nodules throughout the renal tissue (arrowheads) (100× magnification). B, F-PCR analysis for the mutation m.3571insC (MT-ND1) shows peaks for wild-type and mutant alleles in DNA extracted from tissue with oncocytosis. No mutation is detectable in the microdissected normal tissue (0% mutation), whereas only a very small peak is seen for the wild-type allele in the microdissected tumor tissue (93% mutation). Immunohistochemistry for a respiratory chain complex I subunit (NDUFB8) (C) and complex IV subunit (COXIV) (D, overlap shown in E) shows correlation between abundance of mitochondria, as indicated by CIV staining, and absence of CI in multiple nodules throughout the tissue (indicated with white arrows). Normal renal tissue expresses both CI and CIV proteins (central part of images; 200× magnification).
mtDNA mutations specifically occur in oncocytomas
To test if mtDNA mutations are specific for oncocytomas in patients with bilateral and multifocal renal cancer, we sequenced the entire mtDNA in an oncocytoma and a clear cell RCC (ccRCC) that developed in the same patient and were resected in two consecutive surgeries (BMF6) (Supplementary Figure S1). The mtDNA sequence of the ccRCC presented 41 polymorphic nucleotide changes, as compared to the mtDNA reference sequence, and 3 somatic heteroplasmic missense mutations [m.G4244A, MT-ND1, p.S313N, Polyphen-2 score (PSIC): 0.000; m.A11672G, MT-ND4, p.T305A, PSIC: 0.994; m.G12997A, MT-ND5, p.A221T, PSIC:0.906]. On the other hand, the sequence of the renal oncocytoma confirmed the presence of all polymorphisms, but was wild-type for the missense mutations found in the ccRCC. Instead, this sample carried a homoplasmic frameshift mutation in MT-ND4 (m.10952insC, p.L65Pfs*45) as well as a heteroplasmic nonsense mutation in MT-ND5 (m.G12923A, p.W196X), that were absent in the ccRCC and patient blood DNA (Supplementary Figure S1). These results underline the strong correlation between disruptive mtDNA mutations and the occurrence of the oncocytoma tumor type.
mtDNA sequencing in non-oncocytic tumors
Chromophobe RCC can share histological similarities with oncocytomas. In particular the eosinophilic variant of chromophobe RCC closely resembles renal oncocytoma (34). Therefore, we also sequenced the entire mitochondrial genome in a total of 12 sporadic chromophobe RCC (2 classic type, 5 eosinophilic, 4 clear and eosinophilic and one not classified) to test for the presence of mtDNA mutations in this kidney tumor phenotype. Six of these samples turned out to be wild-type, 2 samples carried base changes that are known to be polymorphic, 3 samples carried base changes in the variable D-loop region or a tRNA, while one sample acquired a heteroplasmic missense mutation (50% mutant allele) with a predicted high pathogenic potential (m.G15045A – MT-CYB p.R100Q – Polyphen2 PSIC:1.000) and another sample presented a novel heteroplasmic base change (25% mutant allele) in the MT-RNR2 gene (Supplementary Table S2). All of these mutations were somatic. Overall, sporadic chromophobe RCC only rarely acquired pathogenic mtDNA mutations.
In addition to these sporadic tumors, a patient presented to our institute with bilateral and multifocal chromophobe RCC, and six resected tumors were available for analysis. Notably, all tumors of this patient presented with somatic mtDNA changes. Three tumors carried homoplasmic frameshift insertions (m.3571insC – MT-ND1, m.14623insC – MT-ND6), one tumor had a heteroplasmic frameshift deletion (m.4611delA – MT-ND2) and two tumors showed mutations in the MT-CO1 (m.G6955A) or the D-Loop region (m.549delC, m.T531C) (Supplementary Table S2). Histologic analysis confirmed the eosinophilic chromophobe phenotype of these samples, which therefore represent an exception to the correlation between mtDNA mutations and the oncocytoma phenotype.
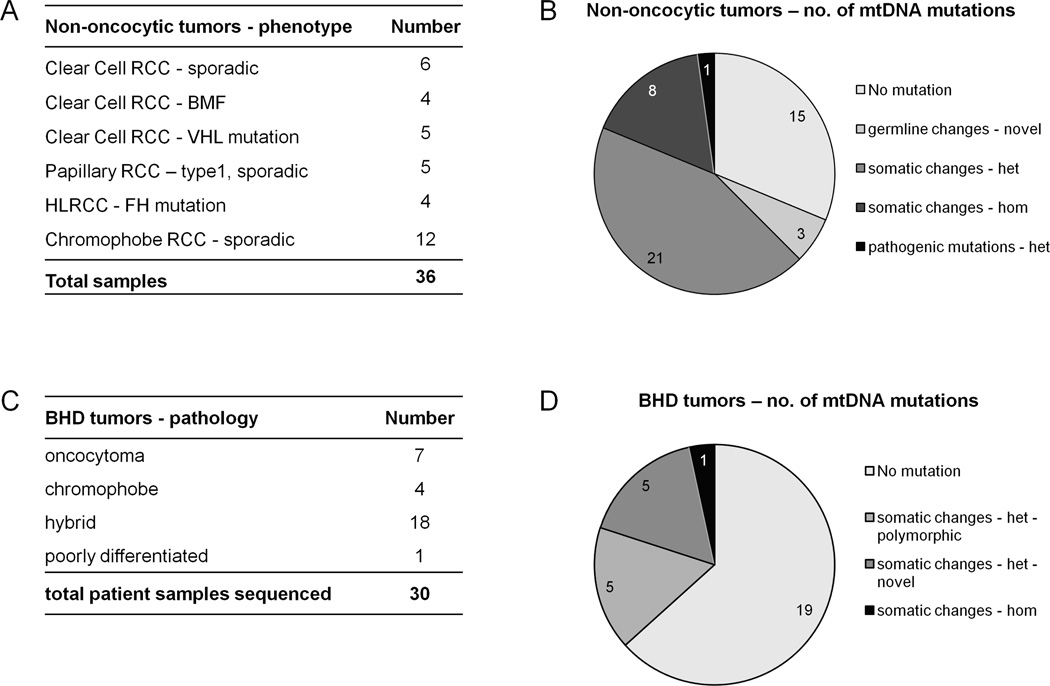
Moreover, we sequenced the entire mtDNA in 24 additional non-oncocytic tumors of different histologies (summarized in Figure 4 and Supplementary Table S3). Eleven of these renal tumor samples displayed a wild-type mitochondrial genome, while the remaining samples carried somatic base changes. In these 13 tumors we found a total of 23 base changes, made up of 5 homoplasmic and 18 heteroplasmic mutations (Supplementary Table S3). Two of the homoplasmic mutations had a strongly predicted pathogenic potential, as analyzed by PolyPhen-2 (m.G3916A – PSIC=0.99; m.G14607A – PSIC=1.000), whereas two missense mutations were predicted to have a low impact on protein function (m.G9387A – PSIC=0.665; m.G12895A – PSIC=0.0290) or tRNA secondary structure (m.A12242C – MT-TS2). Two samples carried heteroplasmic frameshift mutations affecting MT-ND1 (m.3571insC) and MT-ND4 (m.11038delA), which were present in the tumor samples at 85% and 50%, respectively (Supplementary Table S3).
Figure 4.
Summary of phenotypes (A, C) and classification of mtDNA base changes (B, D) in non-oncocytic renal tumors (A,B) and BHD tumors (C,D) showing that mtDNA mutations with pathogenic potential are rare in these samples. het: heteroplasmic; hom: homoplasmic.
Overall, even though mtDNA mutations could be found, non-oncocytic tumors most often carried wild-type mtDNA or presented somatic, mostly heteroplasmic, mutations with low or intermediate predicted pathogenic potential. These data, together with the sequencing data on BMF oncocytomas, again lead to a strong correlation between the oncocytoma phenotype and pathogenic mtDNA mutations (p=0.0005; Exact Fisher’s test).
mtDNA sequencing in BHD tumors
To investigate whether syndromic oncocytic tumors follow the same molecular mechanism as solitary and BMF oncocytomas, we sequenced the entire mtDNA in seven microdissected oncocytic tumors that developed in three BHD patients. Five of these samples did not carry any mtDNA mutations, while the other two samples presented heteroplasmic missense or frameshift mutations (m.G5400A, m.3571insC) at heteroplasmy levels that can be considered below threshold for a phenotypic effect (50% deduced from electropherogram and 30% by F-PCR, respectively) (Supplementary Table S4).
Genetic analysis of FLCN in these samples has been carried out previously (35) and a somatic “second hit” mutation in FLCN was found in three of these tumors. The presence of heteroplasmic somatic mtDNA mutations was not significantly related to the absence of somatic “second hit” mutations in FLCN (p=0.147), ruling out the possibility that mtDNA mutations might serve as a “second hit” event in a cell heterozygous for FLCN expression.
In addition to BHD oncocytomas, we analyzed 23 tumors not classified as oncocytomas from 11 BHD patients, including 4 chromophobe and 18 hybrid oncocytic tumors as well as one poorly differentiated RCC of papillary type (Figure 4). Fourteen of these tumors did not present any somatic nucleotide changes in the mtDNA, while 5 tumors presented heteroplasmic changes, known to be polymorphic, 3 tumors carried novel heteroplasmic somatic mutations and one tumor displayed a heteroplasmic and a homoplasmic somatic missense mutation, predicted to be probably damaging (summarized in Figure 4 and Supplementary Table S4).
Genetic analysis of FLCN in these samples has been carried out previously and in this report [(35); Supplementary Table S4]. In summary, in 19 of 23 tumors a somatic “second hit” mutation of FLCN was detected. Again, the presence of somatic mtDNA nucleotide changes in the tumor was not related to the mutation status of FLCN (p=0.412).
Overall, tumors developed by patients diagnosed with BHD often display oncocytic features. However, in contrast to BMF oncocytomas, these neoplasms carry mostly a wild-type mitochondrial genome. The presence of potentially pathogenic mtDNA mutations is significantly higher in BMF oncocytomas than in tumors with any mutations in FLCN (p<0.0001). Accordingly, IHC staining for CI (NDUFB8), CII (SDHA) and CIV (COXIV) clearly demonstrates that these respiratory chain complexes are present in BHD tumor tissues (Figure 2B,D, F) and furthermore, CI activity assays show that NADH-ubiquinone oxidoreductase activity is not significantly impaired in BHD tumors (Figure 2G).
mtDNA sequences of biopsies are predictive of the tumor mtDNA sequence
Reliable preoperative diagnosis of oncocytoma through imaging techniques and histological analysis of biopsy samples is often inconclusive (5). Since here we find a very strong correlation between the occurrence of pathogenic mtDNA mutations and the benign oncocytoma phenotype in non-syndromic renal tumors, we wanted to test whether genetic analysis of mtDNA in biopsy samples was feasible and predictive of the tumor genotype.
Therefore we performed mock biopsies on two tumor specimens immediately after surgical removal (patients BMF6 and BMF8), collected 2 and 3 biopsies, respectively, from different areas of the tumor and extracted DNA from each sample. PCR amplification was performed on non-diluted DNA. We obtained PCR amplicons with all primers, covering the entire mtDNA and hence were able to sequence the entire mitochondrial genome in the biopsies and compare it to the corresponding tumor tissue. The tumor of patient BMF6 presented 41 polymorphisms and 3 heteroplasmic missense mutations, but no clear pathogenic mutations, and was later diagnosed as ccRCC. The tumor of patient BMF8, diagnosed as a chromophobe RCC, carried 38 polymorphisms and a heteroplasmic insertion in the variable D-Loop region. We detected all nucleotide changes of the tumors in the mtDNA sequences from biopsies, including the heteroplasmic changes (Supplementary Figure S2). These data confirm that the entire mtDNA can be sequenced on limited material, such as biopsy specimens, and that the sequences obtained are representative of the tumor DNA sequence.
Discussion
Renal tumors present a number of different histologies, are caused by mutations in different genes, and follow different clinical courses. Therefore, reliable diagnostic criteria to distinguish between benign and malignant neoplasms, are important for proper patient management. The mitochondrial genome has been analyzed in a variety of tumors, but the role that mtDNA mutations may play in tumorigenesis and in shaping tumor phenotype is still largely debated [e.g.(18, 33, 36)]. Yet, oncocytomas in different organs and, in particular, renal oncocytomas, have been associated with mtDNA mutations (10, 11, 13). We show here that bilateral and multifocal renal oncocytomas also carry somatic pathogenic mitochondrial DNA mutations. Our findings underscore the association between the oncocytoma phenotype and mtDNA mutations, exemplified by one patient who developed a clear cell RCC that carried only a few heteroplasmic missense mutations and an oncocytoma that presented with a homoplasmic and a heteroplasmic frameshift mutation. All of the disruptive mutations found in BMF oncocytoma samples affected CI of the respiratory chain and were homoplasmic or present at heteroplasmy levels that are considered above threshold for a phenotypic effect, based on the previously described oncojanus function of mtDNA-encoded genes (15). On the other hand, one interesting renal oncocytoma sample presented a heteroplasmic tRNA mutation in MT-TS1 that affected the anticodon of the tRNA. Since this alteration could cause misincorporation of amino acids into all mitochondria-encoded proteins, the mutation is predicted to have a dominant negative effect. Thus, in agreement with previous studies (11, 13), we found that all BMF oncocytomas lack a properly assembled NADH-ubiquinone oxidoreductase with dramatically reduced CI enzyme activity.
The pathogenesis of renal oncocytosis, characterized by numerous tiny oncocytic lesions in the kidneys, has not been clearly identified (26). Notably, BMF oncocytomas that developed in patients described in this study carried the same disruptive mtDNA mutation in multiple oncocytomas from both kidneys. The mutation, however, was absent from the blood and normal kidney DNAs of these patients. The methodology used to measure heteroplasmy levels of mtDNA mutations enabled us to obtain a precise measurement of the percentage of mutations and were sensitive enough to detect heteroplasmy levels of 1% [F-PCR, (31)] or even 0.1% (castPCR – manufacturer’s instructions and data not shown). The absence of the mutations from the non-affected tissue together with the presence of the same mutation in contralateral kidneys supports the acquisition of the mtDNA mutation during early embryonic kidney development and implies that this condition was most likely not inherited. Single cells and groups of cells scattered throughout the kidney parenchyma that resemble oncocytic histology, i.e. oncocytosis, carry mtDNA mutations. These cells most likely give rise to oncocytomas, suggesting that the mtDNA mutation might represent the primary hit in these tumors and will eventually account for the benign tumor phenotype (15). mtDNA mutations may provide the cell with an initial growth advantage, according to the Warburg effect (33), but it is expected that additional hits affecting tumor suppressors or oncogenes are necessary to trigger tumor growth initiating from these cells.
Bilateral and multifocal renal tumors have been frequently described in BHD syndrome caused by germline mutations of FLCN (37). These tumors are most frequently chromophobe and hybrid oncocytic tumors, but oncocytomas occur at a low frequency (38). BHD tumors have been shown to present molecular features distinct from sporadic oncocytomas and chromophobe RCC (39). Here we show that BHD patients’ tumors do not carry pathogenic mtDNA mutations, which remains in stark contrast to the presence of disruptive mutations in all analyzed sporadic and BMF renal oncocytomas. Nevertheless, these data are consistent with a previously published report describing a parotid oncocytoma that developed in a BHD syndrome patient, in which no mtDNA mutations were found (40). In sporadic oncocytomas it is thought that mtDNA mutations lead to respiratory deficiency which triggers a compensatory effect that induces mitochondrial biogenesis [for a review see (41)]. However, it has not been ruled out that a defect in mitochondrial turnover pathways may account for the accumulation of defective mitochondria in oncocytomas. On the other hand, FLCN mutations in BHD tumors may account for the increased mitochondria and therefore phenocopy sporadic oncocytomas, but without pathogenic mtDNA mutations. Mitochondrial and OXPHOS-related genes are highly expressed in BHD tumors (39) and a role for FLCN in the regulation of mitochondrial biogenesis through PGC1a, without impairment of mitochondrial function, has recently been demonstrated (27). For that reason, should mtDNA mutations be considered as diagnostic markers for multifocal oncocytomas, it would be important to exclude the possibility that a patient is affected with BHD syndrome.
Several previous studies have shown that many tumors (21), and in particular renal tumors carry somatic alterations of the mitochondrial genome that were, however, mostly heteroplasmic and had no or only a low pathogenic potential (17, 18). Accordingly, here we show in a series of chromophobe, papillary and clear cell RCCs that many of these tumors present mostly heteroplasmic somatic mtDNA mutations. Only tumor samples from one patient with BMF eosinophilic chromophobe RCC showed clearly pathogenic mutations. However, this patient presents with an unusual family history that may suggest a hereditary component and needs to be further investigated. Recently, a comprehensive genomic analysis of chromophobe RCC revealed a significant presence of mtDNA mutations in chromophobe RCC of the eosinophilic subtype (18). Renal oncocytomas have been shown to express low levels of CI genes, but high levels of CIV genes, as compared to chromophobe RCC (14). Our results strongly suggest that this molecular phenotype is due to the presence of mtDNA mutations in oncocytomas and the lack of such mutations in non–eosinophilic chromophobe RCC. The association between mtDNA mutations and eosinophilic chromophobe RCC, however, warrants further investigation.
Various imaging techniques as well as percutaneous biopsies are often performed to diagnose renal masses and facilitate the management of patients (5). However, oncocytomas are often difficult to distinguish from other types of renal tumors on imaging and through biopsies (5). Genetic studies could enhance the diagnosis of oncocytoma, when histological analyses are inconclusive. Genetic testing of biopsies has been shown to be feasible (42, 43). Here we show that it is possible to sequence the entire mitochondrial genome on DNA extracted from tumor biopsies and that the mitochondrial sequences of the biopsies correspond to the tumor sequence. Since it is known that tumors are heterogeneous in their molecular landscape (36), different heteroplasmy levels of somatic mtDNA mutations might be found in different biopsies. However, oncocytomas are most often homoplasmic for pathogenic mtDNA mutations and consequently, every biopsy specimen should present the mutation. For that reason, sequencing of a biopsy sample, possibly in addition to the histologic analysis, could aid in the reliable diagnosis of renal oncocytomas and predict a benign phenotype of the tumor (15).
In conclusion, here we show that, similar to previously reported sporadic oncocytomas (11, 13), disruptive mtDNA mutations affecting complex I of the respiratory chain are hallmarks of BMF renal oncocytomas. Those tumors may arise from kidney oncocytosis, where all oncocytic cells and foci carry a mtDNA mutation. On the other hand, BHD-related renal tumors follow a different pathogenic process and mostly do not present mtDNA mutations. The same holds true for non-oncocytic renal tumors of different histologies. Hence, we demonstrate a role for mtDNA mutations in the development of renal oncocytomas with wild-type FLCN, and propose that mtDNA sequencing, feasible on biopsy samples, or the analysis of respiratory chain complexes, should be further investigated as reliable diagnostic markers for renal oncocytomas.
Supplementary Material
Acknowledgements
The present work was funded in part by Fondazione Italiana Ricerca sul Cancro (FIRC) “Fellowship for abroad – 2011” to ML and was supported by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E (LSS). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
We would like to thank Dr. Hisashi Hasumi, Dr. Yukiko Hasumi, Dr. Masaya Baba and Dr. Christopher Ricketts for helpful discussions and advice.
Footnotes
Disclosure/Conflict of Interest
The authors declare no conflict of interest.
Supplementary information is available at Modern Pathology’s website
References
- 1.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Rodriguez LJ. Appendix 1. Basic properties of mitochondria. Methods in cell biology. 2007;80:809–812. doi: 10.1016/S0091-679X(06)80040-3. [DOI] [PubMed] [Google Scholar]
- 3.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocrine reviews. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 4.Linehan WM, Bratslavsky G, Pinto PA, et al. Molecular diagnosis and therapy of kidney cancer. Annu Rev Med. 2010;61:329–343. doi: 10.1146/annurev.med.042808.171650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahni VA, Silverman SG. Biopsy of renal masses: when and why. Cancer Imaging. 2009;9:44–55. doi: 10.1102/1470-7330.2009.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trpkov K, Yilmaz A, Uzer D, et al. Renal oncocytoma revisited: a clinicopathological study of 109 cases with emphasis on problematic diagnostic features. Histopathology. 2010;57:893–906. doi: 10.1111/j.1365-2559.2010.03726.x. [DOI] [PubMed] [Google Scholar]
- 7.Yusenko MV. Molecular pathology of renal oncocytoma: a review. Int J Urol. 2010;17:602–612. doi: 10.1111/j.1442-2042.2010.02574.x. [DOI] [PubMed] [Google Scholar]
- 8.Kurelac I, MacKay A, Lambros MB, et al. Somatic complex I disruptive mitochondrial DNA mutations are modifiers of tumorigenesis that correlate with low genomic instability in pituitary adenomas. Hum Mol Genet. 2013;22:226–238. doi: 10.1093/hmg/dds422. [DOI] [PubMed] [Google Scholar]
- 9.Bonora E, Porcelli AM, Gasparre G, et al. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res. 2006;66:6087–6096. doi: 10.1158/0008-5472.CAN-06-0171. [DOI] [PubMed] [Google Scholar]
- 10.Gasparre G, Porcelli AM, Bonora E, et al. Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc Natl Acad Sci U S A. 2007;104:9001–9006. doi: 10.1073/pnas.0703056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayr JA, Meierhofer D, Zimmermann F, et al. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res. 2008;14:2270–2275. doi: 10.1158/1078-0432.CCR-07-4131. [DOI] [PubMed] [Google Scholar]
- 12.Simonnet H, Demont J, Pfeiffer K, et al. Mitochondrial complex I is deficient in renal oncocytomas. Carcinogenesis. 2003;24:1461–1466. doi: 10.1093/carcin/bgg109. [DOI] [PubMed] [Google Scholar]
- 13.Gasparre G, Hervouet E, de Laplanche E, et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma. Hum Mol Genet. 2008;17:986–995. doi: 10.1093/hmg/ddm371. [DOI] [PubMed] [Google Scholar]
- 14.Yusenko MV, Ruppert T, Kovacs G. Analysis of differentially expressed mitochondrial proteins in chromophobe renal cell carcinomas and renal oncocytomas by 2-D gel electrophoresis. Int J Biol Sci. 2010;6:213–224. doi: 10.7150/ijbs.6.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasparre G, Kurelac I, Capristo M, et al. A mutation threshold distinguishes the antitumorigenic effects of the mitochondrial gene MTND1, an oncojanus function. Cancer Res. 2011;71:6220–6229. doi: 10.1158/0008-5472.CAN-11-1042. [DOI] [PubMed] [Google Scholar]
- 16.Meierhofer D, Mayr JA, Fink K, et al. Mitochondrial DNA mutations in renal cell carcinomas revealed no general impact on energy metabolism. Br J Cancer. 2006;94:268–274. doi: 10.1038/sj.bjc.6602929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy A, Wilhelm M, Sukosd F, Ljungberg B, Kovacs G. Somatic mitochondrial DNA mutations in human chromophobe renal cell carcinomas. Genes Chromosomes Cancer. 2002;35:256–260. doi: 10.1002/gcc.10118. [DOI] [PubMed] [Google Scholar]
- 18.Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gobbo S, Eble JN, Delahunt B, et al. Renal cell neoplasms of oncocytosis have distinct morphologic, immunohistochemical, and cytogenetic profiles. Am J Surg Pathol. 2010;34:620–626. doi: 10.1097/PAS.0b013e3181d73293. [DOI] [PubMed] [Google Scholar]
- 20.McMahon S, Laframboise T. Mutational patterns in the breast cancer mitochondrial genome, with clinical correlates. Carcinogenesis. 2014 doi: 10.1093/carcin/bgu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larman TC, DePalma SR, Hadjipanayis AG, et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A. 2012;109:14087–14091. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadav N, Chandra D. Mitochondrial DNA mutations and breast tumorigenesis. Biochim Biophys Acta. 2013;1836:336–344. doi: 10.1016/j.bbcan.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warfel KA, Eble JN. Renal oncocytomatosis. J Urol. 1982;127:1179–1180. doi: 10.1016/s0022-5347(17)54285-8. [DOI] [PubMed] [Google Scholar]
- 24.Tickoo SK, Reuter VE, Amin MB, et al. Renal oncocytosis: a morphologic study of fourteen cases. Am J Surg Pathol. 1999;23:1094–1101. doi: 10.1097/00000478-199909000-00013. [DOI] [PubMed] [Google Scholar]
- 25.Adamy A, Lowrance WT, Yee DS, et al. Renal oncocytosis: management and clinical outcomes. J Urol. 2011;185:795–801. doi: 10.1016/j.juro.2010.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda N, Tanaka A, Ohe C, et al. Review of renal oncocytosis (multiple oncocytic lesions) with focus on clinical and pathobiological aspects. Histol Histopathol. 2011;27:1407–1412. doi: 10.14670/HH-27.1407. [DOI] [PubMed] [Google Scholar]
- 27.Hasumi H, Baba M, Hasumi Y, et al. Regulation of mitochondrial oxidative metabolism by tumor suppressor FLCN. J Natl Cancer Inst. 2012;104:1750–1764. doi: 10.1093/jnci/djs418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubino F, Piredda R, Calabrese FM, et al. HmtDB, a genomic resource for mitochondrion-based human variability studies. Nucleic Acids Res. 2012;40:D1150–D1159. doi: 10.1093/nar/gkr1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz-Pesini E, Lott MT, Procaccio V, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurelac I, Lang M, Zuntini R, et al. Searching for a needle in the haystack: comparing six methods to evaluate heteroplasmy in difficult sequence context. Biotechnol Adv. 2012;30:363–371. doi: 10.1016/j.biotechadv.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Baba M, Furihata M, Hong SB, et al. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst. 2008;100:140–154. doi: 10.1093/jnci/djm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasparre G, Porcelli AM, Lenaz G, Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thoenes W, Storkel S, Rumpelt HJ, et al. Chromophobe cell renal carcinoma and its variants--a report on 32 cases. The Journal of pathology. 1988;155:277–287. doi: 10.1002/path.1711550402. [DOI] [PubMed] [Google Scholar]
- 35.Vocke CD, Yang Y, Pavlovich CP, et al. High frequency of somatic frameshift BHD gene mutations in Birt-Hogg-Dube-associated renal tumors. J Natl Cancer Inst. 2005;97:931–935. doi: 10.1093/jnci/dji154. [DOI] [PubMed] [Google Scholar]
- 36.He Y, Wu J, Dressman DC, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nickerson ML, Warren MB, Toro JR, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2:157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 38.Pavlovich CP, Walther MM, Eyler RA, et al. Renal tumors in the Birt-Hogg-Dube syndrome. Am J Surg Pathol. 2002;26:1542–1552. doi: 10.1097/00000478-200212000-00002. [DOI] [PubMed] [Google Scholar]
- 39.Klomp JA, Petillo D, Niemi NM, et al. Birt-Hogg-Dube renal tumors are genetically distinct from other renal neoplasias and are associated with up-regulation of mitochondrial gene expression. BMC Med Genomics. 2010;3:59. doi: 10.1186/1755-8794-3-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pradella LM, Lang M, Kurelac I, et al. Where Birt-Hogg-Dube meets Cowden syndrome: mirrored genetic defects in two cases of syndromic oncocytic tumours. Eur J Hum Genet. 2013;21:1169–1172. doi: 10.1038/ejhg.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasparre G, Romeo G, Rugolo M, Porcelli AM. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta. 2011;1807:633–642. doi: 10.1016/j.bbabio.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Roca CA, Lacroix L, Massard C, et al. Sequential research-related biopsies in phase I trials: acceptance, feasibility and safety. Ann Oncol. 2012;23:1301–1306. doi: 10.1093/annonc/mdr383. [DOI] [PubMed] [Google Scholar]
- 43.Maki J, Robinson K, Reguly B, et al. Mitochondrial genome deletion aids in the identification of false- and true-negative prostate needle core biopsy specimens. Am J Clin Pathol. 2008;129:57–66. doi: 10.1309/UJJTH4HFEPWAQ78Q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.