Abstract
Arthropod cuticles have, in addition to chitin, many structural proteins belonging to diverse families. Information is sparse about how these different cuticular proteins contribute to the cuticle. Most cuticular proteins lack cysteine with the exception of two families (CPAP1 and CPAP3), recently described, and the one other that we now report on that has a motif of 16 amino acids first identified in a protein, Bc-NCP1, from the cuticle of nymphs of the cockroach, Blaberus craniifer (Jensen et al., 1997). This motif turns out to be present as two or three copies in one or two proteins in species from many orders of Hexapoda. We have named the family of cuticular proteins with this motif CPCFC, based on its unique feature of having two cysteines interrupted by five amino acids (C-X(5)-C). Analysis of the single member of the family in Anopheles gambiae (AgamCPCFC1) revealed that its mRNA is most abundant immediately following ecdysis in larvae, pupae and adults. The mRNA is localized primarily in epidermis that secretes hard cuticle, sclerites, setae, head capsules, appendages and spermatheca. EM immunolocalization revealed the presence of the protein, generally in endocuticle of legs and antennae. A phylogenetic analysis found proteins bearing this motif in 14 orders of Hexapoda, but not in some species for which there are complete genomic data. Proteins were much longer in Coleoptera and Diptera than in other orders. In contrast to the 1 and occasionally 2 copies in other species, a dragonfly, Ladona fulva, has at least 14 genes coding for family members. CPCFC proteins were present in four classes of Crustacea with 5 repeats in one species, and motifs that ended C-X(7)-C in Malacostraca. They were not detected, except as obvious contaminants, in any other arthropod subphyla or in any other phylum.
The conservation of CPCFC proteins throughout the Pancrustacea and the small number of copies in individual species indicate that, when present, these proteins are serving important functions worthy of further study.
Keywords: Cuticle, EM immunolocalization, in situ hybridization, arthropod phylogeny, RT-qPCR
Graphical Abstract
1. Introduction
Over a dozen families of cuticular proteins (CPs) have been described. One (CPR) has well over 100 genes in several species (Cornman et al., 2008; Futahashi et al., 2008; Cornman, 2009; Willis, 2010; Willis et al., 2012; Ioannidou et al., 2014; Neafsey et al., 2015). Additional data on temporal and spatial expression (both in terms of tissue distribution and location within the cuticle) have also been published. Early papers are reviewed in Willis et al. (2012), more recent ones are Nor et al. (2014; 2015), Pesch et al. (2015) and Vannini et al. (2014a,b). An unusual family that generally has only one member in a species (and very rarely more than two) was named CPCFC by Willis et al. (2012) because of a motif of C-X(5)-C (two cysteines interrupted by five amino acids). The “type specimen” for CPCFC is Bc-NCP1, isolated from nymphal cuticle of the cockroach, Blaberus craniifer (Jensen et al., 1997) [GenBank: P80674]. The paper describing that sequence established the fundamental property of the family: a 16 amino acid motif, here repeated 3 times, that ends C-X(5)-C. The final motif is at the carboxy-terminus of the protein. In addition, Jensen et al. (1997) speculate, after ruling out a role in cross-linking via quinones: “It is more likely that the three cysteine-containing loops in Bc-NCP1 are involved in some sort of specific interaction or binding, either to metal ions or to other proteins.”
Now we describe, in detail, expression and localization of one member of that family, AgamCPCFC1, in Anopheles gambiae. We conclude with an analysis of the phylogenetic distribution of members of that family in many orders of Pancrustacea (Hexapoda + Crustacea). Our analysis revealed consistent variants of CPCFC proteins in different orders. The wide-spread distribution of this family represents the second time a motif identified in a few cuticular protein sequences (5 in the case of the R&R Consensus in the CPR family (Rebers and Riddiford, 1988), one sequence here (Jensen et al., 1997) turns out to have been conserved in CPs found throughout arthropods (reviewed in Willis 2010; Willis et al., 2012).
2. Materials and methods
2.1. Anopheles rearing
An. gambiae (G3 strain) were obtained as newly hatched first instar larvae from the breeding facility at the University of Georgia Entomology Department. They were raised at 27 °C under a 12:12 photoperiod and fed ground Koi Food Staple Diet (Foster and Smith Aquatics, Rhinelander, WI USA).
2.2. RT-qPCR
An. gambiae larvae, pupae and adults were carefully timed relative to a molt, placed in TRIzol® and immediately frozen. RNA was prepared following the manufacturer's instructions. Superscript III First Strand Synthesis Kit (Invitrogen) with oligo (dT)20 primers was used for cDNA production, and RT-qPCR was carried out with Bio-Rad's CFX Connect Real Time system. Additional details are in Supplementary File 1 that provides MIQE information in a format recommended by Bustin et al. (2013). Calculations were carried out with LinRegPCR software (Ruijter et al., 2009).
The primers used were located near the end of the coding region and extended into the 3’UTR with an amplification product of 103 nt (Supplementary Files 2,3). Before use, the primers were checked on genomic DNA for amplification kinetics against two single copy genes, RpS7 [GenBank:AGAP010592] and the epidermal chitin synthase [GenBank:AGAP001748], to assure that they were only amplifying a single gene. RpS7 was run on every plate with every cDNA preparation, but was not used to normalize values. Rather, we calculate N0, described as R0 in Togawa et al. (2008), basing values on concentrations of RNA determined with NanoDrop N-1000 (Thermo Scientific). This was necessary because we have failed to find housekeeping genes with consistent expression across the range of developmental stages we studied. Figures showing the variable values obtained with the RpS7 primers and CPCFC1 data normalized to RpS7 are in Supplementary File 4.
2.3. In situ hybridization
In situ hybridization was carried out on 4 μm paraffin sections of paraformaldehyde fixed An. gambiae of different developmental stages prepared by the Histology Laboratory of the University of Georgia College of Veterinary Medicine. DIG-labeled anti-sense probe preparation and hybridization followed the methods described in earlier publications from our laboratory (Vannini et al., 2014a,b). The primers used and resulting probes are shown in Supplementary Files 2 and 3, respectively. We used one probe directed against the coding region and another against the 3’UTR. Identical patterns of hybridization were found (Supplementary File 5). Probes were also designed based on the sense strands of both antisense probes. They validated the specificity of the technique (Supplementary File 6). Anatomical nomenclature is based on Harbach and Knight (1980).
2.4. Cloning and expression of AgamCPCFC1
The coding sequence for almost all of the mature form of AgamCPCFC1 was cloned into Lucigen Expresso™ T7 Cloning and Expression System with an N-His tag. Primers are given in Supplementary File 2. They cover the entire coding sequence of the mature protein except for the regions coding for the first four and last three amino acids (Supplementary File 3B).
The expressed protein was solubilized in 3M urea, 10 mM DTT (dithiothreitol), purified with a Talon Imac Metal Affinity Resin packed into a BioRad column, eluted with 1M imidazole and sent to Harlan Bioproducts for antibody production in rabbits, using their 112 day protocol.
2.5 EM immunocytochemistry
Legs and antennae with Johnston's organs were dissected from precisely aged pharate and post-eclosion adults and fixed in 4% parformaldehyde, 0.3% glutaraldehyde + 4% sucrose in phosphate buffer (pH 7.4). Further details about processing and embedding in LR White resin (Electron Microscopy Sciences) and subsequent processing are given in Vannini et al. (2014a,b). Anti-AgamCPCFC1 and secondary antibodies (goat-anti-rabbit, conjugated to 5 nm gold particles, Sigma) were diluted 1:5,000 and 1:50, respectively. We found only an occasional gold particle on sections incubated with hybridization buffer rather than the primary antibody. We used a JEM-1210 transmission electron microscope (JEOL USA) at 120kV. The images were captured with an XR41C Bottom-MountCCD Camera (Advanced Microscopy Techniques).
2.6. Phylogenetic analysis via BLAST searches
BLAST searches (tblastn) for CPCFC family members were carried out at http://blast.ncbi.nlm.nih.gov/Blast.cgi using either the first motif from Blaberus craniifer Bc-NCP1 [GenBank:P80674.1] or its entire sequence. We used default settings except for turning off filtering and masking of low complexity regions. We searched EST and TSA databases. We only included in our analyses (with one exception) sequences that had a signal peptide and a stop codon and at least two occurrences of the 16- amino-acid CPCFC motif. We omitted all sequences that came from the 1KITE - 1K Insect Transcriptome Evolution project submitted in January, 2014, because we found a small number of cases with identical sequences in two or more orders. At the time of writing this paper these data were under review and revision, which may resolve the inconsistencies that we observed. We used the phylogenetic nomenclature of von Reumont et al. (2012) and Misof et al. (2014) as well as many of the sequences produced in their analyses.
Additional searches were done with wgs (whole-genome shotgun contigs) using Odonata (taxid:6961) as the search term. These could not produce complete sequences unless the region coding for the entire protein was in a single exon, something we have not yet seen for CPCFC genes. Nonetheless, we got provocative results for Ladona fulva.
3. Results and discussion
3.1. Genomic structure
AgamCPCFC1 [GenBank:AGAP007980] is coded by a gene with three exons, the first of which codes for only 5 amino acids (Supplementary File 3A). Such a short first exon is a common feature of CPs in other families (Willis et al., 2010). The sequence is certain to be correct; for there are 4 ESTs with 100% sequence identity and an additional 50 with 99% identity, all covering the entire coding sequence. These ESTs came from the Celera Anopheles gambiae EST project with directional cloning on mixed sex adults, using strain RSP-ST (Reduced susc. to Permethrin).
The ortholog in Drosophila melanogaster has only two exons, and the first also codes for only 5 amino acids (Supplementary File 3D).
3.2. Temporal expression of transcripts
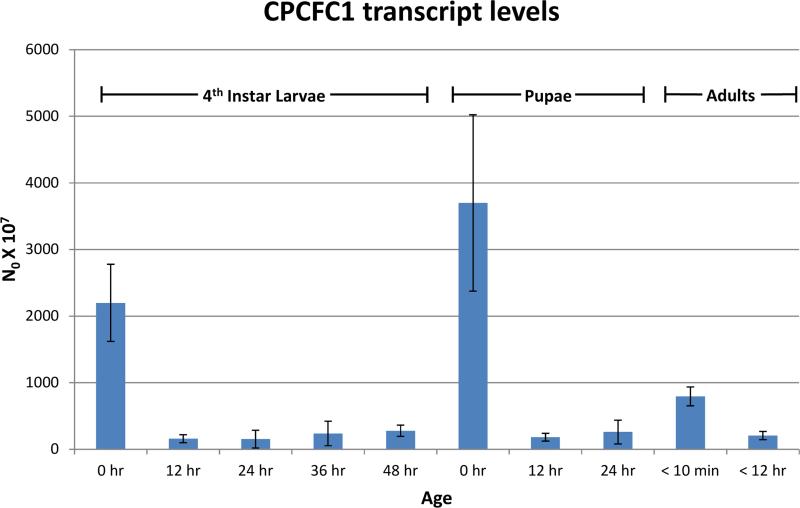
RT-qPCR was used to learn when mRNA from AgamCPCFC1 was present. Highest levels were found immediately after ecdysis to fourth instar larvae, to pupae and to adults. Far lower levels of transcripts were detected in intermolt and pharate periods (Fig. 1).
Fig. 1.
RT-qPCR analysis of AgamCPCFC1 transcripts in Anopheles gambiae. L48 and P24 are actually pharates of the next stage. See Text and Supplementary File 1 for methods.
3.3. Anatomical location of transcripts for AgamCPCFC1
We carried out in situ hybridization to learn where the mRNA for AgamCPCFC1 was localized. We used two different antisense probes, one designed in the coding region, the other in the 3’UTR (Supplementary File 3A). In successive sections, hybridization patterns were identical with the two probes (Supplementary File 5). We selected animals at developmental stages where our RT-qPCR data indicated that mRNA was likely to be present, namely pharate and newly eclosed animals. Sense controls for both probes showed no specific hybridization (Supplementary File 6).
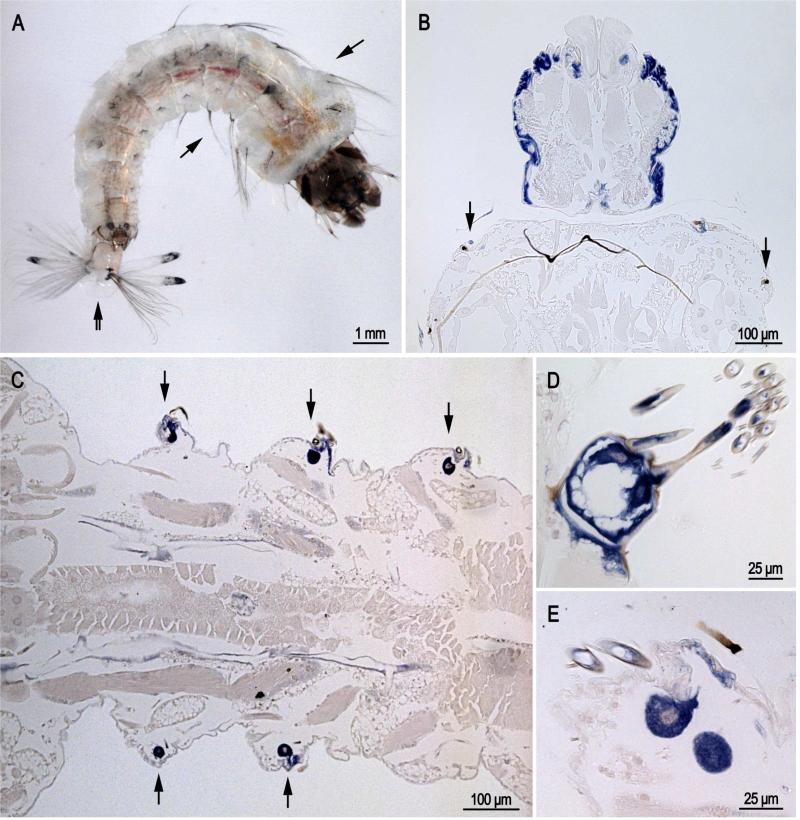
Transcripts were found in epidermis of larvae, pupae and adults underlying cuticle destined to be highly sclerotized, i.e. hard cuticle. Thus in sections of larvae (Fig. 2), probe was found in the head capsule (Fig. 2B), in cells that secrete lateral setae (arrows in Fig. 2A-C) and in the cells that form the grid and brush at the posterior end (Fig. 2D). Our slides of larvae had animals at different developmental ages, thus it was not unexpected that we found many sections without labeled cells in the head capsule.
Fig. 2.
In situ hybridization of AgamCPCFC1 on sections of 4th instar larvae. A. Photograph of larva with arrows showing location of lateral setae on thorax and abdomen and a double arrow indicating the grid and fringe at the posterior end. B. Head capsule and bit of prothoracic segment. Note the presence of hybridization in the small cells that form setae at the anterior edge of the prothorax. C. Section of the abdomen showing cells that are forming setae. D. Grid and accompanying fringe at posterior end of a larva. E. Section showing cells secreting large and small setae. (B,D 3’ probe; C,E coding region probe).
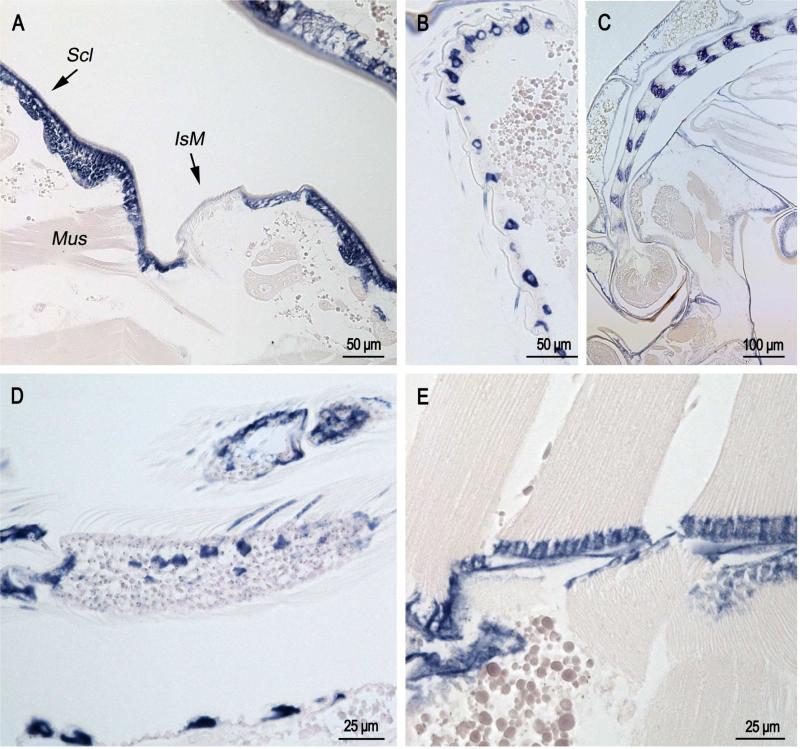
In sections of pupae that were less than one hour after eclosion (Fig. 3), label was present in cells that form bristles on the pupal abdomen (Fig. 3B); it was also present in the developing antennae (Fig. 3C) and adult scales that surprisingly are already forming (Fig. 3D). Label was found in epidermis underlying abdominal sclerites but not intersegmental membranes (Fig. 3A) with the exception of places where muscle is inserting into the intersegmental membrane (Mus in Fig. 3A)
Fig. 3.
In situ hybridization of AgamCPCFC1 on sections of pupae less than 1 hour after pupation. A. Section of abdomen showing epidermal hybridization in sclerites (Scl) and only in intersegmental membrane (IsM) where muscles (Mus) are inserting into the cuticle. B. Lateral surface of pupal abdomen with setae-forming cells. C. Developing antenna in pupa. Structure was recognized because it is similar to that shown in Fig. 76a of Harbach and Knight (1980). D. Limb with developing scales showing hybridization. E. Muscle insertion zone with strong hybridization. (B,D 3’ probe; A,C,E coding region probe.)
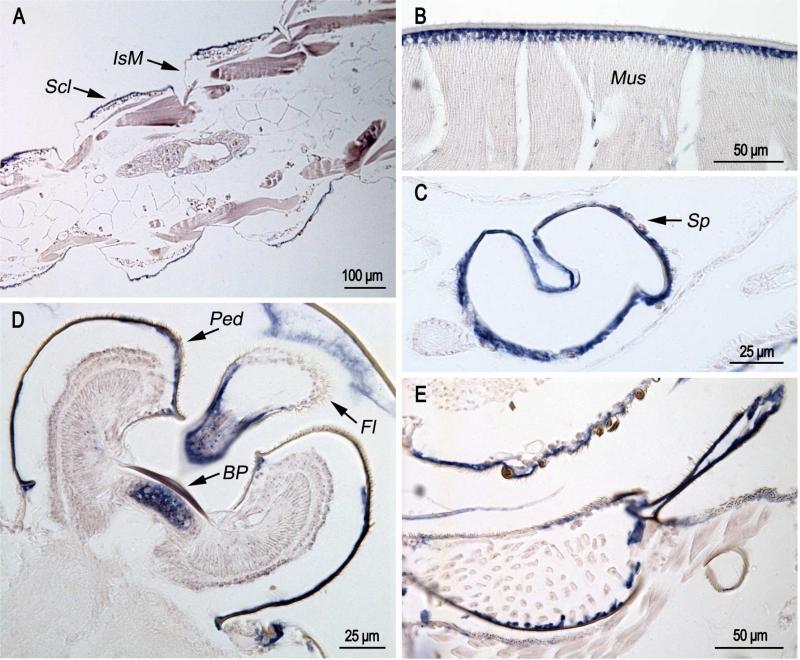
In pharate adults (Fig. 4), hybridization of the probe was found in sclerites (Fig. 4A), in muscle attachment zones (Fig. 4B), and in epidermis of Johnston's organ (JO) both beneath the basal plate and under the pedicel that surrounds the organ (Fig. 4D). It was also present in the epidermis of the flagellum (Fig. 4D), spermatheca (Fig. 4C) and the cervical sclerite (Fig. 4E). Just as in the pupa, CPCFC1 transcript was not found in intersegmental membranes (Fig. 4A).
Fig. 4.
In situ hybridization of AgamCPCFC1 on sections of pharate adults Animals were fixed 24 hours after pupation, which are a few hours before ecdysis to the adult. A. Hybridization to epidermis of sclerites (Scl), but not intersegmental membranes (IsM). B. Hybridization in muscle attachment region. C. Hybridization in spermatheca (Sp). D. Hybridization under basal plate (BP) of Johnston's organ, the surrounding pedicel (Ped) and the flagellum (Fl). E. Hybridization to part of cervical sclerite. (D,E 3’ probe; A,B,C coding region probe.)
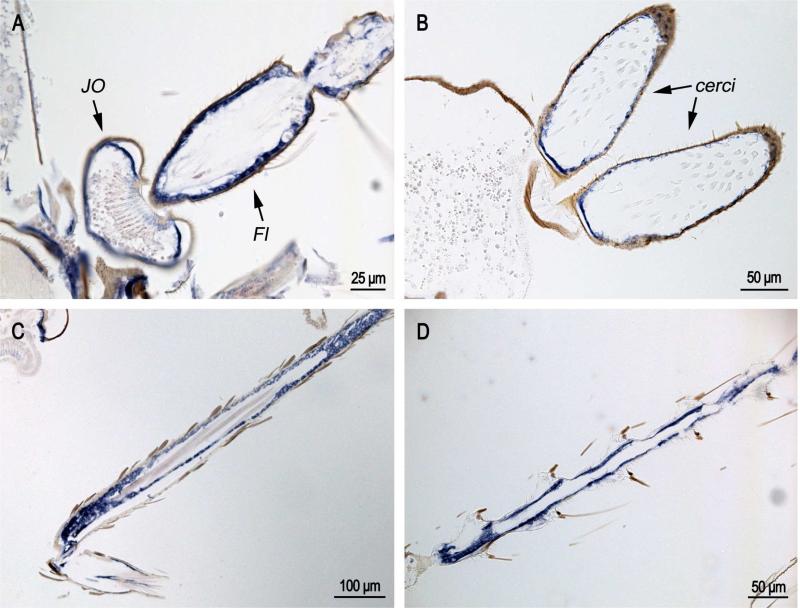
In recently eclosed adults (Fig. 5), CPCFC1 transcript was once again detected in JO and the flagellum of the antennae (Fig. 5A), the male cerci (Fig. 5B), and other appendages (Fig. 5C,D).
Fig. 5.
In situ hybridization of AgamCPCFC1 on adults less than 12 hours after eclosion. A. Antenna with Johnston's organ (JO) and flagellum (Fl) showing strong hybridization. B. cerci at the terminal end of the male abdomen. C and D. Hybridization in appendages. (All coding region probe.)
3.4. Localization of AgamCPCFC1 protein within the cuticle
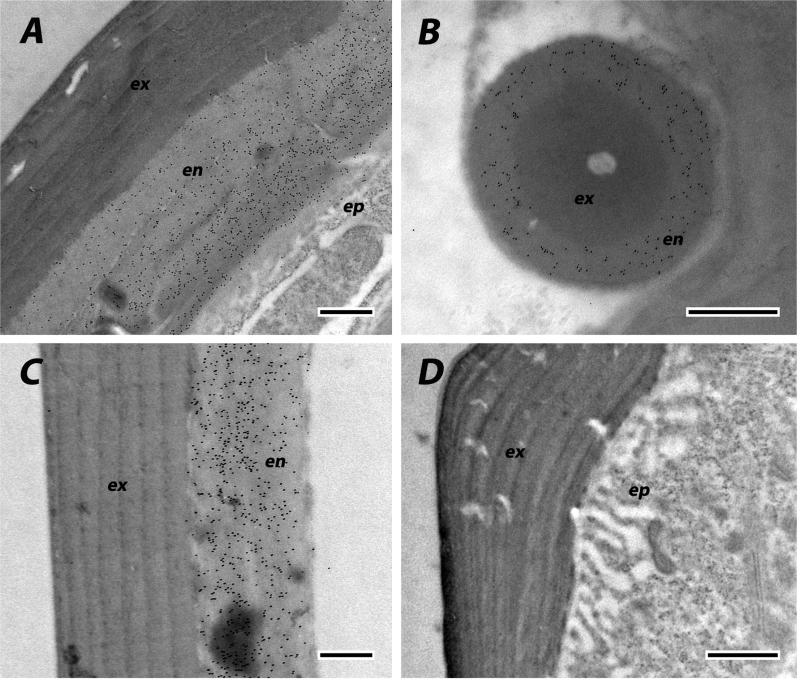
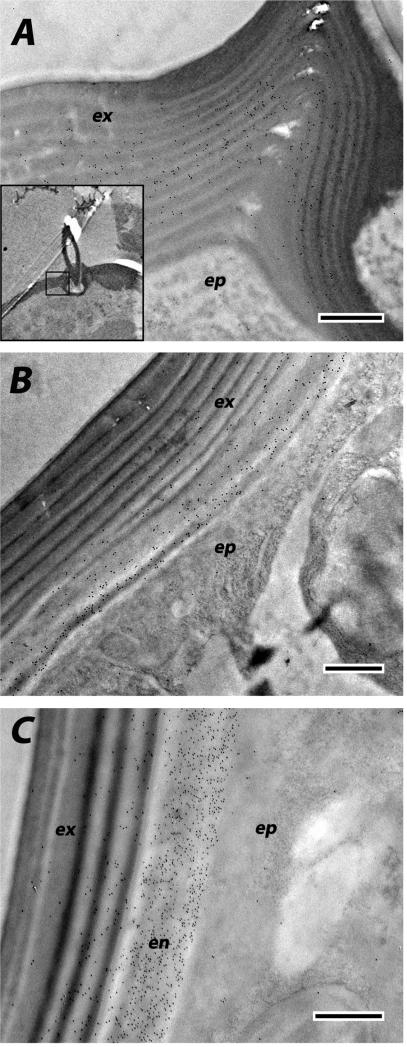
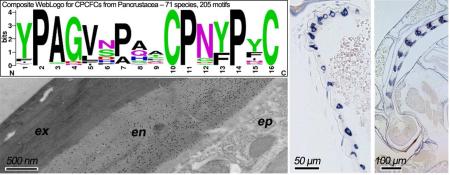
We used EM immunolocalization in order to learn where CPCFC1 was within the cuticle. EM sections were treated with a polyclonal antibody (Ab) that had been raised against most of the mature form of CPCFC1 (Supplementary File 3B). The specificity of the antibody is shown in a Western blot of proteins isolated from adult legs (Supplementary File 3C). Ab binding to EM sections was visualized with a colloidal-gold- labeled secondary antibody against rabbit IgG. We examined structures where the transcript, as visualized with in situ hybridization, was abundant: legs and the antenna. We use the term exocuticle for cuticle formed prior to ecdysis, with endocuticle secretion beginning after ecdysis. In adult legs fixed within a day of eclosion or on Day 8 of the adult stage, the presence of AgamCPCFC1 was strong, exclusively in the endocuticle of both the leg and its apodemes (Figs. 6 A-C). In most regions of the legs of pharate adults (P24), when, by definition, no endocuticle is present, no trace of AgamCPCFC1 was found (Fig. 6D). But in other regions of the pharate adult leg, we did find evidence for AgamCPCFC1 in exocuticle, both in regions with well-formed lamellae and in not yet organized regions next to the epidermal cells. This was most noticeable at the base of the leg and near a joint (Fig. 7A). We also saw label in the pedicel of pharate adults (Fig. 7B) and flagellum of newly emerged adults (Fig. 7C), once again, where endocuticle should not yet be present (Fig. 7B). Absence of an antigen in the cuticle might just mean that it has been masked during the sclerotization process. Hence it would be premature to conclude that except for an occasional region, AgamCPCFC1 is confined to the endocuticle. The higher levels of transcript right after a molt rather than immediately before (Fig. 1), however, are consistent with the endocuticle being the primary destination of the protein.
Fig. 6.
EM Immunolocalization of AgamCPCFC1 on legs from adults of various ages. In these sections label is restricted to endocuticle. A. Leg from adult one day after eclosion. B. Apodeme from same animal. Exocuticle is interior in the apodemes. C. Section of leg from animal 8 days after eclosion. D. Pharate adult with only exocuticle and no labeling visible. ex, exocuticle; en, endocuticle; ep, epidermis. Scale bars are 500 nm.
Fig. 7.
EM immunolocalization of AgamCPCFC1 in both exo- and endo-cuticle. A. Leg of a pharate adult (P24) showing areas of lamellar exocuticle with labeling near a joint. Insert lower power of relevant region. B. Labeling in exocuticle of P24 pedicel. C. Both exo- and endo-cuticle labeled in flagellum of adult <12 h after eclosion. Abbreviations as in Fig. 6. Scale bars are 500 nm.
3.5. Phylogenetic distribution of CPCFC genes in Hexapoda
RNAseq technology has provided a plethora of sequences from diverse arthropods, available as TSA (Transcriptome Shotgun Assembly) that greatly expanded the number of sequences available from ESTs or genomic data. These new data provided a rich source of CPCFCs including some from minor orders. Searches were carried out with blastp and tblastn (see Methods) and we found 72 complete sequences distributed across the Hexapoda (Table 1; Supplementary File 7). We required that a sequence be complete with a signal peptide and a stop codon in order to be included in the analysis, a stringent criterion especially for sequences obtained with Pyrosequencing (454), where we found occasional frame shifts recognized because parts of the protein resided in two different reading frames. No attempt was made to reconcile these. Further details on search strategies are described in Section 2.6.
TABLE 1.
CHARACTERISTICS OF CPCFC FAMILY MEMBERS IN HEXAPODA (signals removed)
| amino acids | |||||
|---|---|---|---|---|---|
| Order/Species | total mature length | to start of motif 1 | between motif 1-2 | between motif 2-3 | final C to end |
| Collembola | |||||
| Orchesella cincta | 53 | 3 | 18 | 0 | |
| Onychiurus arcticus | 55 | 4 | 19 | 0 | |
| Archaeognatha | |||||
| Lepismachilis y-signata | 121 | 4 | 26 | 42 | 1 |
| Odonata | |||||
| Enallagma hageni | 96 | 5 | 29 | 14 | 0 |
| Enallagma hageni | 145 | 6 | 64 | 43 | |
| Orthoptera | |||||
| Teleogryllus commodus | 90 | 6 | 20 | 16 | 0 |
| Gryllotalpa sp. | 92 | 6 | 22 | 16 | 0 |
| Blattodea | |||||
| Blaberus craniifer | 87 | 4 | 22 | 12 | 1 |
| Blattella germanica | 87 | 4 | 22 | 12 | 1 |
| Phthiraptera | |||||
| Pediculus humanus corporis | 154+ | 6 | 82 | 21 | end missing |
| Hemiptera | |||||
| Macrosiphum euphorbiae | 128 | 4 | 60 | 15 | 1 |
| Acyrthosiphon pisum | 128 | 4 | 60 | 15 | 1 |
| Kerria lacca | 119 | 5 | 23 | 42 | 1 |
| HOLOMETABOLA | |||||
| Hymenoptera | |||||
| Cephus cinctus | 151 | 6 | 54 | 43 | 0 |
| Megaloptera | |||||
| Corydalinae sp. | 94 | 2 | 22 | 21 | 1 |
| Neuroptera | |||||
| Chrysopa pallens | 94 | 4 | 24 | 18 | 0 |
| Coleoptera | |||||
| Tribolium castaneum | 158 | 14 | 111 | 1 | |
| Tribolium castaneum | 184 | 14 | 137 | 1 | |
| Dendroctonus frontalis | 180 | 14 | 129 | 4 | |
| Dendroctonus ponderosae | 178 | 9 | 129 | 3 | |
| Pissodes strobi | 195 | 17 | 144 | 2 | |
| Pissodes strobi | 302 | 47 | 222 | 1 | |
| Rhynchophorus ferrugineus | 188 | 18 | 137 | 1 | |
| Anthonomus grandis | 162 | 14 | 114 | 2 | |
| Agrilus planipennis | 165 | 14 | 115 | 1 | |
| Onthophagus taurus | 206 | 14 | 159 | 1 | |
| Diaprepes abbreviatus | 158 | 14 | 111 | 1 | |
| Colaphellus boyringi | 171 | 14 | 124 | 1 | |
| Lepidoptera | |||||
| Bombyx mori | 72 | 2 | 37 | 1 | |
| Spodoptera litura | 72 | 2 | 37 | 1 | |
| Ostrinia furnacalis | 74 | 2 | 39 | 1 | |
| Ostrinia nubilalis | 74 | 2 | 39 | 1 | |
| Antheraea assama | 76 | 2 | 42 | 0 | |
| Antheraea assama | 77 | 1 | 44 | 0 | |
| Antheraea yamamai | 76 | 2 | 42 | 0 | |
| Athetis lepigone | 70 | 2 | 35 | 1 | |
| Agrotis segetum | 72 | 2 | 37 | 1 | |
| Papilio polytes | 74 | 2 | 39 | 1 | |
| Papilio xuthus | 74 | 2 | 39 | 1 | |
| Danaus plexippus | 74 | 2 | 39 | 1 | |
| Heliconius melpomene | 74 | 2 | 39 | 1 | |
| Heliconius melpomene | 74 | 2 | 39 | 1 | |
| Heliconius erato | 74 | 2 | 39 | 1 | |
| Mamestra brassicae | 72 | 2 | 37 | 1 | |
| Siphonaptera | |||||
| Oropsylla silantiewi | 110 | 6 | 35 | 20 | 1 |
| Diptera | |||||
| Anopheles gambiae | 150 | 9 | 72 | 21 | 0 |
| Anopheles darlingi | 149 | 9 | 71 | 21 | 0 |
| Anopheles sinensis | 159 | 9 | 77 | 21 | 0 |
| Anopheles funestus | 148 | 9 | 70 | 21 | 0 |
| Anopheles quadrimaculatus | 152 | 9 | 74 | 20 | 0 |
| Aedes aegypti | 190 | 9 | 111 | 21 | 1 |
| Chironomus riparius | 144 | 7 | 66 | 23 | 0 |
| Sitodiplosis mosellana | 148 | 9 | 68 | 22 | 0 |
| Sitodiplosis mosellana | 131 | 6 | 62 | 15 | 0 |
| Culicoides sonorensis | 165 | 5 | 86 | 22 | 4 |
| Drosophila ananassae | 146 | 9 | 65 | 23 | 1 |
| Drosophila yakuba | 147 | 9 | 66 | 23 | 1 |
| Drosophila grimshawi | 147 | 9 | 66 | 23 | 1 |
| Drosophila melanogaster | 147 | 9 | 66 | 23 | 1 |
| Drosophila erecta | 149 | 9 | 68 | 23 | 1 |
| Drosophila persimilis | 152 | 9 | 71 | 23 | 1 |
| Drosophila simulans | 147 | 9 | 66 | 23 | 1 |
| Drosophila sechellia | 147 | 9 | 66 | 23 | 1 |
| Drosophila mojavensis | 146 | 9 | 65 | 23 | 1 |
| Drosophila willistoni | 146 | 9 | 65 | 23 | 1 |
| Drosophila virilis | 147 | 9 | 66 | 23 | 1 |
| Ceratitis capitata | 137 | 9 | 58 | 21 | 1 |
| Teleopsis dalmanni | 193 | 9 | 113 | 21 | 1 |
| Corethrella appendiculata | 175 | 9 | 95 | 20 | 1 |
| Glossina morsitans morsitans | 133 | 9 | 54 | 21 | 1 |
| Musca domestica | 139 | 5 | 63 | 21 | 1 |
| Bactrocera dorsalis | 145 | 9 | 65 | 22 | 1 |
| Bactrocera cucurbitae | 136 | 9 | 57 | 21 | 1 |
The complete sequences identified were sufficient to gain insight about the CPCFC family. With but two exceptions, the original Blaberus protein (Bc-NCP1) and AgamCPCFC1, the proteins discussed are only putative cuticular proteins. Bc-NCP1 was isolated from clean nymphal cuticle, and we presented immunological evidence for the presence of AgamCPCFC1 in the cuticle. All of the sequences we report have signal peptides, establishing that they are secreted. One incomplete sequence from Pediculus humanus is presented (in different or red type) in Table 1 and Supplementary File 7, but data from it were not used in the numerical analyses.
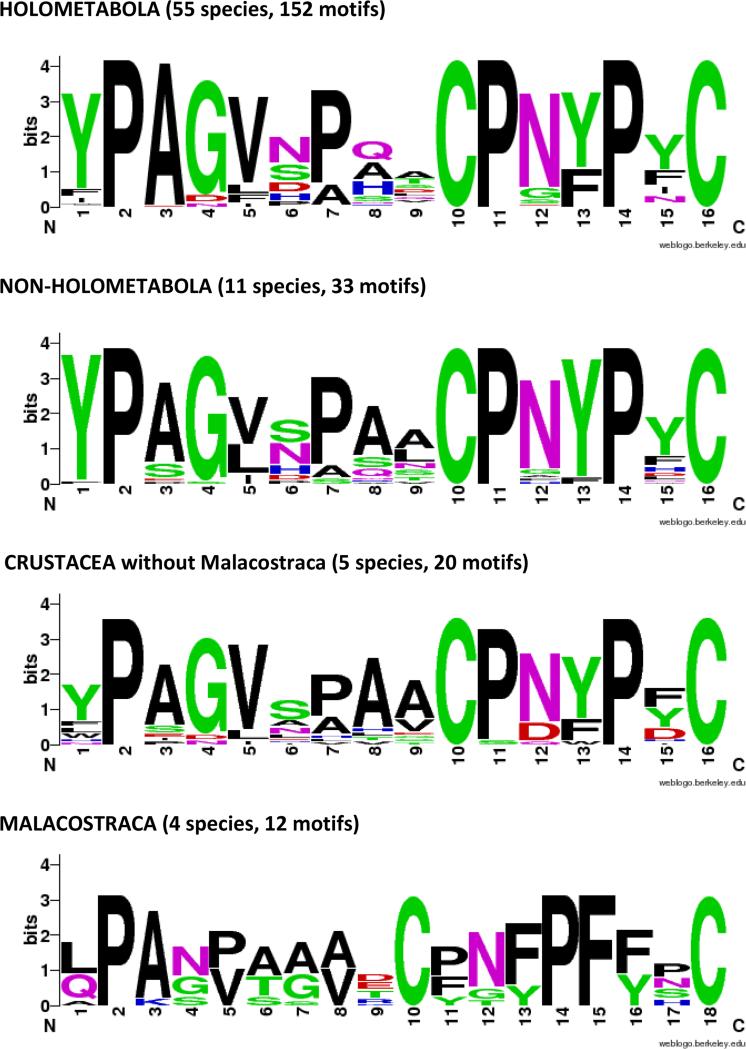
The diagnostic feature of this family is the presence of a 16 amino acid motif, first identified by Jensen et al. (1997). WebLogos (Crooks et al., 2004) based on motifs from holo- and non-holometabolous hexapods and diverse Crustacea are given in Fig. 8. They show that in addition to the two cysteines that provided the name for this family, there are three prolines, in positions 2, 11, 14, that are universal across the Hexapoda. Several other residues are highly conserved, making this an easily recognized and highly conserved motif.
Fig. 8.
WebLogos constructed for CPCFC motifs highlighted in Supplementary Files 7 and 9.
Additional consistent features are evident, but we acknowledge that these conclusions are preliminary and may well be revised as more sequences become available. The most common protein structure of the CPCFC family had three copies of the motif, but sequences from three orders, Collembola, Coleoptera and Lepidoptera, had only two. One of the two sequences from the Odonata also had only two motifs (Table 1). Most species have only a single copy of the gene. The presence of two genes in the coleopteran Tribolium castaneum led to the speculation that where only two motifs were present, there would be two genes. Yet we have identified only 2/10 species of Coleoptera and 2/14 species of Lepidoptera with two copies of CPCFC. There was one dipteran and one odonate with two CPCFC genes (Table 1, Supplementary File 7). An intriguing exception in another odonate, Ladona fulva, is discussed below.
The most surprising phylogenetic finding was that the family was almost completely absent from Hymenoptera with only one complete sequence identified from Cephus cinctus, a sawfly. This is despite the abundance of sequence information for this order, with data from many species and complete genomes for three species of Nasonia and Apis dorsata and Apis mellifera, the latter with a recently updated proteome (Elsik et al. 2014).
SignalP (Petersen et al., 2011) was used to predict the signal peptides shown in Supplementary Files 7 and 9. The first amino acid in Bc-NCP1 is glutamine (Q), which was present as a pyroglutamate residue (Jensen et al., 1997). An initial Q was present, after the signal peptide was removed, in many of the sequences. In addition, we noticed that many of the retrieved sequences had a Q close to the end of the signal peptide. In most cases, the SignalP result showed that this could follow an alternative splice site. The signal for these sequences was modified (bold in Supplementary File 7) to move the Q into the mature protein resulting in 6/12 non-holometabola sequences beginning in this manner, providing further evidence for the conservation of the entire protein sequence. In the Holometabola, Q was less common. Instead, in the Lepidoptera, arginine (R) was the first amino acid in 13/16 sequences, and in the Diptera it was lysine (K) in 22/28. Except for the Coleoptera, there are fewer than 10 amino acids from the start of the mature protein to the start of the first motif. Generally there are zero or one amino acids after the final cysteine at the carboxy- terminus, but occasionally more (Table 1).
Another generalization is that the mature protein, with one exception, does not exceed 130 amino acids except in the Coleoptera and Diptera that have all family members over that length. The lepidopteran sequences are more comparable in length to members of the non-holometabolous orders (Table 1). There also appear to be amino acids immediately adjacent to the 16-amino- acid-motifs that differ between the different motifs within a sequence and among different orders. For example, almost all of the lepidopteran sequences have arginine-glutamic acid (RE) immediately upstream of the first motif, while this was not seen in any of the dipteran sequences, all with longer stretches before the first motif and alanine-glutamine (AQ) most frequently immediately upstream from the first motif (Supplementary File 7). Whether these differences represent something functional or result from a chance event in evolution remains to be learned.
While we have focused our discussion on the number and placement of the CPCFC 16-amino-acid-motif within the protein, it is apparent that the rest of the protein must be conferring important functional properties. This is clearest in the three major Holometabola orders, Coleoptera, Lepidoptera and Diptera. Extensions of the amino-terminus and the regions between motifs are populated by the acidic amino acids, glutamine (Q ) or asparagine (N), with fairly evenly spaced aromatic residues tyrosine (Y), tryptophan (W), or phenylalanine (F) (Supplementary File 7).
In addition to the presence of only two copies of the CPCFC motif in Coleoptera and Lepidoptera, there are other features of the long sequences from these groups and from the Diptera that enable one to assign a sequence to the correct order.
The generalizations presented here are certain to change as data on more species become available. For example, a tblastn search for whole genome sequences (WGS) in just the Odonata revealed evidence for 14 distinct CPCFC genes in Ladona fulva. None were complete, for the start of the signal peptides was missing, something not unexpected since the first exon is generally very short and would not been continuous with the presumed second exon, which in these genes had the rest of the coding region. All ended with stop codons. These 14 genes were distributed across 10 contigs. Ten sequences had three motifs, and 4 had two (Supplementary File 8). Three with two motifs were unusual because the final motif was not near the C-terminus, but from 63-84 amino acids away. Possibly as whole genome sequences become available for other species, more examples will be found with more than two CPCFC genes. Another generalization that is upset by Ladona CPCFCs is that the length of the proteins from the first motif to the end exceeds 131 amino acids in 7 of the sequences, excluding the two with unusual carboxy-termini. Hence, unless an intron interrupts what we have interpreted as a continuous second exon, the Coleoptera and Diptera will not be the only orders with long proteins. The one exception noted above to a non-Holometabola sequence with greater than 140 amino acids interestingly is one of the two sequences from another odonate, Enallagma hageni (Table 1).
3.6. Phylogenetic distribution of CPCFC genes in Crustacea
While the available data are far more limited in the Crustacea, we found representatives of CPCFC in four of the six classes: Ostracoda, Malacostraca, Maxillopoda, and Remipedia (Table 2, Supplementary File 9). Variation among groups was informative. A large number of hits that were not examined further were to sequences that had only one of the motifs. The barnacle (Amphibalanus amphitrite) had five motifs, and that was the only sequence in Crustacea that was longer than 100 amino acids. Remipedia, the group reported by von Reumont et al. (2012) to be most closely related to the hexapods, had two sequences from one species, Speleonectes, one with two motifs, one with three. The more basal group (Ostracoda) had two sequences, both with two motifs. Most intriguing were the 6 members of this family in Malacostraca. All had a variant on the basic motif, namely C-X(7)-C, present twice in each sequence. This variant was not found in any other group of arthropods. Since Jensen et al. (1997) suggested that the motif functions to bind metals, it would be interesting to learn if some unusual metal is used by members of this order.
TABLE 2.
CHARACTERISTICS OF CPCFC FAMILY MEMBERS IN CRUSTACEA (signals removed)
| amino acids | |||||||
|---|---|---|---|---|---|---|---|
| Class/Species | total length | to start of motif 1 | between motif 1-2 | Between motif 2-3 | Between motif 3-4 | Between motif 4-5 | final C to end |
| Ostracoda | |||||||
| Cypridininae sp. | 91 | 3 | 27 | 29 | |||
| Cypridininae sp. | 92 | 4 | 27 | 29 | |||
| Malacostraca | |||||||
| Melita plumulosa mira | 65 | 2 | 28 | ALL Malacostraca are C-X(7)-C | 2 | ||
| Hyalella azteca | 73 | 9 | 29 | 2 | |||
| Hyalella azteca | 72 | 9 | 28 | 2 | |||
| Procambarus clarkii | 47 | 4 | 10 | 0 | |||
| Petrolisthes cinctipes | 48 | 4 | 10 | 1 | |||
| Petrolisthes cinctipes | 48 | 4 | 10 | 1 | |||
| Maxillopoda | |||||||
| Amphibalanus amphitrite | 156 | 4 | 21 | 17 | 17 | 17 | 0 |
| Calanus finmarchicus | 81 | 4 | 13 | 15 | 1 | ||
| Eucyclops serrulatus | 79 | 3 | 13 | 15 | 0 | ||
| Remipedia | |||||||
| Speleonectes cf. tulumensis | 56 | 1 | 14 | 9 | |||
| Speleonectes cf. tulumensis | 76 | 2 | 10 | 15 | 1 | ||
The conservation of CPCFC proteins across the arthropods and the somewhat consistent differences among members of different orders suggest that these proteins must be playing a significant role in the cuticle. Their absence in some Hymenoptera indicates that whatever that role is, it is not irreplaceable.
3.7. Is CPCFC1 found outside Arthropoda?
We wondered if the CPCFC motif so highly conserved in Crustacea and Hexapoda could be found in other groups. They were, and while details are in Supplementary File 10, a summary is given below:
BLAST searches (tblastn, against EST or TSA entries, excluding Arthropoda) turned up five hits. One hit was to a sequence from a Homo sapiens brain cDNA library [GenBank:HY131203.1]. The sequence is not present in the database of Homo sapiens proteins, not surprisingly, because it has a 100% match to a protein from the cockroach, Blatella germanica [GenBank:GBID01001268.1].
We also got hits to two plants, Karelinia caspia (Asteraceae, a daisy, [GenBank:GANI01023091.1]) and Humulus lupulus (common hop, [GenBank: GAAW01027316.1]). TSA entries from another animal, Hynobius chinensis (Chinese salamander, [GenBank:GAQK01079415.1]), also had a CPCFC sequence.
We found a perfect match for the daisy; indeed, the daisy sequence completed an abbreviated sequence for the silverleaf (sweet potato, tobacco) whitefly Bemisia tabaci. The hop was clearly contaminated by a fruit fly, probably in the genus Bactrocera, and the salamander sequence was very close to a chironomid.
A final case of contamination was in Daphnia pulex, the only sequence identified for the crustacean class Branchiopoda. Searches of ESTs for CPCFC in Crustacea result in top hits to Daphnia pulex, but exclusively to library 12, the one where the Daphnia had been exposed to Chaoborus americanus in order to monitor the transcriptional response to this predatory midge (Table S10 in Colbourne et al., 2011). Thus it is not surprising that when the complete Daphnia sequence [GenBank:FE342003.1] is itself used in a BLAST search against ESTs, instead of linking to other Crustacea, the top match is to a different midge, Corethrella appendiculata [GenBank:GANO01004087.1], followed by various mosquitoes.
4. Conclusions
A new family of cuticular proteins, CPCFC, has members widely dispersed among the Pancrustacea. Members are generally present in 1-2 copies per species, with a protein having two to three copies of the 16 amino acid CPCFC motif that ends C-X(5)-C. A notable exception was seen in the dragonfly, Ladona fulva, where 14 genes, each with 2 or 3 CPCFC motifs, were found.
Experimental work with the An. gambiae family member, AgamCPCFC1, revealed that the mRNA is most abundant immediately following a molt; transcripts are found predominantly in epidermis secreting hard cuticle, and the protein has been localized mainly in endocuticle. Available information on phylogenetic distribution and protein characteristics revealed that CPCFC is distributed throughout the Hexapoda and in several classes of Crustacea. Amino acid sequences in two Holometabola orders, Coleoptera and Diptera, were longer than in the other orders. All sequences found in the Malacostraca had a motif that ended C-X(7)-C, rather than C-X(5)-C.
Supplementary Material
CPCFC HIGHLIGHTS.
New cuticular protein family described, characterized by a 16 amino acid motif ending C-X(5)-C.
In Anopheles gambiae, transcripts localized primarily in epidermis underlying hard cuticle.
Proteins localized primarily in endocuticle.
Family members identified in 14 orders of Hexapoda and 4 classes of Crustacea.
Acknowledgements
We thank Drs. Reben Rhaman and Sheng-Cheng Wu for producing the AgamCPCFC1 protein used for antibody generation. We also thank Dr. Mark R. Brown and Anne Robertson for maintaining the mosquito facility from which the animals were obtained, MR Brown for help interpreting mosquito structures, and Dr. Michael Strand for access to his Leica photomicroscope and Jena Johnson for training in its use. Dr. Neal Dittmer alerted us to the presence of two CPCFC genes in Tribolium; Dr. Hugh Robertson found the Cephus sequence; Dr. Michael Pfrender supplied information about Daphnia and Drs. Bernhard Misof and Karen Meusemann provided guidance about the 1KITE sequences. We thank Mary B. Ard of the Electron Microscopy Laboratory at the University of Georgia College of Veterinary Medicine for technical support. Drs. Yihong Zhou and John S. Willis and three anonymous reviewers provided helpful comments on the MS. This research was funded by a grant from the U.S. National Institutes of Health R01AI055624.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare that they have no competing interests.
References
- Bustin SA, Benes V, Garson J, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley G, Wittwer CT, Schjerling P, Day PJ, Abreu M, Aguado B, Beaulieu JF, Beckers A, Bogaert S, Browne JA, Carrasco-Ramiro F, Ceelen L, Ciborowski K, Cornillie P, Coulon S, Cuypers A, De Brouwer S, De Ceuninck L, De Craene J, De Naeyer H, De Spiegelaere W, Deckers K, Dheedene A, Durinck K, Ferreira-Teixeira M, Fieuw A, Gallup JM, Gonzalo-Flores S, Goossens K, Heindryckx F, Herring E, Hoenicka H, Icardi L, Jaggi R, Javad F, Karampelias M, Kibenge F, Kibenge M, Kumps C, Lambertz I, Lammens T, Markey A, Messiaen P, Mets E, Morais S, Mudarra-Rubio A, Nakiwala J, Nelis H, Olsvik PA, Perez-Novo C, Plusquin M, Remans T, Rihani A, Rodrigues-Santos P, Rondou P, Sanders R, Schmidt-Bleek K, Skovgaard K, Smeets K, Tabera L, Toegel S, Van Acker T, Van den Broeck W, Van der Meulen J, Van Gele M, Van Peer G, Van Poucke M, Van Roy N, Vergult S, Wauman J, Tshuikina-Wiklander M, Willems E, Zaccara S, Zeka F, Vandesompele J. The need for transparency and good practices in the qPCR literature. Nat. Methods. 2013;10:1063–1067. doi: 10.1038/nmeth.2697. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Caceres CE, Carmel L, Casola C, Choi JH, Detter JC, Dong Q, Dusheyko S, Eads BD, Frohlich T, Geiler-Samerotte KA, Gerlach D, Hatcher P, Jogdeo S, Krijgsveld J, Kriventseva EV, Kultz D, Laforsch C, Lindquist E, Lopez J, Manak JR, Muller J, Pangilinan J, Patwardhan RP, Pitluck S, Pritham EJ, Rechtsteiner A, Rho M, Rogozin IB, Sakarya O, Salamov A, Schaack S, Shapiro H, Shiga Y, Skalitzky C, Smith Z, Souvorov A, Sung W, Tang Z, Tsuchiya D, Tu H, Vos H, Wang M, Wolf YI, Yamagata H, Yamada T, Ye Y, Shaw JR, Andrews J, Crease TJ, Tang H, Lucas SM, Robertson HM, Bork P, Koonin EV, Zdobnov EM, Grigoriev IV, Lynch M, Boore JL. The ecoresponsive genome of Daphnia pulex. Science. 2011;311:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Togawa T, Dunn WA, He N, Emmons AC, Willis JH. Annotation and analysis of a large cuticular protein family with the R&R Consensus in Anopheles gambiae. BMC Genomics. 2008;9:22. doi: 10.1186/1471-2164-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS. Molecular evolution of Drosophila cuticular protein genes. PLoS ONE. 2009;4:e8345. doi: 10.1371/journal.pone.0008345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, Childers CP, de Graaf DC, Debyser G, Deng J, Devreese B, Elhaik E, Evans JD, Foster LJ, Graur D, Guigo R, Hoff KJ, Holder ME, Hudson ME, Hunt GJ, Jiang H, Joshi V, Khetani RS, Kosarev P, Kovar CL, Ma J, Maleszka R, Moritz RF, Munoz-Torres MC, Murphy TD, Muzny DM, Newsham IF, Reese JT, Robertson HM, Robinson GE, Rueppell O, Solovyev V, Stanke M, Stolle E, Tsuruda JM, Vaerenbergh MV, Waterhouse RM, Weaver DB, Whitfield CW, Wu Y, Zdobnov EM, Zhang L, Zhu D, Gibbs RA. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics. 2014;15:86. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futahashi R, Okamoto S, Kawasaki H, Zhong YS, Iwanaga M, Mita K, Fujiwara H. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1138–1146. doi: 10.1016/j.ibmb.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Harbach RE, Knight KL. Taxonomist's glossary of mosquito anatomy, first ed. Plexus Publishing, Inc.; Marlton, New Jersey: 1980. [Google Scholar]
- Ioannidou ZS, Theodoropoulou MC, Papandreou NC, Willis JH, Hamodrakas SJ. CutProtFam-Pred: detection and classification of putative structural cuticular proteins from sequence alone, based on profile hidden Markov models. Insect Biochem. Mol. Biol. 2014;52:51–59. doi: 10.1016/j.ibmb.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen UG, Rothmann A, Skou L, Andersen SO, Roepstorff P, Hojrup P. Cuticular proteins from the giant cockroach, Blaberus craniifer. Insect Biochem. Mol. Biol. 1997;27:109–120. doi: 10.1016/s0965-1748(96)00074-4. [DOI] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, Niehuis O, Petersen M, Izquierdo-Carrasco F, Wappler T, Rust J, Aberer AJ, Aspöck U, Aspöck H, Bartel D, Blanke A, Berger S, Böhm A, Buckley TR, Calcott B, Chen J, Friedrich F, Fukui M, Fujita M, Greve C, Grobe P, Gu S, Huang Y, Jermiin LS, Kawahara AY, Krogmann L, Kubiak M, Lanfear R, Letsch H, Li Y, Li Z, Li J, Lu H, Machida R, Mashimo Y, Kapli P, McKenna DD, Meng G, Nakagaki Y, Navarrete-Heredia JL, Ott M, Ou Y, Pass G, Podsiadlowski L, Pohl H, von Reumont BM, Schütte K, Sekiya K, Shimizu S, Slipinski A, Stamatakis A, Song W, Su X, Szucsich NU, Tan M, Tan X, Tang M, Tang J, Timelthaler G, Tomizuka S, Trautwein M, Tong X, Uchifune T, Walzl MG, Wiegmann BM, Wilbrandt J, Wipfler B, Wong TK, Wu Q, Wu G, Xie Y, Yang S, Yang Q, Yeates DK, Yoshizawa K, Zhang Q, Zhang R, Zhang W, Zhang Y, Zhao J, Zhou C, Zhou L, Ziesmann T, Zou S, Li Y, Xu X, Zhang Y, Yang H, Wang J, Wang J, Kjer KM, Zhou X. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, Amon J, Arca B, Arensburger P, Artemov G, Assour LA, Basseri H, Berlin A, Birren BW, Blandin SA, Brockman AI, Burkot TR, Burt A, Chan CS, Chauve C, Chiu JC, Christensen M, Costantini C, Davidson VL, Deligianni E, Dottorini T, Dritsou V, Gabriel SB, Guelbeogo WM, Hall AB, Han MV, Hlaing T, Hughes DS, Jenkins AM, Jiang X, Jungreis I, Kakani EG, Kamali M, Kemppainen P, Kennedy RC, Kirmitzoglou IK, Koekemoer LL, Laban N, Langridge N, Lawniczak MK, Lirakis M, Lobo NF, Lowy E, MacCallum RM, Mao C, Maslen G, Mbogo C, McCarthy J, Michel K, Mitchell SN, Moore W, Murphy KA, Naumenko AN, Nolan T, Novoa EM, O'Loughlin S, Oringanje C, Oshaghi MA, Pakpour N, Papathanos PA, Peery AN, Povelones M, Prakash A, Price DP, Rajaraman A, Reimer LJ, Rinker DC, Rokas A, Russell TL, Sagnon N, Sharakhova MV, Shea T, Simao FA, Simard F, Slotman MA, Somboon P, Stegniy V, Struchiner CJ, Thomas GW, Tojo M, Topalis P, Tubio JM, Unger MF, Vontas J, Walton C, Wilding CS, Willis JH, Wu YC, Yan G, Zdobnov EM, Zhou X, Catteruccia F, Christophides GK, Collins FH, Cornman RS, Crisanti A, Donnelly MJ, Emrich SJ, Fontaine MC, Gelbart W, Hahn MW, Hansen IA, Howell PI, Kafatos FC, Kellis M, Lawson D, Louis C, Luckhart S, Muskavitch MA, Ribeiro JM, Riehle MA, Sharakhov IV, Tu Z, Zwiebel LJ, Besansky NJ. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347:1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh MY, Kramer KJ, Muthukrishnan S, Kanost MR, Beeman RW, Arakane Y. Two major cuticular proteins are required for assembly of horizontal laminae and vertical pore canals in rigid cuticle of Tribolium castaneum. InsectBiochem. Mol. Biol. 2014;53C:22–29. doi: 10.1016/j.ibmb.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Noh MY, Muthukrishnan S, Kramer KJ, Arakane Y. Tribolium castaneum RR-1 cuticular protein TcCPR4 is required for formation of pore canals in rigid cuticle. PLoS Genet. 2015;11:e1004963. doi: 10.1371/journal.pgen.1004963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch YY, Riedel D, Behr M. Obstructor-A organizes matrix assembly at the apical cell surface to promote enzymatic cuticle maturation in Drosophila. J. Biol. Chem. 2015;290:10071–10082. doi: 10.1074/jbc.M114.614933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Rebers JE, Riddiford LM. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J. Mol. Biol. 1988;203:411–423. doi: 10.1016/0022-2836(88)90009-5. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Dunn WA, Emmons AC, Nagao J, Willis JH. Developmental expression patterns of cuticular protein genes with the R&R Consensus from Anopheles gambiae. Insect Biochem. Mol. Biol. 2008;38:508–519. doi: 10.1016/j.ibmb.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Augustine Dunn W, Reed TW, Willis JH. Changes in transcript abundance for cuticular proteins and other genes three hours after a blood meal in Anopheles gambiae. Insect Biochem. Mol. Biol. 2014a;44:33–43. doi: 10.1016/j.ibmb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Reed TW, Willis JH. Temporal and spatial expression of cuticular proteins of Anopheles gambiae implicated in insecticide resistance or differentiation of M/S incipient species. Parasit. Vectors. 2014b;7:24. doi: 10.1186/1756-3305-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reumont BM, Jenner RA, Wills MA, Dell'ampio E, Pass G, Ebersberger I, Meyer B, Koenemann S, Iliffe TM, Stamatakis A, Niehuis O, Meusemann K, Misof B. Pancrustacean phylogeny in the light of new phylogenomic data: support for Remipedia as the possible sister group of Hexapoda. Mol. Biol. Evol. 2012;29:1031–1045. doi: 10.1093/molbev/msr270. [DOI] [PubMed] [Google Scholar]
- Willis JH. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol. 2010;40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, Papandreou NC, Iconomidou VA, Hamodrakas SJ. Cuticular Proteins. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. Academic Press; San Diego: 2012. pp. 134–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.