Summary
Amyotrophic lateral sclerosis (ALS) is a clinical syndrome named for its neuropathological hallmark: degeneration of motor neurons in the spinal anterior horn and motor cortex and loss of axons in the lateral columns of the spinal cord. The signature neuropathological molecular signature common to almost all sporadic ALS and most familial ALS is TDP-43 immunoreactive neuronal cytoplasmic inclusions. The neuropathological and molecular neuropathological features of ALS variants primarly lateral sclerosis and progressive muscular atrophy are less certain, but also appear to share the primary features of ALS. A number of genetic causes including mutations in SOD1, FUS, and C9orf72 comprise a disease spectrum and all demonstrate distinctive molecular and neuropathological signatures. Neuropathology will continue to play to a key role in solving the puzzle of ALS pathogenesis.
Keywords: ALS, neuropathology, TDP-43, motor neuron degeneration, C9orf72
Introduction
The first case reports of amyotrophic lateral sclerosis (ALS) date back to Charles Bell in 18241. While a variety of other clinical descriptions followed throughout the 1850s2-4, the correlations between key clinical features of progressive muscle atrophy and muscle spasticity and key neuropathologic features of loss of anterior horn cells and sclerosis in the lateral columns of the spinal cord were first made by Charcot in the 1860’s5, and thus he named the clinical disease by its distinctive neuropathology.6 Significant subsequent contributions included the observation of loss of the giant cells of Betz, best summarized by Brodmann in 19097, of eosinophilic inclusions now called Bunina bodies in 19628, the discovery of ubiquitinated cytoplasmic inclusions in 1988,9, 10 and the discovery that the ubiquitinated inclusions are comprised primarily of TDP-43 in 200611, 12. The association between ALS and frontotemporal lobar dementia (FTLD) has taken three decades to establish13, 14. With advances in genetics beginning in 1993, distinctive neuropathology is being identified in the genetic forms, the main ones being SOD115, TDP-4316, FUS17, 18, and C9orf72 repeat expansions19, 20.
Classic ALS Neuropathology
Gross
In the majority of ALS brains, no gross abnormalities are observed. The spinal cord often reveals atrophy of the anterior nerve roots21. Some cases exhibit atrophy of the precentral gyrus21. In patients with dementia, atrophy of the frontal and/or temporal cortex may be seen21-24, the atrophy being greatest in brains from patients with overlap ALS-frontotemporal dementia (FTD). In addition to these grey matter abnormalities, white matter reduction is also observed, particularly, but not exclusively, in the corticospinal tract25-27.
Microscopic
Microscopic changes include neuronal and axon loss. There is loss of myelinated axons in the lateral and anterior columns of the spinal cord and decreases in size of anterior horn of the spinal cord, best shown by myelin stains such as luxol fast blue (Figure 1A, B) 21. There is degeneration and loss of the large motor neurons in the anterior horn of the spinal cord, lower cranial motor nuclei of the brainstem, and Betz cells in the motor cortex, best seen with routine stains such as hematoxylin and eosin (H&E)21, 28-30 (Figure 1C, D and Figure 2A-F). Morphometric studies of the spinal anterior horn have shown a global reduction of all neurons in the anterior horn, not just the large alpha motor neurons31. There is evidence of reduction in neuron size as well as loss and atrophy of nerve fibers. Other pathological features of ALS include vacuolization, large empty spaces near neurons, and spongiosis, microscopic holes resulting in a sponge-like appearance (Figure 2G,H).
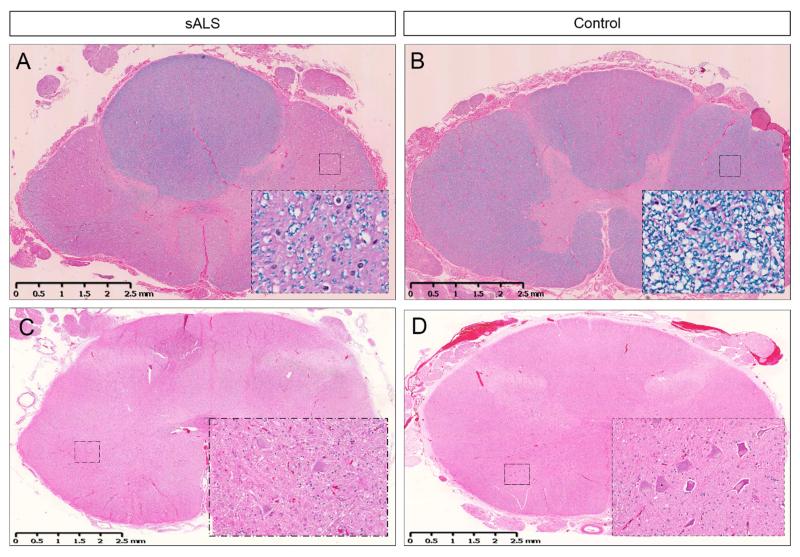
Figure 1. Amyotrophic lateral sclerosis (ALS).
Lateral sclerosis is shown in the thoracic spinal cord in sporadic ALS (A) and compared to control (B). The inserts show loss of myelin in the white matter tracts. Loss of motor neurons is shown in the lumbar spinal cord in sporadic ALS (C) and compared to control (D). The inserts show the motor neurons in the anterior horns under higher power. [Luxol fast blue with hematoxylin and eosin (A, B); hematoxylin and eosin (C, D)]
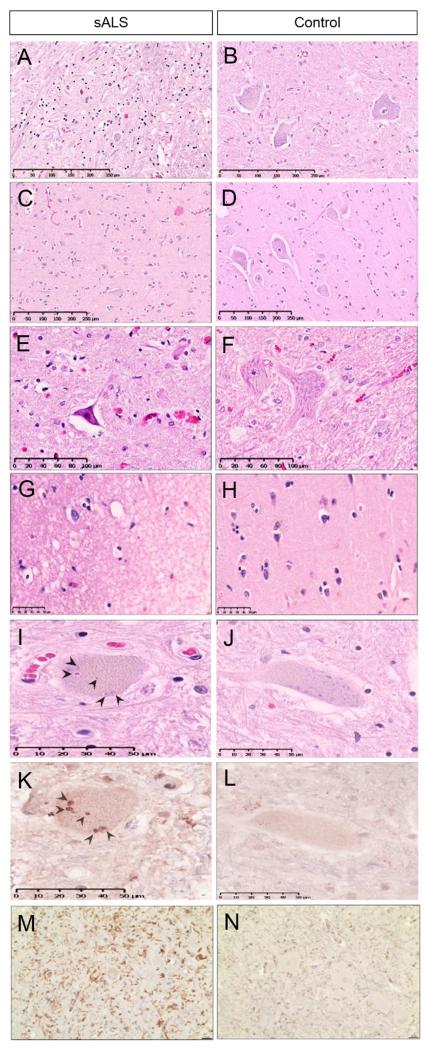
Figure 2. Classic ALS neuropathology.
Loss of motor neurons is shown in an anterior horn of the spinal cord (A) and motor cortex (C) of ALS and compared to control (B, D). Shrinkage and contraction of motor neuron in ALS (E) is compared to control (F). Vacuolization and spongiosis in motor cortex is shown in ALS (G) and compared to control (H). Bunina bodies are seen in the cytoplasm of motor neurons of ALS (I) and compared to control (J). Bunina bodies are positive for cystatin c in ALS (K) and compared to the effect of the stain in control (L). Microglial activation is shown by IBA1 in the anterior horn of the spinal cord in ALS (M) but not control (N). [Hematoxylin and eosin (A-J)]
Bunina bodies are small (3-6 microns), round to oval shaped eosinophilic intracellular inclusions in the motor neurons of the spinal cord and brain stem of both sporadic and familial ALS patients, best seen with H&E32,33 (Figure 2I, J). They are most frequently found in the cytoplasm of motor neurons, but can occasionally be found in dendrites34. Their number per neuron is highly variable, and they can sometimes make chains and clusters. They are rarely seen in Betz cells, neurons of oculomotor nuclei, and Onuf’s nuclei35-37. Immunohistochemically, Bunina bodies are positive for cystatin c (Figure 2K, L) and transferrin and partially colocalize with peripherin38, 39, but they are negative for a variety of proteins commonly associated with neurodegeneration, including tau, alpha and beta tubulin, synaptophysin, amyloid precursor protein, glial fibrillary acidic protein, alpha synuclein, and p6238, 40-42. Interestingly, it remains controversial whether or not Bunina bodies are positive for ubiquitin10, 43. Their biological significance is unknown.
The complexity of glial cells is now well established and recent studies have shown they are crucial in the biology of ALS neurodegeneration44. Reactive astrogliosis surrounds degenerating motor neurons in ALS patients and ALS-animal models45-47. Reactive astrocytes show increased immunoreactivity for GFAP and the calcium binding protein S100β and express inflammatory makers such as COX-2, inducible nitric oxide synthase (iNOS) and neuronal NOS. Increase in GFAP-immunoreactive astrocytes is particularly notable in the grey matter of the spinal cord ventral horn, where normally astrocytes express GFAP at low levels. Cytoplasmic hyaline inclusions and markers of oxidative and nitrative stress accompany astrocyte pathology48, 49.
Activation of microglia is a critical aspect of the glial neuropathology45. It has been correlated with severity of UMN degeneration in ALS (reviewed in Lasiene & Yamanaka, 201150). Activated microglia, responding to neuronal distress, release a variety of proinflammatory cytokines, leading to a higher degree of inflammation in the brains of ALS patients. These proinflammatory molecules include but are not limited to: tumor necrosis factor-α, interleukin-1β, and ionized calcium-binding adapter molecule 2 (Figure 2M, N). Microglia also release reactive oxygen species such as superoxide and nitric oxide, as well as chemokines (monocyte chemoattractant protein 1, macrophage colony stimulating factor) and neurotrophic factors (insulin-like growth factor-1)51. It seems neuroinflammation is a two edged sword, some of it protects against neurodegeneration and some of it drives it44-46.
Molecular Neuropathology: Inclusions and Proteinopathies
Ubiquitin
Some of the most important progress in understanding ALS biology has been driven by key neuropathological discoveries. This began in 1988, when Leigh et al.9, 52 and Lowe et al.10 independently discovered ubiquitin-positive skein-like or dense, round structures in the cytoplasm of anterior horn cells in both familial and sporadic ALS (Figure 3 A-D), inclusions that are not detected by H&E and other routine staining methods. Such inclusions were later identified in FTLD and became the cornerstone of distinguishing FTLD with ubiquinated inclusions (FTLD-U) from FTLD with tau (FTLD-tau) and other inclusions 53-55. In both ALS and FTLD-U, the ubiquitin-positive inclusions have been observed in neurons of the frontal cortex, temporal cortex, hippocampus and striatum32, 56-59. Although they are most commonly found in neurons, they have occasionally been seen in glial cells60. They are negative for proteins commonly associated with neurodegenerative inclusions, such as tau and alpha-synuclein56, 60.
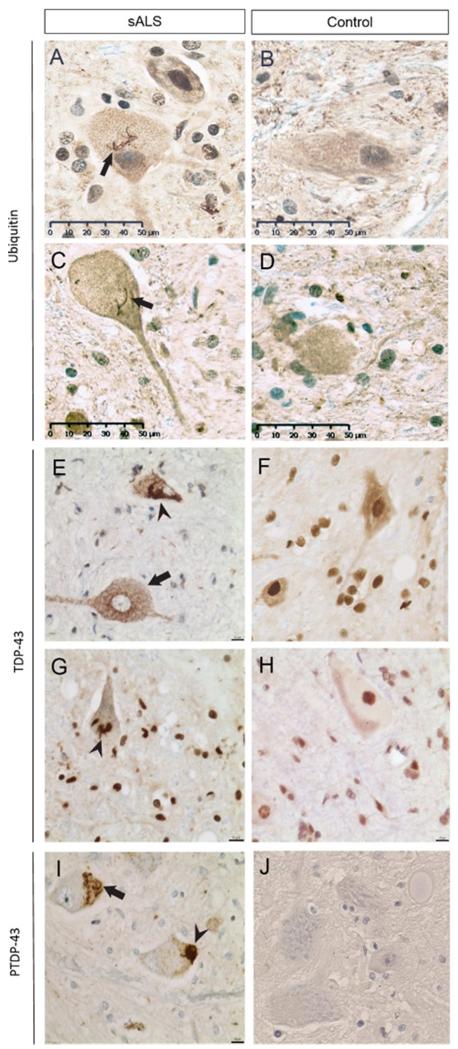
Figure 3. Inclusions in ALS neuropathology.
Ubiquitin skein-like inclusions (arrows) are shown in spinal motor neurons of the lumbar anterior horn (A) and Betz cells of the motor cortex (C) in ALS but not control (B, D). TDP-43 inclusions are shown to be diffuse (arrow) and skein-like (arrow-head) and there is nuclear clearing in the spinal motor neurons of ALS (E), features not seen in controls. Note the normal nuclear TDP-43 in control (F). TDP-43 dense round inclusions (arrowhead) are shown in the motor cortex of ALS (G) but not in controls, which show normal nuclear TDP-43 (H). Phospho-TDP-43 staining shows skein-like (arrow) and dense round (arrow-head) inclusions in ALS lower motor neurons (I), which are not seen in controls. Note pTDP is not seen in normal nuclei (unlike TDP-43) (J).
TDP-43
The presence of ubiquitin-positive inclusions suggested a problem with some other protein(s). Eighteen years later, TDP-43 was identified as the main component of ubiquitinated inclusions in both ALS and FTD patients11, 12. This connected ALS and FTLD-U as TDP-43 proteinopathies61. (Table 1) TDP-43 is a heterogeneous nuclear ribonucleoprotein and has many different cellular functions, including mRNA stability62, 63, mRNA processing64, 65, mRNA transport and translation66, 67 and negative regulation of alternative splicing68. Under normal conditions, TDP-43 is expressed in many tissues including the nuclei of neurons and glial cells. In sporadic and most familial ALS as well as FTLD-TDP-43 (now renamed from FTLD-U), there is loss of nuclear TDP-43 and formation of pathological aggregates in the cytoplasm (Figure 3E-H)69. The mechanism behind this redistribution is poorly understood, and could be either the translocation of TDP-43 from the nuclei to the cytoplasm, or an impaired TDP-43 cytoplasm-to-nucleus shuttling process12, 70-72. With the use of immunoblot analysis, extracted material from brains of patients with FTD-TDP-43, FTD-ALS, and ALS were found to be phosphorylated TDP-43 band at 45 kDa11, 73. This suggested a post-translational modification to TDP-43. Indeed, antibodies against TDP-43 phosphopeptides stain ubiquitinated TDP-43 positive inclusions in FTD/ALS patients (Figure 3I, J)74, 75.
TABLE 1.
ALS PROTEINOPATHIES: MAIN MOLECULAR NEUROPATHOLOGICAL FEATURES
| Proteinopathy | Phenotype | Gene | Main Molecular Features1 | ||||
|---|---|---|---|---|---|---|---|
| Motor cortex (UMN) |
Spinal anterior horn or brain stem motor nuclei (LMN) |
Fronto-temporal regions |
Miscellaneous (cerebellum, hippocampus) |
Descending axonal pathways (e.g. lateral columns) |
|||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
NCI = nuclear cytoplasmic inclusions; GCI = glial cytoplasmic inclusions
No primary FTD phenotypes have been defined by SOD1 pathology.
Included here for comparison–no primary ALS phenotypes have been defined by tau+ neuropathology.
There are different kinds of TDP-43 inclusions including fine skeins, coarse skeins, dot-like, and dense round inclusions (Figure 3 E, G, I). Fine and coarse inclusions are seen with a similar frequency in lower and upper motor neurons, while dot-like and round inclusions are seen more frequently in the motor neurons of the anterior horn76 (Figure 3E-H). In some cases, there is evidence of TDP-43 proteinopathy such as nuclear clearing and/or diffuse or granular cytoplasmic TDP-43 despite the absence of frank cytoplasmic inclusions (Figure 3E)12, 77.
It is now apparent that TDP-43 inclusions are not pathognomonic for ALS or FTLD-TDP-43, since inclusions are also observed in Alzheimer’s disease78-80, Lewy body diseases79-82, Guamanian Parkinsonism dementia complex79, 80, 83 and post-traumatic encephalopathy and neurodegeneration84. TDP-43 is present in the mesiotemporal lobe structure in about 30% of the people 65 years or older, regardless of mental illness status85, indicating the aggregation and misfolding of TDP-43 may be caused by processes normally associated with aging.
Sequential Changes and Neuropathological Staging
Cellular and microscopic
Although it is known that motor neurons degenerate and die in ALS, it is not clear how this degeneration is initiated and progresses or the exact morphological stages of cell death. It is commonly believed that motor neuron death in ALS closely resembles apoptosis, although the evidence for this is incomplete86. Martin postulated three stages of neuronal death in motor neurons of ALS patients: chromatolysis (dissolution of the Nissl bodies in the cell body of a neuron), somatodendritic attrition, and finally apoptosis87. This process of neuronal death is accompanied by morphologic findings such as cytoplasmic and nuclear condensation and darkness, DNA fragmentation in the presence of DNA fragmentation activation factor, as well as a lack of appreciable vacuolar and edematous cytoplasm in dying neurons. Based on these observations he concluded that apoptosis plays a role in neuronal death in ALS cases. Elevated levels of pro-apoptotic proteins Bax and Bak and decreased levels of anti-apoptotic protein Bcl-2 in vulnerable CNS regions in ALS patients compared with controls were also observed, further strengthening the link between motor neuron death and apoptosis87. A related question about ALS regards the spread of the disease and the cell-to-cell spread of disease. A neuropathological correlate of this is that lower motor neuron loss is greatest at the region of onset and decreased outward88,89. Recently, necroptosis, a form of programmed necrosis involving receptor interacting protein 1 and the mixed lineage kinase domain-like protein, has been postulated to be an important driver of motor neuron death90, although the neuropathological underpinnings remain to be established.
Anatomical distribution of pathological changes
Staging of ALS neuropathology, similar to Alzheimer’s and Parkinson’s diseases, has been proposed by Braak et al91 and Brettschneider et al.92. In this, stage 1 disease is characterized by mild burden of pTDP-43 pathology involving motor cortex, brainstem motor nuclei, and spinal motoneurons. Stage 2 disease involves mild-moderate burden of pTDP-43 with dissemination into prefrontal neocortex (middle frontal gyrus), reticular formation, and precerebellar nuclei. Stage 3 disease involves moderate burden of pTDP-43 with dissemination into basal ganglia and prefrontal and postcentral neocortex and striatum. Stage 4 disease involves severe burden of pTDP-43 including the hippocampal formation. In the spinal cord, severity of pTDP-43 pathology in lamina IX motor nuclei and neuronal loss correlated closely with gray and white matter oligodendroglial involvement and was linked to onset of disease88. Interestingly, pTDP-43 pathology sometimes included Onuf’s nucleus and neurons of Clarke’s column but rarely in the intermediolateral nucleus. Gray matter oligodendroglial pTDP-43 inclusions were present in areas devoid of neuronal pTDP-43 aggregates and neuronal loss and suggested involvement is an early event. This staging classification is based on neuropathology, not clinical disease severity (all nervous systems were from patients who died from end-stage disease), and clinical-pathological correlations remain to be established.
Familial ALS: Genetics and Associated Pathology
Approximately 90% of all ALS cases occur sporadically, with no associated family history. The remaining 10% of ALS cases are familial (FALS), and are usually the result of dominantly inherited autosomal mutations. The most common mutations occur in SOD1, TDP-43, FUS, and C9orf72, although several other genes have been identified (reviewed in Renton et al., 201493). The reader is also referred to the chapter titled “Familial ALS” in this issue. Each genetic cause correlates with a relatively distinctive neuropathological signature.
SOD1
Mutations in the superoxide dismutase-1 (SOD1) gene account for 20% of all FALS cases. Mutations throughout the gene have been linked to ALS. In general, updating of SOD1 neuropathology is critically needed105, but it is clear SOD1 ALS patients demonstrate more severe lower motor neuron degeneration than upper motor neuron degeneration. Upper motor neuron degeneration is suggested to be a distal axonopathy94. Anterior horn motor neurons also exhibit Lewy body-like inclusions (LBLIs), which consist of a hyalinized, poorly stainable substance (Figure 4A) that by immunohistochemistry are positive for SOD1, ubiquitin, phosphorylated neurofilaments, and various chaperone proteins96, 97, but negative for TDP-43, p-TDP-43 and FUS98. Isotype specific antibodies can uniquely detect misfolded SOD1 in spinal cord motor neurons of patients with SOD1 mutations (Figure 4B, C). This misfolded SOD1 is absent in the Betz cells in the motor cortex (Figure 4D). Based on the morphology of the motor neurons and the fact that they are TDP-43 negative95, neuropathology suggest molecular mechanisms of SOD1 mutant FALS may be distinct from sporadic ALS. But, importantly, misfolded SOD1 aggregates have been reported in sporadic ALS as well as mutant SOD1 familial ALS99-101, thus suggesting SOD1 protein misfolding may indeed play a role in sporadic disease, although such findings remain controversial102-104.
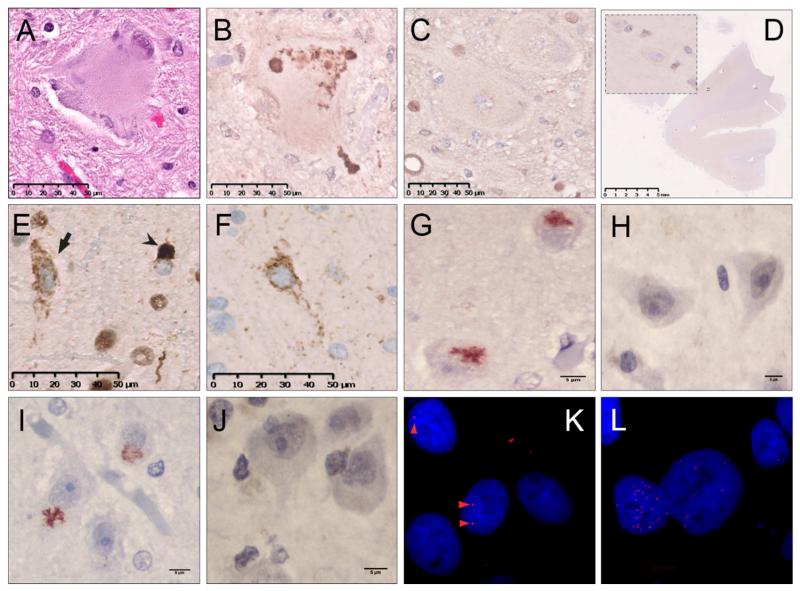
Figure 4. Neuropathology of familial ALS.
Lewy body-like inclusion in a spinal motor neuron from a nervous system of a patient with an SOD1 A4V mutation (A). A subsequent histological section showing co-localization of misfolded SOD1 and the Lewy body-like inclusion (B). Misfolded SOD1 is not seen in spinal motor neurons of controls (C) or in the motor cortex of SOD1 patients (D). Skein-like inclusions (arrow) and dense round inclusions (arrow-head) in spinal motor neurons of lumbar spinal cord of a nervous system of a patient with repeat expanded C9orf72 using anti-TDP antibody (E). Same as (E), using an anti-phospho-TDP-43 antibody (F). Poly GA dipeptide repeat proteins in the hippocampus of a nervous system from a patient with a repeat expansion in C9orf72 (G) that are not seen in sporadic ALS (H). PolyGP dipeptide repeat proteins in the hipppcampus of a nervous system from a patient with repeat expansion in C9orf72 (I) are not seen in sporadic ALS (J). RNA foci from the sense (K) and antisense (L) directions from cultured fibroblasts of a patient with repeat expansion in C9orf72 using fluorescent in situ hybridization.
TDP-43
The discovery of TDP-43 proteinopathy in 2006 was quickly followed by the identification of mutations in the TARDBP gene that encodes it. Mutations are responsible for 2-5% of FALS cases. Approximately 30 mutations have been identified throughout TARDBP106, 107, nearly all of them in the glycine-rich domain, which is responsible for regulating gene expression and protein-protein interactions108. Importantly, the TDP-43 and pTDP-43 proteinopathy that are observed in sporadic ALS are also observed in familial ALS caused by TARDBP mutations. In a neuropathologic study of patients with the Gly298Ser TDP-43 mutation, inclusions were observed in various locations throughout the CNS, including the substantia nigra, dentate gyrus, cingulate gyrus, amygdala, and the frontal and temporal cortices. The quantity of TDP-43 pre-inclusions in FALS patients with this mutation appears to be greater than in SALS patients109.
FUS/TLS
In 2009, mutations in the RNA-binding protein fused in sarcoma/translocated in sarcoma (FUS/TLS) in a subset of FALS patients were identified17, 18. FUS mutations are responsible for 5% of FALS cases, and are distinct from TDP-43 proteinopathies, but follow a similar motif wherein a protein involved in RNA metabolism is mislocalized from the nucleus and aggregates in the cytoplasm of neurons. Mutant FUS forms large, ubiquitinated, TDP-43 negative neuronal cytoplasmic inclusions (NCI) and occasional neuronal intranuclear inclusions (NII) in the spinal cords and brains of affected patients110. These inclusions take the form of fine and coarse granules, as well as filaments, and can be seen in neurons and glia. They are thought to interfere with RNA processing111 and cause the formation of cytoplasmic stress granules112. In contrast with TDP-43 NCIs, cytoplasmic FUS aggregates and non-pathogenic nuclear FUS are not exclusive, and can be observed in the same cell. There is one report that FUS immunoreactive NCIs may be present in sporadic ALS113, but the specificity of the antibodies was not proven and there has been no further confirmation. Bunina bodies are absent following H&E staining, however basophilic cytoplasmic inclusions are present. Specific FUS-ALS mutations may cause distinctive severity and neuropathology114—the p.P525L FUS mutation, with early-onset, has basophilic inclusions and round FUS-positive NCI, while the p.R521C mutation has tangle-like NCI and numerous cytoplasmic inclusions in oligodendroglia. FUS proteinopathy is now understood to account for cases of FTLD-U that are TDP-43-negative115 and thus cause an ALS-FTLD spectrum116.
C9orf72
In 2011, abnormally expanded GGGGCC hexanucleotide repeats in C9orf72 were identified as the most common genetic cause of FALS and FTD19, 20. This not only linked ALS and FTD at the genetic level, but connected them to the repeat expansion diseases. C9orf72 neuropathology displays the signature ubiquitin-positive, TDP-43-positive immunoreactive aggregates in neuronal cytoplasm, and thus is a TDP-43 proteinopathy (Figure 4E F). But it is unique among the TDP-43 proteinopathies in several respects. Most of the ubiquitinated inclusions in C9orf72-ALS are p62-positive, but TDP-43-negative117, 118. Nucleoporin 62 (p62) is a component of the nuclear envelope and thought to be involved in mRNA and protein trafficking into and out of the nucleus119. Another signature is the production of dipeptide repeats proteins (DPRs) resulting from repeat-associated non-ATG (RAN) translation, which occurs bidirectionally. DPRs translated from the sense strand are poly Gly-Ala (Figure 4G,H), poly Gly-Pro (Figure 4I, J), and poly Gly-Arg. DPRs translated from the antisense strand are poly Gly-Pro (also coded by sense), poly Ala-Pro, and poly Pro-Arg. All have been observed in CNS material of C9orf72 cases, although DPRs originating from the sense strand seem to be more frequent than antisense-related dipeptides120, 128. DPRs colocalize with p62 but not TDP-43129, 130. DPR aggregates can be seen in different parts of CNS including, frontal, occipital, temporal, and motor cortex as well as subcortical areas, midbrain, cerebellum and spinal cord but TDP-43 pathology may not correlate better with disease stage131. Another signature of C9-ALS neuropathology is foci of RNA of the expanded repeat (Figure 4K, L). These are detected by fluorescent in-situ hybridization (FISH)120, and are a feature of several of the repeat expansion diseases121. The RNA foci are bidirectionally transcribed and both sense- and antisense-directed expansions are seen125, 126. The foci are in multiple cell types including motor neurons, microglia, and astrocytes20 and in multiple regions of the nervous system including frontal cortex, motor cortex, hippocampus, cerebellum, and spinal cord, as well as lymphoblasts, fibroblasts, and IPSC-derived neurons122,123, 124. RNA foci seem to accumulate in cells with TDP-43 protein abnormalities127.
ALS variants
There are many clinical variants of ALS that appear to be distinctive and a key debate is whether these are distinct disease entities with different biologies, or ends of a continuum. The neuropathological evidence is scarce but suggests the latter, that they share a similar neuropathology and differences are based on the anatomical distribution of the pathological burden rather than biological differences. Separate from this debate about ALS are the important genetic syndromes that affect the motor system but are clearly different from ALS—since these may be confusing, they are reviewed here briefly for clarification.
Primary Lateral Sclerosis (PLS)
PLS is characterized by its upper motor neuron pattern with little or no apparent lower motor neuron involvement.132, 133,134. In this issue, a detailed description can be found in the chapter titled “Primary Lateral Sclerosis.” By some estimates, PLS is approximately 0.5% as prevalent as ALS. The most commonly reported differences in neuropathology between ALS and PLS lie in which regions have greater demyelination. PLS reportedly shows the greatest demyelination near the corpus callosum, whereas in ALS demyelination occurs most in the superior frontal gyrus135. But, PLS neuropathology does show changes in lower motor neurons and these changes are of the same molecular pattern as is seen in typical ALS disease including TDP-43 pathology, at least in some cases 133,136,137. Further study is needed to characterize the hallmarks of ALS, including TDP-43 and Bunina bodies, in PLS.
Progressive Muscular Atrophy (PMA)
PMA is characterized by it lower motor neuron pattern with little or no upper motor neuron involvement138. The reader is also referred to the chapter in this issue titled “Progressive Muscular Atrophy.” ALS and PMA also share similar genetic mutations in familial cases139. Despite the predominant involvement of lower motor neurons clinically, neuropathological studies have shown degeneration of the corticospinal tract even in the absence of upper motor neurons symptoms or signs140. PMA neuropathology may show abnormalities of the UMN by way of CD68 staining of the descending corticospinal tract, abnormalities identified in 50% of patients with clinically isolated LMN disease140. Distinct pathological change is identified in the motor and extra-motor areas of the brains as well as the spinal cords of patients whose disease was clinically limited to the LMN and these changes seem independent of progression rate141 Importantly, as is seen in ALS, inclusions positive for ubiquitin, TDP-43, and FUS are frequently present140, 142. Thus, the strongest evidence points to PMA being part of disease spectrum, not a different disease. .
Overlap FTD
The clinical overlap of ALS and FTD143, 144 is mirrored at the neuropathological level. Nearly all sporadic ALS and most FALS (except SOD1 and FUS) show TDP-43 proteinopathy, whereas only about 50% of FTLD is a TDP-43 proteinopathy. Most of the remaining cases of FTLD are considered as tauopathies and a small percentage are FUS proteinopathies. The discovery of repeat expanded C9orf72 revealed a common genetic link between ALS and FTD and highlighted the fact that both ALS and FTD are phenotypes of disease, as well as diseases. C9-ALS and C9-FTD share many pathological markers (as outlined above), and it remains to be shown whether or not C9-ALS, C9-ALS/FTD, and C9-FTD are different neuropathologically—the assumption is that they are not129..
Spinal Muscular Atrophy (SMA)
Spinal muscular atrophy (SMA), like PMA, is also characterized by degeneration of lower motor neurons 145 and result from homozygous mutations in the SMN1 gene146. SMA affects infants, juveniles, and young adults and is the leading genetic cause of infant death147. A detailed review of SMA is discussed in Chapter 13 of this issue. Affected individuals exhibit muscle weakness and atrophy of muscle fibers145. There is also extensive motor neuron loss, gliosis, neuronophagia, and chromatolysis in the anterior horn145,148. Reduction in the number of Betz cells in the motor cortex has also been observed148. TDP-43 proteinopathy does not appear to contribute to SMA biology, at least in mouse models149.
Hereditary Spastic Paraparesis (HSP)
HSP is a group of genetically heritable diseases that present with late onset slowly progressive spasticity of the lower limbs. The neuropathological findings are almost entirely limited to the pyramidal tracts of the spinal cord, and most significantly affect the longest ascending and descending axons. This axonal degeneration is particularly noticeable in the lumbar region of the spinal cord. Some degeneration of the anterior corticospinal and spinocerebellar tracts is also observed, as well as occasional loss of the cells in the anterior horn150, 151. Only one mutation of HSP has been studied neuropathologically and did show evidence of TDP-43 proteinopathy 152.
ALS Neuropathology: Future Directions and Final Remarks
The clinical syndrome called ALS is actually named by its neuropathology, amyotrophy and lateral sclerosis. Now, with the rapid progress in our understanding of phenotypes, genetics, and molecular biology and with the availability of new microscopic technologies including immunohistochemistry, immunofluorescence, and in situ hybridization, we are beginning to appreciate the extraordinary microscopic and molecular complexity underlying ALS neuropathology and its importance in unraveling the mystery of disease biology.
KEY Points.
-
*
ALS has a distinctive and complex neuropathology, from which its name is derived.
-
*
Many developments in ALS research have been driven by key neuropathological insights, such as the identification of ubiquitinated cytoplasmic inclusions that led to the identification of TDP-43 in ALS.
-
*
New microscopic and visualization techniques are allowing researchers an unprecedented view of the inner workings of the disease at the gross, cellular, and molecular levels
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
“The Authors have nothing to disclose.”
References
- 1.Tyler HR, Shefner J. Amyotrophic lateral sclerosis. Handb Clin Neurol. 1991;15:169–215. [Google Scholar]
- 2.Aran F. Recherches sur une maladie non encore décrite du système musculaire (atrophie musculaire progressive) Arch Gen Med. 1850;24:172. [Google Scholar]
- 3.Cruveilhier J. Sur la paralysie musculaire progressive atrophique. Arch Gen Med. 1853;91:561–603. [Google Scholar]
- 4.Duchenne de Boulogne G. Recherches électro-physiologiques et thérapeutiques. Comp Rend Seances Acad Sci. 1851;32:506. [Google Scholar]
- 5.Charcot J-M, Joffroy A. Deux cas d’atrophie musculaire progressive: avec lésions de la substance grise et des faisceaux antérolatéraux de la moelle épinière. Masson; 1869. [Google Scholar]
- 6.Charcot J. De la sclérose latérale amyotrophique. Prog Med. 1874;2:341–453. [Google Scholar]
- 7.Brodmann K. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Barth: 1909. [Google Scholar]
- 8.Bunina T. Zhurnal nevropatologii i psikhiatrii imeni SS Korsakova. Vol. 62. Moscow, Russia: 1961. On intracellular inclusions in familial amyotrophic lateral sclerosis; pp. 1293–1299. 1952. [PubMed] [Google Scholar]
- 9.Leigh P, Anderton B, Dodson A, Gallo J-M, Swash M, Power D. Ubiquitin deposits in anterior horn cells in motor neurone disease. Neuroscience letters. 1988;93:197–203. doi: 10.1016/0304-3940(88)90081-x. [DOI] [PubMed] [Google Scholar]
- 10.Lowe J, Lennox G, Jefferson D, et al. A filamentous inclusion body within anterior horn neurones in motor neurone disease defined by immunocytochemical localisation of ubiquitin. Neuroscience letters. 1988;94:203–210. doi: 10.1016/0304-3940(88)90296-0. [DOI] [PubMed] [Google Scholar]
- 11.Arai T, Hasegawa M, Akiyama H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and biophysical research communications. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 12.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 13.Hudson AJ. Amyotrophic lateral sclerosis and its association with dementia, parkinsonism and other neurological disorders: a review. Brain. 1981;104:217–247. doi: 10.1093/brain/104.2.217. [DOI] [PubMed] [Google Scholar]
- 14.Kiernan JA, Hudson AJ. Frontal lobe atrophy in motor neuron diseases. Brain. 1994;117(Pt 4):747–757. doi: 10.1093/brain/117.4.747. [DOI] [PubMed] [Google Scholar]
- 15.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. 1993 doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 16.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 19.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellison D, Love S, Chimelli LMC, et al. Neuropathology: a reference text of CNS pathology. Elsevier Health Sciences; 2012. [Google Scholar]
- 22.Chang J, Lomen-Hoerth C, Murphy J, et al. A voxel-based morphometry study of patterns of brain atrophy in ALS and ALS/FTLD. Neurology. 2005;65:75–80. doi: 10.1212/01.wnl.0000167602.38643.29. [DOI] [PubMed] [Google Scholar]
- 23.Abrahams S, Goldstein L, Suckling J, et al. Frontotemporal white matter changes in amyotrophic lateral sclerosis. J Neurol. 2005;252:321–331. doi: 10.1007/s00415-005-0646-x. [DOI] [PubMed] [Google Scholar]
- 24.Murphy JM, Henry RG, Langmore S, Kramer JH, Miller BL, Lomen-Hoerth C. Continuum of frontal lobe impairment in amyotrophic lateral sclerosis. Archives of Neurology. 2007;64:530–534. doi: 10.1001/archneur.64.4.530. [DOI] [PubMed] [Google Scholar]
- 25.Kassubek J, Unrath A, Huppertz HJ, et al. Global brain atrophy and corticospinal tract alterations in ALS, as investigated by voxel-based morphometry of 3-D MRI. Amyotrophic Lateral Sclerosis. 2005;6:213–220. doi: 10.1080/14660820510038538. [DOI] [PubMed] [Google Scholar]
- 26.Roccatagliata L, Bonzano L, Mancardi G, Canepa C, Caponnetto C. Detection of motor cortex thinning and corticospinal tract involvement by quantitative MRI in amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis. 2009;10:47–52. doi: 10.1080/17482960802267530. [DOI] [PubMed] [Google Scholar]
- 27.Kiernan J, Hudson J. Frontal lobe atrophy in motor neuron diseases. Brain. 1994;117:747–757. doi: 10.1093/brain/117.4.747. [DOI] [PubMed] [Google Scholar]
- 28.Hammer RP, Tomiyasu U, Scheibel AB. Degeneration of the human Betz cell due to amyotrophic lateral sclerosis. Experimental neurology. 1979;63:336–346. doi: 10.1016/0014-4886(79)90129-8. [DOI] [PubMed] [Google Scholar]
- 29.Nihei K, McKee AC, Kowall NW. Patterns of neuronal degeneration in the motor cortex of amyotrophic lateral sclerosis patients. Acta neuropathologica. 1993;86:55–64. doi: 10.1007/BF00454899. [DOI] [PubMed] [Google Scholar]
- 30.Dickson D, Weller RO. Neurodegeneration: the molecular pathology of dementia and movement disorders. John Wiley & Sons; 2011. [Google Scholar]
- 31.Stephens B, Guiloff RJ, Navarrete R, Newman P, Nikhar N, Lewis P. Widespread loss of neuronal populations in the spinal ventral horn in sporadic motor neuron disease. A morphometric study. Journal of the Neurological Sciences. 2006;244:41–58. doi: 10.1016/j.jns.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Piao YS, Wakabayashi K, Kakita A, et al. Neuropathology with clinical correlations of sporadic amyotrophic lateral sclerosis: 102 autopsy cases examined between 1962 and 2000. Brain pathology. 2003;13:10–22. doi: 10.1111/j.1750-3639.2003.tb00002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomonaga M, Saito M, Yoshimura M, Shimada H, Tohgi H. Ultrastructure of the Bunina bodies in anterior horn cells of amyotrophic lateral sclerosis. Acta neuropathologica. 1978;42:81–86. doi: 10.1007/BF00690971. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda S, Ishizu H, Kawai K, Otsuki S. Bunina bodies in dendrites of patients with amyotrophic lateral sclerosis. Acta medica Okayama. 1990;44:41–45. doi: 10.18926/AMO/30462. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto K, Mizuno Y, Fujita Y. Bunina bodies in amyotrophic lateral sclerosis. Neuropathology. 2008;28:109–115. doi: 10.1111/j.1440-1789.2007.00873.x. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki S, Maruyama S. Immunocytochemical and ultrastructural studies of the motor cortex in amyotrophic lateral sclerosis. Acta neuropathologica. 1994;87:578–585. doi: 10.1007/BF00293318. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto K, Hirai S, Ishiguro K, Kawarabayashi T, Takatama M. Light and electron microscopic and immunohistochemical observations of the Onuf’s nucleus of amyotrophic lateral sclerosis. Acta neuropathologica. 1991;81:610–614. doi: 10.1007/BF00296370. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto K, Hirai S, Amari M, Watanabe M, Sakurai A. Bunina bodies in amyotrophic lateral sclerosis immunostained with rabbit anti-cystatin C serum. Neuroscience letters. 1993;162:125–128. doi: 10.1016/0304-3940(93)90576-7. [DOI] [PubMed] [Google Scholar]
- 39.Mizuno Y, Fujita Y, Takatama M, Okamoto K. Peripherin partially localizes in Bunina bodies in amyotrophic lateral sclerosis. Journal of the Neurological Sciences. 2011;302:14–18. doi: 10.1016/j.jns.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 40.Mizuno Y, Amari M, Takatama M, Aizawa H, Mihara B, Okamoto K. Transferrin localizes in Bunina bodies in amyotrophic lateral sclerosis. Acta neuropathologica. 2006;112:597–603. doi: 10.1007/s00401-006-0122-4. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki S, Komori T, Iwata M. Neuronal inclusions in sporadic motor neuron disease are negative for alpha-synuclein. Neuroscience letters. 2006;397:15–19. doi: 10.1016/j.neulet.2005.11.056. [DOI] [PubMed] [Google Scholar]
- 42.Mizuno Y, Amari M, Takatama M, Aizawa H, Mihara B, Okamoto K. Immunoreactivities of p62, an ubiqutin-binding protein, in the spinal anterior horn cells of patients with amyotrophic lateral sclerosis. Journal of the neurological sciences. 2006;249:13–18. doi: 10.1016/j.jns.2006.05.060. [DOI] [PubMed] [Google Scholar]
- 43.Murayama S, Mori H, Ihara Y, Bouldin TW, Suzuki K, Tomonaga M. Immunocytochemical and ultrastructural studies of lower motor neurons in amyotrophic lateral sclerosis. Annals of neurology. 1990;27:137–148. doi: 10.1002/ana.410270208. [DOI] [PubMed] [Google Scholar]
- 44.Boillée S, Vande Velde C, Cleveland Don W. ALS: A Disease of Motor Neurons and Their Nonneuronal Neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 45.McGeer PL, McGeer EG. Inflammatory processes in amyotrophic lateral sclerosis. Muscle & Nerve. 2002;26:459–470. doi: 10.1002/mus.10191. [DOI] [PubMed] [Google Scholar]
- 46.Boillée S, Yamanaka K, Lobsiger CS, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 47.Yamanaka K, Chun SJ, Boillee S, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nature neuroscience. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiffer D, Fiano V. Astrogliosis in ALS: possible interpretations according to pathogenetic hypotheses. Amyotrophic Lateral Sclerosis. 2004;5:22–25. doi: 10.1080/14660820310016822. [DOI] [PubMed] [Google Scholar]
- 49.Barbeito LH, Pehar M, Cassina P, et al. A role for astrocytes in motor neuron loss in amyotrophic lateral sclerosis. Brain Research Reviews. 2004;47:263–274. doi: 10.1016/j.brainresrev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Lasiene J, Yamanaka K. Glial cells in amyotrophic lateral sclerosis. Neurol Res Int. 2011;2011:718987. doi: 10.1155/2011/718987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henkel J, Beers D, Zhao W, Appel S. Microglia in ALS: The Good, The Bad, and The Resting. J Neuroimmune Pharmacol. 2009;4:389–398. doi: 10.1007/s11481-009-9171-5. [DOI] [PubMed] [Google Scholar]
- 52.Leigh P, Whitwell H, Garofalo O, et al. Ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis morphology, distribution, and specificity. Brain. 1991;114:775–788. doi: 10.1093/brain/114.2.775. [DOI] [PubMed] [Google Scholar]
- 53.Bergmann M, Kuchelmeister K, Schmid K, Kretzschmar H, Schröder R. Different variants of frontotemporal dementia: a neuropathological and immunohistochemical study. Acta neuropathologica. 1996;92:170–179. doi: 10.1007/s004010050505. [DOI] [PubMed] [Google Scholar]
- 54.Ikeda K, Akiyama H, Arai T, Ueno H, Tsuchiya K, Kosaka K. Morphometrical reappraisal of motor neuron system of Pick’s disease and amyotrophic lateral sclerosis with dementia. Acta neuropathologica. 2002;104:21–28. doi: 10.1007/s00401-001-0513-5. [DOI] [PubMed] [Google Scholar]
- 55.Jackson M, Lennox G, Lowe J. Motor neurone disease-inclusion dementia. Neurodegeneration. 1996;5:339–350. doi: 10.1006/neur.1996.0046. [DOI] [PubMed] [Google Scholar]
- 56.Okamoto K, Murakami N, Kusaka H, et al. Ubiquitin-positive intraneuronal inclusions in the extramotor cortices of presenile dementia patients with motor neuron disease. J Neurol. 1992;239:426–430. doi: 10.1007/BF00856806. [DOI] [PubMed] [Google Scholar]
- 57.Okamoto K, Hirai S, Yamazaki T, Sun X, Nakazato Y. New ubiquitin-positive intraneuronal inclusions in the extra-motor cortices in patients with amyotrophic lateral sclerosis. Neuroscience letters. 1991;129:233–236. doi: 10.1016/0304-3940(91)90469-a. [DOI] [PubMed] [Google Scholar]
- 58.Wightman G, Anderson V, Martin J, et al. Hippocampal and neocortical ubiquitin-immunoreactive inclusions in amyotrophic lateral sclerosis with dementia. Neuroscience letters. 1992;139:269–274. doi: 10.1016/0304-3940(92)90569-s. [DOI] [PubMed] [Google Scholar]
- 59.Kawashima T, Kikuchi H, Takita M, et al. Skein-like inclusions in the neostriatum from a case of amyotrophic lateral sclerosis with dementia. Acta neuropathologica. 1998;96:541–545. doi: 10.1007/s004010050932. [DOI] [PubMed] [Google Scholar]
- 60.Arai T, Nonaka T, Hasegawa M, et al. Neuronal and glial inclusions in frontotemporal dementia with or without motor neuron disease are immunopositive for p62. Neuroscience letters. 2003;342:41–44. doi: 10.1016/s0304-3940(03)00216-7. [DOI] [PubMed] [Google Scholar]
- 61.Geser F, Lee VMY, Trojanowski JQ. Amyotrophic lateral sclerosis and frontotemporal lobar degeneration: A spectrum of TDP-43 proteinopathies. Neuropathology. 2010;30:103–112. doi: 10.1111/j.1440-1789.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ayala YM, De Conti L, Avendaño-Vázquez SE, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. The EMBO journal. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Volkening K, Leystra-Lantz C, Yang W, Jaffee H, Strong MJ. Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS) Brain research. 2009;1305:168–182. doi: 10.1016/j.brainres.2009.09.105. [DOI] [PubMed] [Google Scholar]
- 64.Buratti E, De Conti L, Stuani C, Romano M, Baralle M, Baralle F. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS journal. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- 65.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proceedings of the National Academy of Sciences. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Godena VK, Romano G, Romano M, et al. TDP-43 regulates Drosophila neuromuscular junctions growth by modulating Futsch/MAP1B levels and synaptic microtubules organization. PloS one. 2011;6:e17808. doi: 10.1371/journal.pone.0017808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang I, Wu LS, Chang HY, Shen CKJ. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. Journal of neurochemistry. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 68.Buratti E, Baralle FE. Characterization and Functional Implications of the RNA Binding Properties of Nuclear Factor TDP-43, a Novel Splicing Regulator ofCFTR Exon 9. Journal of Biological Chemistry. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 69.Giordana MT, Piccinini M, Grifoni S, et al. TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol. 2010;20:351–360. doi: 10.1111/j.1750-3639.2009.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thorpe JR, Tang H, Atherton J, Cairns NJ. Fine structural analysis of the neuronal inclusions of frontotemporal lobar degeneration with TDP-43 proteinopathy. Journal of neural transmission. 2008;115:1661–1671. doi: 10.1007/s00702-008-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Geser F, O’Dwyer L, Hardiman O, et al. On the development of markers for pathological TDP-43 in amyotrophic lateral sclerosis with and without dementia. Progress in neurobiology. 2011;95:649–662. doi: 10.1016/j.pneurobio.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mackenzie IR, Neumann M, Baborie A, et al. A harmonized classification system for FTLD-TDP pathology. Acta neuropathologica. 2011;122:111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davidson Y, Kelley T, Mackenzie IR, et al. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta neuropathologica. 2007;113:521–533. doi: 10.1007/s00401-006-0189-y. [DOI] [PubMed] [Google Scholar]
- 74.Hasegawa M, Arai T, Nonaka T, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Annals of neurology. 2008;64:60–70. doi: 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Braak H, Ludolph A, Thal DR, Del Tredici K. Amyotrophic lateral sclerosis: dash-like accumulation of phosphorylated TDP-43 in somatodendritic and axonal compartments of somatomotor neurons of the lower brainstem and spinal cord. Acta neuropathologica. 2010;120:67–74. doi: 10.1007/s00401-010-0683-0. [DOI] [PubMed] [Google Scholar]
- 76.Mori F, Tanji K, Zhang H-X, et al. Maturation process of TDP-43-positive neuronal cytoplasmic inclusions in amyotrophic lateral sclerosis with and without dementia. Acta neuropathologica. 2008;116:193–203. doi: 10.1007/s00401-008-0396-9. [DOI] [PubMed] [Google Scholar]
- 77.Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends in neurosciences. 1998;21:516–520. doi: 10.1016/s0166-2236(98)01276-4. [DOI] [PubMed] [Google Scholar]
- 78.Amador-Ortiz C, Lin WL, Ahmed Z, et al. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Annals of neurology. 2007;61:435–445. doi: 10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Higashi S, Iseki E, Yamamoto R, et al. Concurrence of TDP-43, tau and α-synuclein pathology in brains of Alzheimer’s disease and dementia with Lewy bodies. Brain research. 2007;1184:284–294. doi: 10.1016/j.brainres.2007.09.048. [DOI] [PubMed] [Google Scholar]
- 80.Uryu K, Nakashima-Yasuda H, Forman MS, et al. Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. Journal of neuropathology and experimental neurology. 2008;67:555. doi: 10.1097/NEN.0b013e31817713b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakashima-Yasuda H, Uryu K, Robinson J, et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta neuropathologica. 2007;114:221–229. doi: 10.1007/s00401-007-0261-2. [DOI] [PubMed] [Google Scholar]
- 82.Yokota O, Davidson Y, Arai T, et al. Effect of topographical distribution of α-synuclein pathology on TDP-43 accumulation in Lewy body disease. Acta neuropathologica. 2010;120:789–801. doi: 10.1007/s00401-010-0731-9. [DOI] [PubMed] [Google Scholar]
- 83.Hasegawa M, Arai T, Akiyama H, et al. TDP-43 is deposited in the Guam parkinsonism–dementia complex brains. Brain. 2007;130:1386–1394. doi: 10.1093/brain/awm065. [DOI] [PubMed] [Google Scholar]
- 84.McKee AC, Gavett BE, Stern RA, et al. TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. J Neuropathol Exp Neurol. 2010;69:918–929. doi: 10.1097/NEN.0b013e3181ee7d85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geser F, Robinson JL, Malunda JA, et al. Pathological 43-kDa transactivation response DNA-binding protein in older adults with and without severe mental illness. Archives of neurology. 2010;67:1238–1250. doi: 10.1001/archneurol.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sathasivam S, Ince P, Shaw P. Apoptosis in amyotrophic lateral sclerosis: a review of the evidence. Neuropathology and applied neurobiology. 2001;27:257–274. doi: 10.1046/j.0305-1846.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 87.Martin LJ. Neuronal death in amyotrophic lateral sclerosis is apoptosis: possible contribution of a programmed cell death mechanism. Journal of Neuropathology & Experimental Neurology. 1999;58:459–471. doi: 10.1097/00005072-199905000-00005. [DOI] [PubMed] [Google Scholar]
- 88.Brettschneider J, Del Tredici K, Irwin DJ, et al. Sequential distribution of pTDP-43 pathology in behavioral variant frontotemporal dementia (bvFTD) Acta neuropathologica. 2014;127:423–439. doi: 10.1007/s00401-013-1238-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ravits J, Laurie P, Fan Y, Moore DH. Implications of ALS focality Rostral–caudal distribution of lower motor neuron loss postmortem. Neurology. 2007;68:1576–1582. doi: 10.1212/01.wnl.0000261045.57095.56. [DOI] [PubMed] [Google Scholar]
- 90.Re DB, Le Verche V, Yu C, et al. Necroptosis drives motor neuron death in models of both sporadic and familial ALS. Neuron. 2014;81:1001–1008. doi: 10.1016/j.neuron.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Braak H, Brettschneider J, Ludolph AC, Lee VM, Trojanowski JQ, Del Tredici K. Amyotrophic lateral sclerosis—a model of corticofugal axonal spread. Nature Reviews Neurology. 2013;9:708–714. doi: 10.1038/nrneurol.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brettschneider J, Del Tredici K, Toledo JB, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Annals of neurology. 2013;74:20–38. doi: 10.1002/ana.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nature neuroscience. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ince PG, Tomkins J, Slade JY, Thatcher NM, Shaw PJ. Amyotrophic lateral sclerosis associated with genetic abnormalities in the gene encoding Cu/Zn superoxide dismutase: molecular pathology of five new cases, and comparison with previous reports and 73 sporadic cases of ALS. Journal of Neuropathology & Experimental Neurology. 1998;57:895–904. doi: 10.1097/00005072-199810000-00002. [DOI] [PubMed] [Google Scholar]
- 95.Mackenzie IRA, Bigio EH, Ince PG, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Annals of Neurology. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 96.Mizusawa H, Matsumoto S, Yen S-H, Hirano A, Rojas-Corona R, Donnenfeld H. Focal accumulation of phosphorylated neurofilaments within anterior horn cell in familial amyotrophic lateral sclerosis. Acta neuropathologica. 1989;79:37–43. doi: 10.1007/BF00308955. [DOI] [PubMed] [Google Scholar]
- 97.Okamoto Y, Shirakashi Y, Ihara M, et al. Colocalization of 14-3-3 proteins with SOD1 in Lewy body-like hyaline inclusions in familial amyotrophic lateral sclerosis cases and the animal model. PloS one. 2011;6:e20427. doi: 10.1371/journal.pone.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakamura S, Wate R, Kaneko S, et al. An autopsy case of sporadic amyotrophic lateral sclerosis associated with the I113T SOD1 mutation. Neuropathology. 2014;34:58–63. doi: 10.1111/neup.12049. [DOI] [PubMed] [Google Scholar]
- 99.Bosco DA, Morfini G, Karabacak NM, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nature neuroscience. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Forsberg K, Jonsson PA, Andersen PM, et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PloS one. 2010;5:e11552. doi: 10.1371/journal.pone.0011552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Forsberg K, Andersen PM, Marklund SL, Brännström T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta neuropathologica. 2011;121:623–634. doi: 10.1007/s00401-011-0805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu HN, Sanelli T, Horne P, et al. Lack of evidence of monomer/misfolded superoxide dismutase-1 in sporadic amyotrophic lateral sclerosis. Annals of neurology. 2009;66:75–80. doi: 10.1002/ana.21704. [DOI] [PubMed] [Google Scholar]
- 103.Kerman A, Liu H-N, Croul S, et al. Amyotrophic lateral sclerosis is a non-amyloid disease in which extensive misfolding of SOD1 is unique to the familial form. Acta neuropathologica. 2010;119:335–344. doi: 10.1007/s00401-010-0646-5. [DOI] [PubMed] [Google Scholar]
- 104.Brotherton TE, Li Y, Cooper D, et al. Localization of a toxic form of superoxide dismutase 1 protein to pathologically affected tissues in familial ALS. Proceedings of the National Academy of Sciences. 2012;109:5505–5510. doi: 10.1073/pnas.1115009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ince PG, Highley JR, Kirby J, et al. Molecular pathology and genetic advances in amyotrophic lateral sclerosis: an emerging molecular pathway and the significance of glial pathology. Acta neuropathologica. 2011;122:657–671. doi: 10.1007/s00401-011-0913-0. [DOI] [PubMed] [Google Scholar]
- 106.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yokoseki A, Shiga A, Tan C-F, et al. TDP-43 mutation in familial amyotrophic lateral sclerosis. Annals of Neurology. 2008;63:538–542. doi: 10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]
- 108.Pesiridis GS, Lee VM-Y, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Human Molecular Genetics. 2009;18:R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Verbeeck C, Deng Q, Dejesus-Hernandez M, et al. Expression of Fused in sarcoma mutations in mice recapitulates the neuropathology of FUS proteinopathies and provides insight into disease pathogenesis. Molecular neurodegeneration. 2012;7:53. doi: 10.1186/1750-1326-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takanashi K, Yamaguchi A. Aggregation of ALS-linked FUS mutant sequesters RNA binding proteins and impairs RNA granules formation. Biochemical and biophysical research communications. 2014;452:600–607. doi: 10.1016/j.bbrc.2014.08.115. [DOI] [PubMed] [Google Scholar]
- 112.Vance C, Scotter EL, Nishimura AL, et al. ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Human molecular genetics. 2013;22:2676–2688. doi: 10.1093/hmg/ddt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deng HX, Zhai H, Bigio EH, et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Annals of neurology. 2010;67:739–748. doi: 10.1002/ana.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mackenzie IR, Ansorge O, Strong M, et al. Pathological heterogeneity in amyotrophic lateral sclerosis with FUS mutations: two distinct patterns correlating with disease severity and mutation. Acta neuropathologica. 2011;122:87–98. doi: 10.1007/s00401-011-0838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mackenzie IR, Neumann M, Bigio EH, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta neuropathologica. 2010;119:1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Snowden JS, Hu Q, Rollinson S, et al. The most common type of FTLD-FUS (aFTLD-U) is associated with a distinct clinical form of frontotemporal dementia but is not related to mutations in the FUS gene. Acta neuropathologica. 2011;122:99–110. doi: 10.1007/s00401-011-0816-0. [DOI] [PubMed] [Google Scholar]
- 117.Pikkarainen M, Hartikainen P, Alafuzoff I. Neuropathologic features of frontotemporal lobar degeneration with ubiquitin-positive inclusions visualized with ubiquitin-binding protein p62 immunohistochemistry. Journal of Neuropathology & Experimental Neurology. 2008;67:280–298. doi: 10.1097/NEN.0b013e31816a1da2. [DOI] [PubMed] [Google Scholar]
- 118.King A, Al-Sarraj S, Shaw C. Frontotemporal lobar degeneration with ubiquitinated tau-negative inclusions and additional α-synuclein pathology but also unusual cerebellar ubiquitinated p62-positive, TDP-43-negative inclusions. Neuropathology. 2009;29:466–471. doi: 10.1111/j.1440-1789.2008.00966.x. [DOI] [PubMed] [Google Scholar]
- 119.Bullock TL, Clarkson WD, Kent HM, Stewart M. The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2) J Mol Biol. 1996;260:422–431. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- 120.Zu T, Liu Y, Bañez-Coronel M, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proceedings of the National Academy of Sciences. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Human molecular genetics. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mackenzie IR, Frick P, Neumann M. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta neuropathologica. 2014;127:347–357. doi: 10.1007/s00401-013-1232-4. [DOI] [PubMed] [Google Scholar]
- 123.Gendron TF, Bieniek KF, Zhang YJ, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta neuropathologica. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mori K, Arzberger T, Grasser FA, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta neuropathologica. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 125.Lagier-Tourenne C, Baughn M, Rigo F, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proceedings of the National Academy of Sciences. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mizielinska S, Lashley T, Norona FE, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta neuropathologica. 2013;126:845–857. doi: 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cooper-Knock J, Higginbottom A, Stopford MJ, et al. Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta neuropathologica. 2015:1–13. doi: 10.1007/s00401-015-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mori K, Weng S-M, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 129.Al-Sarraj S, King A, Troakes C, et al. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta neuropathologica. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 130.Boxer AL, Mackenzie IR, Boeve BF, et al. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. Journal of Neurology, Neurosurgery & Psychiatry. 2011;82:196–203. doi: 10.1136/jnnp.2009.204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mackenzie IR, Frick P, Neumann M. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta neuropathologica. 2014;127:347–357. doi: 10.1007/s00401-013-1232-4. [DOI] [PubMed] [Google Scholar]
- 132.Ravits J. Focality, stochasticity and neuroanatomic propagation in ALS pathogenesis. Exp Neurol. 2014;262(Pt B):121–126. doi: 10.1016/j.expneurol.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 133.Kosaka T, Fu YJ, Shiga A, et al. Primary lateral sclerosis: upper-motor-predominant amyotrophic lateral sclerosis with frontotemporal lobar degeneration--immunohistochemical and biochemical analyses of TDP-43. Neuropathology. 2012;32:373–384. doi: 10.1111/j.1440-1789.2011.01271.x. [DOI] [PubMed] [Google Scholar]
- 134.Pringle CE, Hudson AJ, Munoz DG, Kiernan JA, Brown WF, Ebers GC. PRIMARY LATERAL SCLEROSIS: CLINICAL FEATURES, NEUROPATHOLOGY AND DIAGNOSTIC CRITERIA. Brain. 1992;115:495–520. doi: 10.1093/brain/115.2.495. [DOI] [PubMed] [Google Scholar]
- 135.Kolind S, Sharma R, Knight S, Johansen-Berg H, Talbot K, Turner MR. Myelin imaging in amyotrophic and primary lateral sclerosis. Amyotrophic Lateral Sclerosis & Frontotemporal Degeneration. 2013;14:562–573. doi: 10.3109/21678421.2013.794843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kosaka T, Fu YJ, Shiga A, et al. Primary lateral sclerosis: Upper-motor-predominant amyotrophic lateral sclerosis with frontotemporal lobar degeneration–immunohistochemical and biochemical analyses of TDP-43. Neuropathology. 2012;32:373–384. doi: 10.1111/j.1440-1789.2011.01271.x. [DOI] [PubMed] [Google Scholar]
- 137.Dickson DW, Josephs KA, Amador-Ortiz C. TDP-43 in differential diagnosis of motor neuron disorders. Acta neuropathologica. 2007;114:71–79. doi: 10.1007/s00401-007-0234-5. [DOI] [PubMed] [Google Scholar]
- 138.SWANK RL, Putnam TJ. Amyotrophic lateral sclerosis and related conditions: a clinical analysis. Archives of Neurology & Psychiatry. 1943;49:151–177. [Google Scholar]
- 139.van Blitterswijk M, Vlam L, van Es MA, et al. Genetic Overlap between Apparently Sporadic Motor Neuron Diseases. PLoS ONE. 2012;7:e48983. doi: 10.1371/journal.pone.0048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ince P, Evans J, Knopp M, et al. Corticospinal tract degeneration in the progressive muscular atrophy variant of ALS. Neurology. 2003;60:1252–1258. doi: 10.1212/01.wnl.0000058901.75728.4e. [DOI] [PubMed] [Google Scholar]
- 141.Geser F, Stein B, Partain M, et al. Motor neuron disease clinically limited to the lower motor neuron is a diffuse TDP-43 proteinopathy. Acta Neuropathol. 2011;121:509–517. doi: 10.1007/s00401-011-0797-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Riku Y, Atsuta N, Yoshida M, et al. Differential motor neuron involvement in progressive muscular atrophy: a comparative study with amyotrophic lateral sclerosis. BMJ Open. 2014;4 doi: 10.1136/bmjopen-2014-005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 144.Lomen-Hoerth C, Murphy J, Langmore S, Kramer J, Olney R, Miller B. Are amyotrophic lateral sclerosis patients cognitively normal? Neurology. 2003;60:1094–1097. doi: 10.1212/01.wnl.0000055861.95202.8d. [DOI] [PubMed] [Google Scholar]
- 145.Lunn MR, Wang CH. Spinal muscular atrophy. The Lancet. 371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 146.Alías L, Bernal S, Fuentes-Prior P, et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Human genetics. 2009;125:29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- 147.Zhang Z, Pinto AM, Wan L, et al. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19348–19353. doi: 10.1073/pnas.1319280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Araki S, Hayashi M, Tamagawa K, et al. Neuropathological analysis in spinal muscular atrophy type II. Acta Neuropathologica. 2003;106:441–448. doi: 10.1007/s00401-003-0743-9. [DOI] [PubMed] [Google Scholar]
- 149.Turner BJ1, Bäumer D, Parkinson NJ, Scaber J, Ansorge O, Talbot K. BMC: TDP-43 expression in mouse models of amyotrophic lateral sclerosis and spinal muscular atrophy. Neurosci. 2008 Oct 28;9:104. doi: 10.1186/1471-2202-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Behan WM, Maia M. Strümpell’s familial spastic paraplegia: genetics and neuropathology. Journal of Neurology, Neurosurgery & Psychiatry. 1974;37:8–20. doi: 10.1136/jnnp.37.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Harding A. Hereditary spastic paraplegias. Seminars in neurology. 1993:333. doi: 10.1055/s-2008-1041143. [DOI] [PubMed] [Google Scholar]
- 152.Martinez-Lage Maria, et al. TDP-43 Pathology in a Case of Hereditary Spastic Paraplegia with a NIPA1/SPG6 Mutation. Acta Neuropathol. 2012 Aug;124(2):285–291. doi: 10.1007/s00401-012-0947-y. [DOI] [PMC free article] [PubMed] [Google Scholar]