Abstract
Progress in defining the genetics of autoimmune disease has been dramatically enhanced by large scale genetic studies. Genome-wide approaches examining hundreds or for some diseases thousands of cases and controls have been implemented using high throughput genotyping and appropriate algorithms to provide a wealth of data over the last decade. These studies have identified hundreds of non-HLA loci as well as further defining HLA region variations that predispose to different autoimmune diseases. These studies to identify genetic risk loci are also complemented by progress in gene expression studies including definition of expression quantitative trait loci (eQTL), various alterations in chromatin structure including histone marks, DNase I sensitivity, repressed chromatin regions as well as transcript factor binding sites. Integration of this information can partially explain why particular variations can alter proclivity to autoimmune phenotypes. Despite our incomplete knowledge base with only partial definition of hereditary factors and possible functional connections, this progress has and will continue to facilitate a better understanding of critical pathways and critical changes in immunoregulation. Advances in defining and understanding functional variants potentially can lead to both novel therapeutics and personalized medicine in which therapeutic approaches are chosen based on particular molecular phenotypes and genomic alterations.
Keywords: Genetics, Autoimmune Diseases, HLA, Susceptibility Loci, Functional Variants, Immunoregulation
1. Introduction
Our understanding of which genes predispose to different autoimmune diseases has expanded rapidly over the last decade. This progress has been mostly due to genome-wide association studies (GWAS) and the development of various technical and analytic tools. However, despite this progress less than half of the heritability of most autoimmune diseases can be explained and nearly half of this identified genetic risk is due to variations within HLA. The actual functional variants that underlie statistically significant associations are with some notable exceptions are still largely unknown. In the following perspective, I will review some of the more salient advances in the field, provide examples to illustrate specific points, indicate where knowledge is sparse, and discuss the potential for future advances that I believe could further define the pathogenesis and perhaps enable application to diagnoses and therapy. More detailed aspects of the genetics for a variety of autoimmune diseases in presented by experts in the field in other sections of this special issue of the journal. A general paradigm for GWAS and sequence variant studies is shown in Figures 1 and discussed in subsequent sections.
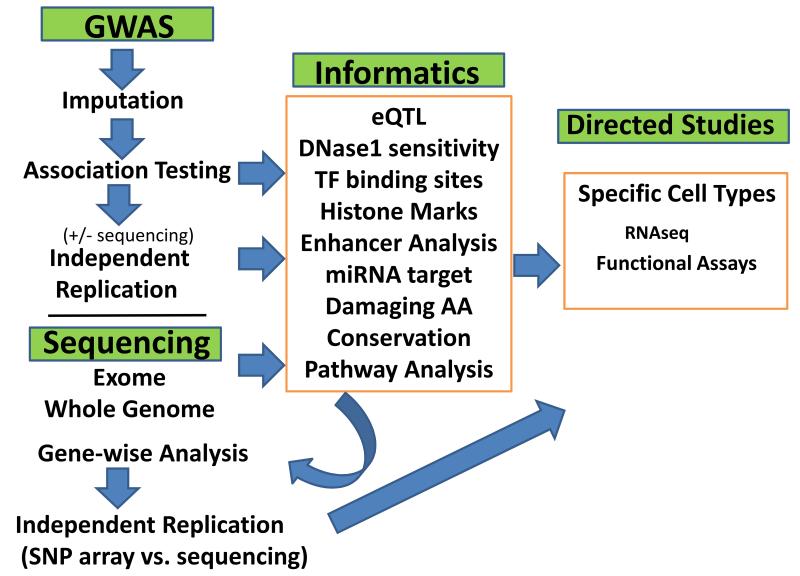
Figure 1.
Diagram of general scheme for genetic studies of complex autoimmune diseases. GWAS studies can greatly benefit from imputation and replication studies for loci discovered in the discovery phase. For some studies replication studies are limited to those loci (genes) that are also part of pathways for genes identified as significantly associated with the disease in previous studies. This is also proposed for sequencing studies to identify less common variants in which power issues may be partially addressed by limiting replication analyses based on pathway information.
1.1 Heritability
Epidemiological studies of most autoimmune diseases including rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), type 1 diabetes (T1D), multiple sclerosis (MS), and primary biliary cirrhosis (PBC) show that there is strong heritability. These include studies showing increased concordance in monozygotic compared to dizygotic twin as well as studies showing increased risk to siblings of proband cases compared to the general population. Although most of these studies are not truly population based and may have biased results, there are some caveats worth noting. First, some autoimmune diseases have much higher sibling relative risk rates than other diseases (e.g. SLE [1, 2], T1D [3], celiac disease [4], and PBC [5, 6] compared to others e.g. RA [2], and MS [7]). Second, although concordance of disease is much higher in monozygotic compared with dizygotic twins for many autoimmune diseases, the overall monozygotic concordance of disease is usually substantially less than 50% [8]. This indicates that stochastic factors including environmental variables are a strong component and although genetics can be very useful in identifying important factors in etiopathogenesis it can only partially predict phenotype. Although some specific environmental factors have been identified (e.g. smoking and rheumatoid arthritis [9, 10]) it is also possible that most of the incomplete concordance is simply chance or indefinable events.
1.2 General Considerations for Identifying Genetic Loci for Autoimmune Disease Susceptibility
The major advance in identifying genetic loci that predispose to autoimmune diseases has been GWAS. Although some non-major histocompatibility (HLA in humans) loci were identified prior to GWAS using linkage or candidate gene studies, and other methodologies including admixture mapping have also enabled identification of a modicum of risk loci, the exponential increase in loci (over 200 for some autoimmune diseases) has been the direct result of GWAS. The basis of GWAS is the technology enabling efficient and accurate genotyping of single base polymorphisms (SNPs) and large collaborative studies such as HapMap [11, 12] defining large numbers (hundreds of thousands) of SNPs in different populations. The success of GWAS is in large part due to practical advantage in conducting case/control design, namely the ability to recruit large numbers of cases and population controls as opposed to the difficulty in recruiting families: power for any association study is largely based on numbers. A critical aspect for these studies has been the ability to adequately control for population substructure differences using statistical methodology. Most commonly this is done by logistic regression using relevant principal components defined by principal component analyses or similar methods [13-15]. In some studies only continental population differences are accounted for, but for the most part type 2 errors (false positives) due to unrecognized stratification differences in case and control populations have been minimized. In fact, many studies have used publically available control genotypes rather than specific matched collections of controls. It is also worth noting that it may be possible to increase power (decrease Type 1 errors, false negatives) to ascertain risk variants by limiting studies to more homogenous populations and additional considerations of population substructure is discussed in subsequent sections (see sections 3.3 and 4). However, GWAS is largely applicable to those loci that fulfill the common variant (>0.05 minor allele frequency) common disease hypothesis since this methodology relies on linkage disequilbrium (LD) between the marker (SNP) detected and the actual disease causing variant(s). The commonly used genotyping platforms (Illumina and Affymetrix chip arrays) and clustering algorithms are also most accurate for minor allele frequencies (MAFs) >0.05, although there have been some improvements in software to enable higher accuracy genotyping.
Another important aspect of current GWAS and other association studies is the ability to accurately impute much of the sequence difference that is not directly genotyped using current genotyping platforms. Imputation is based on sophisticated algorithms that enable under certain conditions the accurate estimation of the sequence variants between genotyped markers based on reference genomes that contain the missing information [16-19]. It relies on the shared loci (almost all of the SNPs in commonly used genotyping platforms are shared with the reference genomes) and the patterns of variation (http://www.1000genomes.org). With the completion of phase 3 of the 1000 genome sequencing study there is now sequence data for multiple populations that can be used to inform imputation(http://www.1000genomes.org/) [20]. This information can suggest candidate non-synonymous damaging variants and in others provides candidate non-coding variants that might be suggestive based on expression quantitative traits, binding sites for transcription factors, DNase sensitiviy or other features (e.g. histone marks) that have been elucidated in particular tissues or cell lines (discussed further in Section 6). Importantly, imputation methods can also be used to determine HLA classical determinants and amino acids.
1.3 Phenotype
A critical aspect for any association study is defining the phenotype of cases. Theoretically, the better the definition of phenotype the greater the likelihood that type 1 errors will be minimized since inclusion of unaffected individuals or those with a different autoimmune disease may increase heterogeneity and dilute the risk of variants for a particular phenotype. However, there may be a practical trade-off between including cases that may not have all the information (e.g. particular autoantibodies or even age of onset) that could increase the power to detect loci due to greater sample size as compared to more rigid criteria. For some autoimmune diseases particular features clearly increase the relative risk of many of the susceptibility loci. A strong example is for SLE where cases with anti-double strand (DS) DNA show substantially higher odds ratios for several lupus susceptibility alleles than those that are anti-DS DNA negative [21]. Similarly, some RA studies show stronger associations when the cases are limited to those with anti-cyclic citrullinated peptide antibodies (anti-CCP) [22]. However, in other studies when cases are limited to anti-CCP positive RA there are fewer susceptibility loci associated with disease, probably mostly due to decreased power as a function of a smaller sample size [23]. Similarly, if for example an SLE study was restricted to only those that manifested the same 4 out of 11 American College of Rheumatology criteria it is likely that many of the identified loci would have been missed due to weaker signals from decreased sample sizes.
The decision on phenotypic definition is analogous to the classic fight between the lumpers and the splitters in the definition of mammalian species [24] and was advanced many years ago with respect to the nosology of genetic disease [25]. However, for some autoimmune diseases the definition of “subtypes” may be crucial. In collaborative work by this author, the study of myasthenia gravis, it was critical to divide the disease by age-of-onset and the lack of thymomas. Here, there are major differences in which HLA genes are important in susceptibility when early onset (<45 years of age) compared to late onset (>50 years of age) are compared ([26, 27] and Seldin, Gregersen, Hammarstrom, unpublished data). In addition, the highest risk non-HLA gene, TNIP1, observed in early onset myasthenia gravis (EOMG) [26] has no effect in late onset myasthenia gravis (LOMG) (Seldin, Gregersen, Hammarstrom, unpublished data).
As the field progresses there are likely to be more studies that examine endophenotypes including the most prominent disease manifestations, disease severity and morbidity. In SLE there is a study suggesting specific loci that predispose to nephropathy [28]. Such studies could also include those individuals that have very poor outcomes or more severe phenotypes.
2. The Major Histocompatibility Complex Region
With rare exception, genes in the human major histocompatibility complex (MHC), HLA, are the strongest risk genes for each autoimmune disease. Several general points are notable: 1) often there is more than one association signal from the HLA region with the second and even third signals sometimes stronger than any of the individual signals from loci outside this region; 2) recent studies suggest that the most of the strongest signals are from classical HLA determinants (not from single SNPs); 3) in some studies epistasis between HLA determinants has been shown or suggested and 4) different HLA specificities are associated with different autoimmune diseases.
For most autoimmune diseases where pathology is characterized/driven by autoantibodies the strongest associations are with MHC class II genes (HLA D region specificities) (e.g. SLE, RA, T1D, and celiac disease [29-31]). For those that are not characterized by autoantibodies (e.g. ankylosing spondylitis, psoriasis, Behcet’s) most are strongly associated with class I genes (HLA-A, HLA-B and HLA-C) [32-34]. However, this is not always the case. The major exception for autoantibody associated disease is the early onset form of myasthenia gravis (EOMG) a disease characterized by anti-acetylcholinesterase receptor antibodies. Here the association is clearly much stronger with the class I HLA-B8 than the DRB1 determinant (DRB1*03) that is in linkage disequilbrium with HLA-B8. For diseases not characterized by autoantibodies, converse examples are the finding that a class II gene (DRB1*11) has been strongly associated with ulcerative colitis and Crohn’s disease [29]. Recently specific amino acid residues in DQA1 were found to be strongly associated with idiopathic achalasia, an enigmatic disease that is now thought to be an autoimmune process [35]. In this context, it may be useful to note that there is evidence for potential role for CD8 cells (that recognize antigen in the context of class I) in other processes than cell mediated cytotoxicity: CD8 cells producing IFN-γ are critical to inducing class switching in B cells or helper function in particular circumstances [36, 37].
Although the specific associations are diverse, overall, these studies are highly suggestive that specific classical determinants or specific amino acids are associated primarily with specific diseases. Thus, the predominant association for RA have the “shared epitope” in which DRB1 determinants that share particular amino acid residues in the third hypervariable region explain the strongest component of the HLA association with this disease [38] but the same amino acids are not very strongly associated with most other autoimmune diseases. In contrast to the strong RA association with the shared epitope (mostly particular DRB1*04 determinants), the strongest MS association is with DRB1*1501, SLE with DRB1*0301, and DRB1*08 with PBC [29, 39, 40] . These associations are often stronger than specific MHC SNPs associated in GWAS and studies using conditional analyses have provided additional support for these specific genes and amino acid polymorphisms for some of these diseases. The overall genetics of HLA, even for the predominant HLA association is much more complicated at least for some autoimmune diseases including T1D. Extensive studies of T1D associated haplotypes show data consistent with a complex relationships among DRB1, DQA1 and DQB1alleles [30]. Here specific combinations show very strong susceptibility or protective effects depending both on haplotypic combinations and particular determinants can in some cases be part of both (conferring increased odds ratios in some contexts but decreased in others).
It is further notable that the D region amino acids residues that have been implicated (e.g. specific amino acids in DRB1 in RA and ulcerative colitis, and DQA1 and achalasia) are critical for either antigen binding or peptide exchange chaperone HLA-DM in the context of antigen binding [35, 41, 42]. These findings further suggest that unknown and elusive antigens are driving these autoimmune diseases, similar to that involved in the pathogenesis of celiac disease where specific DQ amino acids participate in binding gluten peptides [43] or microbes in the seronegative spondyloarthropathies such as ankylosing spondylitis and Reiter’s syndrome [44-46]. Other postulates on the functional relationship of HLA class II genes have been advanced including molecular mimicry and in some studies have supported an alternative postulate that in RA the shared epitope amino acids bind and trigger signal transduction via cell surface calreticulin (CRT) an innate immune receptor [47].
As mentioned above, multiple different MHC associations can be observed in many autoimmune diseases including many secondary loci. These include both class 1 and DPB1 determinants [40] as well as class III for some autoimmune diseases [30]. For RA, a study has attributed all MHC risk to 5 different amino acids including 3 DRB1 amino acids, and one HLA-B amino acid and one DPB1 amino acid [42]. It is also worth noting that the effect-size of the MHC associations varies greatly. For EOMG (HLA-B8), celiac disease (DRB1*03) and ankylosing spondylitis (HLA-B28) and certain T1D haplotypes the odds ratio are >5. For most other autoimmune diseases the odds ratio is ~2.0. A GRR of > 1.6 is rare for non-MHC region associations. Thus, the especially strong associations of HLA determinants suggest that recognition of particular antigens is the likely driver of the initiating event(s) in autoimmune disease.
3. Non-HLA Susceptibility Loci
Several hundred non-HLA loci have been identified that contribute to the association of one or more autoimmune diseases. Most are relatively high frequency perhaps as a reflection of the sensitivity of the methods to detect these loci (i.e. GWAS as discussed above). The vast majority of these loci are located either within or close to genes that participate in immune system response. These include genes that are components of antigen processing, presentation, recognition, differentiation, signaling and/or some form of immune-regulation including particular cytokines, interleukins or interferons. The genes are often primarily expressed in T cells, B cells or dendritic cells. Most have relatively low odds ratios (<1.2) although some have GRR’s >1.5. Most of the SNP variants that show association are non-coding SNPs and many are located between genes and a few in genomic regions that are devoid of genes in what is sometimes termed as gene deserts. A few such as the PTPN22 association are nonsynonymous amino acid differences (e.g. PTPN22 R620W) with functional effects [48-53]. Fine mapping, sequencing, expression studies and a modicum of functional tests have in some cases identified strong candidate variants and potential mechanisms but these are the exception. For some, even with strong associations, it is not clear whether the closest gene is necessarily the gene involved in the functional variation. A possible example of this is the association of SLE (and several other autoimmune diseases including RA) with a number of SNPs within the STAT4 gene [54]. The strongest associated SNPs are all located in an intronic region of STAT4 and thus far studies have not linked these SNPs to any function. Since STAT1 is located only ~10 kb downstream from these SNPs it is certainly possible that the association is due to some regulatory function with respect to STAT1 rather than STAT4. This is a reasonable hypothesis even though the association is much weaker with STAT1, since recombination distance (not directly related to physical distance) does not obviate a possible functional link which, in this example, can still be a cis regulatory mechanism. Although Occam’s razor favors that most of the associations close to or within a reasonable candidate gene are due to a functional connection to this gene, it is possible that some of the identified variants regulate genes in disparate locations (i.e. trans-regulation could account for some of the associations).
For some genes multiple haplotypes are associated with disease including specific TNFAIP3 and TNIP1 haplotypes with SLE [55, 56]. It is also possible that many associations with particular loci actually reflect multiple different functional lower frequency variants that are on the same haplotype. However, efforts to find such rare or uncommon variants within the high frequency haplotypes have thus far not proven to be generally successful and my personal viewpoint is that most of these associations are actually due to common variants, albeit not necessarily the sentinel variant identified in a GWAS study. Although fine mapping and sequencing of genomic regions is sometimes undertaken to further define the genomic variation that explain the association of common variants, much/most of the information on this variation can now probably be obtained using the imputation methods described above. The potential value of sequencing efforts or dense genotyping is further discussed in Section 3.3 and may yet be important for ascertaining rare variants that may also underlie susceptibility to autoimmune diseases.
3.1. Overlap and Differences in non-HLA Genetics of Different Autoimmune Diseases
(For this section, I will confine the discussion to studies in European populations since there are substantial differences among different ancestries that will be discussed in a subsequent section.) Although some non-MHC loci appear unique or relatively unique to specific autoimmune diseases such as the SpB transcription factor for PBC [57], in general there are many non-MHC loci that are shared between different autoimmune diseases [54][58]. Although many of the loci overlap there may be substantial differences in effect size. For example among the genetic loci implicated for both SLE and RA are associations with STAT4 and PTPN22. However, the odds ratio is substantially higher for STAT4 in SLE than in RA [54] and the opposite appears to be the case for PTPN22 [59, 60]. PTPN22 is also a shared susceptibility gene for many autoimmune diseases. This includes a high odds ratio (>2) in T1D [48], and lower odds ratios although still over 1.5 in several other diseases including EOMG [26] but only a minimal effect or no effect in PBC, celiac disease and MS. Interestingly, although the implicated (and in this case a nonsynonymous amino acid variant) also appears to be associated with Crohn’s disease for this disease it has a protective rather than disease promoting effect [61]. There are multiple other examples of differences in apparently shared genetic loci. For EOMG the highest odds ratio for non-HLA genes is observed for TNIP1 and although this gene is also implicated in SLE the odds ratio is much smaller and there are apparently two haplotypes (rather than one in EOMG) that are implicated [26, 56]. IRF5 has a high odds ratio in SLE but moderate odds ratio in PBC [57, 62, 63] and a low odds ratio RA [58]. ITGAM is another strong (high odds ratio) SLE susceptibility locus that is not associated with RA or other autoimmune diseases (including MS, T1D, and celiac disease) except systemic sclerosis where it has only a modest effect size [64, 65].
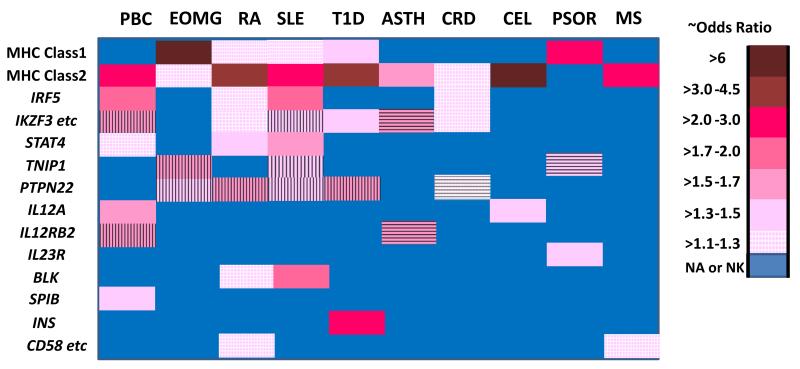
Often for a given autoimmune diseases when sample sizes are increased additional loci are identified that are shared with other autoimmune diseases but many of these have very marginal effect sizes and odds ratios <1.1. Some examples of differences for gene associated loci with relatively high odds ratios in at least one autoimmune disease but show different effects with other autoimmune diseases is depicted in Figure 2. Thus, for non-HLA variants there appears to be a complex pattern of gene effect sizes that is not readily discernible. Why are the odds ratios of these shared susceptibility genes so different between the different autoimmune diseases? Although some differences are due to variability/noise that can plague studies with relatively low sample sizes, it appears that many/most of these differences are real. Going forward it is important to determine how these differences relate to the interplay between the genetic risk factors and how these differential effects can alter the predisposition to different autoimmune diseases.
Figure 2.
Comparison of effect size for gene associated SNPs in multiple different autoimmune diseases. Disease abbreviations that are not clear from the main text include: asthma (ASTH), Crohn’s disease (CRD), celiac disease (CEL) and psoriasis (PSOR). Asthma is included for comparison of autoimmune conditions with another immunologically mediated disease. The approximate odds ratios (ORs) are indicated by the color code legend and for each the OR is indicated in the positive direction (regardless of minor allele frequency). Horizontal or vertical lines are shown over the color coded ORs when the opposite SNP/haplotype is associated with different diseases (e.g. for the IKZF3 etc. in which the asthma association is clearly the opposite of those for PBC and SLE. The ORs are derived from studies of European ancestry subject sets and are preferentially chosen from larger studies or meta-analyses [26, 48, 54, 56, 57, 59-61, 94, 142, 148, 163-173]. For some of the genes for which the legend key indicates no association or not known (NA or NK) there may be limited data that suggests possible association that does not meet the standard criterion (p < 5 ×10−8). Where different SNPs show disparate ORs the association the highest OR is shown. The gene list was selected based on inclusion of most genes with at least moderate ORs (OR> 1.1) in at least one of the autoimmune diseases and is in part biased to emphasize the differences in effect sizes. IKZF3 etc represents a gene cluster that includes IKZF3, ORMDL3, ZPBP2, and GSDMB. CD58 etc. represents a gene cluster including CD58, CD2, and IGSF2.
3.2. Epistasis between non-HLA variants and HLA
There are very few examples of epistasis between different susceptibility variants in any autoimmune disease. Perhaps the strongest examples are those between the non-HLA susceptibility gene ERAP1 and HLA in three diseases: ankylosing spondylitis (HLA-B27) [66]; psoriasis (HLA-C6) [67], and Behcet’s disease (HLA-B51) [68]. ERAP1 codes an aminopeptidase that functions to trim peptides for loading onto MHC class I determinants [69]. For these diseases the ERAP1 variants are only important in the context of these particular HLA determinants and ERAP1 variants have little or no effect in those patients that do not have these HLA determinants. Interestingly, while the same ERAP1 variants show the same effect in ankylosing spondylitis and psoriasis, the opposite effect in seen in Behcet’s disease (c ) suggesting that different requirements for the ERAP1 endoplasmic reticulum processing of the specific protein antigens that are part of triggering the immune responses that are critical to pathogenesis of these diseases.
3.3. Rare/Uncommon variants
Only a small number of rare (minor allele frequency <0.01%) or uncommon (<0.05%) variants have been identified for common complex autoimmune diseases. Some of those identified for common complex autoimmune disease have been based on examining candidate genes that were known to be causal for rare Mendelian diseases with features of autoimmunity. This includes TREX1 that causes autoimmune polyglandular syndrome type 1 [70]. For this gene, rare variants with high effect size has been identified in studies of SLE [71, 72]. A modicum of other rare variants have been identified for autoimmune diseases based on either inclusion in the IMMUNOCHIP array (dense SNP variants for regions based mostly on genes identified by GWAS) including several for celiac disease [73]. However, in general these studies have been hampered by limited database of known variants (early versions of 1000 genome and other sequence information) and it is possible that more success will be realized in the near future for autoimmune diseases. Recent success identifying rare variants using exon chips has been reported for type 2 diabetes, a common non-autoimmune disease [74, 75] and there are multiple ongoing studies using exon chips to try to uncover rare/uncommon variants in autoimmune diseases. A limited number of studies have also examined specific immune system genes by sequencing and some larger studies have used pooling approaches to look for whole exome variants. These and a single study that included a systematic sequencing of >800 immune related genes in psoriasis cases and controls have thus far met with limited success [76].
The overall status of efforts to identify rare variants in autoimmune disease has recently been reviewed [77]. As pointed out by these authors it is too early to make conclusions as to the role of rare variants. Systematic sequencing of exomes is limited and whole genome sequencing of large numbers of cases for specific autoimmune disease is just now on the horizon. Part of the difficulty in these studies is that the power to detect statistically significant associations for rare variants requires either prodigiously large numbers of samples (100s of thousands) or some method to limit the variants under consideration (i.e. those more likely to be functional) and/or applying some type of burden or collapsing algorithm to examine composite effects of multiple rare variants together. The numerous methods and algorithms for these analyses include several software packages that include different types of burden and collapsing tests (Variable Threshold, Madson Browning, KBAC, SKAT, SKAT0, VAAST) that use different algorithms to group together sequence variants to test for evidence of association [78-83]. Some of these methods have been assimilated in a variety of software packages many of which enable incorporation of covariates including population substructure (http://varianttools.sourceforge.net/, http://csg.sph.umich.edu/kang/epacts/). Recent simulation studies suggest that SKAT0 and a generalized version of the C-alpha test may be more powerful than the other methods [84]. For exonic variants, algorithms such as SIFT, Polyphen and MutationTaster are popularly used to identify more likely causal variants and other algorithms use sequence conservation [85-87]. However, these are imperfect and some known causal variants for at least Mendelian diseases would be missed by these approaches. Compilations of functional and potentially functional coding variants can be found using different software [e.g. Annovar (http://annovar.openbioinformatics.org/en/latest/)[88]] and Ensemble (http://uswest.ensembl.org/Homo_sapiens/Info/Index)] [89].
For non-coding variation the situation is even more difficult although there are several search engines to look for critical variations in regulatory sequences including micro-RNAs, transcription binding sites, DNase1 hypersensitivity and expression quantitative trait loci (eQTL) [88-92]. (These issues are discussed further in Section 6.)
4. Ancestry makes a difference
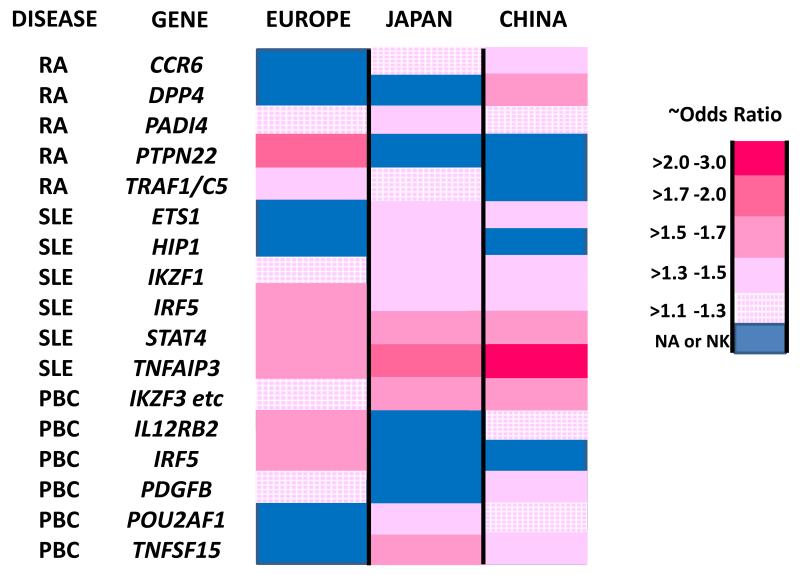
There are substantial differences in the loci defined for autoimmune diseases in disparate population groups. These differences are most apparent comparing results from one continental population to another continental population. The largest amount available data are from studies of European populations and East Asian populations [93-96]. The differences are partially explained by the frequency of particular variants in these different populations. Thus, the PTPN22 R602W variation that appears to account for most of the association of this gene with multiple different autoimmune diseases in European populations is virtually absent in other ethnicities including East Asian population groups [97, 98]. A converse example is the strong association of PADI4 with RA in East Asian populations that apparently plays a much smaller role in European populations consistent with the near absence of the strongest associated SNPs [94, 99]. Examples showing comparisons of associations and odds ratios in European, Japanese and Chinese RA cohorts are shown in Figure 3.
Figure 3.
Comparison of effect sizes for gene polymorphsims in three different population groups in three different autoimmune diseases. The approximate odds ratios (ORs) are indicated by the color code legend and for each the OR is indicated in the positive direction (regardless of minor allele frequency). The ORs were preferentially chosen from larger studies or meta-analyses ([57, 94, 96, 98, 99, 148, 170, 171, 174-179]. The genes selected were chosen on the bases of showing an OR of >1.3 in at least one of the populations chosen. For some of the genes for which the legend key indicates no association or not known (NA or NK) there may be limited data that suggests possible association that does not meet the standard genome-wide criteria for statistical significance.
It should also be noted that when the frequency of an allelic variant is substantially lower in one population group, that even if the odds ratios are in reality similar, the sample sizes might need to be very large to detect statistically significant association for this population group. Thus, there is an association of PADI4 and RA in European derived populations when the sample size is very large. However, the odds ratios are small (OR =~1.1) compared to that observed in Japanese RA (1.36) and is apparently due to the absence of the more strongly associated haplotype [94, 99].
There are also differences within continents. For example, the common variants of PADI4 that are associated with RA in Japanese are not associated in Chinese cohorts [96]. Within Europe there is a substantial difference in frequency in the afore-noted PTPN22 variant with a notable difference between northern and southern population groups: highest frequency in Finland (0.155) [100] and lowest in Italy (0.021) [48]. This emphasizes the importance of controlling for population substructure and using Cochran–Mantel–Haenszell corrections to account for the differences in allele frequencies when meta-analyses are performed in disparate population groups (reviewed in [15]).
It is also worth briefly noting that the striking geographic distribution of PTPN22 suggests that it may have been under natural selection. In fact, other evidence suggests this is the case and suggest that many of the genes associated with SLE have natural selection signals [101]. Other autoimmune susceptibility genes also show evidence for natural selection. In celiac disease a non-synonymous variant in SH2B3 that shows a strong signal for positive selection in Europe and suggestive data that it might trace to the Justinan plague in the Eastern Roman Empire 541-542 AD [102]. Thus, some of the proclivity to autoimmunity may be the flip side of variants that are advantageous in the context of particular infectious agents and in part may underlie a portion of the differences in susceptibility gene variant frequencies between different population groups.
A difficulty in comparing studies in different ancestries is that for most susceptibility loci the actual causal variants are not known. Differences between populations can be the result of different haplotype structure rather than the absence or lack of effect of a particular causal variant (if unknown) or alternatively different causal variants of the same gene in different population groups. This is particularly a problem when candidate polymorphisms are selected based on a different population group. It can be addressed, at least with respect to common variant hypothesis, if sufficient SNPs marking any common haplotype in the study population are included in the analyses. Some have advocated studying multiple populations to enable finer mapping of causal variants. However, this pre-supposes that the same causal variants are operative in different population groups. Meta-analyses using trans-ethnic population samples have uncovered some novel variants in for example RA [94], presumably due to increased sample sizes but this does not negate the observation of differences in the strength of association in different population groups.
It is worth emphasizing that differences in disease phenotypes are also associated with ancestry. There are examples of both differences in the frequency of a particular autoimmune disease as well as differences in detected susceptibility associated variants. While some differences in disease frequency may be due to differences in environmental factors including presumed exposure to different infectious agents others are clearly due to genetics. One of the best examples is the low frequency of T1D in Native American population groups that has been attributed to the near absence of particular HLA determinants. More recently, in mixed population groups such as in the United States, ancestry informative markers (AIMs) or sufficient numbers of unselected independent markers (low intermarker LD) have been used to examine ancestry relationships between and within continents as well as admixture and autoimmune disease. Evidence for increased frequency of SLE has been associated with both West African ancestry and Native American ancestry [103-105]. Several studies of SLE have used this approach to examine whether endophenotypes of this disease are more likely to be associated with particular ancestry [106-109]. Some of these studies suggest substantial differences based on ancestry that cannot be accounted for by environmental covariates, albeit these covariates are imperfectly measured. Future studies may define the underlying variations that presumably can explain why particular ancestries predispose to particular disease manifestations (e.g. mucocutaneous manifestations were higher in northern compared with southern European ancestry and the opposite was found for anti-ds DNA in SLE) [106, 107].
5. Missing Heritability
For most autoimmune diseases, estimates suggest that over 50% of the genetic loci contributing to heritability are not yet elucidated. Many explanations that are not mutually exclusive are possible to account for this missing heritability including: 1) very large numbers of common low risk variants that can only be defined with huge numbers of cases and controls (e.g. sample sizes >>100,000); 2) large numbers of rare variants that have not been detected due to insufficient sequencing, sample size and appropriate models; 3) copy number repeats and/or small insertions or deletions that have not been adequately examined; 4) better definition of disease phenotypes (i.e. reduce disease heterogeneity) is necessary to better identify loci; 5) epistasis between two or more loci that may be difficult to ascertain; and 6) epistasis between different environmental factors and genetic loci (that can only be identified in the context of the environmental factors). As large scale sequencing becomes increasingly practical and perhaps applied systematically to large populations many of these issues should be addressable. In addition, some studies have suggested that a sequence driven search might identify additional genetic factors. This approach, PheWas [110] relies on looking at medical records and laboratory studies to see if particular variants are associated with any of the phenotypes. For example if there is a large database of sequence available it is possible that particular variants might be associated with a specific autoantibody. It is doubtful that ICD codes that are generally used in such efforts would be sufficiently informative to detect many novel associations although the method has shown some promise [110, 111]. Alternatively, it might be possible to selectively study individuals who have sequence variants that are predicted to have a strong functional effect.
6. Towards Functional Studies: Coding Variants, Gene Expression, and Epigenetics
Relatively few non-synonymous variants have been identified in non-MHC genetic loci contributing to the susceptibility to autoimmune diseases. These include PTPN22, TNIP1, ITGAM, and SH2B3 [26, 48, 59, 102, 112]. Although Koch’s postulates cannot be tested, for some variants there is compelling evidence that the variant changes a functional aspect(s) of the immune response. For PTPN22 the R620W variant has been extensively studied and clearly has a major effect on increasing T cell and B cell expansion, dendritic cell hyper-responsiveness and B cell activation on engagement of the antigen receptor [49-53]. However, even here there is still substantial uncertainty with regards the actual mechanism although a gain of function appears to be most likely and there is evidence that the effect is likely to be on peripheral negative selection of T cells and an independent effect on B cells [113, 114].
Most of the confirmed loci that contribute to the genetics of autoimmune disease are non-coding. To connect these to actual mechanisms is difficult and requires extensive circumstantial evidence. In contrast to Mendelian developmental defects that are usually coding, and where model organisms can frequently be used to easily provide evidence for functional defects, defining the physiologic role for non-coding variants in a complex disease may require multiple lines of evidence. As an initial step, many studies have examined gene expression and aspects of changes in DNA (methylation, histone marks) in various cell types. These studies can be either gene specific or genome-wide, and in general include: 1) studies comparing cells from cases compared to controls; 2) normal cells or tissues comparing those with a particular variant(s) to those without. Public databases can provide a valuable tool as will be discussed below.
Genome-wide expression studies have been performed in a variety of cells and tissues. Several approaches are possible. Many studies have used unseparated peripheral blood mononuclear studies may be difficult to interpret even when differential white blood cell counts are used in efforts to adjust based on cell type. Although some findings such as the type 1 (alpha) interferon signal in SLE (comparing cases with controls) are robust [115-119], it is likely that determining most of the functional variations will require finer examination of particular cell types and perhaps in the context of particular activating stimulation. Other studies have examined sorted cell populations and although these may be partially affected by cell separation procedures they are more likely to provide relevant information. For RA, synovial cells have also been studied [120-123], but for most diseases it is difficult to study regional lymphoid or parenchymal tissue that might enrich for particularly relevant subsets of cells.
There are also methodological issues that lead to substantial difficulty in obtaining accurate scans of gene expression. The use of chip gene arrays is likely to be replaced by RNA sequencing studies that may also have the potential to ascertain differences in splice form variants [124, 125] and preferential allele expression when coupled with genomic sequence. Nevertheless, many of these studies have provided some clues to the pathways that are involved in the pathogenesis of individual autoimmune diseases. However, connecting the patterns of gene expression differences to the constellation of genetic factors that are defined by association studies (i.e. hereditary factors) is a critical step that may be hard to bridge.
One of the bridging approaches to look at the functional significance of sequence variants is to utilize eQTL. A number of studies have performed quantitative tests in cells or cell lines derived from different tissues to find polymorphisms (usually SNPs) that are associated with measurements of expression (eQTL). Other studies have examined methylation, DNase1 hypersensitvity and various histone modifications. These studies as well as eQTL can potentially identify specific variants that include those that are also identified by various association studies (GWAS or rare variant analyses) or are in strong linkage disequilibrium with these variants. Clues are often available in silico from public data bases of the larger projects, most notably the integrated encyclopedia of DNA elements in the human genome (Encode) project [126] and compilations of eQTL data. The Encyclopedia of DNA elements (Encode) data compiled and available via the UCSC Genome Browser provide data from multiple different cell lines and a number of tissues. This data provides Dnase sensitivity, information from sequences derived from chromatin immunoprecipitation (ChIP-seq), histone marks and transcription binding sites from a number of cell lines and tissues. A caveat for this approach is that for some genetic variants the right cell population at the right activation state may not be represented in the Encode or other available studies. Thus although this is a good starting point it may have both false negatives and even false positives.
Other tools also include Genevar that can be used to evaluate the measured effect of variants on DNA methylation (http://www.sanger.ac.uk/resources/software/genevar/) [127, 128] and eQTLs (http://www.bios.unc.edu/research/genomic_software/seeQTL/) [129]. The University of Chicago eQTL browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/) also provides another tool to identify expression quantitative trait loci (eQTLs) amongst candidate variants. It is worth noting that a good method for searching eQTL, and transcription factor binding sites including the Encode data is the RegulomeDB [90].
Other potentially useful approaches to finding putative important non-coding variations include searching for conserved motifs (e.g. SinBaD [92]) and using software that can ascertain whether variants affect splicing (e.g. Ensemble [89]). In addition, another resource, PolymiRTS [91], can be used to predict if 3’UTR variants are or are likely to alter micro-RNA (miR) target sites that may also be important in regulating genes or whether miR themselves have variants. Variants in other non-coding RNAs including long non-coding RNAs(lncRNAs) are also possible functional candidates as has been recently discussed [130, 131].
It is also worth commenting that a recent study has developed and used a fine-mapping algorithm, Probabilistic Identification of Causal SNPs (PICs) to identify candidate functional variants at risk loci for autoimmune diseases (http://www.broadinstitute.org/pubs/finemapping/?q=pics) [132]. Together with data from mapping RNA and chromatin in both resting and stimulated CD4(+) T-cells, regulatory T cells, CD8(+) T cells, B cells, and monocytes this study provided data to suggest that most putative causal variants (~60%) map to immune-cell enhancers. However, only 10-20% directly altered recognizable TF binding motifs. Thus, much work remains to further understand the regulatory mechanisms. This reviewer would caution that although the PICs algorithm is useful especially in the context of meta-analyses, when primary data is available the use of conditional analyses can be extremely valuable in evaluating the potential importance of particular SNPs in accounting for association signals.
Another approach to further defining function is the use of animal models and in particular those in the mouse. Many studies have used mouse knockouts to help understand what genes do. This has evolved to using mouse other manipulations in which specific base pairs are changed to study a particular variant (knockin mice). A mouse homologue of the PTPN22 with a variant analogous to R620W variant (PEP R619W) has shown immune cell differences with respect to T cell expansion and transitional, germinal center, and age-related B cell expansion [113]. These mice developed autoantibodies and features of systemic autoimmunity. In addition the defective B cell selection and B lineage–restricted variant expression and was sufficient to promote autoimmunity. However, for studying human gene interactions the differences between mouse and human genes may make such knockin studies difficult or impossible for some/many variants.
Evolving methods to introduce human genes more efficiently into mouse stem cells could in the future enable more physiologic studies of putative human functional variants. Mice with the entire human immunoglobulin gene region have been generated [133] and introduction of multiple human genes in a particular pathway is not beyond the realm of feasibility. Of course such manipulations would have to include specific variants of these genes to mimic conditions similar to those in humans with particular autoimmune diseases. In addition, it is not clear that appropriate stimuli could be used to initiate a particular autoimmune phenotype that could depend on some specific antigenic challenge.
Finally, it should also be noted that there are also epigenetic changes and RNA editing variations that are not due to hereditary factors that may be important in the immunopathogenesis of some autoimmune diseases. Studies of identical twins concordant or discordant for particular autoimmune disease may help identify some of these non-hereditary changes that are part of immunopathogenses of autoimmune disease. RNA sequencing studies have the potential to define RNA editing variations but this may depend on identifying the right cell subtype in which critical (phenotype altering) somatic variations have occurred.
6.1 Pathways
A variety of databases can be queried to ascertain whether loci identified for an autoimmune disease are likely by some statistical measure to be members of particular biologic pathways. In general, these approaches use independent signals (no LD) from association results (often from meta-analyses to query membership in gene sets from a variety of databases often including BioCarta (http://www.biocarta.com/genes/index.asp), KEGG [134], Pathway Interaction Database (PID) (http://pid.nci.nih.gov/userguide/introduction.shtml), Gene Ontology [135] and Reactome [136]. An example would be using the MSigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp) curation of some of these gene sets. A subsequent step uses a program (examples include using the i-GSEA4GWAS web server [137], Different software including Paris [138, 139], and Inrich [140] can be used to identified genes within some distance of the independent signals (e.g. 20 kb of the SNP or a border defined by a 2 log fall off from the peak association) and represent each gene by the strongest association. The gene sets (“pathway categories”) can then assessed for enrichment of signals using a permutation procedure that can also correct for bias from variations in gene size and gene set size. Often, a false discovery rate (FDR) is used to correct for multiple testing based on the distributions of enrichment scores generated by the permutations. Sometimes, the concordance of results between different methods is used as another way of filtering “significant” results. For autoimmune diseases, these studies are usually performed with and without the HLA signal to determine whether the pathways are dependent on HLA, since HLA is part of numerous gene pathways. A study of MS used a protein interaction data base to limit pathway analyses to only gene sets with evidence of physical interaction using a protein-interaction-network-based pathway analysis [141], although it is not clear if this approach is substantially better than those described above.
Such studies have been performed in many autoimmune diseases and including RA [142-144], SLE [144], MS [141], T1D [145], Celiac [146], PBC [147, 148] and Crohn’s disease [149]. These and other studies have highlighted particular pathways of which, not surprisingly, many are shared among different autoimmune diseases. Several of these studies have led to finding additional susceptibility loci by focusing additional genotyping on candidate genes which failed to reach statistical significance in previous studies but were “overrepresented” in pathways.
A caveat of these studies is of course that it is unknown how well the gene sets actually correspond to physiologic pathways. The improved curating and knowledge of these gene sets is ongoing that should facilitate these analyses. Future considerations might include ways of categorizing specific components of these pathways that currently are not distinguished including whether there is up or down regulation/signaling predicted for the different variants and perhaps further the ability to define diseases based on systems biology approaches.
6.2. Implications for Diagnosis and Therapy
Theoretically, genetic risk factors can distinguish who is at high risk for a particular autoimmune disease. Studies of several diseases including RA, MS and PBC have used various genetic risk models to ask whether the combination of loci defined by GWAS studies can identify individuals at high risk for MS, RA, and PBC [150-152]. These studies have shown some promise, however, it is clear that our current state of knowledge of the genetics for most of these autoimmune diseases is still insufficient to meaningfully assist in diagnostic criteria. Furthermore, it is not clear whether incomplete penetrance will severely limit the usefulness of applying genetic information for complex diseases as opposed to Mendelian disorders. It is also possible that the genetic information could meaningfully complement other diagnostic information including serology, gene expression profiles and perhaps environmental risk factors. Theoretically, such information might also assist in determining who is more likely to have disease progression.
Another arena that will clearly receive more attention is studies to evaluate whether particular therapeutic agents are more likely to be effective in the context of different arrays of risk alleles. For SLE several directed studies and a GWAS have examined whether genotypes could in retrospective analyses account for responders or non-responders to TNF blockade. Although several loci have been suggested by such studies none have replicated across studies where examined and at this time none have been confirmed [153-158]. Sample size issues have limited the power of such studies. Other efforts (e.g. in RA and PBC) have suggested using gene pathway information to identify potential therapeutic agents for autoimmune disease [94, 148].
The identification of more risk loci and definition of actual causal variants together with the anticipated large scale implementation of individual sequencing and integration with medical records may enable the large amount of data necessary for evaluating whether more optimal therapeutic decisions can be developed based on genetics. However, actionable genetics for complex diseases such as most autoimmune disease is not yet on the horizon as compared with the beginning of implementation of such decisions for pediatric Mendelian conditions [159-162].
7. Final comments: What to make of it all
Some in the scientific community have questioned whether the myriad of loci identified in GWAS studies has been useful. While GWAS has not solved the problem of what causes autoimmunity it is my contention that it has had an important impact on the field and the identification of these loci will continue to be one of the major drivers in furthering our understanding of what causes these phenotypes. Like most investigative results, more questions are raised from these studies. What are the real functional variants? How do they really alter responses? In which cell populations and at what stage do the genetic variations matter? Why do some variants have a much larger impact in one autoimmune disease than another? These will not be easily solvable but as reviewed here, there is definite progress.
The extensive variation in the human genome has partly been shaped by natural selection including many variants that are theorized to have favored survival to various infections. In depth studies that define functional variation of the loci that predispose to or accelerate autoimmunity may also further our understanding of immune responses to infectious agents. Likewise, ongoing studies directed at defining host responses to infectious agents may also provide needed clues for further unraveling the enigmatic autoimmune diseases.
As discussed in this perspective, the increased understanding of the genetics of autoimmune disease may have an impact in early diagnoses and/or in a decision making tree for therapeutic intervention. At the least, our increased knowledge of the dictionary of relevant gene variations and pathways should focus functional studies. These may also lead to the development of novel therapeutics. Ongoing functional studies will undoubtedly, although perhaps slowly, increase our knowledge of not only pathogenesis but normal biology and immunoregulation.
Hundreds of variations in HLA and non-HLA genes predispose to autoimmunity
For non-coding variations alterations in chromatin structure provide valuable clues
Expression quantitative traits can help identify causal variants
Pathway analyses can facilitate further understanding of autoimmune genetics
Functional studies and accounting for missing heredity are critical to advances
Acknowledgments
This work was supported by R01DK091823.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures
The authors have no conflict of interest to declare
References
- [1].Harley IT, Kaufman KM, Langefeld CD, Harley JB, Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10:285–90. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sundquist K, Martineus JC, Li X, Hemminki K, Sundquist J. Concordant and discordant associations between rheumatoid arthritis, systemic lupus erythematosus and ankylosing spondylitis based on all hospitalizations in Sweden between 1973 and 2004. Rheumatology (Oxford) 2008;47:1199–202. doi: 10.1093/rheumatology/ken184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mehers KL, Gillespie KM. The genetic basis for type 1 diabetes. British medical bulletin. 2008;88:115–29. doi: 10.1093/bmb/ldn045. [DOI] [PubMed] [Google Scholar]
- [4].Korponay-Szabo I, Kovacs J, Lorincz M, Torok E, Goracz G. Families with multiple cases of gluten-sensitive enteropathy. Zeitschrift fur Gastroenterologie. 1998;36:553–8. [PubMed] [Google Scholar]
- [5].Selmi C, Invernizzi P, Keefe EB, Coppel RL, Podda M, Rossaro L, et al. Epidemiology and pathogenesis of primary biliary cirrhosis. J Clin Gastroenterol. 2004;38:264–71. doi: 10.1097/00004836-200403000-00013. [DOI] [PubMed] [Google Scholar]
- [6].Jones DE, Watt FE, Metcalf JV, Bassendine MF, James OF. Familial primary biliary cirrhosis reassessed: a geographically-based population study. J Hepatol. 1999;30:402–7. doi: 10.1016/s0168-8278(99)80097-x. [DOI] [PubMed] [Google Scholar]
- [7].Nielsen NM, Westergaard T, Rostgaard K, Frisch M, Hjalgrim H, Wohlfahrt J, et al. Familial risk of multiple sclerosis: a nationwide cohort study. Am J Epidemiol. 2005;162:774–8. doi: 10.1093/aje/kwi280. [DOI] [PubMed] [Google Scholar]
- [8].Bogdanos DP, Smyk DS, Rigopoulou EI, Mytilinaiou MG, Heneghan MA, Selmi C, et al. Twin studies in autoimmune disease: genetics, gender and environment. J Autoimmun. 2012;38:J156–69. doi: 10.1016/j.jaut.2011.11.003. [DOI] [PubMed] [Google Scholar]
- [9].Padyukov L, Silva C, Stolt P, Alfredsson L, Klareskog L. A gene-environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum. 2004;50:3085–92. doi: 10.1002/art.20553. [DOI] [PubMed] [Google Scholar]
- [10].Heliovaara M, Aho K, Aromaa A, Knekt P, Reunanen A. Smoking and risk of rheumatoid arthritis. J Rheumatol. 1993;20:1830–5. [PubMed] [Google Scholar]
- [11].Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].The International HapMap Project Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- [13].Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- [14].Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–63. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–50. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- [17].Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, et al. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–50. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chung SA, Taylor KE, Graham RR, Nititham J, Lee AT, Ortmann WA, et al. Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet. 2011;7:e1001323. doi: 10.1371/journal.pgen.1001323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Viatte S, Plant D, Bowes J, Lunt M, Eyre S, Barton A, et al. Genetic markers of rheumatoid arthritis susceptibility in anti-citrullinated peptide antibody negative patients. Ann Rheum Dis. 2012;71:1984–90. doi: 10.1136/annrheumdis-2011-201225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cui J, Taylor KE, Lee YC, Kallberg H, Weinblatt ME, Coblyn JS, et al. The influence of polygenic risk scores on heritability of anti-CCP level in RA. Genes Immun. 2014;15:107–14. doi: 10.1038/gene.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Simpson G. The Principles of Classification and a Classification of Mammals. Bulletin of the AMNH. 1945;85:23. [Google Scholar]
- [25].McKusick VA. On lumpers and splitters, or the nosology of genetic disease. Perspectives in biology and medicine. 1969;12:298–312. doi: 10.1353/pbm.1969.0039. [DOI] [PubMed] [Google Scholar]
- [26].Gregersen PK, Kosoy R, Lee AT, Lamb J, Sussman J, McKee D, et al. Risk for myasthenia gravis maps to a (151) Pro-->Ala change in TNIP1 and to human leukocyte antigen-B*08. Ann Neurol. 2012 doi: 10.1002/ana.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Maniaol AH, Elsais A, Lorentzen AR, Owe JF, Viken MK, Saether H, et al. Late onset myasthenia gravis is associated with HLA DRB1*15:01 in the Norwegian population. PLoS One. 2012;7:e36603. doi: 10.1371/journal.pone.0036603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chung SA, Brown EE, Williams AH, Ramos PS, Berthier CC, Bhangale T, et al. Lupus nephritis susceptibility loci in women with systemic lupus erythematosus. J Am Soc Nephrol. 2014;25:2859–70. doi: 10.1681/ASN.2013050446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rioux JD, Goyette P, Vyse TJ, Hammarstrom L, Fernando MM, Green T, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A. 2009;106:18680–5. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Noble JA, Erlich HA. Genetics of type 1 diabetes. Cold Spring Harbor perspectives in medicine. 2012;2:a007732. doi: 10.1101/cshperspect.a007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kaukinen K, Partanen J, Maki M, Collin P. HLA-DQ typing in the diagnosis of celiac disease. Am J Gastroenterol. 2002;97:695–9. doi: 10.1111/j.1572-0241.2002.05471.x. [DOI] [PubMed] [Google Scholar]
- [32].Schlosstein L, Terasaki PI, Bluestone R, Pearson CM. High association of an HL-A antigen, W27, with ankylosing spondylitis. N Engl J Med. 1973;288:704–6. doi: 10.1056/NEJM197304052881403. [DOI] [PubMed] [Google Scholar]
- [33].de Menthon M, Lavalley MP, Maldini C, Guillevin L, Mahr A. HLA-B51/B5 and the risk of Behcet’s disease: a systematic review and meta-analysis of case-control genetic association studies. Arthritis Rheum. 2009;61:1287–96. doi: 10.1002/art.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, Jenisch S, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–51. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gockel I, Becker J, Wouters MM, Niebisch S, Gockel HR, Hess T, et al. Common variants in the HLA-DQ region confer susceptibility to idiopathic achalasia. Nature genetics. 2014;46:901–4. doi: 10.1038/ng.3029. [DOI] [PubMed] [Google Scholar]
- [36].Maggi E, Giudizi MG, Biagiotti R, Annunziato F, Manetti R, Piccinni MP, et al. Th2-like CD8+ T cells showing B cell helper function and reduced cytolytic activity in human immunodeficiency virus type 1 infection. J Exp Med. 1994;180:489–95. doi: 10.1084/jem.180.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mohr E, Cunningham AF, Toellner KM, Bobat S, Coughlan RE, Bird RA, et al. IFN-{gamma} produced by CD8 T cells induces T-bet-dependent and -independent class switching in B cells in responses to alum-precipitated protein vaccine. Proc Natl Acad Sci U S A. 2010;107:17292–7. doi: 10.1073/pnas.1004879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–13. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- [39].Invernizzi P. Human leukocyte antigen in primary biliary cirrhosis: an old story now reviving. Hepatology. 2011;54:714–23. doi: 10.1002/hep.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Invernizzi P, Ransom M, Raychaudhuri S, Kosoy R, Lleo A, Shigeta R, et al. Classical HLA-DRB1 and DPB1 alleles account for HLA associations with primary biliary cirrhosis. Genes Immun. 2012;13:461–8. doi: 10.1038/gene.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Achkar JP, Klei L, de Bakker PI, Bellone G, Rebert N, Scott R, et al. Amino acid position 11 of HLADRbeta1 is a major determinant of chromosome 6p association with ulcerative colitis. Genes Immun. 2012;13:245–52. doi: 10.1038/gene.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nature genetics. 2012;44:291–6. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Qiao SW, Iversen R, Raki M, Sollid LM. The adaptive immune response in celiac disease. Seminars in immunopathology. 2012;34:523–40. doi: 10.1007/s00281-012-0314-z. [DOI] [PubMed] [Google Scholar]
- [44].Ebringer RW, Cawdell DR, Cowling P, Ebringer A. Sequential studies in ankylosing spondylitis. Association of Klebsiella pneumoniae with active disease. Ann Rheum Dis. 1978;37:146–51. doi: 10.1136/ard.37.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Schwimmbeck PL, Yu DT, Oldstone MB. Autoantibodies to HLA B27 in the sera of HLA B27 patients with ankylosing spondylitis and Reiter’s syndrome. Molecular mimicry with Klebsiella pneumoniae as potential mechanism of autoimmune disease. J Exp Med. 1987;166:173–81. doi: 10.1084/jem.166.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kvien TK, Glennas A, Melby K, Granfors K, Andrup O, Karstensen B, et al. Reactive arthritis: incidence, triggering agents and clinical presentation. J Rheumatol. 1994;21:115–22. [PubMed] [Google Scholar]
- [47].de Almeida DE, Ling S, Holoshitz J. New insights into the functional role of the rheumatoid arthritis shared epitope. FEBS letters. 2012;585:3619–26. doi: 10.1016/j.febslet.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–8. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- [49].Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–9. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- [50].Zhang J, Zahir N, Jiang Q, Miliotis H, Heyraud S, Meng X, et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet. 2011;43:902–7. doi: 10.1038/ng.904. [DOI] [PubMed] [Google Scholar]
- [51].Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179:4704–10. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- [52].Arechiga AF, Habib T, He Y, Zhang X, Zhang ZY, Funk A, et al. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009;182:3343–7. doi: 10.4049/jimmunol.0713370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bottini N, Peterson EJ. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annual review of immunology. 2014;32:83–119. doi: 10.1146/annurev-immunol-032713-120249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008 doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Adrianto I, Wang S, Wiley GB, Lessard CJ, Kelly JA, Adler AJ, et al. Association of two independent functional risk haplotypes in TNIP1 with systemic lupus erythematosus. Arthritis Rheum. 2012;64:3695–705. doi: 10.1002/art.34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu X, Invernizzi P, Lu Y, Kosoy R, Lu Y, Bianchi I, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet. 2010;42:658–60. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Associations between interferon regulatory factor 5 polymorphisms and rheumatoid arthritis: a meta-analysis. Molecular biology reports. 2013;40:1791–9. doi: 10.1007/s11033-012-2233-4. [DOI] [PubMed] [Google Scholar]
- [59].Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chung SA, Criswell LA. PTPN22: its role in SLE and autoimmunity. Autoimmunity. 2007;40:582–90. doi: 10.1080/08916930701510848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Diaz-Gallo LM, Espino-Paisan L, Fransen K, Gomez-Garcia M, van Sommeren S, Cardena C, et al. Differential association of two PTPN22 coding variants with Crohn’s disease and ulcerative colitis. Inflammatory bowel diseases. 2011;17:2287–94. doi: 10.1002/ibd.21630. [DOI] [PubMed] [Google Scholar]
- [62].Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–32. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hirschfield GM, Liu X, Han Y, Gorlov IP, Lu Y, Xu C, et al. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet. 2010;42:655–7. doi: 10.1038/ng.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee YH, Bae SC. Association between the functional ITGAM rs1143679 G/A polymorphism and systemic lupus erythematosus/lupus nephritis or rheumatoid arthritis: an update meta-analysis. Rheumatology international. 2014;35:815–23. doi: 10.1007/s00296-014-3156-2. [DOI] [PubMed] [Google Scholar]
- [65].Anaya JM, Kim-Howard X, Prahalad S, Chernavsky A, Canas C, Rojas-Villarraga A, et al. Evaluation of genetic association between an ITGAM non-synonymous SNP (rs1143679) and multiple autoimmune diseases. Autoimmun Rev. 2011;11:276–80. doi: 10.1016/j.autrev.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLAB27 in disease susceptibility. Nat Genet. 2011;43:761–7. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2012;45:202–7. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A. 2006;103:9202–7. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–8. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- [71].Namjou B, Kothari PH, Kelly JA, Glenn SB, Ojwang JO, Adler A, et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 2011;12:270–9. doi: 10.1038/gene.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–7. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- [73].Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nature communications. 2015;6:5897. doi: 10.1038/ncomms6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, Stringham HM, et al. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Tang H, Jin X, Li Y, Jiang H, Tang X, Yang X, et al. A large-scale screen for coding variants predisposing to psoriasis. Nat Genet. 2014;46:45–50. doi: 10.1038/ng.2827. [DOI] [PubMed] [Google Scholar]
- [77].Massey J, Eyre S. Rare variants and autoimmune disease. Briefings in functional genomics. 2014;13:392–7. doi: 10.1093/bfgp/elu011. [DOI] [PubMed] [Google Scholar]
- [78].Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. American journal of human genetics. 2012;91:224–37. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS genetics. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS genetics. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, et al. Pooled association tests for rare variants in exon-resequencing studies. American journal of human genetics. 2010;86:832–8. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nature reviews Genetics. 2010;11:459–63. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hu H, Huff CD, Moore B, Flygare S, Reese MG, Yandell M. VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol. 2013;37:622–34. doi: 10.1002/gepi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Clarke GM, Rivas MA, Morris AP. A flexible approach for the analysis of rare variants allowing for a mixture of effects on binary or quantitative traits. PLoS Genet. 2013;9:e1003694. doi: 10.1371/journal.pgen.1003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- [86].Adzhubei I, Jordan DM, Sunyaev SR, Haines Jonathan L, et al. Current protocols in human genetics / editorial board. 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods. 2010;7:575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- [88].Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, et al. Ensembl 2013. Nucleic acids research. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lehmann KV, Chen T. Exploring functional variant discovery in non-coding regions with SInBaD. Nucleic Acids Res. 2013;41:e7. doi: 10.1093/nar/gks800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–7. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- [94].Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lee HS, Kim T, Bang SY, Na YJ, Kim I, Kim K, Kim JH, Chung YJ, Shin HD, Kang YM, Shim SC, Suh CH, Park YB, Kim JS, Kang C, Bae SC. Ethnic specificity of lupus-associated loci identified in a genome-wide association study in Korean women. Ann Rheum Dis. 2014;73:1240–5. doi: 10.1136/annrheumdis-2012-202675. [DOI] [PubMed] [Google Scholar]
- [96].Jiang L, Yin J, Ye L, Yang J, Hemani G, Liu AJ, et al. Novel risk loci for rheumatoid arthritis in Han Chinese and congruence with risk variants in Europeans. Arthritis & rheumatology (Hoboken, NJ. 2014;66:1121–32. doi: 10.1002/art.38353. [DOI] [PubMed] [Google Scholar]
- [97].Gregersen PK, Lee HS, Batliwalla F, Begovich AB. PTPN22: setting thresholds for autoimmunity. Seminars in immunology. 2006;18:214–23. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- [98].Yamamoto K, Okada Y, Suzuki A, Kochi Y. Genetics of rheumatoid arthritis in Asia-present and future. Nature reviews. 2015 doi: 10.1038/nrrheum.2015.7. [DOI] [PubMed] [Google Scholar]
- [99].Ikari K, Kuwahara M, Nakamura T, Momohara S, Hara M, Yamanaka H, et al. Association between PADI4 and rheumatoid arthritis: a replication study. Arthritis Rheum. 2005;52:3054–7. doi: 10.1002/art.21309. [DOI] [PubMed] [Google Scholar]
- [100].Seldin MF, Shigeta R, Laiho K, Li H, Saila H, Savolainen A, et al. Finnish case-control and family studies support PTPN22 R620W polymorphism as a risk factor in rheumatoid arthritis, but suggest only minimal or no effect in juvenile idiopathic arthritis. Genes Immun. 2005;6:720–2. doi: 10.1038/sj.gene.6364255. [DOI] [PubMed] [Google Scholar]
- [101].Ramos PS, Shaftman SR, Ward RC, Langefeld CD. Genes associated with SLE are targets of recent positive selection. Autoimmune diseases. 2014;2014:203435. doi: 10.1155/2014/203435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhernakova A, Elbers CC, Ferwerda B, Romanos J, Trynka G, Dubois PC, et al. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet. 2010;86:970–7. doi: 10.1016/j.ajhg.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Molokhia M, McKeigue P. Risk for rheumatic disease in relation to ethnicity and admixture. Arthritis Res. 2000;2:115–25. doi: 10.1186/ar76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Molokhia M, Hoggart C, Patrick AL, Shriver M, Parra E, Ye J, et al. Relation of risk of systemic lupus erythematosus to west African admixture in a Caribbean population. Hum Genet. 2003;112:310–8. doi: 10.1007/s00439-002-0883-3. [DOI] [PubMed] [Google Scholar]
- [105].Seldin MF, Qi L, Scherbarth HR, Tian C, Ransom M, Silva G, et al. Amerindian ancestry in Argentina is associated with increased risk for systemic lupus erythematosus. Genes Immun. 2008 doi: 10.1038/gene.2008.25. [DOI] [PubMed] [Google Scholar]
- [106].Chung SA, Tian C, Taylor KE, Lee AT, Ortmann WA, Hom G, et al. European population substructure is associated with mucocutaneous manifestations and autoantibody production in systemic lupus erythematosus. Arthritis Rheum. 2009;60:2448–56. doi: 10.1002/art.24707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Richman IB, Chung SA, Taylor KE, Kosoy R, Tian C, Ortmann WA, et al. European population substructure correlates with systemic lupus erythematosus endophenotypes in North Americans of European descent. Genes Immun. 2010;11:515–21. doi: 10.1038/gene.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Richman IB, Taylor KE, Chung SA, Trupin L, Petri M, Yelin E, et al. European Genetic Ancestry Protects against the Development of Renal Disease in Systemic Lupus Erythematosus. Arthritis Rheum. 2012 doi: 10.1002/art.34567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Sanchez E, Rasmussen A, Riba L, Acevedo-Vasquez E, Kelly JA, Langefeld CD, et al. Impact of genetic ancestry and sociodemographic status on the clinical expression of systemic lupus erythematosus in American Indian-European populations. Arthritis Rheum. 2012;64:3687–94. doi: 10.1002/art.34650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Hebbring SJ, Schrodi SJ, Ye Z, Zhou Z, Page D, Brilliant MH. A PheWAS approach in studying HLA DRB1*1501. Genes Immun. 2013;14:187–91. doi: 10.1038/gene.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–4. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- [113].Dai X, James RG, Habib T, Singh S, Jackson S, Khim S, et al. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest. 2013;123:2024–36. doi: 10.1172/JCI66963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wu DJ, Zhou W, Enouz S, Orru V, Stanford SM, Maine CJ, et al. Autoimmunity-associated LYP W620 does not impair thymic negative selection of autoreactive T cells. PLoS One. 2014;9:e86677. doi: 10.1371/journal.pone.0086677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Baechler EC, Gregersen PK, Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Current opinion in immunology. 2004;16:801–7. doi: 10.1016/j.coi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- [116].Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferoninducible gene expression signature in peripheral blood cells of patients with severe lupus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2610–5. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Baechler EC, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, Novitzke J, et al. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Molecular medicine (Cambridge, Mass. 2007;13:59–68. doi: 10.2119/2006-00085.Baechler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Niewold TB. Interferon alpha as a primary pathogenic factor in human lupus. J Interferon Cytokine Res. 2011;31:887–92. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Niewold TB, Adler JE, Glenn SB, Lehman TJ, Harley JB, Crow MK. Age- and sex-related patterns of serum interferon-alpha activity in lupus families. Arthritis Rheum. 2008;58:2113–9. doi: 10.1002/art.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Badot V, Galant C, Nzeusseu Toukap A, Theate I, Maudoux AL, Van den Eynde BJ, et al. Gene expression profiling in the synovium identifies a predictive signature of absence of response to adalimumab therapy in rheumatoid arthritis. Arthritis Res Ther. 2009;11:R57. doi: 10.1186/ar2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Galligan CL, Baig E, Bykerk V, Keystone EC, Fish EN. Distinctive gene expression signatures in rheumatoid arthritis synovial tissue fibroblast cells: correlates with disease activity. Genes Immun. 2007;8:480–91. doi: 10.1038/sj.gene.6364400. [DOI] [PubMed] [Google Scholar]
- [122].Devauchelle V, Marion S, Cagnard N, Mistou S, Falgarone G, Breban M, et al. DNA microarray allows molecular profiling of rheumatoid arthritis and identification of pathophysiological targets. Genes Immun. 2004;5:597–608. doi: 10.1038/sj.gene.6364132. [DOI] [PubMed] [Google Scholar]
- [123].Verweij CL. Transcript profiling towards personalised medicine in rheumatoid arthritis. The Netherlands journal of medicine. 2009;67:364–71. [PubMed] [Google Scholar]
- [124].Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome research. 2012;22:2008–17. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Hooper JE. A survey of software for genome-wide discovery of differential splicing in RNA-Seq data. Human genomics. 2014;8:3. doi: 10.1186/1479-7364-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics (Oxford, England) 2010;26:2474–6. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. American journal of human genetics. 2013;93:876–90. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Xia K, Shabalin AA, Huang S, Madar V, Zhou YH, Wang W, et al. seeQTL: a searchable database for human eQTLs. Bioinformatics (Oxford, England) 2012;28:451–2. doi: 10.1093/bioinformatics/btr678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, et al. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015 doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- [131].Hrdlickova B, Kumar V, Kanduri K, Zhernakova DV, Tripathi S, Karjalainen J, et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome medicine. 2014;6:88. doi: 10.1186/s13073-014-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2014;518:337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Macdonald LE, Karow M, Stevens S, Auerbach W, Poueymirou WT, Yasenchak J, et al. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5147–52. doi: 10.1073/pnas.1323896111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhang K, Cui S, Chang S, Zhang L, Wang J. i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 2010;38:W90–5. doi: 10.1093/nar/gkq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Yaspan BL, Bush WS, Torstenson ES, Ma D, Pericak-Vance MA, Ritchie MD, et al. Genetic analysis of biological pathway data through genomic randomization. Hum Genet. 2011;129:563–71. doi: 10.1007/s00439-011-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Yaspan BL, Veatch OJ, Haines Jonathan L, et al. Current protocols in human genetics / editorial board. 2011. Strategies for pathway analysis from GWAS data. Chapter 1:Unit1 20. [DOI] [PubMed] [Google Scholar]
- [140].Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–9. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Sclerosis IM. Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. American journal of human genetics. 2013;92:854–65. doi: 10.1016/j.ajhg.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–8. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Zhang MM, Jiang YS, Lv HC, Mu HB, Li J, Shang ZW, et al. Pathway-based association analysis of two genome-wide screening data identifies rheumatoid arthritis-related pathways. Genes Immun. 2014;15:487–94. doi: 10.1038/gene.2014.48. [DOI] [PubMed] [Google Scholar]
- [144].Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Molecular biology reports. 2012;39:10627–35. doi: 10.1007/s11033-012-1952-x. [DOI] [PubMed] [Google Scholar]
- [145].Evangelou M, Smyth DJ, Fortune MD, Burren OS, Walker NM, Guo H, et al. A method for gene-based pathway analysis using genomewide association study summary statistics reveals nine new type 1 diabetes associations. Genet Epidemiol. 2014;38:661–70. doi: 10.1002/gepi.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Kumar V, Gutierrez-Achury J, Kanduri K, Almeida R, Hrdlickova B, Zhernakova DV, et al. Systematic annotation of celiac disease loci refines pathological pathways and suggests a genetic explanation for increased interferon-gamma levels. Hum Mol Genet. 2015;24:397–409. doi: 10.1093/hmg/ddu453. [DOI] [PubMed] [Google Scholar]
- [147].Kar SP, Seldin MF, Chen W, Lu E, Hirschfield GM, Invernizzi P, et al. Pathway-based analysis of primary biliary cirrhosis genome-wide association studies. Genes Immun. 2013 doi: 10.1038/gene.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Cordell H, Han Y, Li Y, Mells G, Hirschfield G, Greene C, et al. An international genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and highlights pathogenic pathways for drug targeting. Nat Genet. doi: 10.1038/ncomms9019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Carbonetto P, Stephens M. Integrated enrichment analysis of variants and pathways in genome-wide association studies indicates central role for IL-2 signaling genes in type 1 diabetes, and cytokine signaling genes in Crohn’s disease. PLoS Genet. 2013;9:e1003770. doi: 10.1371/journal.pgen.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Wang JH, Pappas D, De Jager PL, Pelletier D, de Bakker PI, Kappos L, et al. Modeling the cumulative genetic risk for multiple sclerosis from genome-wide association data. Genome medicine. 2009;3:3. doi: 10.1186/gm217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Yarwood A, Han B, Raychaudhuri S, Bowes J, Lunt M, Pappas DA, et al. A weighted genetic risk score using all known susceptibility variants to estimate rheumatoid arthritis risk. Ann Rheum Dis. 2015;74:170–6. doi: 10.1136/annrheumdis-2013-204133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Tang R, Chen H, Miao Q, Bian Z, Ma W, Feng X, et al. The cumulative effects of known susceptibility variants to predict primary biliary cirrhosis risk. Genes Immun. 2015 doi: 10.1038/gene.2015.2. [DOI] [PubMed] [Google Scholar]
- [153].Cui J, Saevarsdottir S, Thomson B, Padyukov L, van der Helm-van Mil AH, Nititham J, et al. PTPRC rheumatoid arthritis risk allele is also associated with response to anti-TNF therapy. Arthritis Rheum. 2010 doi: 10.1002/art.27457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Liu C, Batliwalla F, Li W, Lee A, Roubenoff R, Beckman E, et al. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol Med. 2008;14:575–81. doi: 10.2119/2008-00056.Liu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Suarez-Gestal M, Perez-Pampin E, Calaza M, Gomez-Reino JJ, Gonzalez A. Lack of replication of genetic predictors for the rheumatoid arthritis response to anti-TNF treatments: a prospective case-only study. Arthritis Res Ther. 2010;12:R72. doi: 10.1186/ar2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Krintel SB, Palermo G, Johansen JS, Germer S, Essioux L, Benayed R, et al. Investigation of single nucleotide polymorphisms and biological pathways associated with response to TNFalpha inhibitors in patients with rheumatoid arthritis. Pharmacogenetics and genomics. 2012;22:577–89. doi: 10.1097/FPC.0b013e3283544043. [DOI] [PubMed] [Google Scholar]
- [157].Plant D, Bowes J, Potter C, Hyrich KL, Morgan AW, Wilson AG, et al. Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum. 2011;63:645–53. doi: 10.1002/art.30130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Umicevic Mirkov M, Cui J, Vermeulen SH, Stahl EA, Toonen EJ, Makkinje RR, et al. Genome-wide association analysis of anti-TNF drug response in patients with rheumatoid arthritis. Ann Rheum Dis. 2013;72:1375–81. doi: 10.1136/annrheumdis-2012-202405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England journal of medicine. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. Jama. 2014;312:1880–7. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, Petrikin JE, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Science translational medicine. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, et al. Genetics of type 1 diabetes: what’s next? Diabetes. 2010;59:1561–71. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–55. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of Systemic Lupus Erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008 doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- [166].Hoppenbrouwers IA, Aulchenko YS, Janssens AC, Ramagopalan SV, Broer L, Kayser M, et al. Replication of CD58 and CLEC16A as genome-wide significant risk genes for multiple sclerosis. J Hum Genet. 2009;54:676–80. doi: 10.1038/jhg.2009.96. [DOI] [PubMed] [Google Scholar]
- [167].Lee YH, Rho YH, Choi SJ, Ji JD, Song GG, Nath SK, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases--a meta-analysis. Rheumatology (Oxford) 2007;46:49–56. doi: 10.1093/rheumatology/kel170. [DOI] [PubMed] [Google Scholar]
- [168].Chen YF, Chang JS. PTPN22 C1858T and the risk of psoriasis: a meta-analysis. Molecular biology reports. 2012;39:7861–70. doi: 10.1007/s11033-012-1630-z. [DOI] [PubMed] [Google Scholar]
- [169].Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Sigurdsson S, Goring HH, Kristjansdottir G, Milani L, Nordmark G, Sandling JK, et al. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum Mol Genet. 2008;17:872–81. doi: 10.1093/hmg/ddm359. [DOI] [PubMed] [Google Scholar]
- [171].Juran BD, Hirschfield GM, Invernizzi P, Atkinson EJ, Li Y, Xie G, et al. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Ramos PS, Criswell LA, Moser KL, Comeau ME, Williams AH, Pajewski NM, et al. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet. 2011;7:e1002406. doi: 10.1371/journal.pgen.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–90. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Kochi Y, Okada Y, Suzuki A, Ikari K, Terao C, Takahashi A, et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet. 2010;42:515–9. doi: 10.1038/ng.583. [DOI] [PubMed] [Google Scholar]
- [175].Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Yang W, Tang H, Zhang Y, Tang X, Zhang J, Sun L, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am J Hum Genet. 2013;92:41–51. doi: 10.1016/j.ajhg.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Dong M, Li J, Tang R, Zhu P, Qiu F, Wang C, et al. Multiple Genetic Variants Associated with Primary Biliary Cirrhosis in a Han Chinese Population. Clinical reviews in allergy & immunology. 2015 doi: 10.1007/s12016-015-8472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Okada Y, Shimane K, Kochi Y, Tahira T, Suzuki A, Higasa K, et al. A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus eyrthematosus in Japanese. PLoS Genet. 2012;8:e1002455. doi: 10.1371/journal.pgen.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Nakamura M, Nishida N, Kawashima M, Aiba Y, Tanaka A, Yasunami M, et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet. 2012;91:721–8. doi: 10.1016/j.ajhg.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]